Abstract
Pea seed-borne mosaic potyvirus (PSbMV) isolates are divided into pathotypes P-1, P-2, and P-4 according to their infection profile on a panel of Pisum sativum lines. P. sativum PI 269818 is resistant to P-1 and P-2 isolates and is susceptible to P-4 isolates. Resistance to P-1 is inherited as a single recessive gene, denoted sbm-1, and the pathogenicity determinant has previously been mapped to the virus-coded protein VPg. In the cultivar Bonneville, a second recessive gene, sbm-2, confers specific resistance to P-2. By exchanging cistrons between a P-2 and a P-4 isolate, the P3-6k1 cistron was identified as the PSbMV host-specific pathogenicity determinant on Bonneville. Exchange of P3-6k1 did not affect infection on PI 269818, and infection of Bonneville was not altered by substitution of the VPg cistron, indicating that P3-6k1 and VPg are independent determinants of pathotype-specific infectivity. On PI 269818 the pathogenicity determinant of both P-1 and P-2 mapped to the N terminus of VPg. This suggests that VPg from the P-1 and P-2 isolates are functionally similar on this host and that resistance to P-1 and P-2 in PI 269818 may operate by the same mechanism. Identification of VPg–sbm-1 and P3-6k1–sbm-2 as independent pairs of genetic interactors between PSbMV and P. sativum provides a simple explanation of the three known pathotypes of PSbMV. Furthermore, analysis of β-glucuronidase-tagged P-2 virus indicated that sbm-2 resistance affected an early step in infection, implying that the P3-6k1 region plays a critical role in potyvirus replication or cell-to-cell movement.
The genus Potyvirus of the family Potyviridae belongs to the picornavirus supergroup of animal and plant viruses (19) and contains 30% of all known plant viruses (40). The potyvirus genome is an approximately 10-kb polyadenylated RNA molecule that is covalently linked at the 5′ terminus to a tyrosine residue of the virus-coded protein VPg (22). The RNA contains a single open reading frame encoding a polyprotein that is cleaved into 8 to 10 mature proteins by three virus-encoded proteinases (Fig. 1A) (reviewed in reference 32).
FIG. 1.
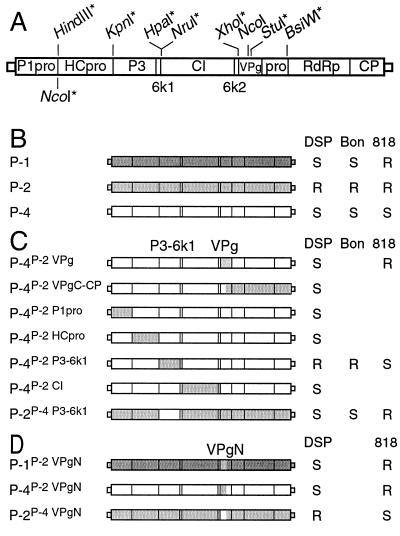
(A) Cistron map of the PSbMV genome, with an indication above the map of the restriction sites used to assemble recombinant full-length clones. The NcoI site indicated below the map was used to insert the GUS coding region in tagged constructs. All restriction sites marked with an asterisk were introduced by site-directed mutagenesis. (B) Diagrammatic representation of PSbMV pathotypes P-1 (dark grey), P-2 (light grey), and P-4 (white) and their infection profiles (R, resistant; S, susceptible) on P. sativum genotypes DSP, Bonneville (Bon), and PI 269818 (818). (C) Chimeras of PSbMV pathotypes P-2 and P-4 and their infection profiles on P. sativum genotypes DSP, Bon, and 818. (D) PSbMV chimeras with N-terminal VPg substitutions between pathotypes P-1, P-2, and P-4 and their infection profiles on P. sativum genotypes DSP and PI 269818.
A large number of naturally occurring dominant and recessive potyvirus resistance genes have been identified (28). The nature of these resistances and the relationship between resistances within a host species or between different plant species are generally not known. Interestingly, a high proportion of potyvirus resistance genes are inherited as recessive characters (9, 28). Recessive resistance is often strain specific, and in three studies this has led to identification of VPg as the host-specific pathogenicity determinant (15, 23, 37). Through analysis of chimeric and mutant viruses, VPg was found to determine long-distance movement of tobacco etch virus (TEV) in Nicotiana tabacum cultivar V20 (37), cell-to-cell movement and, to some extent, replication of tobacco vein mottling virus (TVMV) in N. tabacum cultivar TN86 homozygous for the resistance gene va (23), and replication of pea seed-borne mosaic virus (PSbMV) in P. sativum line PI 269818 carrying the resistance gene sbm-1 (15). One interpretation of resistance observed in plants carrying recessive resistance genes is that these plants lack a function essential for a particular step in infection (9, 31). In the above examples, these host factors would be needed for long-distance movement, cell-to-cell movement, and replication, implying that VPg has a role in each of these different processes. The first evidence that recessive resistance may actually be caused by the lack of a host factor came from a recent study demonstrating that va resistance in N. tabacum is caused by loss of a large chromosomal fragment (24). The recessive gene mlo, which confers resistance against powdery mildew in barley, is, however, an example demonstrating that recessive resistance can also be caused by lack of a host factor that suppresses a resistance response (3).
