Abstract
Immunotoxins are widely applied for cancer therapy. However, bacterial expression of immunotoxins usually leads to the formation of insoluble and non-functional recombinant proteins. This study was aimed to improve soluble expression of a novel anti-HER2 immunotoxin under the regulation of the trc promoter in Escherichia coli by optimization of the cultivation conditions using response surface methodology (RSM). To conduct RSM, four cultivation variables (i.e., inducer concentration, post-induction time, post-induction temperature, and medium recipe), were selected for statistical characterization and optimization using the Box-Behnken design and Design Expert software. Based on the developed model using the Box-Behnken design, the optimal cultivation conditions for soluble expression of anti-HER2 immunotoxin were determined to be 0.1 mM IPTG for induction in the LB medium at 33 °C for 18 h. The expressed immunotoxin was successfully purified using affinity chromatography with more than 90% purity and its bioactivity was confirmed using cell-based ELISA. Technical approach developed in this study can be generally applied to enhance the production yield and quality of recombinant proteins using E. coli as the gene expression system.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13568-024-01765-6.
Keywords: Immunotoxin, Response surface methodology (RSM), Cultivation optimization, Protein solubility, Escherichia coli
Introduction
Immunotoxins are protein molecules widely used for cancer therapy these days (Xing et al. 2021). Successful biological action of immunotoxins relies on their effective attachment to the target cell and subsequent translocation of the toxin’s bioactive fragment into the cytosol of the target cell. Because of their high cytotoxicity and potential inhibition of protein synthesis in eukaryotic cells, immunotoxins are often expressed in bacterial hosts for subsequent medical administration.
Expression of immunotoxins in E. coli usually leads to the formation and accumulation of insoluble aggregates with no biological function, i.e., inclusion bodies (Akbari et al. 2017; Kim et al. 2020). This presents a major technological challenge that requires a laborious and time-consuming process for protein refolding. The retrieval of soluble proteins from pre-existing inclusion bodies frequently results in reduced protein yield and biological activity, substantially increasing the production expenses associated with downstream separation processing. Alternatively, the soluble expression of foreign proteins in E. coli can be enhanced by the following bioprocessing and/or genetic strategies, including modulation and optimization of cultivation conditions (e.g., temperature, medium composition, and aeration, etc.), co-expression with chaperons, genetic modification of host strains, and modifying the level of inducer and the promoter strength (Rosano et al. 2019).
Different promoters have been genetically explored for heterologous gene expression in E. coli. Among them, the T7 promoter appears to be stronger than the common promoters, such as tac, lac, and trp (Rosano et al. 2019). The T7 promoter is one of the most common E. coli promoters widely used to produce recombinant proteins, although its induction with isopropyl D-1β-thiogalactopyranoside (IPTG) usually leads to high level expression of foreign proteins and correspondingly formation of inclusion bodies (Li and Rinas 2020).
On the other hand, the trc promoter, a hybrid fusion of the lac and trp promoters, offers more robust regulatory capabilities and higher promoter strength than the native lac promoter (Samuelson 2011). Previous studies demonstrated that the trc promoter, though exhibiting a lower promoter strength compared to the T7 one, can potentially generate a higher quantity of soluble recombinant protein. Lee et al. reported metabolic engineering of E. coli and improvement of 2′-Fucosyllactose titers and yields following the decrease in promoter strength of α-1,2 fucosyltransferase. In their study, the trc promoter replaced the T7 promoter led to a considerable increase in the soluble expression levels of α-1,2 fucosyltransferase and subsequently improvement of ′-Fucosyllactose production in E. coli (Lee et al. 2021). Mayer and coworkers also reported that, upon the overproduction of human proteins for X-ray crystallography and NMR studies, incorporating the trc promoter, instead of the T7 promoter, with proper optimization of culture conditions can effectively increase the solubility and yield of the heterologous proteins (Mayer et al. 2004). Therefore, proper integration of genetic and bioprocessing strategies appears to be crucial for more efficient soluble expression of recombinant proteins.
Traditionally, optimizing cultivation conditions for recombinant protein expression involves altering one variable at a time (Bezerra et al. 2008). However, this approach is not only time-consuming but also leads to misinterpretation of results due to potential uncharacterized interactions among several explanatory variables (e.g., temperature, pH of the culture medium, and inducer concentration). Several alternative methods exist to analyze multiple variables simultaneously, such as full factorial and fractional factorial design (Lee et al. 2006). Response Surface Methodology (RSM) is a suitable approach for effectively determining the interactions among experimental/operating variables and relying on the simultaneous variation of multiple factors in the designated design, resulting in more reliable prediction of the optimal conditions (Liu et al. 2018). Box-Behnken design and central composite design are both widely used in RSM for experimental design and optimization. Box-Behnken Design involves three levels of each factor and requires fewer runs and delivers maximal information compared to central composite design. Its robustness against experimental error and noise makes it suitable for systems with potential variability, especially in well-defined design spaces with linear relationships between factors and response (Beg and Akhter 2021).
In our previous study (Shariaty Vaziri et al. 2023), a novel anti-HER2 immunotoxin containing a modified Pseudomonas toxin (PE35KDEL) was developed. This fusion protein (i.e., scFv-PE35KDEL) was expressed using the pET28a vector containing the T7 promoter, resulting in significant formation of inclusion bodies with only 10% of the resulting proteins being soluble. Adjusting the culture conditions (e.g., temperature and inducer concentration) barely improved the soluble expression of the recombinant protein (Shariaty Vaziri et al. 2023). As mentioned above, the trc promoter has been successfully applied to enhance soluble expression of several recombinant proteins in E. coli (Mayer et al. 2004; Lee et al. 2021). Therefore, in the current study, the immunotoxin was expressed under the regulation of the trc promoter. Moreover, based on the previous studies (Muntari et al. 2012; Shariati et al. 2022), the cultivation conditions, such as inducer concentration, culture temperature, growth medium, and induction timing, were optimized using Box–Behnken design with RSM to improve the soluble expression of this immunotoxin.
Materials and methods
Bacterial strains, plasmids, and oligonucleotides
A list of bacterial strains, plasmids, and oligonucleotides used in this study is provided in Table 1. Standard recombinant DNA technologies were applied for molecular cloning (Hofnung 1993). Phusion HF was obtained from New England Biolabs (Ipswich, MA, USA). All synthesized oligonucleotides and gBlocks were obtained from Integrated DNA Technologies (Coralville, IA, USA). E. coli HI-Control 10G chemically competent cell (Lucigen, Middleton, WI, USA) was used as the host for plasmid cloning and propagation. E. coli BL21(DE3) was used as the expression host to produce recombinant anti-HER2 immunotoxin.
Table 1.
