Abstract
We have used vaccinia virus as a vector to clone a 22.5-kbp cDNA that represents the 5′ and 3′ ends of the human coronavirus 229E (HCoV 229E) genome, the HCoV 229E replicase gene, and a single reporter gene (coding for green fluorescent protein [GFP]) located downstream of a regulatory element for coronavirus mRNA transcription. When RNA transcribed from this cDNA was transfected into BHK-21 cells, a small percentage of cells displayed strong fluorescence. A region of the mRNA encoding GFP was amplified by PCR and shown to have the unique mRNA leader-body junction indicative of coronavirus-mediated transcription. These data show that the coronavirus replicase gene products suffice for discontinuous subgenomic mRNA transcription.
Coronaviruses are enveloped positive-strand RNA viruses with a genome size of approximately 30 kb. More than two-thirds of the genome encodes an RNA-dependent RNA-replicase, which is expressed from the viral genomic RNA (13, 21). The replicase gene is comprised of two large overlapping open reading frames (ORFs), ORF1a and ORF1b, which are translated as two polyprotein precursors, pp1a and pp1ab. The larger protein, pp1ab, is expressed by programmed (−1) ribosomal frameshifting. Extensive processing of the polyproteins by virus-encoded proteinases leads to the formation of a replication-transcription complex in the cytoplasm of the infected cell (31).
A key feature of coronaviruses is their unique transcription strategy. This strategy leads to the synthesis of a nested set of 3′ coterminal subgenomic mRNAs, encoding mainly structural proteins. It has been shown that the synthesis of each subgenomic mRNA involves a discontinuous step by which the so-called 3′ body sequence is fused to the genomic 5′ leader sequence (22). This process most probably occurs during the synthesis of subgenomic, negative-strand templates (19, 28). The fusion of leader and body sequences during discontinuous transcription is determined, at least in part, by cis-acting elements, termed transcription-associated sequences (TAS). These elements are located at the 5′ end of the genome and at 3′ proximal sites corresponding to the individual transcription units (5).
Until recently, the study of coronavirus transcription was essentially restricted to the analysis of defective RNA templates that depend upon transcriptional functions provided by a helper virus (15). Nevertheless, some general characteristics of coronavirus transcription have been revealed. Thus, it has been shown that coronavirus-specific transcripts can be generated from defective RNA templates that contain one or several TAS elements (15, 29). Also, mutagenesis of TAS elements in defective RNAs has revealed that TAS base pairing plays an important role in coronavirus discontinuous transcription (27). However, the use of defective RNAs to study coronavirus transcription has focused attention on the template RNA rather than the viral gene products that provide transcriptional functions.
Our limited knowledge of coronavirus replicase proteins has been obtained mainly by biochemical and genetic studies. For example, the in vitro analysis of recombinant proteins has shown that coronavirus replicase polypeptides have proteinase (31), nucleoside triphosphatase (NTPase) (9), and RNA duplex unwinding (20) activities. At the same time, genetic analysis of temperature-sensitive mutants that are defective in RNA synthesis has been used to correlate a limited number of specific mutations in replicase polypetides with defects in positive- and negative-strand RNA synthesis (S. Siddell, D. Sawicki, Y. Meyer, V. Thiel, and S. Sawicki, submitted for publication). These studies need to be continued and extended, but it is also clear that we need to complement them with a reverse genetic approach. Indeed, several different reverse genetic methods, including targeted RNA recombination (7, 10, 12) and the construction of infectious cDNA copies of both human and porcine coronavirus genomes (2, 24, 30), have been developed for coronaviruses in the last few years. Unfortunately, these systems are all dependent on the rescue of recombinant coronaviruses and, therefore, are not particularly suitable for the analysis of defective genomes encoding mainly replicative and transcriptional functions.
The analysis of replication and transcription in many positive-strand RNA viruses has been greatly facilitated by the use of synthetic RNAs that, once introduced in susceptible cells, are able to replicate autonomously; i.e., so-called replicons (1, 6, 8, 14). This approach has been used to study, for example, the replication and transcription of arteriviruses, a family of RNA viruses that are grouped together with the coronaviruses in the order Nidovirales. It has been demonstrated that base pairing between TAS elements guides the process of arterivirus discontinuous transcription (28) and that the arterivirus replicase, in the absence of any further structural or nonstructural proteins, is sufficient for genome replication and subgenomic mRNA transcription (17). In a particularly elegant study, Tijms et al. (26) have shown that the arterivirus zinc finger-containing papain-like proteinase nsp1 has an essential role in subgenomic mRNA synthesis.
