Abstract
Rapid on-site cytopathology evaluation (ROSE) has been considered an effective method to increase the diagnostic ability of endoscopic ultrasound-guided fine needle aspiration (EUS-FNA); however, ROSE is unavailable in most institutes worldwide due to the shortage of cytopathologists. To overcome this situation, we created an artificial intelligence (AI)-based system (the ROSE-AI system), which was trained with the augmented data to evaluate the slide images acquired by EUS-FNA. This study aimed to clarify the effects of such data-augmentation on establishing an effective ROSE-AI system by comparing the efficacy of various data-augmentation techniques. The ROSE-AI system was trained with increased data obtained by the various data-augmentation techniques, including geometric transformation, color space transformation, and kernel filtering. By performing five-fold cross-validation, we compared the efficacy of each data-augmentation technique on the increasing diagnostic abilities of the ROSE-AI system. We collected 4059 divided EUS-FNA slide images from 36 patients with pancreatic cancer and nine patients with non-pancreatic cancer. The diagnostic ability of the ROSE-AI system without data augmentation had a sensitivity, specificity, and accuracy of 87.5%, 79.7%, and 83.7%, respectively. While, some data-augmentation techniques decreased diagnostic ability, the ROSE-AI system trained only with the augmented data using the geometric transformation technique had the highest diagnostic accuracy (88.2%). We successfully developed a prototype ROSE-AI system with high diagnostic ability. Each data-augmentation technique may have various compatibilities with AI-mediated diagnostics, and the geometric transformation was the most effective for the ROSE-AI system.
Subject terms: Cancer, Cancer imaging
Introduction
Endoscopic ultrasound-guided fine needle aspiration (EUS-FNA) has recently become widely used, and a high diagnostic ability for pancreatic solid lesions has been reported1–3. However, there is significant variability in the diagnostic ability of EUS-FNA among different facilities, and a certain number of patients require repetitive EUS-FNA due to a lack of adequate tissue4. Several factors, such as the number of needle passes, needle type, needle size, and endoscopists’ experience, are associated with the diagnostic ability of EUS-FNA3. Repetitive EUS-FNA lead to the physical and financial burden on patients, and methods to improve the diagnostic ability of EUS-FNA are necessary.
Rapid on-site cytopathology evaluation (ROSE) is performed on-site by a cytopathologist. It is an effective method to increase the diagnostic ability of EUS-FNA by providing immediate feedback on the characteristics and adequacy of the samples5,6. However, ROSE is unavailable in most institutions worldwide due to the shortage of cytopathologists7.
Recently, image recognition using a convolutional neural network (CNN) or transformer based on artificial intelligence (AI) has been used in various fields, including gastroenterology8. The replacement of manual ROSE with AI for ROSE will reduce the work of cytopathologists and the difference in diagnostic ability between the facilities. There are reports using deep learning in the pathological classification of pancreatic solid masses by a relatively small sample size of cytopathological slides from EUS-FNA9,10. Notably, many training images are required to create AI models with high accuracy; however, the collection and annotation of training images require great effort from medical specialists. The comprehensiveness of the data is vital in establishing efficient machine-learning models. Therefore, the differences between technicians and facilities during specimen processing pose a significant barrier to generalizability. In recent years, data-augmentation with various techniques has been reported as a valuable method for generating many image datasets from a small image dataset for training AI11,12. Data-augmentation may provide a solution to guarantee data comprehensiveness. However, to our knowledge, no study has compared the effectiveness of various data-augmentation techniques for optimal image datasets of AI training in the gastroenterological or pathological fields. We developed a computer-assisted diagnostic system for ROSE using the transformer-based AI (ROSE-AI system); however, the performance depends on the training images augmented by various data-augmentation techniques. This study aimed to verify the beneficial effects of the ROSE-AI system, which was trained by the augmented images, and to compare the effectiveness of various data-augmentation techniques.
Methods
Training data for the ROSE AI system
The ROSE-AI system was trained using slide images from patients who underwent EUS-FNA for solid pancreatic masses at Okayama University Hospital between April 2019 and September 2022. We collected EUS-FNA slides from patients with pancreatic and non-pancreatic cancer. All non-cancer patients were confirmed to have pathological findings of no malignancy by two experienced certified cytopathologists and underwent at least 6 months of follow-up with no rapid progression of pancreatic disease. Clinicopathological data, such as age, sex, size, and location of the lesions, were obtained from medical records. All patients provided written informed consent for the EUS-FNA.