Bacterial strains, plasmids, and primers used in this study
| Name | Description or relevant genotype | Source |
|---|---|---|
| E. coli host strains | ||
| HI-Control 10G | mcrA, ∆(mrr-hsdRMS-mcrBC), endA1, recA1,ϕ80dlacZ∆M15, ∆lacX74, araD139, ∆(ara leu)7697, galU, galK, rpsL (StrR), nupG, λ−, tonA, Mini-F lacIq1 (GentR) | Lucigen |
| BL21(DE3) | F– ompT gal dcm lon hsdSB(rB–mB–) λ(DE3 [lacI lacUV5-T7p07 ind1 sam7 nin5]) [malB+]K−12(λS) | Lab stock |
| Plasmids | ||
| pTrc99a | pBR322 ori, Ampr | Amann et al. (1988) |
| pTrc99a-HER2 | pBR322 ori, Ampr, Ptrc:: scFv-PE35KDEL | This study |
| Primers | ||
| pTrc99a-HER2F | CATCACAAGGATGAGCTTTAAGGCTGTTTTGGCGGATGAGAG | This study |
| pTrc99a-HER2R | TTCCACTAACTGTACTTCCATTTATTCTTCCTCCTTTTTAAAGTTAATGCTAGC | This study |
The synthetic gene of scFv-PE35KDEL encoding anti-HER2 immunotoxin (GenBank accession number: PP759094) was codon-optimized and synthesized by Integrated DNA Technologies (Coralville, IA, USA) in two fragments with 21 bp overlap. These two DNA fragments were Gibson–assembled with the PCR-linearized pTrc99a as the backbone, which was amplified using the primer set pTrc99a-HER2F/pTrc99a-HER2R. It is noteworthy that the original ribosome-binding site (RBS) in pTrc99a was substituted with a strong RBS sequence, AAGGAG. Additionally, transcription of the clone synthetic gene was under the regulation of the trc promoter (Supplementary Fig. 1).
Characterization of expression of anti-HER2 immunotoxin in E. Coli
The overnight-grown seed culture of BL21(DE3) harboring pTrc99a-HER2 was transferred to fresh LB medium with an inoculation ratio of 10% v/v. The resulting culture was kept at 37 ºC until reaching the logarithmic phase. Then, the protein expression was induced by adding 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) with subsequent incubation at 180 rpm at 37 ºC for 3 h (Malekian et al. 2019).
Experimental design for bacterial expression and analysis of recombinant immunotoxin
A three-level four-factor Behnken experimental design was applied to statistically investigate the effects of four independent cultivation variables on the soluble expression of anti-HER2 immunotoxin in E. coli BL21(DE3). The input factors include IPTG concentration (at three levels: 0.1, 0.55 and 1 mM), post-induction time (at three levels: 2, 10 and 18 h), post-induction temperature (at three levels: 23–37 ºC), and medium recipe (at three categories: LB, 2XYT, and TB), whereas the quantitative factor levels were coded as − 1 (low level), 0 (central point level), and 1 (high level) (Table 2). As a result, 45 cultivation runs with three replications (i.e., runs 4, 7, and 19 (for LB), 11, 13, and 28 (for 2xYT), and 3, 29, and 42 (for TB)) at center point were conducted in parallel (Table 3). For LB medium, runs 4, 5, 6, 7, 14,18, 19, 21, 25, 30, 33, 34, 35, 39, and 43 were performed simultaneously from one seed culture (i.e., overnight culture in LB). The same seed culture (i.e., overnight culture in TB) was used to prepare samples of runs 3, 8, 9, 12, 16, 22, 27, 29, 31, 36, 37, 41, 42, 44, and 45 for TB medium. For 2xYT medium, runs 1, 2, 10, 11, 13, 15, 17, 20, 23, 24, 26, 28, 32, 38, and 40 were performed simultaneously from one seed culture (i.e., overnight culture in 2xYT).
Table 2.
The variables of cultivation conditions optimized for soluble expression of anti-HER immunotoxin and their symbols and levels (coded and actual values) used in the experimental design
| Symbol | Variable | Levels | Unit | ||
|---|---|---|---|---|---|
| − 1 | 0 | 1 | |||
| A | Post-induction time | 2 | 10 | 18 | h |
| B | Post-induction temperature | 23 | 30 | 37 | °C |
| C | IPTG concentration | 0.1 | 0.55 | 1 | mM |
| D | Culture medium | LB | 2xYT | TB | - |
Table 3.
Box–Behnken experimental design of 4 factors (A: post-induction time, B: post-induction temperature, C: IPTG concentration and D: culture meium) in 3 levels and corresponding response (soluble anti-HER2 immunotoxin) values
| Run | A (h) | B (°C) | C (mM) | D | Response (soluble immunotoxon (µg/ml)) | Total immunotoxon (µg/ml) |
|---|---|---|---|---|---|---|
| 4 | 10 | 30 | 0.55 | LB | 199.62 | 856.566 |
| 5 | 18 | 30 | 1 | LB | 98.123 | 996.526 |
| 6 | 10 | 23 | 0.1 | LB | 234.928 | 795.566 |
| 7 | 10 | 30 | 0.55 | LB | 185 | 897.311 |
| 14 | 2 | 30 | 1 | LB | 85 | 380.011 |
| 18 | 18 | 30 | 0.1 | LB | 259.535 | 1127.181 |
| 19 | 10 | 30 | 0.55 | LB | 172.212 | 842.26 |
| 21 | 18 | 23 | 0.55 | LB | 120 | 888.344 |
| 25 | 10 | 37 | 1 | LB | 108.271 | 823.12 |
| 30 | 2 | 23 | 0.55 | LB | 170.671 | 496.11 |
| 33 | 10 | 37 | 0.1 | LB | 173.21 | 886.522 |
| 34 | 10 | 23 | 1 | LB | 136.384 | 645.93 |
| 35 | 2 | 37 | 0.55 | LB | 65.743 | 573.077 |
| 39 | 2 | 30 | 0.1 | LB | 161.417 | 609.65 |
| 43 | 18 | 37 | 0.55 | LB | 190.667 | 1469.989 |
| 3 | 10 | 30 | 0.55 | TB | 166.499 | 950.44 |
| 8 | 18 | 37 | 0.55 | TB | 178.667 | 986.127 |
| 9 | 18 | 30 | 1 | TB | 196.321 | 1339.70 |