In this paper, we describe a system that will complement existing methods to analyze coronavirus discontinuous transcription. Also, using this system, we demonstrate that the coronavirus replicase gene products are the only viral proteins required for coronavirus subgenomic RNA synthesis.
Cloning of a recombinant vaccinia virus, HCoV-vec-1.
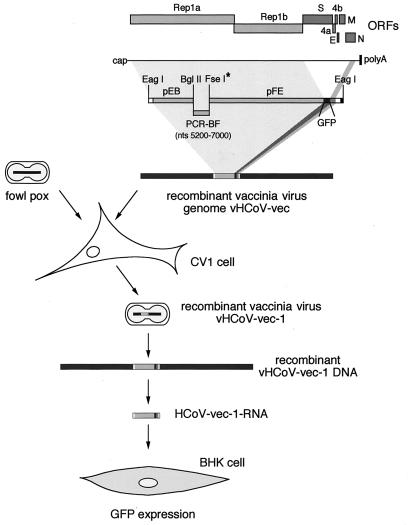
The overall strategy we adopted is illustrated in Fig. 1. Briefly, cDNA fragments were ligated in vitro to produce a 22.5-kbp cDNA. This cDNA represents the 5′-proximal 20.6 kb of the HCoV 229E genome encompassing the entire replicase gene, the gene encoding green fluorescent protein (GFP), the 3′ proximal 1 kb of the genome, and a synthetic poly(A) tail of 42 nucleotides (nt). The start of the GFP ORF is located 1 nt downstream of the ORF1b stop codon and 7 nt downstream of the TAS element that normally controls the discontinuous transcription of mRNA2. The cDNA was ligated in vitro to NotI-cleaved vaccinia virus DNA, and a recombinant vaccinia virus, vHCoV-vec-1, was rescued by transfection into fowlpox-infected CV-1 cells. The recombinant vHCoV-vec-1 DNA was then used as a template for the in vitro transcription of a synthetic HCoV-vec-1 RNA. This RNA was transfected into BHK-21 cells that were subsequently monitored for the expression of GFP.
FIG. 1.
Strategy for the construction of a coronavirus-based vector RNA that mediates the expression of GFP. The structural relationship among the HCoV 229E ORFs, HCoV 229E genomic RNA, plasmid, PCR cDNA fragments, and in vitro RNA transcripts is shown. The position of three silent mutations, which create a unique FseI (∗) site, is depicted in the recombinant cDNA fragments. Relevant restriction sites are indicated. The cDNA fragments pEB, PCR-BF (which represents the HCoV 229E genomic region [nt 5200 to 7000] that is unstable in bacterial cloning systems), and pFE were assembled by in vitro ligation with the restriction sites BglII and FseI. Subsequent ligation of the resulting cDNA with NotI-cleaved vNotI/tk vaccinia virus DNA produced the recombinant vaccinia virus vHCoV-vec DNA. Recombinant vaccina virus vHCoV-vec-1 was recovered and RNA transcripts were produced in vitro by using genomic vHCoV-vec-1 DNA and bacteriophage T7 RNA polymerase. The HCoV-vec-1 RNA was transfected into BHK-21 cells and monitored for the expression of GFP (see text for details).