The potyvirus PSbMV mainly infects P. sativum and, to some extent, other legumes (16). Historically, PSbMV isolates have been grouped as pathotypes P-1, P-2, and P-4, depending on their infection profile on a selection of P. sativum lines (1). Sequences of cDNA clones representing an isolate of each of the PSbMV pathotypes P-1, P-2, and P-4 have been obtained, and DNA constructs expressing infectious mRNA are available (13, 14, 25). These DNA constructs represent a good starting point for a molecular dissection of the observed pathotypes. The P. sativum lines that have been used to differentiate the PSbMV pathotypes are of four categories: (A) susceptible to P-1, P-2, and P-4 (all-susceptible); (B) resistant to P-1, P-2, and P-4 (multiresistant); (C) resistant to P-2; and (D) resistant to P-1 and P-2 (25). Resistances to all three pathotypes of PSbMV in P. sativum are inherited as single recessive genes, designated sbm-1, sbm-2, sbm-3, and sbm-4 (26, 27). The sbm genes were mapped to linkage groups II (sbm-2) and VI (sbm-1, sbm-3, and sbm-4) as part of two clusters of recessive resistance genes affecting several potyviruses (29, 38). Resistance gene sbm-2 was mapped in category C P. sativum cultivar Bonneville (26), which is resistant to P-2 and susceptible to both P-1 and P-4 (1). Gene sbm-1 was mapped in multiresistant category B line PI 193586 (11) and later was shown to cosegregate with sbm-3 and sbm-4 (29). Resistance genes sbm-3 and sbm-4 were both defined in category B line PI 347492 as conferring resistance to P-2 and P-4, respectively (26, 27). A P. sativum category D line, PI 269818, which is resistant to pathotype P-1 and P-2 but susceptible to pathotype P-4 (1), was used to map the P-1/P-4 differentiating host-specific pathogenicity determinant to the VPg cistron (15). In this study, we show that host-specific pathogenicity of PSbMV on sbm-2/sbm-2 plants depends on the P3-6k1 cistron, whereas infection of PI 269818 plants depends on the VPg cistron. Based upon these two independent interactions between viral pathogenicity determinants and recessive resistance genes in P. sativum, we propose a simple model to explain the three known PSbMV pathotypes.
MATERIALS AND METHODS
Construction of chimeric viruses.
Cloned cDNA from virus isolates DPD1, L1, and NY was used to generate infectious clones p35S-P1-IB/IICIVA/VIA, p35S-L1-5, and p35S-NY VA, representing pathotypes P-1, P-2, and P-4, respectively (13, 25). Chimeric constructs and β-glucuronidase (GUS)-tagged derivatives were obtained by standard cloning procedures (36), including Pfu DNA polymerase (Stratagene)-mediated amplification of minor regions and site-directed mutagenesis on single-stranded DNA (Sculptor; Amersham). To stabilize the molecular clones in Escherichia coli, plant introns were inserted at selected sites according to published procedures (13). Seven chimeric viruses, P-4P-2 P1pro, P-4P-2 HCpro, P-4P-2 P3-6k1, P-4P-2 CI, P-4P-2 VPg, P-4P-2 VPgN, and P-4P-2 VPgC-CP, with regions of P-4 replaced by P-2 sequence were generated. In addition we constructed two viruses, P-2P-4 P3-6k1 and P-2P-4 VPgN, with regions of P-2 replaced by P-4 sequence and one virus, P-1P-2 VPgN, with part of the VPg coding region of P-1 replaced by the corresponding P-2 sequence. The following restriction sites (shown in Fig. 1A) were used to generate the recombinant plasmids: HindIII between P1 proteinase (P1pro) and helper component proteinase (HCpro) coding regions, KpnI between HCpro and P3-6k1, HpaI at the 3′ terminus of P3-6k1, NruI at the 5′ terminus of cylindrical inclusion protein (CI), XhoI between 6k2 and VPg, and StuI and NcoI in the center of VPg and BsiWI at the 3′ terminus of VPg. Sites HindIII, KpnI, HpaI, NruI, and XhoI in the P-2 sequence were generated during assembly of P-2 infectious clone p35S-L1-5 (25). StuI and BsiWI were introduced at nucleotide (nt) 6349 to 6354 and 6558 to 6563 of P-2, respectively. Corresponding sites were inserted into the P-4 sequence: HindIII at nt 1234 to 1239, KpnI at nt 2671 to 2676, HpaI and NruI at nt 3868 to 3873, XhoI at nt 5941 to 5946, StuI at nt 6313 to 6318, and BsiWI at nt 6522 to 6527. In the P-1 sequence, XhoI and StuI sites were inserted at nt 6003 to 6008 and nt 6373 to 6378, respectively. All nucleotide changes were silent.