| 12 | 2 | 30 | 0.1 | TB | 155.118 | 477.37 |
| 16 | 2 | 30 | 1 | TB | 80.921 | 550.639 |
| 22 | 18 | 23 | 0.55 | TB | 80 | 972.569 |
| 27 | 2 | 37 | 0.55 | TB | 40.554 | 726.31 |
| 29 | 10 | 30 | 0.55 | TB | 178 | 877 |
| 31 | 10 | 37 | 1 | TB | 97.654 | 993.46 |
| 36 | 10 | 23 | 1 | TB | 72.028 | 758.212 |
| 37 | 2 | 23 | 0.55 | TB | 180.922 | 849.65 |
| 41 | 10 | 23 | 0.1 | TB | 152.623 | 786.12 |
| 42 | 10 | 30 | 0.55 | TB | 156 | 684.883 |
| 44 | 10 | 37 | 0.1 | TB | 76.923 | 459.666 |
| 45 | 18 | 30 | 0.1 | TB | 63.756 | 1074.565 |
| 1 | 10 | 23 | 0.1 | 2xYT | 200.919 | 797.16 |
| 2 | 10 | 37 | 0.1 | 2xYT | 63.93 | 282.69 |
| 10 | 18 | 23 | 0.55 | 2xYT | 205 | 870.436 |
| 11 | 10 | 30 | 0.55 | 2xYT | 191.914 | 684.21 |
| 13 | 10 | 30 | 0.55 | 2xYT | 173 | 665 |
| 15 | 18 | 30 | 1 | 2xYT | 223.746 | 1293.674 |
| 17 | 18 | 30 | 0.1 | 2xYT | 167.631 | 984.713 |
| 20 | 2 | 37 | 0.55 | 2xYT | 22.418 | 407.99 |
| 23 | 2 | 23 | 0.55 | 2xYT | 114.134 | 480.111 |
| 24 | 2 | 30 | 1 | 2xYT | 70.324 | 411.862 |
| 26 | 10 | 37 | 1 | 2xYT | 115.249 | 911.36 |
| 28 | 10 | 30 | 0.55 | 2xYT | 180 | 634.28 |
| 32 | 18 | 37 | 0.55 | 2xYT | 174.668 | 1258.144 |
| 37 | 2 | 23 | 0.55 | TB | 180.922 | 849.65 |
| 40 | 2 | 30 | 0.1 | 2xYT | 152 | 740.11 |
Data analysis of the experimental design was carried out using the Design Expert software (version 13.0.5.0, StatEase Inc., Minneapolis, USA) to develop a model. The good fit of the model was confirmed by measuring the difference between actual and predicted values for each point (i.e. studentized residuals).
After protein expression for each cultivation, bacterial cells were collected by centrifugation at 3,500 g for 20 min, and the resulting cell pellet was resuspended in phosphate-buffered saline (PBS). For cell disruption, the suspension was sonicated for five cycles of 1 min with an interval of 1 min on ice between cycles. The soluble and insoluble fractions of the cell were separated using centrifugation at 7,500 g and 4 °C for 15 min.
To evaluate the expression level and solubility of the expressed anti-HER2 immunotoxin, the soluble fraction of the sonicated culture sample, insoluble and total protein fraction were analyzed using a 12% (w/v) sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The intensity of the corresponding bands, soluble and total proteins, were assessed by Gel Analyzer 19.1 software (Lazar Software, Hungary) and the amount of the target protein was estimated based on the standard protein (i.e., bovine serum albumin) (Table 3). Then, the percentage ratio of the soluble and the insoluble protein to the total protein was determined (Table 2).
The soluble fraction of the culture sample was used to purify anti-HER2 immunotoxin using affinity chromatography under native conditions, as described in our previous study with some modifications (Akbari et al. 2015a). The protein concentration in each purification fraction was determined using the Bradford method (Malekian et al. 2019).
Biological activity of anti-HER2 immunotoxin through cell-based ELISA
To evaluate the bioactivity of the purified anti-HER2 immunotoxin, two cancer cell lines, i.e., the HER2-overexpressing cell line of SK-OV-3 and the HER2-low-expressing cell of MCF-7, were seeded in a 96-well ELISA plate at a cell density of 2 × 105 cells/ml and 37 ºC overnight. The cells were rinsed with PBS and then immobilized with 4% formalin buffer for 20 min. After that, the plate was washed thrice with PBS and then treated with 1% bovine serum albumin in PBS for 2 h at the room temperature. Then, samples containing anti-HER2 immunotoxin in a serial dilution were added to individual wells and incubated for 2 h at 25 ºC with gentle shaking. Four wells were only treated with PBS (i.e., no immunotoxin was added) as negative control. Each well was rinsed for three times with 0.05% Tween 20 in PBS. HRP-conjugated anti-His antibody (1: 50,000 dilution) was added to each well for incubation for 2 h at the room temperature with gentle shaking. After washing three times, OPD substrate was added to each well for incubation for 20 min. The colorimetric reaction was terminated by addition of 10 µl of 2M H2SO4 to each well, and the absorbance was measured at 450 and 490 nm (Akbari et al. 2014).
Results
Small-scale expression of anti-HER2 immunotoxin
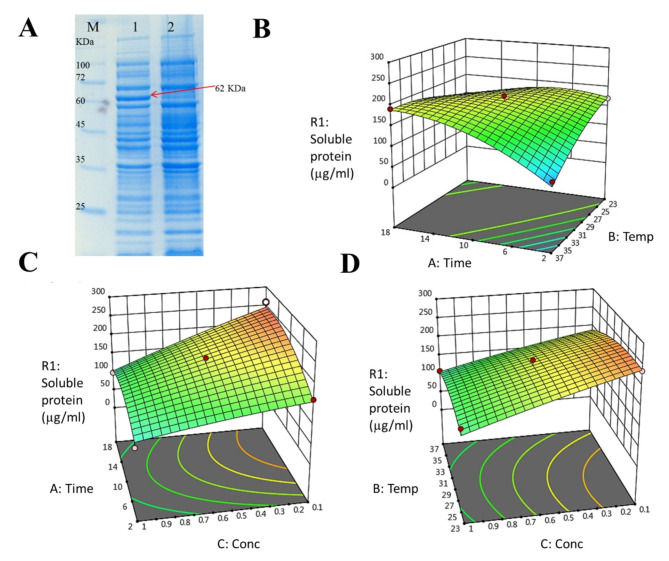
E. coli BL21 (DE3) harboring pTrc99a-HER2 was used to express anti-HER2 immunotoxin while being cultivated at 37 ºC in LB medium for 3 h with a general induction condition at 1 mM IPTG. SDS-PAGE analysis suggested that the full-length immunotoxin was successfully expressed with an envisioned molecular weight of ~ 62 kDa (Fig. 1A). However, most of the expressed immunotoxin appeared to be in the insoluble fraction as inclusion bodies (approximately 95%). The results nevertheless confirmed the proper design of the expression plasmid and the expression strategy.
Fig. 1.
