Abstract
CD4-immunoglobulin G2 (CD4-IgG2) incorporates four copies of the D1D2 domains of CD4 into an antibody-like molecule that potently neutralizes primary human immunodeficiency virus type 1. Here electron microscopy was used to explore the structure and functional valence of CD4-IgG2 in complex with gp120. CD4-γ2, a divalent CD4-immunoglobulin fusion protein, was evaluated in parallel. Whereas CD4-γ2–gp120 complexes adopted a simple Y-shaped structure, CD4-IgG2–gp120 complexes consisted of four gp120s arrayed about a central CD4-IgG2 molecule, a structure more reminiscent of complement C1q. Molecular modeling corroborated the electron microscopy data and further indicated that CD4-IgG2 but not CD4-γ2 has significant potential to cross-link gp120-gp41 trimers on the virion surface, suggesting a mechanism for the heightened antiviral activity of CD4-IgG2.
CD4-immunoglobulin G2 (CD4-IgG2; PRO 542) is a recombinant antibody-like fusion protein wherein the heavy- and light-chain variable domains of human IgG2 have been replaced with the D1D2 domains of human CD4 (2). CD4-γ2 is a homodimer comprising D1D2 genetically fused to the hinge and Fc region of the γ2 heavy chain (2). Unlike monovalent and divalent CD4-based proteins, CD4-IgG2 broadly and potently neutralizes primary human immunodeficiency virus type 1 (HIV-1) isolates (3, 7, 9, 10, 28, 29) and has demonstrated encouraging antiviral activity in humans (14, 25). In this study, we used electron microscopy (EM) and molecular modeling to explore the structures of CD4-IgG2 and CD4-γ2 alone and in complex with gp120. These data were used to estimate the ability of these agents to cross-link gp120s either within or between the trimeric envelope spikes on the virion surface.
Immunoelectron microscopy.
Negative-staining immunoelectron microscopy was performed as described previously (22, 23). Individually, neither CD4-IgG2 nor CD4-γ2 produced interpretable images typical of IgG molecules. Both appeared to be considerably reduced in mass compared to IgG and often lacked distinctive Fc and Fab arms (data not shown), observations that are not unexpected given that the CD4 D1D2 components are more filamentous (narrow) than the IgG Fab arms they replace.
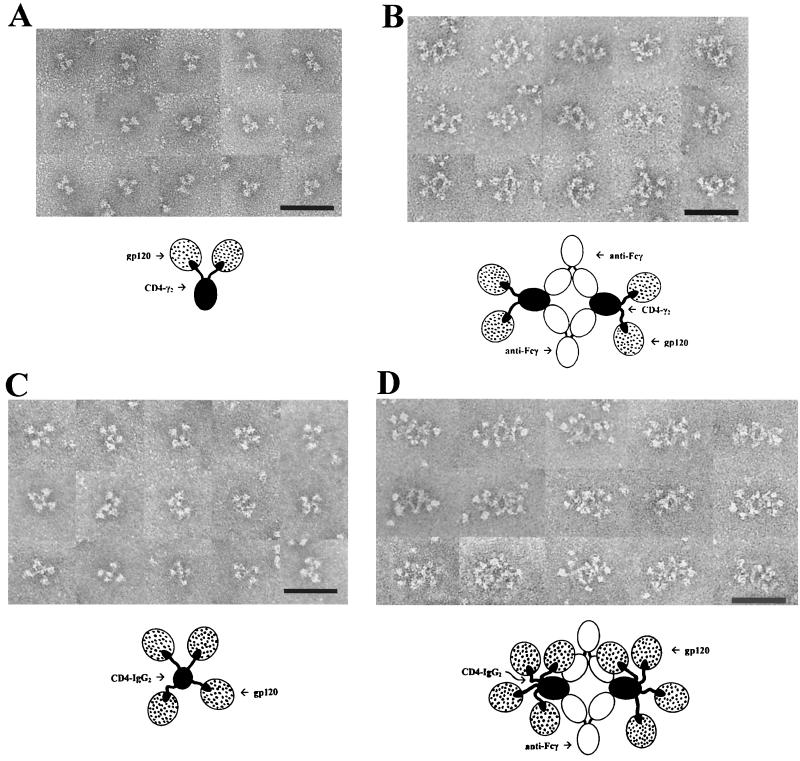
However, interpretable images emerged when both molecules were reacted with recombinant, monomeric HIV-1JR-FL gp120 (27). CD4-γ2–gp120 complexes (Fig. 1A) typically appeared as trilobed structures comprising two rounded structures (putative gp120s) in association with a smaller ovoid structure (putative Fc). The rounded and ovoid structures were sometimes separated by a gap that was often bridged by a thin filament (putative D1D2). The spatial relationships between the structures were variable, suggesting considerable segmental flexibility. To establish the identities of the individual elements of the complexes, they were reacted with a monoclonal antibody (MAb) to human