Abstract
Pure laparoscopic donor hepatectomy (PLDH) has become a standard practice for living donor liver transplantation in expert centers. Accurate understanding of biliary structures is crucial during PLDH to minimize the risk of complications. This study aims to develop a deep learning-based segmentation model for real-time identification of biliary structures, assisting surgeons in determining the optimal transection site during PLDH. A single-institution retrospective feasibility analysis was conducted on 30 intraoperative videos of PLDH. All videos were selected for their use of the indocyanine green near-infrared fluorescence technique to identify biliary structure. From the analysis, 10 representative frames were extracted from each video specifically during the bile duct division phase, resulting in 300 frames. These frames underwent pixel-wise annotation to identify biliary structures and the transection site. A segmentation task was then performed using a DeepLabV3+ algorithm, equipped with a ResNet50 encoder, focusing on the bile duct (BD) and anterior wall (AW) for transection. The model’s performance was evaluated using the dice similarity coefficient (DSC). The model predicted biliary structures with a mean DSC of 0.728 ± 0.01 for BD and 0.429 ± 0.06 for AW. Inference was performed at a speed of 15.3 frames per second, demonstrating the feasibility of real-time recognition of anatomical structures during surgery. The deep learning-based semantic segmentation model exhibited promising performance in identifying biliary structures during PLDH. Future studies should focus on validating the clinical utility and generalizability of the model and comparing its efficacy with current gold standard practices to better evaluate its potential clinical applications.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-73434-4.
Keywords: Artificial intelligence in medicine, Laparoscopic liver donor hepatectomy, Biliary structures
Subject terms: Machine learning, Liver
Introduction
Pure laparoscopic donor hepatectomy (PLDH) has emerged as the standard procurement practice for living donor liver transplantation (LDLT) in expert centers1–4. Given that PLDH influences both the donor and recipient’s postoperative outcomes, the procedure demands a refined technical approach, resulting in a prolonged learning curve5,6. A thorough anatomical understanding of donor bile duct division is critical during PLDH to prevent biliary complications, notably biliary leakage (3.3%) or stricture (1.6%) for donors7–9. To enhance comprehension of biliary structures, preoperative magnetic resonance cholangiopancreatography (MRCP) is deemed essential, while intraoperative image guidance techniques such as cholangiography or indocyanine green (ICG) near-infrared fluorescence method are strongly advised2,10. However, intraoperative cholangiography (IOC) requires exposure to radiation, and ICG method entails infra-red (IR) camera and a device capable of processing IR images11.
Surgical data science (SDS) is an emerging field in data science that seeks to improve surgical outcomes by extracting valuable insights from various digitalized data generated throughout the entire process of surgical care process12,13. In particular, computer vision (CV), a subfield of artificial intelligence (AI), employs digital images or videos to train computers to understand and automate tasks typically performed by human visual system14. Recent studies have demonstrated impressive results, with CV models accurately interpreting anatomical structures, surgical instruments, and surgical procedures15–19.
With the potential of SDS and CV analysis to enhance clinical outcomes, we proposed the utilization of these technologies for providing intraoperative image guidance for better understanding of biliary structure during PLDH. This study aims to develop a deep learning-based semantic segmentation model capable of identifying biliary structures, thereby assisting in determining the optimal transection site.
Methods
Patients
This study is a single-institution retrospective feasibility analysis which includes 30 intraoperative videos of PLDH from Samsung Medical Center, utilizing the intraoperative ICG near-infrared fluorescence method between January 2021 and April 2022. Our center has extensive experience in PLDH, having performed over 600 cases20. The surgical team consists of four experienced donor surgeons, although the videos included in this study were all from procedures performed by a single surgeon (GS. Choi). All donors were injected 0.1 mg/kg of indocyanine green (Dianogreen, Daiichi Sankyo Co, Tokyo, Japan) intraoperatively about 30 min before exposure of the hilar plate21. The biliary structures were clearly visualized by using infra-red endoscopic camera (IR Telescopes 10 mm, Olympus, Tokyo, Japan). The types of bile ducts were classified according to the modified classification system proposed by Huang et al.: type I, normal type; type II, trifurcation of right anterior, right posterior, and left hepatic duct; type III, right posterior duct draining into left hepatic duct; type IV, early branching of right posterior duct from the common hepatic duct; type V, right posterior duct draining into cystic duct; type VI, other types of variation in bile duct anatomy22. For detailed information regarding our bile duct division technique, we refer readers to our previously published paper23. The study was conducted in accordance with the Declaration of Helsinki and the Istanbul Declaration, and was reviewed and approved by the Institutional Review Board (IRB, SMC-2022-07-149-001). Due to the retrospective nature of the study, IRB of Samsung Medical Center waived the need of obtaining informed consent.