A SDS-PAGE analysis of total protein from transformed Escherichia coli BL21(DE3) with scFv-PE35KDEL-pTrc99a (lane 1) and E. coli non-transformed (lane 2) after induction with 1 mM IPTG at 37 ºC in LB medium for 3 h. M: protein marker (PM1500, SMOBIO). The arrow indicates the band of interest. B–D 3D response surface plot representing the interaction between two variables on the soluble expression of anti-HER2 immunotoxin (R1: response) in LB medium. B Interaction between post-induction time and post-induction temperature when IPTG concentration was 0.55 mM, C interaction between post-induction time and IPTG concentration when post-induction temperature was 30 ºC, and D interaction between post-induction temperature and IPTG concentration when post-induction time was 10 h
Correlation of cultivation conditions using the RSM
The effect of the four operating factors (i.e., inducer concentration, temperature, induction timing, and medium composition) of the cultivation on the soluble expression of anti-HER2 immunotoxin was characterized using RSM. A total of 45 cultivations was conducted simultaneously, with soluble expression of anti-HER2 immunotoxin ranging between 22.4 and 260 µg/ml (Table 3). The results suggest the potential importance of optimization of cultivation conditions. To assess the experimental error and model reproducibility, cultivation was further carried out in triplicate under three central point conditions (i.e., 0.55 mM IPTG, 10 h post-induction time, and 30 °C in the culture medium of LB (runs 4, 7, and 19), 2xYT (11, 13, and 28) or TB (3, 29, and 42)). The anti-HER2 immunotoxin concentrations were determined to be 184 ± 14, 182 ± 10 and 167 ± 1 µg/ml, respectively, with the coefficient of variation (i.e., standard deviation/mean×100%) being less than 7.68, 5.26 and 6.59%, for LB, 2xYT, and TB cultures, respectively (Table 3).
The data of anti-HER2 immunotoxin concentrations obtained from the 45 experiments (Table 3) were further analyzed by the Design Expert software (Akbari et al. 2015b). As culture medium (D) is not a quantitative parameter (i.e., it is a categorical or nominal variable), the optimum level of other three quantitative parameters (i.e., A, B and C) for each culture medium was found. Correlation between the level of soluble anti-HER2 immunotoxin expression in each medium culture and the three independent variables was formulated based on the following quadratic regression equations:
Soluble anti-HER2 immunotoxin expression in LB medium (µg/ml) = 141 − 7.84(A: Time) + 10.9(B: Temp) − 110(C: Conc) + 0.784(A: Time)(B: Temp) − 5.90(A: Time)(C: Conc) + 2.67(B: Temp)(C: Conc) − 0.477(A: Time)² − 0.374(B: Temp)² − 20.2(C: Conc)².
Soluble anti-HER2 immunotoxin expression in 2xYT medium (µg/ml) = -261.769 -4.702(A: Time) + 41.040(B: Temp) -411.109(C: Conc) + 0.274(A: Time)(B: Temp) + 9.569(A: Time)(C: Conc) + 13.414(B: Temp)(C: Conc) − 0.117(A: Time)² − 0.920(B: Temp)² − 102.318 (C: Conc)².
Soluble anti-HER2 immunotoxin expression in TB medium (µg/ml) = -21.522–35.427(A: Time) + 26.748(B: Temp) -214.053 C + 1.067(A: Time)(B: Temp) + 14.358(A: Time)(C: Conc) + 8.042(B: Temp)(C: Conc) − 0.176(A: Time)2 − 0.725(B: Temp)² -155.636(C: Conc)².
The fitting quality of the quadratic regression model was determined using analysis of variance (ANOVA) and the P values were applied to evaluate the significance of each coefficient (Table 4). The correlation coefficient (R²) of the regression model was 0.993, suggesting that the model could cover and predict 99.3% of the variation in the response. The F-value of 14.7 with a low probability value (P < 0.0001) indicates that the model is significant in terms of predicting soluble expression of anti-HER2 immunotoxin based on cultivation of the engineered E. coli. On the other hand, the lack of fit for the model was insignificant (P > 0.05) with an F value of 3.79, suggesting that there could be a 8.03% probability for a Lack of Fit F-value to appear as a result of the noise. The results indicate that the model could fit the experimental data properly.
Table 4.
Analysis of variance (ANOVA) and regression coefficients of the developed response surface quadratic model for soluble expression of anti-HER2 immunotoxin regarding the influence of post-induction time (A), post-induction temperature (B), IPTG concentration (C) and culture medium (D)
| Source | Sum of squares | Df | Mean square | F-value | p-value | |
|---|---|---|---|---|---|---|
| Model | 1.36E + 05 | 29 | 4674.13 | 14.74 | < 0.0001 | Significant |
| A-Time | 18089.11 | 1 | 18089.11 | 57.03 | < 0.0001 | Significant |
| B-Temp | 8172.32 | 1 | 8172.32 | 25.77 | 0.0001 | Significant |
| C-Conc | 10199.15 | 1 | 10199.15 | 32.16 | < 0.0001 | Significant |
| D-Culture | 7851.57 | 2 | 3925.79 | 12.38 | 0.0007 | Significant |
| AB | 18882.44 | 1 | 18882.44 | 59.54 | < 0.0001 | Significant |
| AC | 5614.2 | 1 | 5614.2 | 17.7 | 0.0008 | Significant |
| AD | 7915.93 | 2 | 3957.96 | 12.48 | 0.0006 | Significant |
| BC | 7698.9 | 1 | 7698.9 | 24.27 | 0.0002 | Significant |
| BD | 1246.78 | 2 | 623.39 | 1.97 | 0.1745 | Not significant |
| CD | 10989.26 | 2 | 5494.63 | 17.32 | 0.0001 | Significant |
| A² | 2990.26 | 1 | 2990.26 | 9.43 | 0.0078 | Significant |
| B² | 12045.68 | 1 | 12045.68 | 37.98 | < 0.0001 | Significant |
| C² | 3903.63 | 1 | 3903.63 | 12.31 | 0.0032 | Significant |
| ABD | 4052.39 | 2 | 2026.19 | 6.39 | 0.0098 | Significant |
| ACD | 11626.06 | 2 | 5813.03 | 18.33 | < 0.0001 | Significant |
| BCD | 2292.19 | 2 | 1146.09 | 3.61 | 0.0524 | Not significant |
| A²D | 1124.51 | 2 | 562.25 | 1.77 | 0.2036 | Not significant |
| B²D | 1357.49 | 2 | 678.75 | 2.14 | 0.1522 | Not significant |
| C²D | 1410.5 | 2 | 705.25 | 2.22 | 0.1426 | Not significant |
| Residual | 4757.38 | 15 | 317.16 | |||
| Lack of Fit | 3956.16 | 9 | 439.57 | 3.29 | 0.0803 | Not significant |
| Pure Error | 801.22 | 6 | 133.54 | |||
| Cor Total | 1.40E + 05 | 44 |
Evaluation of the effects of cultivation variables for optimization
As illustrated in Table 4, the effects of all four cultivation parameters on soluble expression of anti-HER2 immunotoxin were significant (P < 0.001) though the culture medium (D) had a less significant effect (P = 0.0007). Additionally, the interaction terms of AB, AC, AD, BC, and CD were shown to have a significant impact on the soluble expression of anti-HER2 immunotoxin (P < 0.05), suggesting their potential combinatory (dual interaction) effects on the cultivation performance. However, the interaction of post-induction time and culture medium (BD) was insignificant. Moreover, square interactions as A², B², C², and triple interactions asABD and ACD are other significant model terms (P < 0.01).