IgG Fc (MAb 1302; Chemicon, Temecula, Calif.). This approach lead to the formation of four-membered ring structures with fixed geometries composed of two anti-Fc MAbs cross-linking two CD4-γ2 molecules (Fig. 1B). In these images, gp120 and Fc are identified unequivocally. Up to two gp120 molecules laterally protrude from opposite sides of the complexes and appear to be attached by filamentous arms. Complexes formed in the absence of gp120 did not display the lateral globular structures (data not shown).
FIG. 1.
Electron micrographs and interpretive diagrams of CD4-γ2 and CD4-IgG2 in complex with monomeric recombinant HIV-1JR-FL gp120. Bar = 50 nm. (A) CD4-γ2 and gp120; (B) CD4-γ2, gp120, and anti-Fc MAb; (C) CD4-IgG2 and gp120; (D) CD4-IgG2, gp120, and anti-Fc MAb.
CD4-IgG2–gp120 complexes (Fig. 1C) typically consisted of four irregular spheres (putative gp120s) arrayed about a smaller central structure (putative Fc). The putative gp120s were clearly attached to the central structure by thin strands. These images resemble those obtained for C1q (26), whose central stalk often binds perpendicular to the membrane while the six arms with their globular heads protrude radially. When anti-Fc MAb is added, the many attached gp120s form a crowded image wherein some of the gp120s may be folded back over one another or over the Fc. Nevertheless, at least three gp120s per CD4-IgG2 are seen in most of the complexes (Fig. 1D). Compared with IgG, the Fab arms of CD4-IgG2 appear less compact, consistent with their ability to project radially and independently of one another. In this arrangement, each arm would serve as a flexible tether for gp120. The results demonstrate that CD4-IgG2 can bind four gp120 monomers simultaneously with minimal steric interference between D1D2 domains.
Molecular modeling of CD4-IgG2 and CD4-γ2 in complex with monomeric gp120, trimeric gp120 and arrays of trimeric gp120.
Models were constructed using the SwissPDBviewer (http://www.expasy.ch/spdbv/text/index.htm) and the O Protein Crystallographic Package (http://imsb.au.dk/∼mok/o/) and used to lend support to the EM-based interpretations and investigate the likely mode of binding to viral surfaces. The CD4-IgG2 model was constructed using the constant regions of human IgG1κ Fab 3D6 (Protein Data Bank [PDB] access code 1DFB [12]), the Fc domain of human IgG1 antibody POT (PDB code 1FC1 [4]), and the D1D2 domains of CD4 (PDB code 1CDH [24]). Amino acids and disulfide bonds in the hinge and surrounding regions were converted from IgG1 to IgG2 and structurally refined using O. D1D2 (K1 to F179) was grafted onto A114 of the CH1 domain and T109 of the light chain (Kabat numbering [15]). The CD4-γ2 model was constructed by linking D1D2 to E226 of the γ2 hinge region.
Models of CD4-γ2 and CD4-IgG2 in complex with monomeric gp120 were constructed using the X-ray structure of the gp120 core in complex with D1D2 and Fab 17b (PDB code 1GC1 [16]).