Video segmentation and training dataset
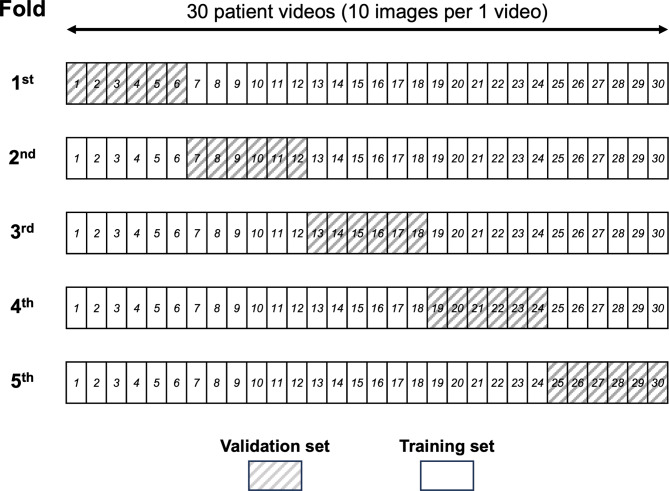
The videos were recorded in MP4 format with a display resolution of 1920 × 1080 pixels and a frame rate of 30 frames per second (fps). Frames were extracted at a rate of 10 fps from each video, capturing the period from bile duct isolation to the opening of anterior wall of right hepatic duct. This was achieved using ffmpeg 4.1 software (www.ffmpeg.org). Frames with obscured fields due to smoke, completely obscured biliary structures by surgical instruments, or camera positioned outside the surgical field were excluded. Finally, 300 images (10 images from each of the 30 intraoperative videos) were selected for the model training and validation. The five-fold cross-validation was performed on 30 videos. For each validation cycle, four out of five groups (24 videos) were used to train the model, while the remaining group (6 videos) was reserved for validation. This process was repeated five times, with each group serving as the validation set once and as part of training set four times (Fig. 1).
Fig. 1.
Schematic representation of five-fold cross-validation. Each row represents one of the five ‘folds’ used in the validation process, with a total of 30 patient videos divided into training and validation sets. The columns represent individual patient videos, each containing 10 images, as indicated by the numbers 1 through 30. Shaded boxes within each fold indicate the videos selected as the validation set for that particular cycle, with the remaining videos used as the training set. Each video serves as part of the validation set once throughout the five cycles, ensuring that every video contributes to the validation of the model, while being used four times in the training set.
Annotation of biliary structure
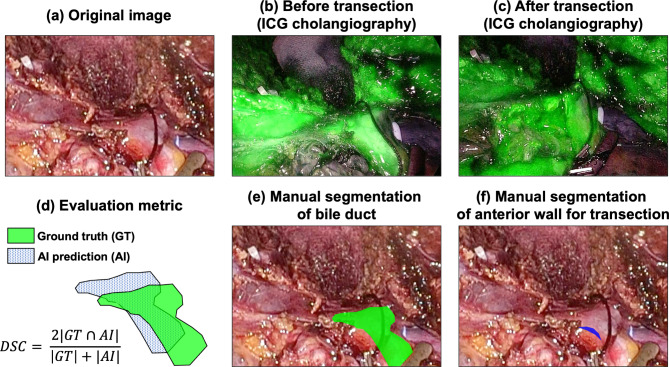
In every intraoperative video included in this study, biliary structures were confirmed using the indocyanine green (ICG) near-infrared fluorescence method (Fig. 2a,b). Pixel-wise labeling of biliary structure and transection site was performed with reference to the ICG fluorescence images from each intraoperative video. Annotations were completed using the Computer Vision Annotation Tool (www.cvat.org, Intel). The proposed model is designed to perform segmentation in two distinct ways. First, segmentation was carried out to mask the entire biliary structure, as predicted with reference to the ICG image (annotated as BD; bile duct, Fig. 2b,e). Second, annotation was performed to mask the anterior wall of the junction of common and right hepatic duct, which represents the area of interest for the operator when opening the bile duct (annotated as AW; anterior wall, Fig. 2c,f). Annotation was performed by fellow surgeon (N. Oh) and confirmed by senior surgeon who experienced more than 300 cases of PLDH (GS. Choi).
Fig. 2.
Ground truth annotation process. This figure illustrates the step-by-step process of creating ground truth annotations for the biliary structure segmentation by referencing indocyanine green (ICG) cholangiography. (a) Actual surgical images extracted from the procedure, (b) Structures of the bile duct extracted from ICG cholangiography, (c) The actual site where the bile duct was transected during surgery, (d) Demonstrates the Dice Similarity Coefficient (DSC), quantitatively showing the level of agreement between the ground truth and the AI-inferred regions, (e) Manual segmentation of the bile duct structure, generated with reference to (b), (f) Segmentation of the anterior wall, representing the proposed area for bile duct transection, created with reference to (c).