The effects of three variables (i.e., IPTG concentration, post-induction time, and post-induction temperature) and their interactions and effects on the soluble expression of anti-HER2 immunotoxin in the LB culture (i.e., the optimal medium) were graphically shown in the 3-dimensional response surface plots (Fig. 1). Figure 1B shows the response surface plot associated with the interaction and effects of post-induction time and post-induction temperature on the soluble expression of anti-HER2 immunotoxin when the IPTG concentration was maintained at its central point level (0.55 mM). Clearly, soluble expression of the protein was enhanced with increasing time and decreasing temperature. As presented in Fig. 1C, soluble expression of anti-HER2 immunotoxin increases remarkably with increasing time and decreasing IPTG concentration when the post-induction temperature was 30 ºC (i.e., central point level). Figure 1D further shows the response surface plot of the effects of post-induction temperature and IPTG concentration on soluble expression of the protein when the post-induction time was 10 h (i.e., central point level). Clearly, soluble expression of the protein was enhanced with decreasing IPTG concentration and temperature. Overall, we found that increasing the post-induction time and decreasing temperature and IPTG concentration would improve soluble protein expression yield in LB medium. As shown in Table 3, among 45 runs the highest yield of total protein (i.e., both soluble and insoluble fractions) production was obtained with 0.55 mM IPTG at 37 ºC for 18 h in LB medium. Run 30 (i.e. 0.55 mM IPTG at 23 ºC for 2 h in LB medium) showed the most proportion of soluble to insoluble immunotoxin (i.e., 34.4/65.6). Run 20 (0.55 mM IPTG at 37 ºC for 2 h in 2xYT medium) exhibited the lowest proportion of soluble to insoluble immunotoxin (i.e., 5.5/94.5).
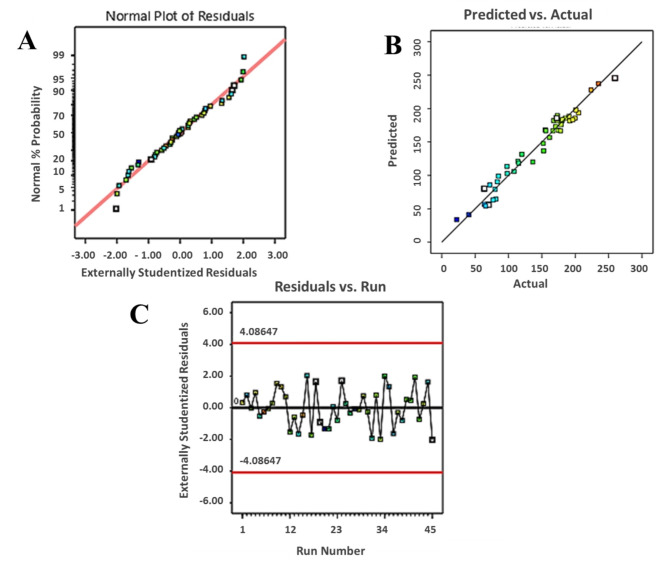
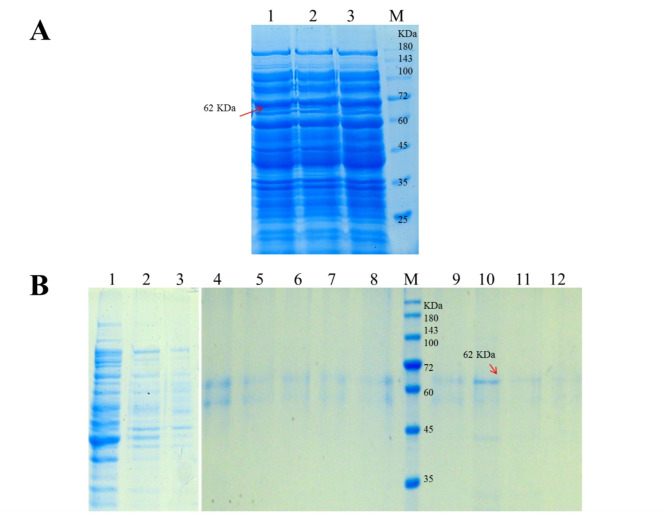
The studentized residuals provide the range of standard deviations associated with the difference between the actual and predicted values. As shown in Fig. 2A, the plot of normal probability for soluble expression of anti-HER2 immunotoxin shows a normal and linear distribution, revealing no outlier run in the considered range. The straight line shows that no response transformation was needed. Figure 2B further shows that the actual and predicted levels for soluble expression of anti-HER2 immunotoxin are in reasonable agreement. Additionally, the plot of residuals versus the experimental runs remains within the boundaries specified in Fig. 2C. These statistical analyses collectively indicate that all data points follow a normal pattern and that the established model is adequate for the prediction of the cultivation performance for soluble expression of anti-HER2 immunotoxin. Finally, according to the model, the optimal solution with the highest soluble expression of the protein (261 µg/ml) would be achieved with 0.1 mM IPTG in LB medium culture at 33 °C for 18 h. To verify the predicted optimum condition, three independent experiments was carried out; in average, 272 ± 7.3 µg/ml of soluble anti-HER2 immunotoxin was expressed and this level was rather close to the predicted optimum (261 µg/ml) (Fig. 3A).
Fig. 2.
Normal (%) probability plot of the ‘studentized’ residuals (A), linear regression plot for the relationship between the predicted and actual values (B) and the residuals versus run number plot (C) for the model of soluble expression of anti-HER2 immunotoxin
Fig. 3.