During the course of our initial studies, we found that one particular region of the HCoV 229E genome (ca. nt 5200 to 7000) was unstable in the form of plasmid cDNA, bacterial artificial chromosome cDNA, or bacteriophage vector cDNA (V. Thiel, unpublished observations). To overcome this problem, we devised a two-phase cloning strategy based upon the optimization of in vitro DNA ligation and the use of vaccinia virus as a eukaryotic cloning vector. First, we constructed the HCoV-vec insert cDNA. Three cDNA fragments were derived from the plasmids pEB and pFE and the reverse transcription-PCR (RT-PCR) product BF. The plasmid pEB is based on pBluescript II KS+ and contains sequences corresponding to nt 1 to 5207 of the HCoV 229E genome preceded by an additional G nucleotide, the sequence for the bacteriophage T7 RNA polymerase promoter, and the restriction sites Bsp120I and EagI. The plasmid pFE is based on pBR322 and contains sequences corresponding to nt 6993 to 20569 of the HCoV 229E genome followed by the GFP gene, the HCoV 229E sequences from nt 26279 to 27277, a synthetic poly(A) tail of 42 nt, and the restriction sites ClaI, Bsp120I, and EagI. The nucleotides at positions 6994, 6997, and 7000 of pFE were mutated from their original sequence to produce an FseI site that is useful for cloning purposes. The RT-PCR product BF was produced with poly(A)-containing RNA from HCoV 229E-infected MRC-5 cells as described previously (25). The RT primer was 5′ CTACTCACGATATCGTAC 3′ (nt 7840 to 7858); the PCR primers were 5′ AGTTGGTGTTATTGCTGATAAGGAC 3′ (nt 5176 to 5200) and 5′ GACATAGGCCGGCCCTGTTGGTTGCACATTTGTTTTGGT 3′ (nt 6968 to 7006). The longer PCR primer contained the nucleotide changes that generate the FseI site. To prepare the DNA fragments EB and BF for in vitro ligation, the plasmid pEB was digested with EagI and BglII, treated with alkaline phospatase, phenol extracted, and ethanol precipitated. The RT-PCR product BF was digested with BglII and precipitated with the DS primer remover reagent (MoBiTec, Göttingen, Germany). Ligation of the DNA fragments EB and BF produced a 7-kbp product, EF, which was digested with FseI and gel purified with QiaexII resin (Qiagen, Hilden, Germany). The fragment EF was ligated to a 15.5-kbp DNA fragment that was derived by digestion of pFE with FseI and EagI, treatment with alkaline phosphatase, and agarose gel purification with QiaexII resin. The products of this reaction were analyzed by pulsed-field gel electrophoresis (PFGE) and are shown in Fig. 2a. The ligation substrates EF (7 kbp) and FE (15.5 kbp), as well as the expected ligation products of 14 kbp (ligation of EF and EF) and 22.5 kbp (ligation of EF and FE), are visible.
FIG. 2.
Cloning of HCoV-vec cDNA in the vaccinia virus genome. (a) Assembly of a 22.5-kbp HCoV-vec cDNA by in vitro ligation. DNA fragments EF and FE were ligated and analyzed by PFGE in comparison with a high-molecular-weight DNA marker (Life Technologies, Karlsruhe, Germany). The ligation substrates and products are illustrated. The relevant restriction sites and the sizes of DNA fragments are shown. AP indicates treatment of DNA fragment ends with alkaline phosphatase. (b) Forced ligation of recombinant vaccinia virus genomes. PFGE analysis of the ligation reaction containing EagI-cleaved, dephosphorylated HCoV-vec insert cDNA and NotI-cleaved vaccinia virus vNotI/tk DNA in the presence of NotI enzyme is shown. The accumulated ligation products, comprised of two vaccinia virus DNA arms and a copy of the HCoV-vec insert cDNA (long/insert/long, long/insert/short, or short/insert/short), as well as relevant substrates and intermediate ligation products, are indicated. DNA bands that are not indicated represent substrates and predicted minor ligation products comprised of pEB, PCR-BF, pFE, and vNotI/tk DNA fragments. (c) Southern blot analysis of 10 random vaccinia virus vHCoV-vec clones. DNA from CV-1 cells infected with vNotI/tk (lane vNot) or recombinant vHCoV-vec clones was digested with HindIII and analyzed by Southern blotting with a random-primed 32P DNA probe representing the HCoV 229E genome between nt 1048 and 20582. Lane M shows a HindIII-digested DNA fragment generated by RT-PCR from HCoV 229E poly(A)-containing RNA. This 19.5-kbp cDNA fragment represents the HCoV genome between nt 1048 and 20582. The two patterns of hybridization (cf. lanes 1 to 6 and 9 with lanes 7, 8, and 10) represent the two possible insert orientations.
In order to produce a recombinant vaccinia virus containing the HCoV-vec DNA, we used a cloning system that has been designed for the insertion of foreign DNA into a single NotI site of the vaccinia virus vNotI/tk genome by in vitro ligation (16). The ligation reaction described above was ligated without further purification to NotI-cleaved vaccinia virus DNA. To eliminate religation of vNotI/tk vector DNA, we added NotI enzyme to the ligation reaction. As illustrated in Fig. 2b, religated vaccinia virus arms (NotI-NotI fusions) were recleaved by the NotI enzyme, whereas ligation products comprised of insert cDNA and vaccinia virus arms (EagI-NotI fusions) were resistant to cleavage. This resulted in an accumulation of ligation products containing the insert DNA. This protocol obviates the need to select for recombinant vaccinia viruses in the subsequent rescue procedure.