Experienced endoscopists or trainees performed EUS-FNA using a curved linear array scanning scope (GF-UCT260; Olympus, Tokyo, Japan, or EG 580 UT; Fujifilm Co., Tokyo, Japan). Under endosonographic guidance, a needle puncture was performed using color Doppler to avoid vessels in the region. The needle type and size were determined at an endo-sonographer’s discretion. For each puncture, a cytopathological slide was prepared from the expressed material, air-dried, and Diff-Quik-stained by cytopathologists. The puncture procedure was repeated until an adequate specimen was obtained using ROSE.
Algorithm and design of the artificial intelligence system
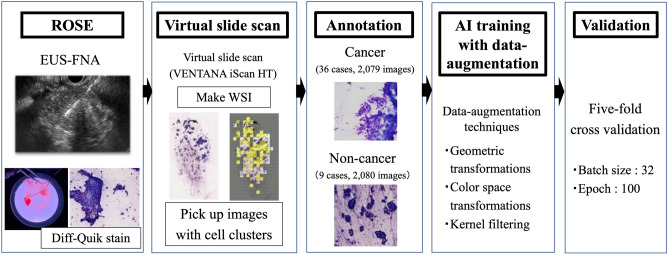
The establishment of the transformer-based AI system is shown in Fig. 1. First, EUS-FNA slides were digitized as whole-slide images (WSI) using a virtual slide scan (VENTANA iScan HT; Roche, Basel, Switzerland). The WSIs were then divided into smaller images (256 × 256 pixels). The divided images in which the ratio of the area with stained cells to the total image area exceeded a threshold (≥ 0.03) were automatically extracted for training data. The divided images were preprocessed for stain color normalization to maintain consistency. The divided images from the WSI were used for AI training to detect cancer cells automatically. The divided images were classified and annotated as those with and without cancer cells.
Figure 1.
Procedures to establish the ROSE-AI system in this study. At first, we collected EUS-FNA slides with Diff-Quik stained. Next, using a virtual slide scan, EUS-FNA slides were digitized as whole-slide images (WSI). The WSIs were then divided into smaller images (256 × 256 pixels), and the divided images were classified and annotated as those with and without cancer cells. Data augmentation was performed to increase the number of original data points. The ROSE-AI system was established using a transformer-based model, and we performed five-fold cross-validation.
The ROSE-AI system was established using a transformer-based model, the Vision transformer13. Transformers are a type of artificial neural network used in deep learning that has been used to analyze visual imagery. In our study, we adopted the Vision transformer model, which resized the input image to 224 × 224 pixels and then divided it into patches of 16 × 16 pixels. This model comprised 12 transformer layers, with a hidden size of 768 and multi-head attention of 12 heads. The total number of parameters was approximately 86 million. Of note, positional encoding was learnable within the model. For the final classification, the last layer of the model was replaced with a fully connected layer, and the original number of classes was modified to fit two classes of this study. This customization allowed the model to be adjusted to fit the specific task of this study. In this study, we performed a five-fold cross-validation due to the difficulty of collecting many images. The images of four-five from each group’s images were used as the training dataset, and the images of one-five from each group were used as the testing dataset. For each fold of the five-fold cross-validation, the dataset was randomly partitioned into training and validation sets in an 8:2 ratio, and each fold was experimented with approximately 3320 test images and 830 validation images. However, a separate test set was held out for final evaluation. The transformer was trained using the dataset and validated using the PyTorch deep learning framework. Notably, all layers of the transformer were fine-tuned from the weights of ImageNet using stochastic gradient descent as an optimizer with a learning rate of 0.0001, 100 epochs, and a batch size 32. The appropriate learning rate was determined through repeated trial and error. The optimal number of epochs with the lowest loss points in the validation set was selected. Finally, a layer-wise relevance propagation (LRP) method was used to visualize and evaluate our AI system14.