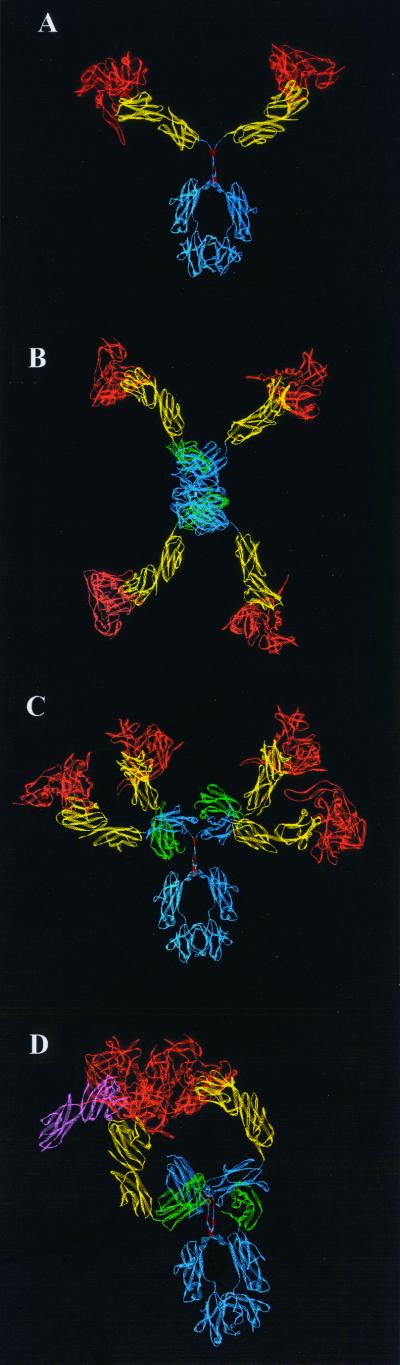
The model of CD4-γ2 in complex with gp120 (Fig. 2A) is consistent with the EM images in that the three lobes correspond to the Ig Fc (blue) and two CD4 D1D2 domain pairs (yellow) in complex with two gp120s (orange). For CD4-IgG2–gp120 complexes, four gp120s could easily be arrayed about a central Fc standing “on end” (e.g., compare Fig. 2B and 1C). In contrast to the more compact structure of intact IgG Fab, the CD4 arms of CD4-IgG2 protrude tangentially from the CL (green) and CH1 domains (blue), providing considerable flexibility with minimal steric inhibition. Thus, CD4-IgG2 appears to be functionally tetravalent. The CD4-IgG2–gp120 model is shown in a side view in Fig. 2C for comparison.
FIG. 2.
Models of monomeric (A to C) and trimeric (D) gp120 cores in complex with CD4-γ2 and CD4-IgG2. (A) CD4-γ2 with monomeric gp120; (B and C) top and side views, respectively, of CD4-IgG2 with monomeric gp120; (D) CD4-IgG2 with trimeric gp120. For clarity, only two of the four D1D2 domains of CD4-IgG2 are shown in panel D. Color codes: blue, Ig heavy chain; red, IgG2 hinge region disulfide bonds; green, light-chain Cκ domain; yellow, D1D2 domains of CD4-IgG2; orange, gp120 core. The pink structure in panel D represents the D1D2 domains docked in the nearest CD4 binding site not occupied by CD4-IgG2 and is included for orientation purposes. Note that the left-hand CD4 arm (yellow) of CD4-IgG2 is unable to extend and maneuver into the requisite docking position (pink CD4) of the left-hand gp120 subunit.
The potential for polyvalent interaction (i.e., cross-linking) of a single anti-gp120 construct with a single gp120 trimer, as would appear on a viral or infected-cell surface, was tested. Trimeric gp120 (orange) was modeled as described previously (17). Upon docking with CD4-IgG2 (Fig. 2D) and CD4-γ2 (data not shown), it was apparent that no amount of flexibility in the hinge, CH1, or CL regions would allow cross-linking of intratrimer gp120s. Intratrimer cross-linking would require rotation of the D1 and D2 domains relative to one another such that significant steric clashes occur (data not shown). Thus, neither molecule appears to be capable of cross-linking intratrimer gp120s, as originally suggested (17).