Deep learning model
The model architecture employed DeepLabv3+ as its foundation, with ResNet50 pre-trained on the ImageNet dataset serving as the encoder24–26. To address the limitation of a small dataset, data augmentation techniques such as geometric transformations (flips, rotations, etc.), color transformations (contrast, saturation, hue, etc.), and Gaussian noise and patch-based zero masking were applied. These augmentation techniques increased the DSC by 1.4% points compared to not using them (Supplementary Table 1). All data were normalized according to the mean and standard deviation of each RGB channel and resized to a pixel size of 256 by 256. The model’s hyperparameter details are provided in Supplementary Table 2.
Computing
We utilized Python as our programming language and Pytorch, an open-source machine learning framework, for segmentation AI modeling. The computational resources employed included an Nvidia GeForce RTXTM 3060 with 12GB of VRAM as the GPU, and an AMD RyzenTM 5 5600X 6-Core Processor @ 3.7 GHz with 32GB of RAM as the CPU.
Evaluation metrics
The model’s performance was assessed using the Dice Similarity Coefficient (DSC) between the manually segmentation and prediction by deep learning model, which calculates the harmonic mean of precision and recall. This metric demonstrates the extent to which the model’s predicted region overlaps with the ground truth image (Fig. 2d). The DSC ranges from 0 to 1, with higher values indicating a closer match between the predicted and ground truth images. In this study, the average DSC was calculated for two classes (BD and AW). The DSC is defined as follows:
DSC (Dice Similarity Coefficient) = 2×TP/(2×TP + FP + FN), precision = TP/(TP + FP), and recall = TP/(TP + FN), where TP (True Positive) denotes cases where both the predicted and ground truth values are positive. FP (False Positive) refers to cases where the predicted value is positive, but the ground truth value is negative. FN (False Negative) represents cases where the predicted value is negative, but the ground truth value is positive.
Results
Patient characteristics in the entire cohort and each validation set are summarized in Table 1. Median age of the patients is 40.5 [29.5, 47.5, IQR], male patients were 16 (53.5%), female patients were 14 (46.7%). Type I bile duct was most common (23/30, 76.7%), type III (3/30, 10%) and IV (3/30, 10%) followed.
Table 1.
Baseline characteristics of entire cohort and each validation set.
| Patient characteristics | Total (N = 30) | 1st val set (n = 6) | 2nd val set (n = 6) | 3rd val set (n = 6) | 4th val set (n = 6) | 5th val set (n = 6) |
|---|---|---|---|---|---|---|
| Age (median, IQR) | 40.5 [29.5, 47.5] | 42.5 [36.8, 45.2] | 41.0 [30.8, 53.5] | 27.0 [26.0, 33.2] | 47.0 [43.0, 49.5] | 40.5 [35.5, 48.5] |
| Sex (%) | ||||||
| M | 16 (53.3) | 2 (33.3) | 5 (83.3) | 6 (100.0) | 1 (16.7) | 2 (33.3) |
| F | 14 (46.7) | 4 (66.7) | 1 (16.7) | 0 (0.0) | 5 (83.3) | 4 (66.7) |
| BMI (median, IQR) | 23.8 [23.0, 27.1] | 23.9 [23.4, 27.6] | 23.2 [22.0, 23.8] | 24.6 [22.3, 26.2] | 26.8 [24.4, 27.4] | 24.9 [21.9, 27.0] |
| Type of bile duct (%) | ||||||
| I | 23 (76.7) | 5 (83.3) | 5 (83.3) | 6 (100.0) | 4 (66.7) | 3 (50.0) |
| II | 0 | 0 | 0 | 0 | 0 | 0 |
| III | 3 (10.0) | 1 (16.7) | 1 (16.7) | 0 (0.0) | 1 (16.7) | 0 (0.0) |
| IV | 3 (10.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (16.7) | 2 (33.3) |
| V | 0 | 0 | 0 | 0 | 0 | 0 |
| VI | 1 (3.3) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (16.7) |
| Number of duct opening (%) | ||||||
| 1 | 24 (80) | 5 (83.3) | 5 (83.3) | 6 (100.0) | 4 (66.7) | 4 (66.7) |
| 2 | 6 (20) | 1 (16.7) | 1 (16.7) | 0 (0.0) | 2 (33.3) | 2 (33.3) |
| Hospitaled day (median, IQR) | 7.0 [6.0, 8.0] | 7.5 [7.0, 8.0] | 7.0 [6.2, 8.5] | 6.5 [5.2, 7.8] | 7.5 [5.5, 8.0] | 6.0 [6.0, 6.0] |
The types of bile ducts: type I, normal type; type II, trifurcation of right anterior, right posterior, and left hepatic duct; type III, right posterior duct draining into left hepatic duct; type IV, early branching of right posterior duct from the common hepatic duct; type V, right posterior duct draining into cystic duct; type VI, other types of variation in bile duct anatomy.