Comparison of ROSE-AI systems with different data-augmentation techniques
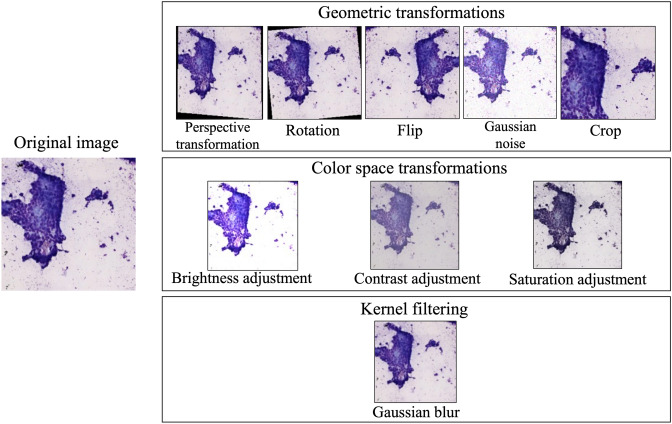
We used three data-augmentation techniques: geometric transformations, color-space transformations, and kernel filtering (Fig. 2). Geometric transformation is a common data-augmentation technique, and it has the advantage of preserving cell morphology even after data-augmentation by rotation, flip and crop. When performing ROSE, factors such as lighting conditions, microscope performance, and staining variations across facilities could potentially affect the images. Therefore, color space transformation ensured generalizability, and kernel filtering was used to adapt minor blurriness in microscope images. Details of the data-augmentation techniques used in this study are shown in Table 1. The data-augmentation formulae used are shown in Supplemental Fig. 115–22. Transformations by data-augmentation are associated with probability and magnitude. Data-augmentation was performed to increase the number of original data points. Each time an image is loaded into memory, the data-augmentation techniques are applied according to their respective probabilities and magnitude. We compared the diagnostic abilities of ROSE-AI systems alone and in combination with the three data-augmentation techniques. All methods were performed according to relevant guidelines and regulations.
Figure 2.
Examples of data augmentation techniques. We used three data augmentation techniques: geometric transformations, color-space transformations, and kernel filtering. Geometric transformations included perspective transformations, rotation, flipping, Gaussian noise, and cropping. Color-space transformations included brightness, contrast, and saturation adjustments. Gaussian blur was applied as Kernel filtering.
Table 1.
List of the data-augmentation techniques used in this study.
| Category | Name of method | Description |
|---|---|---|
| Geometric transformations | Perspective transformation | Transform the image from three-dimensional coordinates to two-dimensional coordinates by magnitude degrees |
| Rotation | Rotate the image by magnitude degrees | |
| Flip | Horizontal and vertical reversal of the image | |
| Gaussian noise | Add noise to the image by magnitude degrees | |
| Crop | A portion of the original image is randomly selected and removed | |
| Color space transformations | Brightness adjustment | Adjust the brightness by magnitude degrees |
| Contrast adjustment | Adjust the contrast by magnitude degrees | |
| Saturation adjustment | Adjust saturation by magnitude degrees | |
| Kernel filtering | Gaussian blur | The image is slightly blurred or smoothed out by magnitude degrees |
Geometric transformations
Geometric transformations include perspective transformations, rotation, flipping, Gaussian noise, and cropping. The perspective transformation was randomly performed with a probability of 50% and a magnitude of 0.2. Rotation was randomly performed with a probability of 100% and a magnitude of -10° to + 10°. For flipping, horizontal and vertical flips were randomly applied with a probability of 50% each. Gaussian noise was applied with a probability of 50%, and the noise followed a Gaussian distribution with a mean of 0 and variance in the range of (10, 50). A crop with a probability of 100% was selected by randomly cutting out a region. In geometric transformations, all these methods were applied simultaneously.
Color-space transformations
Color-space transformations include brightness, contrast, and saturation adjustments. Brightness adjustment was performed with a probability of 100% by randomly adjusting the magnitude between 0.9 and 1.5. Contrast adjustment was applied with a probability of 100% by randomly adjusting the magnitude between 0.9 and 1.3. Saturation adjustment was performed with a probability of 100% by randomly adjusting the magnitude between 0.8 and 1.4. In color space transformations, all these methods were applied simultaneously.
Kernel filtering
Gaussian blur was applied with a probability of 100% using a filter with a kernel size (3,3) and standard deviation ranging (0.01, 2.0).
Combination of data augmentation techniques
Each method's probability and magnitude remained the same as those described above using a combination of data-augmentation techniques.
Statistical analyses
Categorical variables were reported as percentages, whereas continuous variables were reported as median and interquartile range (IQR). The diagnostic ability was determined regarding sensitivity, specificity, and accuracy for diagnosing “Cancer” with a 95% confidence interval (CI). The receiver operating characteristic (ROC) curve and area under the curve (AUC) of the classification accuracy of the ROSE-AI system were described and calculated. Analyses were performed using Mac’s JMP Pro15 software program (SAS Institute Inc., Cary, NC, USA).