Four tagged virus variants, P-2-GUS, P-4-GUS, P-2-GUSP-4 P3-6k1, and P-4-GUSP-2 P3-6k1 were created by insertion of the bacterial GUS coding sequence into infectious full-length cDNA clones between P1pro and HCpro coding regions as described by Dolja et al. (6). An NcoI site was introduced at nt 1339 to 1344 and 1295 to 1300 of P-2 and P-4, respectively, and the GUS coding sequence flanked by NcoI restriction sites (6) was inserted. A nonreplicating construct, P-1-GUSVNN, encoding a defective RNA-dependent RNA polymerase was generated as described for TEV (5) by changing the conserved Gly351-Asp352-Asp353 motif to Val-Asn-Asn.
P. sativum lines and crosses.
Cultivar Dark Skinned Perfection (DSP) was obtained from Danfeldt, Odense, Denmark, cultivar Bonneville and plant introduction line PI 269818 were kindly provided by Karen Keller, USDA Agricultural Research Service, Corvallis, Oregon, and cultivar Fjord was obtained from Svalof Weibull, Hammenhög, Sweden. Cultivar Fjord belongs to category A and is susceptible to the three known pathotypes of PSbMV. Cultivars DSP and Bonneville belong to category C and are resistant only to PSbMV P-2. Resistance gene sbm-2 was originally defined in Bonneville (26). PI 269818 belongs to category D and is resistant to P-1 and P-2, while it is susceptible to P-4.
To test allelism of resistance to P-2 in Bonneville, DSP, and PI 269818, crosses were made by pollinating young emasculated flowers of Bonneville with pollen from DSP and PI 269818 with pollen from Bonneville. In addition, DSP flowers received pollen from all-susceptible cultivar Fjord. For each cross, F1 progeny from different pollination events were manually inoculated with sap from Fjord infected with P-2 isolate L1. Three weeks postinoculation (p.i.), uninoculated upper leaves were tested by enzyme-linked immunosorbent assay (ELISA).
Virus inoculation and detection.
In general, virus infection was established by manual inoculation of cDNA constructs on all-susceptible cultivar Fjord using carborundum as an abrasive (korn grit F400; Dragon). Test plants were inoculated with infected plant material homogenized in 50 mM sodium phosphate, pH 7.0. Plants were analyzed 3 weeks p.i. by ELISA. Readings four times higher than those of mock-inoculated controls were rated positive. ELISA procedures were performed as previously described (18) using a polyclonal antibody against PSbMV coat protein.
To avoid a virus amplification step in susceptible cultivar Fjord, GUS-expressing constructs were inoculated directly on test plants by particle bombardment by using the Helios gene gun (Bio-Rad) as described earlier (30). A virus amplification step may result in part of the virus population having deletions in the GUS sequence (7). Fully expanded leaves of 3-week-old DSP and Fjord plants were inoculated, and GUS activity was visualized in samples of inoculated leaves and upper uninoculated leaves 1, 2, and 3 weeks p.i. Histochemical assays for GUS activity were conducted by the protocol of Hodal et al. (12), except that sorbitol was omitted from the buffer and 0.1% Tween was added. Inoculated and upper uninoculated leaves were assayed 6 and 12 days p.i.
RESULTS
Resistance to P-2 is allelic in Bonneville and DSP but caused by a different gene in PI 269818.
P. sativum cultivar Bonneville is resistant to PSbMV pathotype P-2, while it is susceptible to both P-1 and P-4 (1). Resistance gene sbm-2 was originally defined in Bonneville (26), but this cultivar is no longer commercially available. Cultivar DSP is commercially available and reacts to PSbMV pathotypes in the same manner as Bonneville. To test whether resistance to P-2 is caused by the same allele, the F1 progeny from a cross between DSP and Bonneville was inoculated with PSbMV P-2. Plants from 10 independent pollination events were resistant to P-2, while progeny from crosses between DSP and all-susceptible cultivar Fjord were susceptible. These results suggest that resistance to P-2 in DSP is allelic to and functionally similar to the recessive sbm-2 gene in Bonneville. To test if PI 269818, which is resistant to P-1 and P-2, carries sbm-2, PI 269818 was crossed with Bonneville. F1 progeny of three independent crosses were analyzed for resistance to pathotype P-2 virus. All gave rise to progeny that were systemically infected within 3 weeks p.i., demonstrating that PI 269818 resistance to P-2 is not due to the sbm-2 gene. Thus, according to previous definitions, the genes conferring resistance to P-1 and P-2 in PI 269818 will be designated sbm-1 and sbm-3, although it remains to be shown that the genes conferring resistance to P-1 and P-2 in this host are identical to the genes originally defined in multiresistant category B lines PI 193586 and PI 347492 (11, 26).
VPg is not a determinant of PSbMV virulence in sbm-2/sbm-2 plants.