After incubation for 16 h at 25°C in NotI digestion buffer supplemented with 1 mM ATP, the ligation products were transfected with Lipofectin into CV-1 cells that had been infected with fowlpox virus (multiplicity of infection of 5) 1 h previously. Two hours later, the cells were harvested and replated on 96-well plates with a fourfold excess of noninfected, nontransfected CV-1 cells. Vaccinia virus clones were rescued within 2 weeks from 96-well plates that had shown a cytopathic effect. The rescued clones were plaque purified and analyzed by Southern blotting (3). DNA from vaccinia virus-infected CV-1 cells was digested with HindIII, and the resulting fragments were electrophoresed and transferred to a nylon membrane. Hybridization was done with a 32P multiprime-labeled (Amersham, Freiburg, Germany) DNA probe corresponding to HCoV 229E nt 1048 to 20582. As is shown for 10 representative clones in Fig. 2c (lanes 1 to 10), the pattern of hybdridization confirms the integrity of the inserted HCoV-vec DNA. Furthermore, this result demonstrates that more than 90% of the rescued vaccinia virus clones are indeed recombinant and can be isolated without selection. Finally, the authenticity of the entire 22.5-kbp insert DNA of one recombinant vaccinia virus clone, vHCoV-vec-1, was confirmed by nucleotide sequence analysis (data not shown).
In vitro synthesis and functional analysis of HCoV-vec-1 RNA.
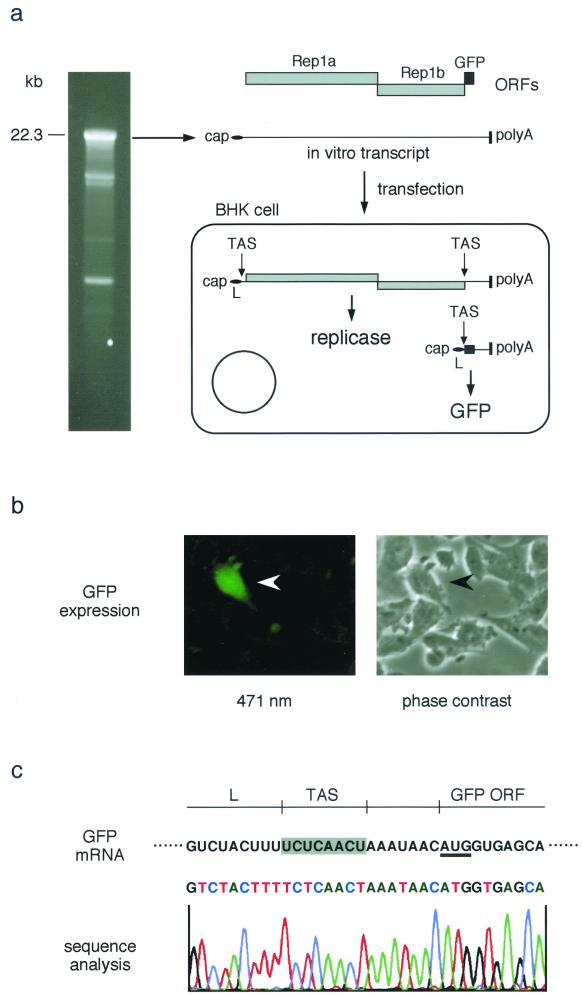
The recombinant vaccinia virus vHCoV-vec-1 was propagated to produce high-titer stocks, the virus was purified, and the genomic DNA was isolated (3). In order to produce a DNA template for the in vitro transcription of HCoV-vec-1 RNA, the HCoV-vec-1 DNA was digested with ClaI, deproteinized by phenol extraction, and ethanol precipitated. Approximately 5 to 10 μg of this DNA was then used to in vitro transcribe HCoV-vec-1 RNA in the presence of m7G(5′)ppp(5′)G (ratio of 1:1 with GTP) by using a bacteriophage T7 polymerase-based system according to the supplier's instructions (RiboMax; Promega, Mannheim, Germany). The reaction was done for 2.5 h at 25°C followed by DNase I treatment and RNA precipitation. As is shown in Fig 3a, this protocol gave both a reasonable amount (approximately 50 μg per reaction) and high proportion of full-length (i.e., 22.3 kb) RNA. When this RNA was transfected into BHK-21 cells by electroporation (3), a small percentage of cells (∼0.1%) displayed strong fluorescence, indicative of GFP expression (Fig. 3b).