Various biochemical, crystallographic, EM, and modeling data suggest that the p17 matrix protein forms trimers that are organized into a paracrystalline icosahedral matrix below the membrane of mature virions (6, 13, 21), forming a mesh of interconnected rings into which the cytoplasmic tail of gp41 may insert (6). This pattern positions the envelope trimers at the vertices of equilateral triangles having a center-to-center distance of 210 Å. The model is consistent with EM data indicating an initial 70 to 80 envelope spikes per virion (pre-gp120 shedding) and similar center-to-center distances (11, 20, 21).
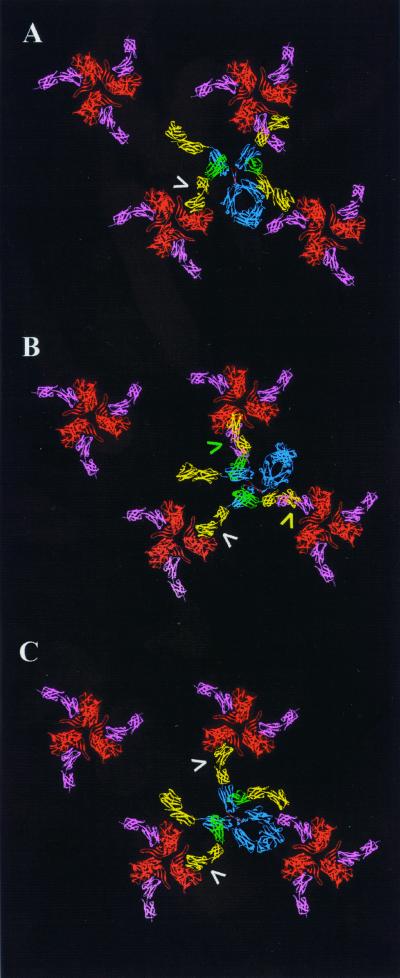
Based on this information, a symmetric assembly of gp120 trimers was constructed to test for the possibility of intertrimer cross-linking. Trimers were first rotated so as to orient the CD4 binding site (occupied by phantom pink CD4 D1D2 molecules [Fig. 3]) toward neighboring trimers. When docked into CD4 binding sites of one such trimer (Fig. 3A, arrow), CD4-IgG2 spanned to two adjacent trimers, but none of the free D1D2 domains (yellow) was in a position compatible with docking. We next rotated each of the trimers 20° clockwise so as to place the C termini of bound D1D2 segments in closest proximity. When an initial D1D2 domain of CD4-IgG2 was fully docked (Fig. 3B, white arrow), two additional CD4s either overshot (green arrow) or undershot (yellow arrow) adjacent trimers by 12 and 23 Å, respectively. In a third model, we rotated each of the gp120 trimers in unison to search for other configurations that could accommodate cross-linking. Figure 3C shows one orientation (45° clockwise with respect to Fig. 3A) favorable for two fully docked CD4 arms. A final model allowed free rotation of the gp120 trimers. The most favorable of the configurations were similar to that in Fig. 3C and allowed two but not three arms of CD4-IgG2 to fully dock (data not shown). It should be noted that our model assumes that the D1D2 domains remain rigid. Any additional segmental flexibility would further increase the potential for cross-linking. Because of its smaller reach (165 versus 220 Å for CD4-IgG2), CD4-γ2 fell 40 to 50 Å short of cross-linking even optimally oriented trimers (data not shown). The implications of these observations are currently being tested through experimental studies of the detailed stoichiometry of virus neutralization by these molecules. In additional studies, we are examining whether cross-linking is possible with anti-gp120 antibodies, which are intermediate in size between CD4-γ2 and CD4-IgG2 and can target additional neutralization epitopes outside the CD4 binding site.