By analyzing chimeras of PSbMV pathotypes P-1 and P-4, VPg has previously been shown to determine pathotype-specific virulence of PSbMV on PI 269818 carrying recessive resistance gene sbm-1 (15). VPg is also a virulence determinant of TVMV and TEV on N. tabacum carrying recessive resistance genes (23, 37). To test if the failure of PSbMV P-2 to infect P. sativum cultivars homozygous for the resistance gene sbm-2 was determined by VPg, a chimera was generated with the VPg cistron from pathotype P-2 isolate L1 replacing the corresponding region in the P-4 isolate NY. The chimeric virus P-4P-2 VPg was inoculated to DSP plants carrying sbm-2 and to all-susceptible cultivar Fjord as a control of inoculum infectivity and inoculation efficiency. At 3 weeks p.i. upper uninoculated leaves were tested for the presence of PSbMV coat protein by ELISA. Leaf extracts of all plants of both DSP and Fjord were positive in ELISA (Fig. 1C). This demonstrated that P-4P-2 VPg infected both DSP and Fjord systemically and suggested that VPg of P-1, P-2, and P-4 isolates DPD1, L1, and NY are functionally identical on DSP. These data do not support the hypothesis that VPg determines PSbMV virulence on P. sativum homozygous for resistance gene sbm-2.
A determinant of PSbMV pathogenicity on sbm-2/sbm-2 plants is located within the P3-6k1 sequence.
To identify the putative viral determinant for virulence on sbm-2/sbm-2 P. sativum plants, a set of P-4 chimeric viruses containing segments of the PSbMV P-2 L1 genome was assembled. Together with P-4P-2 VPg, these constructs represented the entire coding region of PSbMV P-2 L1 (Fig. 1C). Initially, viability of the chimeric viruses was confirmed by inoculating the recombinant cDNA constructs to all-susceptible cultivar Fjord. Sap from Fjord systemically infected with each of the chimeric viruses P-4P-2 P1pro, P-4P-2 HCpro, P-4P-2 P3-6k1, P-4P-2 CI, and P-4P-2 VPgC-CP was used to inoculate DSP plants. For each chimera, six DSP plants were inoculated and at 3 weeks p.i. upper uninoculated leaves were tested by ELISA for the presence of PSbMV coat protein. All DSP plants except those inoculated with P-4P-2 P3-6k1 were positive in ELISA, showing that chimeric viruses P-4P-2 P1pro, P-4P-2 HCpro, P-4P-2 CI, and P-4P-2 VPgC-CP infected DSP systemically and were not affected by sbm-2 resistance. P-4P-2 P3-6k1 was also tested on Bonneville, the cultivar in which sbm-2 was originally defined (26). Twelve plants were inoculated, and none of these were positive in ELISA 3 weeks p.i. These data suggested that properties of P3-6k1 are responsible for the failure of P-2 L1 to infect P. sativum plants homozygous for sbm-2. To test if P3-6k1 functions as a pathogenicity determinant, a chimera of P-2 carrying P3-6k1 of P-4 was constructed. The chimera P-2P-4 P3-6k1 was viable on all-susceptible cultivar Fjord, and sap from systemically infected leaves was inoculated to 12 DSP and 12 Bonneville plants. Three weeks p.i. PSbMV coat protein was detected in uninoculated leaves of all 12 DSP and all 12 Bonneville plants. This demonstrated that P-2P-4 P3-6k1 was infectious on DSP and Bonneville and that the P3-6k1 region determines virulence on P. sativum plants homozygous for resistance gene sbm-2.
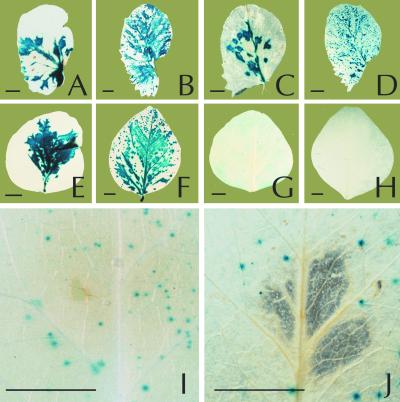
Visualization of infection by GUS-tagged chimeras shows that P-4P-2 P3-6k1 does not spread in inoculated DSP leaves.
To visualize the extent of infection of P-4P-2 P3-6k1 and P-2P-4 P3-6k1, two constructs were generated with GUS inserted between P1pro and HCpro coding sequences. The tagged viruses express the GUS reporter protein as an N-terminal fusion to HCpro. Similarly, the GUS coding sequence was inserted into the P-2 and P-4 full-length clones to obtain P-2-GUS and P-4-GUS. The GUS-tagged full-length cDNA clones were inoculated directly to Fjord and DSP plants by particle bombardment. All four constructs caused infections in both inoculated and upper uninoculated leaves of all-susceptible Fjord plants within 2 weeks p.i. as shown by in situ GUS activity analysis. GUS-stained leaves from plants inoculated with P-4-GUSP-2 P3-6k1 and P-2-GUSP-4 P3-6k1 are shown in Fig. 2A through D. In cultivar DSP, only plants inoculated with constructs containing the P3-6k1 cistron from P-4, P-4-GUS, and P-2-GUSP-4 P3-6k1 (Fig. 2F) resulted in GUS activity in uninoculated leaves, thus confirming data obtained by ELISA with constructs not containing the GUS gene. In inoculated leaves, GUS activity from P-4-GUS and P-2-GUSP-4 P3-6k1 (Fig. 2E) gave rise to extensive blue staining spreading into large areas of the inoculated leaves. In contrast, only small foci of GUS activity were detected in leaves of DSP inoculated with P-2-GUS and P-4-GUSP-2 P3-6k1 (Fig. 2G) and no GUS activity was detected in uninoculated leaves of these plants (Fig. 2H). The small inoculation foci on inoculated leaves were of the same size and intensity as those we have observed upon bombardment with a DNA construct containing only the GUS coding sequence with a PSbMV 5′-untranslated region placed behind the 35S promoter (data not shown) or nonreplicating P-1-GUSVNN encoding a defective RNA-dependent RNA polymerase (Fig. 2I and J). These observations suggest that P-2-GUS and P-4-GUSP-2 P3-6k1 do not spread or replicate to an extent exceeding that of P-1-GUSVNN. Gene sbm-2 may thus cause a block of replication or cell-to cell movement, depending on the nature of the P3-6k1 region.