Val validation, IQR interquartile ratio, M male, F female.
Table 2 represents the DSC values obtained during each validation of the semantic segmentation task for the BD and AW. The mean DSC of the five-fold cross-validation for BD was 0.728 ± 0.01, with the highest DSC of 0.758 achieved on the 1st validation set. The mean precision of BD was 0.713 ± 0.2 and the mean recall was 0.746 ± 0.02. The mean DSC of AW was 0.429 ± 0.06, with the highest DSC of 0.513 in the 1st validation set. The mean precision of AW was 0.366 ± 0.07 and the mean recall was 0.528 ± 0.05. The proposed deep learning model operated at a speed of over 15.3 FPS. Representative images of the semantic segmentation results are presented in Fig. 3 and supplementary Fig. 1.
Table 2.
The performance of the AI model for each validation according to the annotation type AI, artificial intelligence; DSC, Dice similarity coefficient; BD, bile duct; AW, anterior wall; SD, standard deviation.
| Validation set | DSC | Precision | Recall |
|---|---|---|---|
| 1st | |||
| BD | 0.758 | 0.735 | 0.782 |
| AW | 0.513 | 0.482 | 0.579 |
| 2nd | |||
| BD | 0.728 | 0.737 | 0.719 |
| AW | 0.424 | 0.355 | 0.525 |
| 3rd | |||
| BD | 0.726 | 0.734 | 0.719 |
| AW | 0.397 | 0.302 | 0.575 |
| 4th | |||
| BD | 0.723 | 0.677 | 0.777 |
| AW | 0.339 | 0.282 | 0.423 |
| 5th | |||
| BD | 0.706 | 0.683 | 0.731 |
| AW | 0.475 | 0.409 | 0.568 |
| Mean (SD) | |||
| BD | 0.728 (0.01) | 0.713 (0.2) | 0.746 (0.02) |
| AW | 0.429 (0.06) | 0.366 (0.07) | 0.528 (0.05) |
Fig. 3.
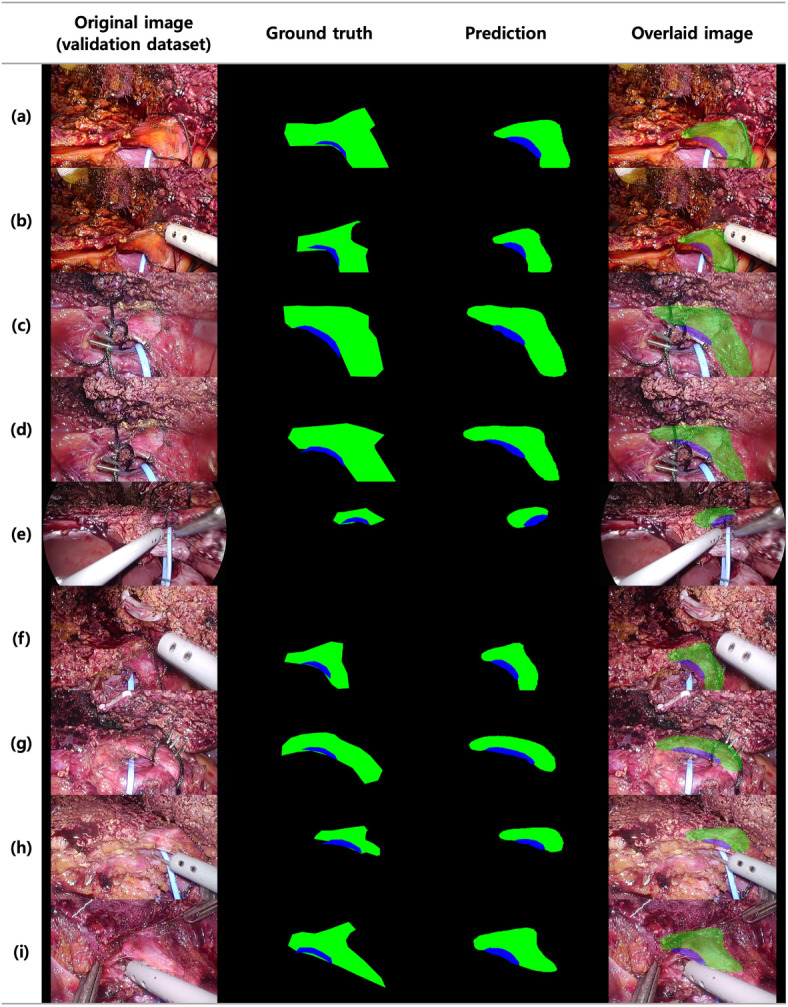
Results of segmentation for bile duct (BD) and anterior wall (AW) in the validation set. From left, the first images are original images of the validation set, the second are the ground truth of the target area (BD, AW) corresponding to the original images, the third are the predicted image by the deep learning model after receiving the original image as an input. The fourth images are the predicted images overlaid onto the original images. Black corresponds to the background, blue to BD, and green to AW. For further insights into the interpretability of these results, please refer to supplementary Fig. 1, which provides heatmap visualizations to enhance the understanding of the segmentation outcomes.