A SDS-PAGE analysis of soluble protein fraction from three repeats (lane 1–3) of experiments under the optimum conditions; M: protein marker (PM1500, SMOBIO). The arrow indicates the band of interest. B SDS–PAGE analysis of the purified anti-HER2 immunotoxin using a Ni–NTA column under native conditions. Lane 1: soluble fraction before loading onto the column; lane 2–5: resin wash with 20 mM imidazole; lane 6–8: resin wash with 40 mM imidazole; lane 9, 10, 11, and 12: eluted fractions with 100, 200, 400, and 600 mM imidazole, respectively; M: protein marker (PM1500, SMOBIO). The arrow indicates the band of interest
Purification and evaluation of the biological activity of anti-HER 2 immunotoxin
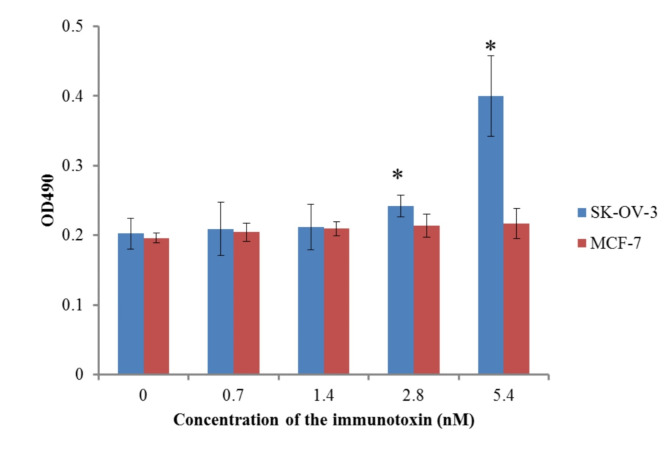
After the cultivation for the expression of anti-HER2 immunotoxin under the optimal condition, the fusion protein was successfully purified by affinity chromatography using a Ni-NTA column under native conditions (Fig. 3B). Out of the 1-L of bacterial culture, approximately 27.2 mg of anti-HER2 immunotoxin (340 µg/ml) was recovered, the percentage yield of purified protein obtained from the soluble fraction was found to be approximately 40%.and the purity was determined to be 90%. Using this purified protein, the biological activity was further evaluated using an ELISA assay. As shown in Fig. 4, the attachment of anti-HER2 immunotoxin to SK-OV-3 cells (HER2-ovrexprssing cell line) was successfully demonstrated in a dose-dependent manner. Anti-HER2 immunotoxin can bind to MCF-7 at a lower level than SK-OV-3, showing the specificity of binding of the protein to HER2. The significant difference in the binding of anti-HER2 immunotoxin to SK-OV-3 and MCF-7 was more observable when a high concentration of the protein was used.
Fig. 4.
Binding assay of anti-HER2 immunotoxin to SK-OV-3 (i.e., HER2-overexpressing cell line) and MCF-7 (i.e., HER2- low expressing cell line) cells using ELISA. The cells were treated with different concentration of anti-HER2 immunotoxin and HRP-anti-His antibody was added for binding detection. Asterisks show the means which were significantly different (P < 0.05) from cells treated without the immunotoxin (negative control). (mean SD, n = 4)
Discussion
The previous study demonstrated that anti-HER2 immunotoxin can be considered as a promising candidate for HER2-targetd cancer therapy (Shariaty Vaziri et al. 2023). However, large-scale production of this immunotoxin was limited by the accumulation of insoluble inclusion bodies upon cultivation of the engineered E. coli strain with a transformed immunotoxin-encoded gene whose expression was regulated by the T7 promoter. One approach to enhance soluble expression of recombinant proteins is to modulate, not necessarily increase, promoter strength. In the present study, it was demonstrated that proper integration of the genetic (i.e., using the trc promoter to replace the T7 one) and bioprocessing (i.e., optimization of cultivation conditions) strategies can potentially lead to a remarkable cultivation performance for more soluble expression of the recombinant anti-HER2 immunotoxin. Tegel et al. evaluated the effects of three different IPTG-inducible promoters (i.e., T7, trc, and lacUV5) for expression of seventeen fusion proteins in E. coli (Tegel et al. 2011). They observed that the use of the most potent promoter (i.e., T7 with the highest promoter strength) often led to an ineffective protein production with a low soluble fraction. Furthermore, such technical issue could be partially alleviated based on the use of the trc or T7 promoter in combination with E. coli Rosetta (DE3) as the expression host, resulting in a higher quantity of soluble protein. Balzer et al. compared three different promoter systems (LacI/PT7lac, LacI/Ptrc, AraC/PBAD) for regulated expression of five genes (Balzer et al. 2013). It was observed that the LacI/PT7lac system, while generating a high-level of mRNA transcript, produced the highest amount of insoluble protein. They also observed that the LacI/Ptrc system had a more effective initiation upon protein expression with a more homogeneous protein expression profile. Instead of modifying promoter strength, Tae Byun et al. improved soluble production of an anti-HER scFv via N-terminal fusion of maltose-binding protein. They used nickel-affinity chromatography to purify the protein, and similar to our study, the obtained protein successfully bound to SK-OV-3 cells (Byun et al. 2023). Cultivation condition can potentially influence both the expression and solubility of recombinant proteins and, therefore, should be properly evaluated. The use of statistical design enables not only determination of the optimal culture condition but also characterization of the interactions amongst numerous experimental parameters and operating variables (Behravan and Hashemi 2021). In this study, RSM was specifically applied to optimize the cultivation performance for soluble expression of anti-HER2 immunotoxin under the regulation of the trc promoter in E. coli and it was also demonstrated satisfactory predictability based on the established model. Similar application of RSM for optimization of recombinant protein production was previously reported (Shafiee et al. 2017). For example, Shafiee et al. applied RSM to optimize the expression of recombinant DT386-BR2 in E. coli Rosetta (DE3). They reported that the induction with 1 mM IPTG for 2 h at 37 °C would result in higher total protein expression of the immunotoxin (Shafiee et al. 2017). All these reports suggest the importance of optimization of induction conditions for bacterial expression of recombinant immunotoxins. In our work, among 45 runs the highest soluble expression of the protein was achieved with 0.1 mM IPTG in LB medium culture at 30 °C for 18 h (i.e., run 18). The developed model predicted that the optimum condition for soluble expression of the protein would be 0.1 mM IPTG in LB medium culture at 33 °C for 18 h which was confirmed by three independent experiments. Medium composition could have remarkable metabolic effects on cell growth and gene expression upon conducting E. coli cultivation (Kram and Finkel 2015). It was previously observed that using the TB medium would result in more production of recombinant DT386-BR2, compared to LB and superbroth (SB) (Shafiee et al. 2017). On the other hand, Heydari et al. used the Taguchi method to optimize the medium composition, in particular carbon and nitrogen sources of chemically defined medium, for the expression of Denileukin diftitox in E. coli (Heydari et al. 2014) Handayani et al. compared four autoinduction media, LB, 2XYT, TB, and Imperial College London (IC), for soluble expression of moloney murine leukemia virus-reverse transcriptase in E. coli BL21 star (DE3). They reported that IC and LB media resulted the highest yield of protein production (Handayani et al. 2024) While nutrient components with high energy and carbon levels could potentially enhance biomass production and recombinant protein expression, their effects on the soluble production of recombinant proteins have been controversial. Some studies showed that the use of culture medium with higher yeast extract and tryptone contents could result in more soluble expression of the target protein (Kanno et al. 2019; Aghdam et al. 2022), while others found the opposite (Kim et al. 2017). Kanno et al. reported that the use of enriched media, such as TB, potentially resulted in more soluble expression of recombinant Zika virus ΔNS1 in E. coli (Aghdam et al. 2022). Though it was observed more soluble expression of anti-HER2 immunotoxin in E. coli grown in a less rich medium (i.e., LB medium), the current results are in agreement with the previous report comparing three media (i.e., LB, TB, 2xYT) for the expression of cis-aconitate decarboxylase in E. coli BL21(DE3) (Kim et al. 2017). Basically, Kim et al. reported that, though a higher cell density could be obtained in TB medium, more enzyme production with a higher yield was obtained in LB medium (Kim et al. 2017). This might be attributable to the metabolic load induced by enhanced cell growth that could promote the formation of inclusion bodies. Our finding showed that post-induction time was also critical for soluble expression of anti-HER2 immunotoxin, especially under a low IPTG induction concentration (i.e., 0.1 mM). Other researchers also manipulated the inducer concentration and induction timing to reduce metabolic load for the producing cells (Aghdam et al. 2022). Reducing the metabolic stress based on the use of a low inducer concentration could potentially mediate a metabolic balance between cell growth and recombinant protein expression in E. coli, improving the solubility and folding of the recombinant protein (Papaneophytou et al. 2013).