FIG. 3.
Expression of GFP by using HCoV-vec-1 RNA (a) The structural relationship among HCoV-vec-1 ORFs, the in vitro-transcribed HCoV-vec-1 RNA, and the intracellular mRNA produced by coronavirus replicase-mediated discontinuous transcription is illustrated together with the predicted intracellular translation products (i.e., the HCoV 229E replicase and GFP). Additionally, 1 μg of capped RNA transcribed from vHCoV-vec-1 DNA in vitro was visualized by ethidium bromide staining after agarose gel electrophoresis. (b) GFP expression analyzed by fluorescence microscopy of BHK-21 cells transfected with HCoV-vec-1 RNA. (c) The TAS region of the intracellular mRNA was sequenced by RT-PCR amplification and cycle sequencing. The sequence corresponding to HCoV 229E leader (L), the TAS region, and the first 10 nt of the GFP-ORF is shown.
To confirm that the observed GFP expression reflects coronavirus-mediated transcription of HCoV-vec-1 RNA, we isolated poly(A)-containing RNA from transfected cells and amplified, by RT-PCR, a DNA fragment that spans the unique mRNA leader-body junction created during coronavirus transcription. The RT primer we used corresponds to HCoV 229E nt 26802 to 26822, and the PCR was done with a primer pair corresponding to HCoV 229E nt 26481 to 26496 and HCoV 229E nt 21 to 39 (i.e., a leader sequence-specific primer). Sequence analysis of the RT-PCR product by using a GFP-specific oligonucleotide (5′ ACGGGCAGCTTGCCGGTGGTGCA 3′) shows, indeed, that coronavirus-specific, subgenomic mRNA synthesis has occurred and the leader-body fusion has taken place at the expected position (Fig. 3c). This result demonstrates conclusively that the coronavirus replicase gene products are the only viral proteins needed to assemble a complex capable of discontinuous transcription and that this complex can be assembled in vertebrate cells of nonhuman origin.
The basis of the approach taken in this study is the use of vaccinia virus as a cloning vector for large cDNA inserts. In this respect, we believe the vaccinia virus system has a number of advantages. First, in this system, we have never observed instability of the cloned insert cDNA, and, irrespective of the size of the cDNA insert, we have not seen any differences in the infectivity, growth, or kinetics of the recombinant vaccinia viruses compared to the parental virus. Second, we have shown that large cDNA fragments, assembled by in vitro ligation, can be efficiently cloned into the vaccinia virus genome. Thus, by incorporating the NotI enzyme in the ligation reactions, more than 90% of recovered vaccinia viruses are recombinant. This protocol facilitates the isolation of recombinant vaccinia virus clones without the need for selection, obviates the need for plasmid intermediates carrying full-length insert cDNAs, and represents a flexible way to introduce defined mutations into large cDNA clones. In the longer term, we predict this system will be useful for the analysis of coronavirus transcription in a variety of eukaryotic cells, in the absence of helper virus components, independent of the virus replication cycle and without the requirement for receptor-mediated infection.
The detection of coronavirus-specific transcripts in this report represents the first direct evidence that the coronavirus replicase proteins suffice for subgenomic RNA synthesis. However, we think it is important to state that our results do not prove that replication of the transfected RNA has occurred, nor do they exclude the possibility that additional viral or host cell proteins may have regulatory roles in coronavirus replication or transcription (4, 11, 18, 23). It is striking, in this respect, that we have observed only a small percentage of green fluorescent cells after electroporation of the HCoV-vec-1 RNA into BHK-21 cells. In contrast, when we use alphavirus-based RNAs, we routinely achieve transfection efficiencies of more than 50%. This could be due to the extraordinary size of the HCoV-vec-1 RNA, which may make it more difficult to transfect by electroporation. Alternatively, the RNA could be subject to degradation before the replicase polyproteins have been translated and an active complex has been formed. Since in natural coronavirus infections, the genomic RNA is initially protected by the nucleocapsid structure, this may represent a fundamental difference compared to the transfection of naked RNA. Further experiments are required to address these questions.