Ethical consideration
This study was approved by our hospital’s Institutional Review Board (approval number: 1908–057).
Results
Patient characteristics of training data
Seventy EUS-FNA slides were collected from 45 patients. A total of 36 cases (55 EUS-FNA slides) of pancreatic cancer and nine cases (six cases of autoimmune pancreatitis, three cases of chronic pancreatitis, and 15 EUS-FNA slides) of non-pancreatic cancer were included. From these slides, we prepared 2,079 divided images with cancer cells and 2,080 with non-cancer cells as the training data. Datasets of images that were increased by data-augmentation were used for the analysis.
Patient characteristics are shown in Table 2. The median age of the patients was 70 years (IQR: 67–77), and 58% were males. Regarding the disease location, the pancreas’s head, body, and tail accounted for 40%, 38%, and 22%, respectively. Among them, 80% of the final cytological diagnoses were adenocarcinomas; the rest were non-malignant. Consistently, the final diagnoses using the tissue pathology were pancreatic cancer (n = 36; 80%), autoimmune pancreatitis (n = 6; 13%), and chronic pancreatitis (n = 3; 7%).
Table 2.
Clinical characteristics of the patients in the present study.
| Parameters | |
|---|---|
| Age, median (IQR), years | 70 (67–77) |
| Sex, male, n (%) | 26 (58) |
| Size of the lesion, median (IQR), cm | 25 (18–36) |
| Location, n (%) | |
| Head | 18 (40) |
| Body | 17 (38) |
| Tail | 10 (22) |
| Cytopathological diagnosis, n (%) | |
| Adenocarcinoma | 36 (80) |
| Non-malignancy | 9 (20) |
| Final diagnosis, n (%) | |
| Pancreatic cancer | 36 (80) |
| Autoimmune pancreatitis | 6 (13) |
| Chronic pancreatitis | 3 (7) |
IQR, Interquartile range.
Comparison of diagnostic ability of the ROSE-AI system trained with different data-augmentation techniques
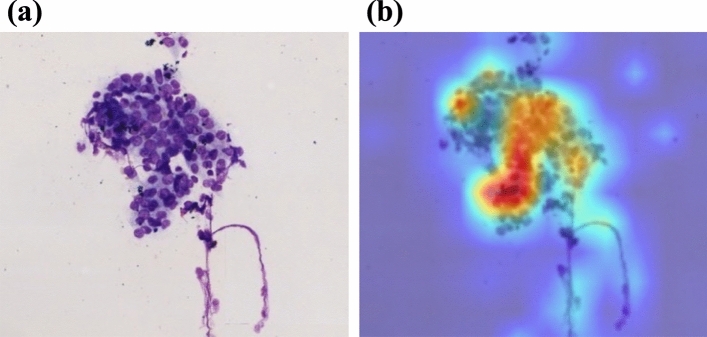
The diagnostic ability of the ROSE-AI system, which automatically evaluates the images of slides obtained by EUS-FNA without data-augmentation, had a sensitivity, specificity, and accuracy of 87.5% (95% CI 86.6–88.8), 79.7% (95% CI 78.5–80.7), and 83.7% (95% CI 82.6–84.7), respectively. Representative images of the ROSE-AI system detecting cancer cells are presented in Fig. 3.
Figure 3.
Representative images of the ROSE-AI system detecting cancer cells. A layer-wise relevance propagation (LRP) method was used to visualize and evaluate the ROSE-AI system. (a) Original image. (b) Enhanced image by LRP method: ROSE-AI system focuses on the red area.
The diagnostic abilities of the ROSE-AI system trained with each data-augmentation technique are shown in Table 3. Using individual data-augmentation technique, the sensitivity, specificity, and accuracy were 91.4% (95% CI 90.4–92.3), 85.0% (95% CI 84.0–85.9), and 88.2% (95% CI 87.2–89.1) in geometric transformations, 87.2% (95% CI 86.1–88.3), 77.8% (95% CI 76.6–78.9), and 82.6% (95% CI 81.3–83.6) in color space transformations, 87.1% (95% CI 84.5–89.2), 76.3% (95% CI 73.8–78.4), and 81.8% (95% CI 79.3–83.9) in kernel filtering, respectively. When multiple data-augmentation techniques were used in combination, the sensitivity, specificity, and accuracy were 86.7% (95% CI 85.5–87.8), 68.7% (95% CI 67.5–69.8), and 77.7% (95% CI 76.5–78.8) in geometric and color space transformations, 84.6% (95% CI 81.9–86.8), 76.2% (95% CI 73.6–78.5) and 80.4% (95% CI 77.8–82.3) in color space transformations and kernel filtering, 91.9% (95% CI 89.9–93.4), 83.5% (95% CI 81.6–84.9), and 87.7% (95% CI 85.8–89.2) in kernel filtering and geometric transformations, 86.8% (95% CI 84.2–89.1), 73.7% (95% CI 71.1–75.9), and 80.4% (95% CI: 77.8–82.3) in geometric and color space transformations and kernel filtering, respectively.