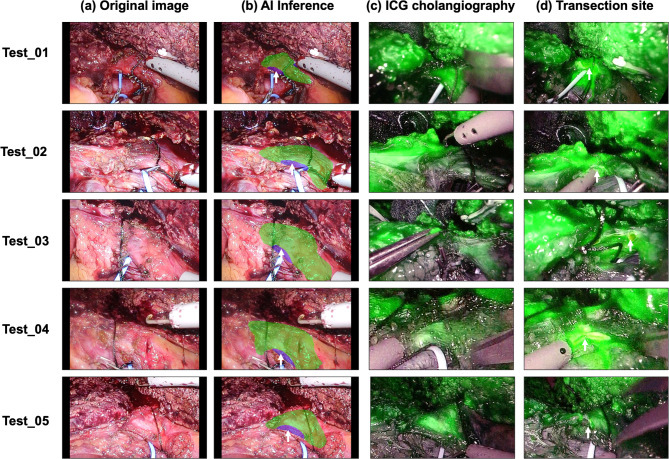
Our model was subsequently tested on five new PLDH cases, using videos that the deep learning model had never encountered previously. The outcomes of these test sets are presented in Fig. 4. The BD and AW prediction made by the proposed model for all five test sets are visualized in Fig. 4b. The actual transection sites during operations are shown in Fig. 4d. In all cases, the actual transection sites were located within the AW proposed by the model. The corresponding five test videos are available for the following links (Test_01, https://youtu.be/Wu7--pndgho; Test_02, https://youtu.be/oNjOpZCZp-M; Test_03, https://youtu.be/ccZogB_H_0k; Test_04, https://youtu.be/wx3Ya9ubQ9I; Test_05, https://youtu.be/JbOzNiDkusc).
Fig. 4.
Comparison between the results inferred by the AI model and ICG cholangiography on the test set. The following five examples showcase the results obtained by the proposed AI model when applied to videos it had never seen during the training process. (a) Original image, (b) The AI model’s predictions for bile duct (BD) in green and anterior wall (AW) in blue. The white arrows point to the actual transection site as confirmed in (c,d) ICG cholangiography, (d) Image after bile duct transection, with white arrows indicating the site where the bile duct was transected.
Discussion
This study was conducted to assess the feasibility of employing an AI model for the delineation of biliary structures and the identification of optimal transection site during PLDH. The proposed AI model predicted biliary structures with a mean DSC of 0.728 ± 0.01 and transection sites with a DSC of 0.429 ± 0.06. Notably, the model performed real-time inference at a speed of 15.3 FPS, demonstrating that deep learning-based real-time recognition of anatomical structures during surgery is feasible approach. To further the research in this field and allow for external validation, we have made the AI model’s codebase available in a public repository (https://github.com/SMC-SSISO/Bile-Duct-Segmentation), inviting collaborators and researchers to engage with our work and contribute to its ongoing development and refinement.
Recent advancements in computer vision analysis for surgical imaging have demonstrated significant progress, particularly in the application of segmentation to guide intraoperative anatomy across various surgical procedures27. The DSC (Dice similarity coefficient) measures the extent to which the predictions made by AI match the actual structures, with values ranging from 0 to 1, as a value closer to 1 signifies a greater performance of the AI’s prediction. Within the realm of surgical imaging segmentation, studies have reported a spectrum of DSC values from 0.58 to 0.79. For instances, research on prostate segmentation during transanal total mesorectal excision (TaTME) reported an average DSC of 0.71 ± 0.04, while studies focusing on the masking of the inferior mesenteric artery (IMA) during colorectal resection yielded a mean DSC of 0.798 ± 0.016119,28. Furthermore, investigations into masking the recurrent laryngeal nerve (RLN) in esophagectomy achieved an average DSC of 0.5815. All these studies utilized the DeepLabV3+ model, as did our study. Our AI model achieved a DSC of 0.728 ± 0.01 for bile duct segmentation, reflecting a comparable level of accuracy to those previously reported researches, despite the constraints of a limited sample size.