FIG. 2.
In situ analysis of P. sativum cultivar Fjord (A through D) and DSP (E through J) inoculated with PSbMV chimera P-2-GUSP-4 P3-6k1 (A, B, E, and F), P-4-GUSP-2 P3-6k1 (C, D, G, H, and J), or replication-defective P-1-GUSVNN (I). Histochemical localization of GUS activity was performed on inoculated leaves (A, C, E, G, I, and J) and uninoculated leaves (B, D, F, and H) 12 days p.i. Bars, 5 mm.
VPg but not the P3-6k1 region determines P-2 infection of PI 269818 plants.
PI 269818 is resistant to PSbMV P-1 and P-2 and is susceptible to P-4 (1). It has been shown that virulence of P-1 and P-4 isolates DPD1 and NY is determined by VPg (15). Further analysis of the VPg cistron demonstrated that the virulence determinant lies within the region encoding the first 122 amino acids of the VPg cistron (2). To test if the failure of P-2 isolate L1 to infect PI 269818 depends on the same region of VPg or on the P3-6k1 region, PI 269818 plants were inoculated with P-4P-2 VPg, P-4P-2 VPgN, P-2P-4 VPgN, P-1P-2 VPgN, P-4P-2 P3-6k1, or P-2P-4 P3-6k1. At least six plants were inoculated with each virus, and 3 weeks p.i. upper uninoculated leaves were tested by ELISA for the presence of PSbMV coat protein. All plants inoculated with P-2P-4 VPgN or P-4P-2 P3-6k1 were positive in ELISA and all plants inoculated with P-4P-2 VPgN, P-1P-2 VPgN, or P-2P-4 P3-6k1 were negative (Fig. 1C and D). Thus, exchange of the P3-6k1 region between P-2 and P-4 did not alter infectivity on PI 269818. P-4P-2 P3-6k1 was infectious on PI 269818 just like P-4, while P-2P-4 P3-6k1 and P-2 both were noninfectious on PI 269818. In contrast, chimeras in which the region encoding the N terminus of VPg was exchanged all had the same infectivity on PI 269818 as the virus donating the VPgN sequence. P-4P-2 VPgN and P-1P-2 VPgN did not infect PI 269818, while P-2P-4 VPgN was infectious on this host. This suggests that while pathotype-specific infectivity of PSbMV on sbm-2/sbm-2 plants depends on properties of the P3-6k1 region, specific infectivity on PI 269818 depends only on the VPgN region.
DISCUSSION
The potyviral VPg cistron has previously been shown to determine host-specific pathogenicity on hosts carrying recessive resistance genes (15, 23, 37). It was logical, therefore, to test the hypothesis that VPg determines host-specific pathogenicity of PSbMV on P. sativum plants homozygous for resistance gene sbm-2. However, the experiments did not support this hypothesis, because exchanging VPg between P-2 isolate L1 and P-4 isolate NY did not affect infectivity on DSP. Instead, the P3-6k1 coding region was found to control infection of P. sativum homozygous for resistance gene sbm-2. Replacing P3-6k1 of P-4 NY with the corresponding region from P-2 L1 prevented infection on DSP and Bonneville, while replacing P3-6k1 of P-2 L1 with the corresponding region from P-4 NY was sufficient to convert P-2 L1 into a fully infectious virus on these two cultivars. The P3 region of isolates L1 and NY are 84% identical, whereas the small 6k1 region is 98% identical (25). The variation is highest in the N-terminal half of P3, but with 52-amino-acid differences between the two isolates in the P3-6k1 region further analysis is required to determine which part of this region affects infectivity on sbm-2 plants.