FIG. 3.
Models of CD4-IgG2 in complex with arrays of gp120 trimers. (A) Docking of CD4-IgG2 with trimers oriented such that the CD4 binding sites project toward neighboring trimers. (B and C) gp120 trimers are rotated 20° and 45° clockwise with respect to those in panel A. Arrows indicate CD4 arms that closely approach (yellow and green) or fully engage (white) a CD4 binding site of gp120. Structures are color coded as described for Fig. 2.
There are numerous uncertainties as to the orientation of gp120 on the viral surface, limiting our ability to draw firm conclusions regarding cross-linking. First, only the CD4-bound conformation of gp120 is known, whereas the target gp120 molecule is in the native configuration. Also, little is known regarding the geometry and flexibility of the gp120-gp41 interaction. Similarly, the rotational and diffusional freedom of the envelope spikes is poorly understood (1, 5, 8, 19, 30). Lastly, cross-linking may be variably affected by host cell proteins incorporated into the viral membrane.
In addition, mechanisms other than trimer cross-linking may contribute to the heightened antiviral activity of CD4-IgG2. First, the higher valence of CD4-IgG2 serves to increase the likelihood that a second gp120 will be engaged before CD4-IgG2 diffuses away from the virion surface following gp120 stripping or dissociation of the CD4-IgG2–gp120 complex (18). Second, the steric effect of CD4-IgG2 binding may significantly hinder gp120 interactions with cell surface CD4 or fusion coreceptors, either directly or, because of its size, indirectly by preventing close apposition of virus and target cell. Last and more conceptually, CD4-IgG2 may cross-link multiple virions and thereby accelerate their clearance. Additional studies are needed to distinguish between these alternatives.
In summary, we have used negative-stain immunoelectron microscopy to demonstrate that CD4-γ2 and CD4-IgG2 are functionally divalent and tetravalent, respectively, in binding monomeric gp120, i.e., there is no steric inhibition between CD4 arms. Molecular models are consistent with the electron microscopic images and have been used to explore the interactions of these molecules with trimeric gp120. The models reveal that CD4-IgG2 but not CD4-γ2 has considerable potential to cross-link envelope trimers on the virion surface.
Acknowledgments
We thank Wayne Hendrickson and John Moore for critical reading of the manuscript.
This work was supported in part by NIH grants R21 AI44291 and R01 AI43084.
REFERENCES
- 1.Akari H, Fukumori T, Adachi A. Cell-dependent requirement of human immunodeficiency virus type 1 gp41 cytoplasmic tail for Env incorporation into virions. J Virol. 2000;74:4891–4893. doi: 10.1128/jvi.74.10.4891-4893.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allaway G P, Davis-Bruno K L, Beaudry G A, Garcia E B, Wong E L, Ryder A M, Hasel K W, Gauduin M C, Koup R A, McDougal J S. Expression and characterization of CD4-IgG2, a novel heterotetramer that neutralizes primary HIV type 1 isolates. AIDS Res Hum Retrovir. 1995;11:533–539. doi: 10.1089/aid.1995.11.533. [DOI] [PubMed] [Google Scholar]
- 3.Allaway G P, Ryder A M, Beaudry G A, Maddon P J. Synergistic inhibition of HIV-1 envelope-mediated cell fusion by CD4-based molecules in combination with antibodies to gp120 or gp41. AIDS Res Hum Retrovir. 1993;9:81–87. doi: 10.1089/aid.1993.9.581. [DOI] [PubMed] [Google Scholar]
- 4.Deisenhofer J. Crystallographic refinement and atomic models of a human Fc fragment and its complex with fragment B of protein A from Staphylococcus aureus at 2.9- and 2.89-Å resolution. Biochemistry. 1981;20:2361–2370. [PubMed] [Google Scholar]