Table 3.
Diagnostic ability of the AI system with each data-augmentation method.
| Sensitivity | Specificity | Accuracy | PPV | NPV | |
|---|---|---|---|---|---|
| No data-augmention | 87.5 | 79.7 | 83.7 | 81.8 | 86.7 |
| G | 91.4 | 85.0 | 88.2 | 86.7 | 91.7 |
| C | 87.2 | 77.8 | 82.6 | 80.0 | 86.1 |
| K | 87.1 | 76.3 | 81.8 | 79.5 | 85.7 |
| G + C | 86.7 | 68.7 | 77.7 | 74.1 | 83.6 |
| C + K | 84.6 | 76.2 | 80.4 | 78.8 | 83.5 |
| K + G | 91.9 | 83.5 | 87.7 | 85.1 | 92.0 |
| G + C + K | 86.8 | 73.7 | 80.4 | 77.4 | 85.1 |
AI, Artificial intelligence.
PPV, Positive predictive value.
NPV, Negative predictive value.
G, Geometric transformations.
C, Color space transformations.
K, Kernel filtering.
The ROC curves for the ROSE-AI system with each data-augmentation technique are shown in Fig. 4. The AUC for the ROSE-AI system without data-augmentation was 0.902. Using individual data-augmentation techniques, the AUCs were 0.954 for geometric transformations, 0.896 for color space transformations, and 0.895 for kernel filtering. Using a combination of data-augmentation techniques, the AUCs were 0.873 for geometric and color space transformations, 0.887 for color space transformations and kernel filtering, 0.946 for kernel filtering and geometric transformations, and 0.881 for geometric and color space transformations and kernel filtering. Therefore, the ROSE-AI system trained only with the augmented data using the geometric transformation technique had the highest diagnostic accuracy.
Figure 4.
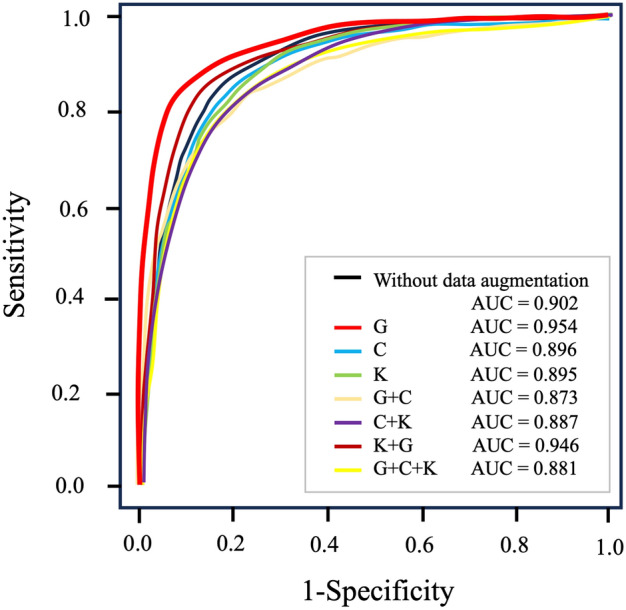
The ROC curves for diagnosing pancreatic cancer by the ROSE-AI system using different data-augmentation techniques. The diagnostic performance for each data augmentation method is shown. The ROSE-AI system trained only with the augmented data using the geometric transformation technique had the highest diagnostic accuracy (AUC = 0.954). ROC, receiver operating characteristic; AUC, area under the curve; G, geometric transformations; C, color space transformations; K, kernel filtering.
Discussion
Recently, computer-aided diagnosis (CAD) using AI models has been developed in various clinical fields to support the shortage of pathologists23,24. This shortage also results in a disincentive for the popularization of ROSE in EUS-FNA7. If the ROSE-AI system is put into practical use and becomes widespread, it will reduce the burden of cytopathologists and the difference in diagnostic ability between facilities. It is also expected to reduce patients’ physical and financial burdens from reexamination and readmission.