The primary goal of employing computer-aided anatomy recognition, as explored in this study, is to augment surgical precision by enabling surgeons to more accurately identify critical anatomical structures. This technological advancement aims to decrease the incidence of adverse events and complications, ultimately improving overall surgical outcomes29. To establish the clinical efficacy of this kind of intraoperative decision support tool, it is crucial to demonstrate that it not only comparable but potentially surpasses the safety and effectiveness of existing techniques. In the context of PLDH, the conventional standard involves using ICG or intraoperative cholangiography (IOC) for bile duct division7. On the other hands, the implementation of a computer vision-based cholangiography system may hold significant potential benefits. If further validated, this approach may eliminate the need for administrating ICG to patients and using IR cameras for its visualization. Currently, IOC requires the use of a C-arm, leading to unwanted radiation exposure for patients. In contrast, a computer vision-based system for cholangiography identification offers the distinct advantage of not subjecting patients to additional radiation or IV ICG. By interfacing the laparoscopic image hub system with a computer, real-time inference results can be visualized directly during surgery. Therefore, future research should focus on comparing the new deep learning-based method of bile duct recognition with these established techniques to justify the integration of new technology into routine surgical practice30.
In this study, a supervised learning approach was utilized to train a computer vision model to segment biliary structure. This approach necessitates the use of precise and consistent ground-truth labels, which are created by human annotators. Particularly in computer vision application, this process involves manual, pixel-by-pixel annotation of regions of interest within raw images. The performance of the computer vision model is heavily reliant on this input data. However, due to inherent human biases, annotations may vary among annotators, and even within the work of a single annotator, inconsistencies may arise from individual errors31. Consequently, it is important to recognize that models trained on these data may also exhibit biases and errors32. To reduce the effects of human bias and error in the data, alternative approaches like unsupervised or self-supervised learning can be considered. These methods enable the model to discern and learn from the intrinsic structure of the data without relying extensively on human-generated labels. This approach can diminish the influence of potential biases and errors, enhancing the model’s objectivity and reliability33.
The limitations of this study include its experimental feasibility nature, which primarily focuses on determining whether a deep learning model can effectively recognize biliary structures, without demonstrating its actual clinical utility. To establish the model’s clinical value, a comparison with the current gold standard practice is necessary. Additionally, the DSC of AW was relatively low compared to BD. This can be attributed to AW’s smaller area and elongated shape, which result in a significantly lower DSC value, even with minor errors. Furthermore, the variability in bile duct anatomy types may affect the model’s performance. It is important to note that surgery is a dynamic procedure with bleeding or bile leaks, which can affect the surgical field and, consequently, the AI model’s performance. The model was trained on 30 surgical videos from a single surgeon in a retrospective study, lacking external validation, which restricts the generalizability of the model. To overcome this limitation and to validate the applicability of this approach, there is a need for a multicenter, multisurgeon international study group. Such a collaborative effort would facilitate the collection of a more diverse and extensive dataset, reflecting a wider range of surgical techniques and patient anatomies.
Conclusion
The deep learning-based semantic segmentation model exhibited promising performance in identifying biliary structure during PLDH. Further study should focus on validating the clinical utility and generalizability of the model and comparing its efficacy with current standard practices to better evaluate its potential clinical applications.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Abbreviations
- CV
Computer vision
- PLDH
Pure laparoscopic donor hepatectomy
- LDLT
Living donor liver transplantation
- ICG
Indocyanine green
- AW
Anterior wall
- BD
Bile duct
- DSC
Dice similarity coefficient
- FPS
Frame per second
- SDS
Surgical data science
Author contributions
Conceptualization: N.O., G.-S.C. Methodology: N.O., G.-S.C., B.K., T.K. Investigation: N.O., B.K., T.K. Resources: N.O., MD, J.R., J.K., G.-S.C. Writing-original draft: N.O., B.K. Writing-review and editing: N.O., B.K., G.-S.C. Supervision: G.-S.C. Funding acquisition: G.-S.C.
Funding
This study was supported by the ‘Bio&Medical Technology Development Program’ of the National Research Foundation (NRF) funded by the Korean government (MSIT) [RS-2023-00222838], Korea Government Grant Program for Education and Research in Medical AI’ through the Korea Health Industry Development Institute (KHIDI), funded by the Korea government (MOE, MOHW), and ‘Future Medicine 2030 Project’ of the Samsung Medical Center [SMX1230771].