- 5.Dubay J W, Roberts S J, Hahn B H, Hunter E. Truncation of the human immunodeficiency virus type 1 transmembrane glycoprotein cytoplasmic domain blocks virus infectivity. J Virol. 1992;66:6616–6625. doi: 10.1128/jvi.66.11.6616-6625.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Forster M J, Mulloy B, Nermut M V. Molecular modelling study of HIV p17gag (MA) protein shell utilising data from electron microscopy and X-ray crystallography. J Mol Biol. 2000;298:841–857. doi: 10.1006/jmbi.2000.3715. [DOI] [PubMed] [Google Scholar]
- 7.Fouts T R, Binley J M, Trkola A, Robinson J E, Moore J P. Neutralization of the human immunodeficiency virus type 1 primary isolate JR-FL by human monoclonal antibodies correlates with antibody binding to the oligomeric form of the envelope glycoprotein complex. J Virol. 1997;71:2779–2785. doi: 10.1128/jvi.71.4.2779-2785.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Freed E O, Martin M A. Domains of the human immunodeficiency virus type 1 matrix and gp41 cytoplasmic tail required for envelope incorporation into virions. J Virol. 1996;70:341–351. doi: 10.1128/jvi.70.1.341-351.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gauduin M-C, Allaway G P, Maddon P J, Barbas C F, Burton D R, Koup R A. Effective ex vivo neutralization of human immunodeficiency virus type 1 in plasma by recombinant immunoglobulin molecules. J Virol. 1996;70:2586–2592. doi: 10.1128/jvi.70.4.2586-2592.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gauduin M C, Allaway G P, Olson W C, Weir R, Maddon P J, Koup R A. CD4-immunoglobulin G2 protects Hu-PBL-SCID mice against challenge by primary human immunodeficiency virus type 1 isolates. J Virol. 1998;72:3475–3478. doi: 10.1128/jvi.72.4.3475-3478.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gelderblom H R, Hausmann E H S, Ozel M, Pauli G, Koch M A. Fine structure of human immunodeficiency virus (HIV) and immunolocalization of structural proteins. Virology. 1987;156:171–176. doi: 10.1016/0042-6822(87)90449-1. [DOI] [PubMed] [Google Scholar]
- 12.He X M, Ruker F, Casale E, Carter D C. Structure of a human monoclonal antibody Fab fragment against gp41 of human immunodeficiency virus type 1. Proc Natl Acad Sci USA. 1992;89:7154–7158. doi: 10.1073/pnas.89.15.7154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hill C P, Worthylake D, Bancroft D P, Christensen A M, Sundquist W I. Crystal structures of the trimeric human immunodeficiency virus type 1 matrix protein: implications for membrane association and assembly. Proc Natl Acad Sci USA. 1996;93:3099–3104. doi: 10.1073/pnas.93.7.3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jacobson M A, Hardy D, Connick E, Watson J, DeBruin M. Phase 1 trial of a single dose of recombinant human interleukin-12 in human immunodeficiency virus-infected patients with 100–500 CD4 cells/μL. J Infect Dis. 2000;182:1070–1076. doi: 10.1086/315819. [DOI] [PubMed] [Google Scholar]
- 15.Kabat E A, Wu T T, Perry H M, Gottesman K S, Foeller C. Sequences of proteins of immunological interest. 5th ed. Bethesda, Md: National Institutes of Health; 1991. [Google Scholar]
- 16.Kwong P D, Wyatt R, Robinson J, Sweet R W, Sodroski J, Hendrickson W A. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;383:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kwong P D, Wyatt R, Sattentau Q J, Sodroski J, Hendrickson W A. Oligomeric modeling and electrostatic analysis of the gp120 envelope glycoprotein of human immunodeficiency virus. J Virol. 2000;74:1962–1972. doi: 10.1128/jvi.74.4.1961-1972.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moore J P, McKeating J A, Weiss R A, Sattentau Q J. Dissociation of gp120 from HIV-1 virions induced by soluble CD4. Science. 1990;250:1139–1142. doi: 10.1126/science.2251501. [DOI] [PubMed] [Google Scholar]