ROSE-AI system is expected to be widely used instead of the conventional effort-consuming ROSE. Previous studies have reported the effectiveness of deep learning in the pathological classification of pancreatic solid masses in EUS-FNA9,10. One study using deep learning methods reported that the diagnostic accuracy was 83.4% in internal validation and 88.7% in external validation using 467 digitized images9. However, in the study, there was an imbalance in the number of images used across the categories due to a shortage of images. Another study reported that the AUC for cancer diagnosis using the AI with deep learning was 0.95810. They also showed generalizable and robust performance on internal datasets, external datasets, and subgroup analysis. In addition, the performance of their system was superior to that of trained endoscopists and comparable to that of cytopathologists on their testing datasets. An AI system with excellent performance does not always lead to general usage clinically in the real world because of its bias due to the insufficient datasets used for the training. Data comprehensiveness is essential to ensure the generalizability of the ROSE-AI system, which requires large quantities of datasets. Obtaining sufficient training data to improve diagnostic ability remains a major challenge in creating the ROSE-AI system. Indeed, pathological re-evaluation requires significant effort when creating a training dataset from EUS-FNA slides.
Data-augmentation has been reported as a valuable technique to compensate for efficient learning in AI training11,12. It has the advantage of increasing the amount of training data by creating new images from the original images. AI training with data-augmentation has been reported as a useful technique for creating AI systems that detect Barrett’s esophagus and colorectal polyps in gastroenterology25,26. However, few reports focus on the differences among data-augmentation techniques in the deep learning approach for CAD. One study compared the data-augmentation techniques of rotation, scaling, and distortion, and the rotation technique was the only method to improve the diagnostic ability for peripheral blood leukocyte recognition in the field of hematology27. Another study optimized data-augmentation and CNN hyperparameters for detecting coronavirus disease 2019 from chest radiographs regarding validation accuracy28. The study evaluated common augmentation techniques in the chest radiograph classification literature (resize value, resize method, rotate, zoom, warp, light, flip, and normalize), recently proposed methods (mixup and random erasing), and combinations of these methods. Individual data-augmentation methods yielded slightly increased task performance, and “Rotate” showed the highest AUC (0.965) performance. In addition, the combination of these optimized methods significantly improved the performance. Therefore, optimal data-augmentation techniques are effective for the efficient training datasets for AI.
To our knowledge, this is the first report to compare various data-augmentation techniques on the AI system of ROSE. Our study demonstrates that data-augmentation influences the performance of an AI system. Data-augmentation may provide a solution to guarantee data comprehensiveness. However, it seems there are compatibilities between various data-augmentation techniques and the contents of AI training. This study also demonstrated that combining various data-augmentation techniques does not necessarily produce synergistic effects. Therefore, we should select the highly effective data-augmentation technique from different techniques. Cytological diagnosis is based on the characteristics of cells or cell clusters, such as nuclear enlargement, variability in nuclear size, and irregularity of nuclear margins. Therefore, techniques such as color space transformations and kernel filtering may render the morphology unclear and work unfavorably in cellular diagnosis. Geometric transformations may be more useful than other data-augmentation techniques in the ROSE-AI system and other morphological diagnostic AI systems, such as endoscopic and computed tomography (CT)/magnetic resonance imaging (MRI) images. One study demonstrated that geometric transformations help detect prostate cancer in diffusion-weighted MRI using a CNN29. However, the study reported that the noise augmentation method's effect is insignificant. There are various techniques, including perspective transformation, rotation, flip, noise, and crop in geometric transformations, and the effectiveness of each technique may vary in terms of improving diagnostic performance. We tried to select the best effective technique from various data-augmentation techniques for the ROSE-AI system. However, because the advances in this field are much faster than we expected, new data-augmentation techniques other than those we used may become more suitable.
Various recommendations have been made for the development of AI systems with a focus on societal implementation30. Efficient and continuous data-collection, annotation, and data-augmentation are essential for creating such high-quality systems. Currently, the construction of large-scale registries and datasets for the development of AI systems is being planned and executed31. By managing the data with considering data augmentation from the collection stage, AI system can be more efficiently developed32. Additionally, flexible annotation of training data is important according to the product concept33. To develop AI systems that can be socially implemented, it is crucial to build a well-coordinated team of specialists, including data managers, programmers, and clinical physicians.