Data availability
The data that support the findings of this study are not openly available due to reasons of sensitivity and are available from the corresponding author upon reasonable request.
Declarations
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Both authors contributed equally to this manuscript.
Contributor Information
Namkee Oh, Email: ngnyou@gmail.com.
Gyu-Seong Choi, Email: med9370@gmail.com.
References
- 1.Rhu, J., Choi, G. S., Kim, J. M., Kwon, C. H. D. & Joh, J. W. Complete transition from open surgery to laparoscopy: 8-year experience with more than 500 laparoscopic living donor hepatectomies. Liver Transplant.28, 1158–1172. 10.1002/lt.26429 (2022). [DOI] [PubMed] [Google Scholar]
- 2.Kwon, C. H. D. et al. Laparoscopic donor hepatectomy for adult living donor liver transplantation recipients. Liver Transplant.24, 1545–1553. 10.1002/lt.25307 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Hong, S. K. et al. Pure laparoscopic donor hepatectomy: A multicenter experience. Liver Transplant.27, 67–76. 10.1002/lt.25848 (2021). [DOI] [PubMed] [Google Scholar]
- 4.Rhu, J., Choi, G. S., Kim, J. M., Joh, J. W. & Kwon, C. H. D. Feasibility of total laparoscopic living donor right hepatectomy compared with open surgery: Comprehensive review of 100 cases of the initial stage. J. Hepatobiliary Pancreat. Sci.27, 16–25. 10.1002/jhbp.653 (2020). [DOI] [PubMed] [Google Scholar]
- 5.Rhu, J., Choi, G. S., Kwon, C. H. D., Kim, J. M. & Joh, J. W. Learning curve of laparoscopic living donor right hepatectomy. Br. J. Surg.107, 278–288. 10.1002/bjs.11350 (2020). [DOI] [PubMed] [Google Scholar]
- 6.Hong, S. K. et al. The learning curve in pure laparoscopic donor right hepatectomy: A cumulative sum analysis. Surg. Endosc.33, 3741–3748. 10.1007/s00464-019-06668-3 (2019). [DOI] [PubMed] [Google Scholar]
- 7.Cherqui, D. et al. Expert consensus guidelines on minimally invasive donor hepatectomy for living donor liver transplantation from innovation to implementation: A joint initiative from the International Laparoscopic Liver Society (ILLS) and the Asian-Pacific Hepato-Pancreato-Biliary Association (A-PHPBA). Ann. Surg.273, 96–108. 10.1097/SLA.0000000000004475 (2021). [DOI] [PubMed] [Google Scholar]
- 8.Rhu, J. et al. A novel technique for bile duct division during laparoscopic living donor hepatectomy to overcome biliary complications in liver transplantation recipients: “Cut and clip” rather than “clip and cut”. Transplantation105, 1791–1799. 10.1097/TP.0000000000003423 (2021). [DOI] [PubMed] [Google Scholar]
- 9.Rhu, J., Choi, G. S., Kim, J. M., Kwon, C. H. D. & Joh, J. W. Risk factors associated with surgical morbidities of laparoscopic living liver donors. Ann. Surg.278, 96–102. 10.1097/SLA.0000000000005851 (2023). [DOI] [PubMed] [Google Scholar]
- 10.Hong, S. K. et al. Optimal bile duct division using real-time indocyanine green near-infrared fluorescence cholangiography during laparoscopic donor hepatectomy. Liver Transplant.23, 847–852. 10.1002/lt.24686 (2017). [DOI] [PubMed] [Google Scholar]
- 11.Mizuno, S. & Isaji, S. Indocyanine green (ICG) fluorescence imaging-guided cholangiography for donor hepatectomy in living donor liver transplantation. Am. J. Transplant.10, 2725–2726. 10.1111/j.1600-6143.2010.03288.x (2010). [DOI] [PubMed] [Google Scholar]
- 12.Maier-Hein, L. et al. Surgical data science for next-generation interventions. Nat. Biomed. Eng.1, 691–696. 10.1038/s41551-017-0132-7 (2017). [DOI] [PubMed] [Google Scholar]
- 13.Maier-Hein, L. et al. Surgical data science—From concepts toward clinical translation. Med. Image Anal.76, 102306. 10.1016/j.media.2021.102306 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ward, T. M. et al. Computer vision in surgery. Surgery169, 1253–1256. 10.1016/j.surg.2020.10.039 (2021). [DOI] [PubMed] [Google Scholar]
- 15.Sato, K. et al. Real-time detection of the recurrent laryngeal nerve in thoracoscopic esophagectomy using artificial intelligence. Surg. Endosc.36, 5531–5539. 10.1007/s00464-022-09268-w (2022). [DOI] [PubMed] [Google Scholar]