Cultivation temperature is one of the most important factors influencing the expression and solubility of recombinant proteins. While the optimal temperature for soluble expression of anti-HER2 immunotoxin was 33 ºC in this study, more soluble expression of recombinant proteins at a temperature around 30 ºC was observed previously (Mühlmann et al. 2017). Another study evaluated the effect of induction conditions (i.e., temperature, time and IPTG cocncetration) on the soluble expression of anti-HER2 scFv in two E. coli strains, BL21(DE3) and SHuffleT7 (Ahmadzadeh et al. 2020). They used altering one variable at a time and did not applied design of experiments such as RSM. They reported the higher level of soluble protein in BL21(DE3) strain was achieved at lower induction temperature (25 ºC) as compared to 37 ºC. However, the soluble expression of anti-HER2 scFv in SHuffle T7 was not significantly influenced by the temperature. It has been perceived that recombinant proteins produced at a low temperature might have higher solubility and bioactivity potentially because of more functional expression of molecular chaperones. However, such temperature effects may not always hold and should be further studied for different recombinant proteins. Some studies reported that higher temperatures promote soluble expression of recombinant proteins. In particular, Vincentelli et al. reported that recombinant protein expression at 37 ºC yielded more soluble protein than 25 ºC and 17 ºC (Vincentelli et al. 2011). Such controversy could be potentially associated with different recombinant protein properties, expression vectors, and expression hosts.
From our study, the Box-Behnken Design can be applied to optimize the multivariable cultivation conditions for soluble expression of other proteins with similar features. More characterization, purification, and functional researches are still required to evaluate proper folding and biological activities of anti-HER2 immmunotoxin. Further in vitro and in vivo studies are needed to demonstrate the clinical application of the immunotoxin.”
In conclusion, RSM was successfully applied to optimize cultivation conditions for soluble expression of anti-HER2 immunotoxin under the regulation of the trc promoter in E. coli such that a sufficient amount of biologically active and soluble recombinant protein could be obtained for subsequent in vitro and in vivo trials. The results suggest that inducer concentration, medium recipe, post-induction temperature, and post-induction time have significant effects on the cultivation performance for soluble expression of anti-HER2 immunotoxin in E. coli. The optimal conditions were determined to be 0.1 mM IPTG for induction of gene expression at 33 °C for 18 h in the LB medium. The approaches documented in this study can be generally applied to enhance the expression yield and solubility of recombinant proteins in E. coli.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We are grateful to thank Mrs Fatemeh Moazen for her excellent technical assistance.
Funding
This study was financially supported by the Isfahan University of Medical Sciences (Grant No. 199495).
Data availability
Data will be shared upon request.
Declarations
Competing interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Vajihe Akbari, Email: v_akbari@pharm.mui.ac.ir, Email: akbarivajihe@yahoo.com.
C. Perry Chou, Email: cpchou@uwaterloo.ca.
References
- Aghdam MA, Tohidkia MR, Ghamghami E, Ahmadikhah A, Khanmahamadi M, Baradaran B, Mokhtarzadeh A (2022) Implementation of a design of experiments to improve periplasmic yield of functional ScFv antibodies in a phage display platform. Adv Pharm Bull 12(3):583–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmadzadeh M, Farshdari F, Nematollahi L, Behdani M, Mohit E (2020) Anti-HER2 scFv expression in Escherichia coli SHuffle®T7 express cells: effects on solubility and biological activity. Mol Biotechnol 62(1):18–30. 10.1007/s12033-019-00221-2 [DOI] [PubMed] [Google Scholar]
- Akbari V, Mir Mohammad Sadeghi H, Jafrian-Dehkordi A, Abedi D, Chou CP (2014) Functional expression of a single-chain antibody fragment against human epidermal growth factor receptor 2 (HER2) in Escherichia coli. J Ind Microbiol Biotechnol 41(6):947–956 [DOI] [PubMed] [Google Scholar]
- Akbari V, Sadeghi HM, Jafarian-Dehkordi A, Abedi D, Chou CP (2015a) Improved biological activity of a single chain antibody fragment against human epidermal growth factor receptor 2 (HER2) expressed in the periplasm of Escherichia coli. Prot Exp Purif 116:66–74 [DOI] [PubMed] [Google Scholar]
- Akbari V, Sadeghi HM, Jafarian-Dehkordi A, Chou CP, Abedi D (2015b) Optimization of a single-chain antibody fragment overexpression in Escherichia coli using response surface methodology. Res Pharm Sci 10(1):75–83 [PMC free article] [PubMed] [Google Scholar]
- Akbari B, Farajnia S, Ahdi Khosroshahi S, Safari F, Yousefi M, Dariushnejad H, Rahbarnia L (2017) Immunotoxins in cancer therapy: review and update. Int Rev Immunol 36(4):207–219 [DOI] [PubMed] [Google Scholar]
- Amann E, Ochs B, Abel K-J (1988) Tightly regulated tac promoter vectors useful for the expression of unfused and fused proteins in Escherichia coli. Gene 69(2):301–315 [DOI] [PubMed] [Google Scholar]
- Balzer S, Kucharova V, Megerle J, Lale R, Brautaset T, Valla S (2013) A comparative analysis of the properties of regulated promoter systems commonly used for recombinant gene expression in Escherichia coli. Microb Cell Fact 12:26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beg S, Akhter S (2021) Box–Behnken designs and their applications in pharmaceutical product development. In: Beg, S. (eds) Design of experiments for pharmaceutical product development. Springer, Singapore. 10.1007/978-981-33-4717-5_7
- Behravan A, Hashemi A (2021) Statistical optimization of culture conditions for expression of recombinant humanized anti-EpCAM single-chain antibody using response surface methodology. Res Pharm Sci 16(2):153–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezerra MA, Santelli RE, Oliveira EP, Villar LS, Escaleira LA (2008) Response surface methodology (RSM) as a tool for optimization in analytical chemistry. Talanta 76(5):965–977 [DOI] [PubMed] [Google Scholar]