This study has some limitations. First, the training and evaluation of the AI system were performed using data collected retrospectively from only one institution. The detailed methods for creating pathological slides vary among facilities. Therefore, it is unclear whether the ROSE-AI system can be used with the same diagnostic ability at other facilities if the training data are collected from only one facility. Second, our study was analyzed using five-fold cross-validation, and we could not perform a validation study using new images. In the future, prospective studies should be conducted to further evaluate the value of the ROSE-AI system in the diagnosis of EUS-FNA in clinical practice. To evaluate the actual usefulness of the ROSE-AI system, comparing the diagnostic abilities of ROSE-AI, endoscopists, and pathologists through external validation is also necessary. Third, we evaluate the small types of pancreatic diseases, including pancreatic cancers, autoimmune pancreatitis, and chronic pancreatitis. Accordingly, evaluation including other pancreatic diseases such as neuroendocrine neoplasms and solid pseudopapillary neoplasms is necessary in the next stage.
In conclusion, the geometric transformation technique is the most useful technique for training the ROSE-AI system. We believe the efficient ROSE-AI system will soon assist endoscopists in performing ROSE where cytopathologists are unavailable.
Supplementary Information
Author contributions
YF and UD: writing, conception, and design of the study. RS, TO, MA, KM, KM, HT, TY, KM, SH, KT, and HI: collecting the training data and annotation. CT, TT, and AO: Data analysis. HK and YK : critically revising the article for important intellectual content. MO: Final approval of the article.
Funding
This work was supported by JSPS KAKENHI Grant Number 23K11932 (granted to D.U.), JSPS KAKENHI Grant Number 24K18948 (granted to Y.F.) and Grant from Okayama prefecture (awarded to Y.F.).
Data availability
The datasets analyzed during the current study are available from the corresponding author on reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-72312-3.
References
- 1.Yoshinaga, S. et al. Safety and efficacy of endoscopic ultrasound-guided fine needle aspiration for pancreatic masses: a prospective multicenter study. Dig. Endosc.32, 114–126 (2020). [DOI] [PubMed] [Google Scholar]
- 2.Dumonceau, J. M. et al. Indications, results, and clinical impact of endoscopic ultrasound (EUS)-guided sampling in gastroenterology: European Society of Gastrointestinal Endoscopy (ESGE) clinical guideline-updated January 2017. Endoscopy49(07), 695–714 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Bang, J. Y., Hawes, R. & Varadarajulu, S. A meta-analysis comparing Procore and standard fine-needle aspiration needles for endoscopic ultrasound-guided tissue acquisition. Endoscopy48, 339–349 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Hawes, R. H. The evolution of endoscopic ultrasound: improved imaging, higher accuracy for fine needle aspiration and the reality of endoscopic ultrasound-guided interventions. Curr. Opin. Gastroenterol.26, 436–444 (2010). [DOI] [PubMed] [Google Scholar]
- 5.Schmidt, R. L., Walker, B. S., Howard, K., Layfield, L. J. & Adler, D. G. Rapid on-site evaluation reduces needle passes in endoscopic ultrasound-guided fine-needle aspiration for solid pancreatic lesions: a risk benefit analysis. Dig. Dis. Sci.58, 3280–3286 (2013). [DOI] [PubMed] [Google Scholar]
- 6.Matynia, A. P. et al. Impact of rapid on-site evaluation on the adequacy of endoscopic-ultrasound guided fine-needle aspiration of solid pancreatic lesions: a systematic review and meta-analysis. J. Gastroenterol. Hepatol.29, 697–705 (2014). [DOI] [PubMed] [Google Scholar]
- 7.Lewin, D. Optimal EUS-guided FNA cytology preparation when rapid on-site evaluation is not available. Gastrointest Endosc.91, 847–848 (2020). [DOI] [PubMed] [Google Scholar]
- 8.Xu, Y. et al. Comparison of diagnostic performance between convolutional neural networks and human endoscopists for diagnosis of colorectal polyp: a systematic review and meta-analysis. PLoS ONE16, e0246892 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin, R. et al. Application of artificial intelligence to digital-rapid on-site cytopathology evaluation during endoscopic ultrasound-guided fine needle aspiration: a proof-of-concept study. J. Gastroenterol. Hepatol.10, 16073 (2022). [DOI] [PubMed] [Google Scholar]
- 10.Zhang, S. et al. A deep learning-based segmentation system for rapid onsite cytologic pathology evaluation of pancreatic masses: A retrospective, multicenter, diagnostic study. EBiomedicine80, 104022 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kebaili, A., Lapuyade-Lahorgue, J. & Ruan, S. Deep learning approaches for data augmentation in medical imaging: a review. J. Imaging9, 9 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chlap, P. et al. A review of medical image data augmentation techniques for deep learning applications. J. Med. Imaging Radiat. Oncol.65, 545–563 (2021). [DOI] [PubMed] [Google Scholar]
- 13.Dosoviskiy, A. et al. An image is worth 16x16 words: transformers for image recognition at scale. Arxiv2010, 11929 (2020). [Google Scholar]
- 14.Chefer Hila, Gur S, Wolf L. Transformer interpretability beyond attention visualization. 2021 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Nashville, TN, USA. 782–91 (2021).