The potyviral P3 protein is released from the polyprotein by two different catalytic events. The HCpro/P3 junction is cleaved autocatalytically by HCpro, liberating the N terminus of P3 (4). The C terminus of P3 is processed by the NIa proteinase, resulting in either a 42-kDa P3-6k1 fusion or a 37-kDa P3 (10). Proteolytic processing between P3 and 6k1 was not required for plum pox virus viability, which could indicate that P3-6k1 is the functional protein (33). Results from localization studies of P3-6k1 and P3 have not been conclusive (20, 34, 35), but despite contrasting results a role in viral replication was proposed (20, 35). This is further supported by the observation that insertional mutations to TVMV P3 resulted in replication-defective mutants (17) and that P3 interacts with the potyviral RNA-dependent RNA polymerase both in vitro and in vivo in the yeast two-hybrid system (21). We found that P-2-GUS and P-4-GUSP-2 P3-6k1 inoculated by particle bombardment to DSP gave rise to small blue inoculation foci of the same size and intensity as a replication-deficient PSbMV (Fig. 2I and J). These observations suggest a role of P3-6k1 in replication or cell-to-cell movement but could also be explained by a failure to suppress host defense, as suggested by Voinnet et al. (39).
The identification of P3-6k1 as a pathogenicity determinant for PSbMV implies that at least two such determinants—the second being VPg—are involved in pathotype-specific resistance in P. sativum. Single cistron exchanges between the three PSbMV cDNA clones demonstrated that the two pathogenicity determinants operated independently (Fig. 1C and D). Since it is possible to explain the observed pattern of infection solely on the basis of P3-6k1 and VPg, we propose a simple model, based on these two viral pathogenicity determinants, to resolve the PSbMV pathotypes (Fig. 3). The model predicts a one-to-one relationship between P. sativum recessive resistance genes and PSbMV cistrons. With this model we propose that recessive resistance reflects a lack of (functional) interaction between the host factor encoded by the resistance gene and the virus-encoded pathogenicity determinant, which prevents multiplication or spread of the virus. Accordingly, P-2-encoded P3-6k1 does not produce a functional interaction with the sbm-2 gene product, while P3-6k1 of both P-1 and P-4 do. In the same manner, P-1- and P-2-encoded VPg do not produce a functional interaction with the sbm-1 gene product in PI 269818, while P-4 VPg does. This differs from the classical gene-for-gene relationship between dominant resistance genes, R-genes, and pathogen avirulence genes involving recognition of the pathogen and induced defense responses. Current models predict that the products of R-genes act as receptors for the products of pathogen avirulence genes, while susceptibility depends on lack of recognition (8).
FIG. 3.
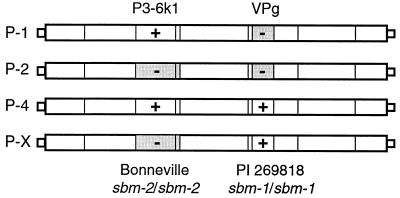
A simple model explaining the infection pattern of PSbMV pathotypes P-1, P-2, and P-4 by properties of the P3-6k1 and VPg regions. According to this model, specific resistance of P. sativum genotypes, like Bonneville homozygous for resistance gene sbm-2, depends on the P3-6k1 region. Specific resistance of genotypes homozygous for sbm-1, here represented by PI 269818, depends on VPg. This model also predicts a fourth pathotype, P-X, that carries a P3-6k1 region with properties similar to those of P3-6k1 of P-2 and therefore does not infect Bonneville but has a VPg region similar to that of VPg of P-4 and therefore is infectious on PI 269818.
The model also predicts the existence of a fourth pathotype, P-X, infectious on sbm-1 plants but noninfectious on sbm-2 plants. To our knowledge, a naturally occurring pathotype with these characteristics has not been described, but the artificially constructed chimera, P-2P-4 VPgN, fulfills the criteria for such a pathotype and shows that its existence, in principle, should be possible.
Only the resistance genes sbm-1 and sbm-2 are incorporated in the proposed model, but as previously explained, two more genes, sbm-3 and sbm-4, have been proposed to explain recessive resistance against PSbMV in P. sativum (26, 27). Further research on the interactions between P. sativum genotypes and PSbMV pathotypes is needed in order to fully elucidate all viral and host determinants involved in the known virus-host pattern of recessive resistance. However, the proposed model provides the first simple explanation to the existence of three PSbMV pathotypes.
ACKNOWLEDGMENTS
We thank Merete Albrechtsen for fruitful discussions and critically reading the manuscript and Jim Carrington for p306GUS, which contains the GUS coding sequence. Expert technical assistance from Birgit Olsen and Heidi Langhoff was much appreciated.
This work was supported by Danish Agricultural and Veterinary Research Council grant 9702802 and EU grant CT97-2356.