Acknowledgments
V.T. and J.H. contributed equally to this work.
We thank V. ter Meulen for support.
This work was supported by the German Research Council (DFG).
REFERENCES
- 1.Agapov E V, Frolov I, Lindenbach B D, Pragai B M, Schlesinger S, Rice C M. Noncytopathic Sindbis virus RNA vectors for heterologous gene expression. Proc Natl Acad Sci USA. 1998;95:12989–12994. doi: 10.1073/pnas.95.22.12989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Almázan F, González J M, Pénzes Z, Izeta A, Calvo E, Plana-Durán J, Enjuanes L. Engineering the largest RNA virus genome as an infectious bacterial artificial chromosome. Proc Natl Acad Sci USA. 2000;97:5516–5521. doi: 10.1073/pnas.97.10.5516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausubel F, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1987. [Google Scholar]
- 4.Baric R S, Nelson G W, Fleming J O, Deans R J, Keck J G, Casteel N, Stohlman S A. Interactions between coronavirus nucleocapsid protein and viral RNAs: implications for viral transcription. J Virol. 1988;62:4280–4287. doi: 10.1128/jvi.62.11.4280-4287.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Vries A F, Horzinek M C, Rottier P J M, de Groot R J. The genome organization of the Nidovirales: similarities and differences between arteri-, toro-, and coronaviruses. Semin Virol. 1997;8:33–47. doi: 10.1006/smvy.1997.0104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Diez J, Ishikawa M, Kaido M, Ahlquist P. Identification and characterization of a host protein required for efficient template selection in viral RNA replication. Proc Natl Acad Sci USA. 2000;97:3913–3918. doi: 10.1073/pnas.080072997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fischer F, Stegen C F, Koetzner C A, Masters P S. Analysis of a recombinant mouse hepatitis virus expressing a foreign gene reveals a novel aspect of coronavirus transcription. J Virol. 1997;71:5148–5160. doi: 10.1128/jvi.71.7.5148-5160.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Herold J, Andino R. Poliovirus RNA replication requires genome circularization through a protein-protein bridge. Mol Cell. 2001;7:581–591. doi: 10.1016/S1097-2765(01)00205-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heusipp G, Harms U, Siddell S G, Ziebuhr J. Identification of an ATPase activity associated with a 71-kilodalton polypeptide encoded in gene 1 of the human coronavirus 229E. J Virol. 1997;71:5631–5634. doi: 10.1128/jvi.71.7.5631-5634.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hsue B, Masters P S. Insertion of a new transcriptional unit into the genome of mouse hepatitis virus. J Virol. 1999;73:6128–6135. doi: 10.1128/jvi.73.7.6128-6135.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim K H, Makino S. Two murine coronavirus genes suffice for viral RNA synthesis. J Virol. 1995;69:2313–2321. doi: 10.1128/jvi.69.4.2313-2321.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuo L, Godeke G J, Raamsman M J B, Masters P S, Rottier P J M. Retargeting of coronavirus by substitution of the spike glycoprotein ectodomain: crossing the host cell species barrier. J Virol. 2000;74:1393–1406. doi: 10.1128/jvi.74.3.1393-1406.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lai M M, Cavanagh D. The molecular biology of coronaviruses. Adv Virus Res. 1997;48:1–100. doi: 10.1016/S0065-3527(08)60286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lohmann V, Korner F, Koch J, Herian U, Theilmann L, Bartenschlager R. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science. 1999;285:110–113. doi: 10.1126/science.285.5424.110. [DOI] [PubMed] [Google Scholar]
- 15.Makino S, Joo M, Makino J K. A system for study of coronavirus mRNA synthesis: a regulated, expressed subgenomic defective interfering RNA results from intergenic site insertion. J Virol. 1991;65:6031–6041. doi: 10.1128/jvi.65.11.6031-6041.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Merchlinsky M, Moss B. Introduction of foreign DNA into the vaccinia virus genome by in vitro ligation: recombination-independent selectable cloning vectors. Virology. 1992;190:522–526. doi: 10.1016/0042-6822(92)91246-q. [DOI] [PubMed] [Google Scholar]