- 15.Soumith, C. PyTorch RandomPerspective. PyTorchhttps://pytorch.org/vision/main/generated/torchvision.transforms.RandomPerspective.html (2017).
- 16.Soumith, C. PyTorch RandomRotation. PyTorchhttps://pytorch.org/vision/main/generated/torchvision.transforms.RandomRotation.html (2017).
- 17.Soumith, C. PyTorch RandomHorizontalFlip. PyTorchhttps://pytorch.org/vision/main/generated/torchvision.transforms.RandomHorizontalFlip.html (2017).
- 18.Soumith, C. PyTorch RandomVerticalFlip. PyTorchhttps://pytorch.org/vision/main/generated/torchvision.transforms.RandomVerticalFlip.html (2017).
- 19.Soumith, C. PyTorch GaussNoise. PyTorchhttps://albumentations.ai/docs/api_reference/augmentations/transforms/#albumentations.augmentations.transforms.GaussNoise (2017).
- 20.Soumith, C. PyTorch RandomCrop. PyTorchhttps://pytorch.org/vision/main/generated/torchvision.transforms.RandomCrop.html (2017).
- 21.Soumith, C. PyTorch ColorJitter. PyTorchhttps://pytorch.org/vision/main/generated/torchvision.transforms.ColorJitter.html (2017).
- 22.Soumith, C. PyTorch GaussianBlur. PyTorchhttps://pytorch.org/vision/0.18/generated/torchvision.transforms.GaussianBlur.html (2017).
- 23.Hermsen, M. et al. Deep learning-based histopathologic assessment of kidney tissue. J. Am. Soc. Nephrol.30, 1968–1979 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sanyal, P., Mukherjee, T., Barui, S., Das, A. & Gangopadhyay, P. Artificial intelligence in cytopathology: a neural network to identify papillary carcinoma on thyroid fine-needle aspiration cytology smears. J. Pathol. Inform.9, 43 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Souza, L. A. et al. Assisting Barrett’s esophagus identification using endoscopic data augmentation based on Generative Adversarial Networks. Comput. Biol. Med.126, 104029 (2020). [DOI] [PubMed] [Google Scholar]
- 26.Adjei, P. E., Lonseko, Z. M., Du, W., Zhang, H. & Rao, N. Examining the effect of synthetic data augmentation in polyp detection and segmentation. Int. J. Comput. Assist. Radiol. Surg.17, 1289–1302 (2022). [DOI] [PubMed] [Google Scholar]
- 27.Nozaka, H. et al. The effect of data augmentation in deep learning approach for peripheral blood leukocyte recognition. Stud. Health Technol. Inform.290, 273–277 (2022). [DOI] [PubMed] [Google Scholar]
- 28.Monshi, M. M. A., Poon, J., Chung, V. & Monshi, F. M. CovidXrayNet: Optimizing data augmentation and CNN hyperparameters for improved COVID-19 detection from CXR. Comput. Biol. Med.133, 104375 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hao, R., Namdar, K., Liu, L., Haider, M. A. & Khalvati, F. A Comprehensive study of data augmentation strategies for prostate cancer detection in diffusion-weighted MRI using convolutional neural networks. J. Digit Imaging34, 862–876 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lococo, F. et al. Implementation of artificial intelligence in personalized prognostic assessment of lung cancer: a narrative review. Cancers16(10), 1832 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shigeki A. Japan medical image database, https://www.radiology.jp/j-mid/ (2020).
- 32.DAMA International. Data Management Body of Knowledge (DMBOK) (2nd ed.). (2017).
- 33.Keisuke, H. et al. Detecting colon polyps in endoscopic images using artificial intelligence constructed with automated collection of annotated images from an endoscopy reporting system. Dig. Endosc.34, 1021–1029 (2022). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets analyzed during the current study are available from the corresponding author on reasonable request.