REFERENCES
- 1.Alconero R, Provvidenti R, Gonsalves D. Three pea seedborne mosaic virus pathotypes from pea and lentil germ plasm. Plant Dis. 1986;70:783–786. [Google Scholar]
- 2.Borgstrøm, B., and I. E. Johansen. Mutations in Pea seed-borne mosaic potyvirus genome-linked protein VPg alter pathotype specific virulence in Pisum sativum. Mol. Plant-Microbe Interact., in press. [DOI] [PubMed]
- 3.Büschges R, Hollricher K, Panstruga R, Simons G, Wolter M, Frijters A, van Daelen R, van der Lee T, Diergaarde P, Groenendijk J, Töpsch S, Vos P, Salamini F, Schulze-Lefert P. The barley Mlo gene: a novel control element of plant pathogen resistance. Cell. 1997;88:695–705. doi: 10.1016/s0092-8674(00)81912-1. [DOI] [PubMed] [Google Scholar]
- 4.Carrington J C, Cary S M, Parks T D, Dougherty W G. A second proteinase encoded by a plant potyvirus genome. EMBO J. 1989;8:365–370. doi: 10.1002/j.1460-2075.1989.tb03386.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carrington J C, Haldeman R, Dolja V V, Restrepo-Hartwig M A. Internal cleavage and trans-proteolytic activities of the VPg-proteinase (NIa) of tobacco etch potyvirus in vivo. J Virol. 1993;67:6995–7000. doi: 10.1128/jvi.67.12.6995-7000.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dolja V V, McBride H J, Carrington J C. Tagging of plant potyvirus replication and movement by insertion of β-glucuronidase into the viral polyprotein. Proc Natl Acad Sci USA. 1992;89:10208–10212. doi: 10.1073/pnas.89.21.10208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dolja V V, Herndon K L, Pirone T P, Carrington J C. Spontaneous mutagenesis of a plant potyvirus genome after insertion of a foreign gene. J Virol. 1993;67:5968–5975. doi: 10.1128/jvi.67.10.5968-5975.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ellis J, Dodds P, Pryor T. Structure, function and evolution of plant disease resistance genes. Curr Opin Plant Biol. 2000;3:278–284. doi: 10.1016/s1369-5266(00)00080-7. [DOI] [PubMed] [Google Scholar]
- 9.Fraser R S S. The genetics of plant-virus interactions: implications for plant breeding. Euphytica. 1992;63:175–185. [Google Scholar]
- 10.García J A, Martín M T, Cervera M T, Riechmann J L. Proteolytic processing of the plum pox potyvirus polyprotein by the NIa protease at a novel cleavage site. Virology. 1992;188:697–703. doi: 10.1016/0042-6822(92)90524-s. [DOI] [PubMed] [Google Scholar]
- 11.Hagedorn D J, Gritton E T. Inheritance of resistance to the pea seed-borne mosaic virus. Phytopathology. 1973;63:1130–1131. [Google Scholar]
- 12.Hodal L, Bochardt A, Nielsen J E, Mattsson O, Okkels F T. Detection, expression and specific elimination of endogenous β-glucuronidase activity in transgenic and non-transgenic plants. Plant Sci. 1992;87:115–122. [Google Scholar]
- 13.Johansen I E. Intron insertion facilitates amplification of cloned virus cDNA in Escherichia coli while biological activity is reestablished after transcription in vivo. Proc Natl Acad Sci USA. 1996;93:12400–12405. doi: 10.1073/pnas.93.22.12400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johansen I E, Keller K E, Dougherty W G, Hampton R O. Biological and molecular properties of a pathotype P-1 and a pathotype P-4 isolate of pea seed-borne mosaic virus. J Gen Virol. 1996;77:1329–1333. doi: 10.1099/0022-1317-77-6-1329. [DOI] [PubMed] [Google Scholar]
- 15.Keller K E, Johansen I E, Martin R R, Hampton R O. Potyvirus genome-linked protein (VPg) determines pea seedborne mosaic virus pathotype-specific virulence in Pisum sativum. Mol Plant-Microbe Interact. 1998;11:124–130. doi: 10.1094/MPMI.1998.11.2.124. [DOI] [PubMed] [Google Scholar]
- 16.Khetarpal R K, Maury Y. Pea seed-borne mosaic virus: a review. Agronomie. 1987;7:215–224. [Google Scholar]
- 17.Klein P G, Klein R R, Rodríguez-Cerezo E, Hunt A G, Shaw J G. Mutational analysis of the tobacco vein mottling virus genome. Virology. 1994;204:759–769. doi: 10.1006/viro.1994.1591. [DOI] [PubMed] [Google Scholar]
- 18.Kohnen P D, Johansen I E, Hampton R O. Characterization and molecular detection of the P4 pathotype of pea seedborne mosaic potyvirus. Phytopathology. 1995;85:789–793. [Google Scholar]
- 19.Koonin E V, Dolja V V. Evolution and taxonomy of positive-strand RNA viruses: implications of comparative analysis of amino acid sequences. Crit Rev Biochem Mol Biol. 1993;28:375–430. doi: 10.3109/10409239309078440. [DOI] [PubMed] [Google Scholar]
- 20.Langenberg W G, Zhang L. Immunocytology shows the presence of tobacco etch virus P3 protein in nuclear inclusions. J Struct Biol. 1997;118:243–247. doi: 10.1006/jsbi.1997.3856. [DOI] [PubMed] [Google Scholar]