- 17.Molenkamp R, van Tol H, Rozier B C, van Der Meer Y, Spaan W J, Snijder E J. The arterivirus replicase is the only viral protein required for genome replication and subgenomic mRNA transcription. J Gen Virol. 2000;81:2491–2496. doi: 10.1099/0022-1317-81-10-2491. [DOI] [PubMed] [Google Scholar]
- 18.Nelson G W, Stohlman S A, Tahara S M. High affinity interaction between nucleocapsid protein and leader/intergenic sequence of mouse hepatitis virus RNA. J Gen Virol. 2000;81:181–188. doi: 10.1099/0022-1317-81-1-181. [DOI] [PubMed] [Google Scholar]
- 19.Sawicki S G, Sawicki D L. A new model for coronavirus transcription. Adv Exp Med Biol. 1998;440:215–219. doi: 10.1007/978-1-4615-5331-1_26. [DOI] [PubMed] [Google Scholar]
- 20.Seybert A, Hegyi A, Siddell S G, Ziebuhr J. The human coronavirus 229E superfamily 1 helicase has RNA and DNA duplex-unwinding activities with 5′-to-3′ polarity. RNA. 2000;6:1056–1068. doi: 10.1017/s1355838200000728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Siddell S G, Snijder E J. Coronaviruses, toroviruses and arteriviruses. In: Mahy B W J, Collier L, editors. Topley & Wilson's microbiology and microbial infections. 9th ed. London, United Kingdom: Arnold; 1998. pp. 463–484. [Google Scholar]
- 22.Spaan W, Delius H, Skinner M, Armstrong J, Rottier P, Smeekens S B, van der Zeijst A, Siddell S G. Coronavirus mRNA synthesis involves fusion of non-contiguous sequences. EMBO J. 1983;2:1839–1844. doi: 10.1002/j.1460-2075.1983.tb01667.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stohlman S A, Baric R S, Nelson G N, Soe L H, Welter L M, Deans R J. Specific interaction between coronavirus leader RNA and nucleocapsid protein. J Virol. 1988;62:4288–4295. doi: 10.1128/jvi.62.11.4288-4295.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thiel V, Herold J, Schelle B, Siddell S G. Infectious RNA transcribed in vitro from a cDNA copy of the human coronavirus genome cloned in vaccinia virus. J Gen Virol. 2001;82:1273–1281. doi: 10.1099/0022-1317-82-6-1273. [DOI] [PubMed] [Google Scholar]
- 25.Thiel V, Rashtchian A, Herold J, Schuster D M, Guan N, Siddell S G. Effective amplification of 20-kb DNA by reverse transcription PCR. Anal Biochem. 1997;252:62–70. doi: 10.1006/abio.1997.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tijms M A, van Dinten L C, Gorbalenya A E, Snijder E J. A zinc finger-containing papain-like protease couples subgenomic mRNA synthesis to genome translation in a positive-stranded RNA virus. Proc Natl Acad Sci USA. 2001;98:1889–1894. doi: 10.1073/pnas.041390398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van der Most R G, de Groot R J, Spaan W J. Subgenomic RNA synthesis directed by a synthetic defective interfering RNA of mouse hepatitis virus: a study of coronavirus transcription initiation. J Virol. 1994;68:3656–3666. doi: 10.1128/jvi.68.6.3656-3666.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Marle G, Dobbe J C, Gultyaev A P, Luytjes W, Spaan W J, Snijder E J. Arterivirus discontinuous mRNA transcription is guided by base pairing between sense and antisense transcription-regulating sequences. Proc Natl Acad Sci USA. 1999;96:12056–12061. doi: 10.1073/pnas.96.21.12056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Marle G, Luytjes W R, van der Most G, van der Straaten T, Spaan W J. Regulation of coronavirus mRNA transcription. J Virol. 1995;69:7851–7856. doi: 10.1128/jvi.69.12.7851-7856.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yount B, Curtis K M, Baric R S. Strategy for systematic assembly of large RNA and DNA genomes: transmissible gastroenteritis virus model. J Virol. 2000;74:10600–10611. doi: 10.1128/jvi.74.22.10600-10611.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ziebuhr J, Snijder E J, Gorbalenya A E. Virus-encoded proteinases and proteolytic processing in the Nidovirales. J Gen Virol. 2000;81:853–879. doi: 10.1099/0022-1317-81-4-853. [DOI] [PubMed] [Google Scholar]