- 16.Kitaguchi, D. et al. Deep learning-based automatic surgical step recognition in intraoperative videos for transanal total mesorectal excision. Surg. Endosc.36, 1143–1151. 10.1007/s00464-021-08381-6 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kitaguchi, D. et al. Real-time automatic surgical phase recognition in laparoscopic sigmoidectomy using the convolutional neural network-based deep learning approach. Surg. Endosc.34, 4924–4931. 10.1007/s00464-019-07281-0 (2020). [DOI] [PubMed] [Google Scholar]
- 18.Anteby, R. et al. Deep learning visual analysis in laparoscopic surgery: A systematic review and diagnostic test accuracy meta-analysis. Surg. Endosc.35, 1521–1533. 10.1007/s00464-020-08168-1 (2021). [DOI] [PubMed] [Google Scholar]
- 19.Kitaguchi, D. et al. Real-time vascular anatomical image navigation for laparoscopic surgery: Experimental study. Surg. Endosc.36, 6105–6112. 10.1007/s00464-022-09384-7 (2022). [DOI] [PubMed] [Google Scholar]
- 20.Rhu, J., Choi, G.-S., Kim, J. M., Kwon, C. H. D. & Joh, J.-W. Risk factors associated with surgical morbidities of laparoscopic living liver donors. Ann. Surg.278, 96–102 (2023). [DOI] [PubMed] [Google Scholar]
- 21.Liu, H. et al. Investigation of the optimal indocyanine green dose in real-time fluorescent cholangiography during laparoscopic cholecystectomy with an ultra-high-definition 4K fluorescent system: A randomized controlled trial. Updates Surg.75, 1903–1910 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oh, N. et al. 3D auto-segmentation of biliary structure of living liver donors using magnetic resonance cholangiopancreatography for enhanced preoperative planning. Int. J. Surg.110, 1975–1982 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rhu, J. et al. A novel technique for bile duct division during laparoscopic living donor hepatectomy to overcome biliary complications in liver transplantation recipients: “Cut and clip” rather than “clip and cut”. Transplantation105, 1791–1799 (2021). [DOI] [PubMed] [Google Scholar]
- 24.Chen, L. C., Zhu, Y., Papandreou, G., Schroff, F. & Adam, H. Encoder–decoder with atrous separable convolution for semantic image segmentation. In Proceedings of the European Conference on Computer Vision (ECCV). https://arxiv.org/abs/1802.02611 (2018).
- 25.Russakovsky, O. et al. ImageNet large scale visual recognition challenge. Int. J. Comput. Vis.115, 211–252. 10.1007/s11263-015-0816-y (2015). [Google Scholar]
- 26.He, K., Zhang, X., Ren, S. & Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognitionhttps://arxiv.org/abs/1512.03385 (2016).
- 27.den Boer, R. B. et al. Computer-aided anatomy recognition in intrathoracic and -abdominal surgery: A systematic review. Surg. Endosc.36, 8737–8752. 10.1007/s00464-022-09421-5 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kitaguchi, D. et al. Computer-assisted real-time automatic prostate segmentation during TaTME: A single-center feasibility study. Surg. Endosc.35, 2493–2499. 10.1007/s00464-020-07659-5 (2021). [DOI] [PubMed] [Google Scholar]
- 29.Hashimoto, D. A., Rosman, G., Rus, D. & Meireles, O. R. Artificial intelligence in surgery: Promises and perils. Ann. Surg.268, 70–76. 10.1097/SLA.0000000000002693 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aeffner, F. et al. The gold standard paradox in digital image analysis: Manual versus automated scoring as ground truth. Arch. Pathol. Lab. Med.141, 1267–1275. 10.5858/arpa.2016-0386-RA (2017). [DOI] [PubMed] [Google Scholar]
- 31.Tajbakhsh, N. et al. Embracing imperfect datasets: A review of deep learning solutions for medical image segmentation. Med. Image Anal.63, 101693. 10.1016/j.media.2020.101693 (2020). [DOI] [PubMed] [Google Scholar]
- 32.Norori, N., Hu, Q., Aellen, F. M., Faraci, F. D. & Tzovara, A. Addressing bias in big data and AI for health care: A call for open science. Patterns (N. Y.)2, 100347. 10.1016/j.patter.2021.100347 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krishnan, R., Rajpurkar, P. & Topol, E. J. Self-supervised learning in medicine and healthcare. Nat. Biomed. Eng.6, 1346–1352. 10.1038/s41551-022-00914-1 (2022). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are not openly available due to reasons of sensitivity and are available from the corresponding author upon reasonable request.