- 19.Murakami T, Freed E O. The long cytoplasmic tail of gp41 is required in a cell type-dependent manner for HIV-1 envelope glycoprotein incorporation into virions. Proc Natl Acad Sci USA. 2000;97:343–348. doi: 10.1073/pnas.97.1.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nermut M V, Hockley D J, Bron P, Thomas D, Zhang W H, Jones I M. Further evidence for hexagonal organization of HIV gag protein in prebudding assemblies and immature virus-like particles. J Struct Biol. 1998;123:143–149. doi: 10.1006/jsbi.1998.4024. [DOI] [PubMed] [Google Scholar]
- 21.Nermut M V, Hockley D J, Jowett J B M, Jones I M, Garreau M, Thomas D. Fullerene-like organization of HIV gag-protein shell in virus-like particles produced by recombinant baculovirus. Virology. 1994;198:288–296. doi: 10.1006/viro.1994.1032. [DOI] [PubMed] [Google Scholar]
- 22.Roux K H. Immunoelectron microscopy of idiotype-anti-idiotype complexes. Methods Enzymol. 1989;178:130–144. doi: 10.1016/0076-6879(89)78010-1. [DOI] [PubMed] [Google Scholar]
- 23.Roux K H. Negative stain immunoelectron microscopic analysis of small macromolecules of immunologic significance. Methods. 1996;10:247. doi: 10.1006/meth.1996.0099. [DOI] [PubMed] [Google Scholar]
- 24.Ryu S E, Truneh A, Sweet R W, Hendrickson W A. Structures of an HIV and MHC binding fragment from human CD4 as refined in two crystal lattices. Structure. 1994;2:59–74. doi: 10.1016/S0969-2126(00)00008-3. [DOI] [PubMed] [Google Scholar]
- 25.Shearer W T, Israel R J, Starr S, Fletcher C V, Wara D, Rathore M, Church J, DeVille J, Fenton T, Graham B, Samson P, Staprans S, McNamara J, Moye J, Madon P J, Olson W C. Recombinant CD4-IgG2 in human immunodeficiency virus type 1-infected children: phase 1/2 study. J Infect Dis. 2000;182:1774–1779. doi: 10.1086/317622. [DOI] [PubMed] [Google Scholar]
- 26.Shelton E, Yonemasu K, Stroud R M. Ultrastructure of the human complement component, Clq (negative staining-glutamine synthetase-biologically active Clq) Proc Natl Acad Sci USA. 1972;69:65–68. doi: 10.1073/pnas.69.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Trkola A, Dragic T, Arthos J, Binley J M, Olson W C, Allaway G P, Cheng-Mayer C, Robinson J, Maddon P J, Moore J P. CD4-dependent, antibody-sensitive interactions between HIV-1 and its co-receptor CCR-5. Nature. 1996;384:184–187. doi: 10.1038/384184a0. [DOI] [PubMed] [Google Scholar]
- 28.Trkola A, Ketas T, Kewalramani V N, Endorf F, Binley J M, Katinger H, Robinsin J, Littman D R, Moore J P. Neutralization sensitivity of human immunodeficiency virus type 1 primary isolates to antibodies and CD4-based reagents is independent of coreceptor usage. J Virol. 1998;72:1876–1885. doi: 10.1128/jvi.72.3.1876-1885.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Trkola A, Pomales A B, Yuan H, Koraber B, Maddon P J, Allaway G P, Katinger H, Barbas C F, Burton D R, Ho D D. Cross-clade neutralization of primary isolates of human immunodeficiency virus type 1 by human monoclonal antibodies and tetrameric CD4-IgG. J Virol. 1995;69:6609–6617. doi: 10.1128/jvi.69.11.6609-6617.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu S, Yuan X, McLane M F, Lee T H, Esex M. Mutations in the cytoplasmic domain of human immunodeficiency virus type 1 transmembrane protein impair the incorporation of Env proteins into mature virions. J Virol. 1993;67:213–221. doi: 10.1128/jvi.67.1.213-221.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]