- 21.Merits A, Guo D, Järvekülg L, Saarma M. Biochemical and genetic evidence for interactions between potato A potyvirus-encoded proteins P1 and P3 and proteins of the putative replication complex. Virology. 1999;263:15–22. doi: 10.1006/viro.1999.9926. [DOI] [PubMed] [Google Scholar]
- 22.Murphy J F, Rychlik W, Rhoads R E, Hunt A G, Shaw J G. A tyrosine residue in the small nuclear inclusion protein of tobacco vein mottling virus links the VPg to the viral RNA. J Virol. 1991;65:511–513. doi: 10.1128/jvi.65.1.511-513.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nicolas O, Dunnington S W, Gotow L F, Pirone T P, Hellmann G M. Variations in the VPg protein allow a potyvirus to overcome va gene resistance in tobacco. Virology. 1997;237:452–459. doi: 10.1006/viro.1997.8780. [DOI] [PubMed] [Google Scholar]
- 24.Noguchi S, Tajima T, Yamamoto Y, Ohno T, Kubo T. Deletion of a large genomic segment in tobacco varieties that are resistant to potato virus Y (PVY) Mol Gen Genet. 1999;262:822–829. doi: 10.1007/s004380051146. [DOI] [PubMed] [Google Scholar]
- 25.Olsen B S, Johansen I E. Nucleotide sequence and infectious cDNA clone of the L1 isolate of pea seed-borne mosaic potyvirus. Arch Virol. 2001;146:15–25. doi: 10.1007/s007050170187. [DOI] [PubMed] [Google Scholar]
- 26.Provvidenti R, Alconero R. Inheritance of resistance to a lentil strain of pea seed-borne mosaic virus in Pisum sativum. J Hered. 1988;79:45–47. [Google Scholar]
- 27.Provvidenti R, Alconero R. Inheritance of resistance to a third pathotype of pea seed-borne mosaic virus in Pisum sativum. J Hered. 1988;79:76–77. [Google Scholar]
- 28.Provvidenti R, Hampton R O. Sources of resistance to viruses in the Potyviridae. Arch Virol Suppl. 1992;5:189–211. doi: 10.1007/978-3-7091-6920-9_17. [DOI] [PubMed] [Google Scholar]
- 29.Provvidenti R, Muehlbauer F J. Evidence of a cluster of linked genes for resistance to pea seedborne mosaic virus and clover yellow vein virus on chromosome 6. Pisum Newsletter. 1990;22:43–45. [Google Scholar]
- 30.Rajamäki M-L, Valkonen J P T. The 6K2 protein and the VPg of potato virus A are determinants of systemic infection in Nicandra physaloides. Mol Plant-Microbe Interact. 1999;12:1074–1081. doi: 10.1094/MPMI.1999.12.12.1074. [DOI] [PubMed] [Google Scholar]
- 31.Revers F, Le Gall O T, Candresse T, Maule A J. New advances in understanding the molecular biology of plant/potyvirus interactions. Mol Plant-Microbe Interact. 1999;12:367–376. [Google Scholar]
- 32.Riechmann J L, Laín S, García J A. Highlights and prospects of potyvirus molecular biology. J Gen Virol. 1992;73:1–16. doi: 10.1099/0022-1317-73-1-1. [DOI] [PubMed] [Google Scholar]
- 33.Riechmann J L, Cervera M T, García J A. Processing of the plum pox virus polyprotein at the P3–6K1 junction is not required for virus viability. J Gen Virol. 1995;76:951–956. doi: 10.1099/0022-1317-76-4-951. [DOI] [PubMed] [Google Scholar]
- 34.Rodríguez-Cerezo E, Shaw J G. Two newly detected nonstructural viral proteins in potyvirus-infected cells. Virology. 1991;185:572–579. doi: 10.1016/0042-6822(91)90527-i. [DOI] [PubMed] [Google Scholar]
- 35.Rodríguez-Cerezo E, Ammar E D, Pirone T P, Shaw J G. Association of the non-structural P3 viral protein with cylindrical inclusions in potyvirus-infected cells. J Gen Virol. 1993;74:1945–1949. doi: 10.1099/0022-1317-74-9-1945. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook J, Fritsch E F, Maniatis T A. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 37.Schaad M C, Lellis A D, Carrington J C. VPg of tobacco etch potyvirus is a host genotype-specific determinant for long distance movement. J Virol. 1997;71:8624–8631. doi: 10.1128/jvi.71.11.8624-8631.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Timmerman G M, Frew T J, Miller A L, Weeden N F, Jermyn W A. Linkage mapping of sbm-1, a gene conferring resistance to pea seedborne mosaic virus, using molecular markers in Pisum sativum. Theor Appl Genet. 1993;85:609–615. doi: 10.1007/BF00220920. [DOI] [PubMed] [Google Scholar]
- 39.Voinnet O, Pinto Y M, Baulcombe D C. Suppression of gene silencing: a general strategy used by diverse DNA and RNA viruses of plants. Proc Natl Acad Sci USA. 1999;96:14147–14152. doi: 10.1073/pnas.96.24.14147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ward C W, Shukla D D. Taxonomy of potyviruses: current problems and some solutions. Intervirology. 1991;32:269–296. doi: 10.1159/000150211. [DOI] [PubMed] [Google Scholar]