- Byun KT, Kim B, Cho J, Lee I, Lee MG, Park D, Kang TB, Won HS, Kim CG (2023) Development of an anti-HER2 single-chain variable antibody fragment construct for high-yield soluble expression in Escherichia coli and one-step chromatographic purification. Biomolecules 13(10):1508. 10.3390/biom13101508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handayani CV, Laksmi FA, Andriani A, Nuryana I, Mubarik NR, Agustriana E, Dewi KS, Purnawan A (2024) Expression of soluble moloney murine leukemia virus-reverse transcriptase in Escherichia coli BL21 star (DE3) using autoinduction system. Mol Biol Rep 51(1):628. 10.1007/s11033-024-09583-6 [DOI] [PubMed] [Google Scholar]
- Heydari M, Robatjazi SM, Zeinoddini M, Darabi E (2014) Optimization of chemically defined cell culture media for recombinant ONTAK immunotoxin production. Iran J Med Microbiol 8(3):51–57 [Google Scholar]
- Hofnung M (1993) A short course in bacterial genetics and a laboratory manual and handbook for Escherichia coli and related bacteria: edited by JH Miller, Cold Spring Harbor Laboratory Press, 1992, Laboratory Manual, p 456. Biochimie 75(6): 501
- Kanno AI, Leite LCC, Pereira LR, de Jesus MJR, Andreata-Santos R, Alves R, Durigon EL, Ferreira LCS, Gonçalves VM (2019) Optimization and scale-up production of Zika virus ∆NS1 in Escherichia coli: application of response surface methodology. AMB Express 10(1):1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Seo HM, Bhatia SK, Song HS, Kim JH, Jeon JM, Choi KY, Kim W, Yoon JJ, Kim YG, Yang YH (2017) Production of itaconate by whole-cell bioconversion of citrate mediated by expression of multiple cis-aconitate decarboxylase (cadA) genes in Escherichia coli. Sci Rep 7:39768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JS, Jun SY, Kim YS (2020) Critical issues in the development of immunotoxins for anticancer therapy. J Pharml Sci 109(1):104–115 [DOI] [PubMed] [Google Scholar]
- Kram KE, Finkel SE (2015) Rich medium composition affects Escherichia coli survival, glycation, and mutation frequency during long-term batch culture. Appl Environ Microbiol 81(13):4442–4450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KM, Rhee CH, Kang CK, Kim JH (2006) Sequential and simultaneous statistical optimization by dynamic design of experiment for peptide overexpression in recombinant Escherichia coli. Appl Biochem Biotechnol 135(1):59–80 [DOI] [PubMed] [Google Scholar]
- Lee JW, Kwak S, Liu JJ, Yun EJ, Jin YS (2021) 2’-Fucosyllactose production in engineered Escherichia coli with deletion of waaF and wcaJ and overexpression of FucT2. J Biotechnol 340:30–38. 10.1016/j.jbiotec.2021.08.007 [DOI] [PubMed] [Google Scholar]
- Li Z, Rinas U (2020) Recombinant protein production associated growth inhibition results mainly from transcription and not from translation. Microb Cell Fact 19:83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Jiang J, Yan F, Xu Y, Yang M, Gao Y, Aihemaiti A, Zou Q (2018) Optimization of simultaneous production of volatile fatty acids and bio-hydrogen from food waste using response surface methodology. RSC Adv 8(19):10457–10464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malekian R, Sima S, Jahanian-Najafabadi A, Moazen F, Akbari V (2019) Improvement of soluble expression of GM-CSF in the cytoplasm of Escherichia coli using chemical and molecular chaperones. Prot Exp Purif 160:66–72 [DOI] [PubMed] [Google Scholar]
- Mayer MR, Dailey TA, Baucom CM, Supernak JL, Grady MC, Hawk HE, Dailey HA (2004) Expression of human proteins at the southeast collaboratory for structural genomics. J Struct Funct Genom 5(1–2):159–165 [DOI] [PubMed] [Google Scholar]
- Mühlmann M, Forsten E, Noack S, Büchs J (2017) Optimizing recombinant protein expression via automated induction profiling in microtiter plates at different temperatures. Microb Cell Fact 16(1):220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muntari B, Amid A, Mel M, Jami MS, Salleh HM (2012) Recombinant bromelain production in Escherichia coli: process optimization in shake flask culture by response surface methodology. AMB Express 2:12. 10.1186/2191-0855-2-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papaneophytou CP, Rinotas V, Douni E, Kontopidis G (2013) A statistical approach for optimization of RANKL overexpression in Escherichia coli: purification and characterization of the protein. Prot Exp Purif 90(1):9–19 [DOI] [PubMed] [Google Scholar]
- Rosano GL, Morales ES, Ceccarelli EA (2019) New tools for recombinant protein production in Escherichia coli: a 5-year update. Prot Sci 28(8):1412–1422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuelson JC (2011) Recent developments in difficult protein expression: a guide to E. Coli strains, promoters, and relevant host mutations. Methods Mol Biol 705:195–209 [DOI] [PubMed] [Google Scholar]
- Shafiee F, Rabbani M, Jahanian-Najafabadi A (2017) Optimization of the expression of DT386-BR2 fusion protein in Escherichia coli using response surface methodology. Adv Biomed Res 6:22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shariati FS, Keramati M, Cohan RA (2022) Indirect optimization of staphylokinase expression level in dicistronic auto-inducible system. AMB Express 12(1):124. 10.1186/s13568-022-01464-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shariaty Vaziri Z, Shafiee F, Akbari V (2023) Design and construction of scFv-PE35KDEL as a novel immunotoxin against human epidermal growth factor receptor 2 for cancer therapy. Biotechnol Lett 45(4):537–550 [DOI] [PubMed] [Google Scholar]
- Tegel H, Ottosson J, Hober S (2011) Enhancing the protein production levels in Escherichia coli with a strong promoter. FEBS J 278(5):729–739 [DOI] [PubMed] [Google Scholar]
- Vincentelli R, Cimino A, Geerlof A, Kubo A, Satou Y, Cambillau C (2011) High-throughput protein expression screening and purification in Escherichia coli. Methods (San Diego Calif) 55(1):65–72 [DOI] [PubMed] [Google Scholar]
- Xing Y, Xu K, Li S, Cao L, Nan Y, Li Q, Li W, Hong Z (2021) A single-domain antibody-based anti-psma recombinant immunotoxin exhibits specificity and efficacy for prostate cancer therapy. Int J Mol Sci 22(11) [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be shared upon request.