Abstract
Background
Depigmentation of specific areas of the skin is a persistent and long-lasting dermatologic disorder known as vitiligo, stemming from the impairment and disruption of melanocytes both structurally and functionally, leading to the loss of pigmentation in those regions.
Aim
Our objective was to identify the pivotal genes and upstream regulators, transcription factors (TFs), microRNAs (miRNAs), and pathways implicated in the pathogenesis of vitiligo.
Methods
An integrated analysis was conducted using microarray datasets on vitiligo obtained from the Gene Expression Omnibus (GEO) database. The functional annotation and potential pathways of differentially expressed genes (DEGs) were additionally investigated through Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses. Various bioinformatics approaches were utilized, making use of publicly accessible databases to identify appropriate TFs and miRNAs.
Results
Our investigation identified TYR, MLANA, TYRP1, PMEL, OCA2, SLC45A2, GPR143, DCT, TRPM1, and EDNRB as the most appropriate genes associated with vitiligo. Our suggestion is that the identified biological processes include developmental pigmentation (GO:0048066) and pigment metabolic processes (GO:0042440) as the most suitable biological processes. In contrast, the KEGG pathways that showed significance in our analysis are Tyrosine metabolism (Path: hsa00350) and Melanogenesis (Path: hsa04916). We hypothesized the involvement of ten TFs and 73 miRNAs in the regulation of genes related to vitiligo.
Conclusion
TYR, MLANA, TYRP1, PMEL, OCA2, SLC45A2, GPR143, DCT, TRPM1, and EDNRB are the top ten genes that are pivotal in the progression and exhibition of vitiligo. The biological, cellular, molecular, and KEGG pathways of those genes has an imperative role in the pathogenesis of vitiligo. TFs and miRNAs that interact with this gene are listed, shedding light on the regulatory mechanisms governing the expression of these key genes in vitiligo.
Keywords: Vitiligo, miRNAs, transcription factors, microarray, DEGs, GEO
Introduction
Patches of depigmented white skin result from the autoimmune destruction of epidermal melanocytes in vitiligo, which is a prevalent disorder.1 The gradual loss of pigmentation in this condition has posed challenges as effective treatments were limited until recent advancements.2 Vitiligo is a complex disorder that stems from a combination of environmental factors and genetic predisposition. It exhibits the hallmarks of a classic polygenic disorder, where each genetic element has a minimal impact on the overall manifestation of the disease.3 Despite being a polygenic disorder, the level of polygenicity in vitiligo is relatively low compared to other complex traits, while its heritability remains considerably high.4 As a result, researchers have found it more manageable to uncover and understand the genetic makeup of vitiligo compared to the genetic complexities seen in most other multifaceted traits.5
The significance of genetic factors in vitiligo was an early point of interest due to the frequent clustering of symptoms within families. Research conducted by Das et al delved into genetic epidemiology, revealing a complex interplay of multifactorial, polygenic inheritance patterns.6 This groundbreaking work brought to light the concept of vitiligo as a “complex disease”, a notion that continues to shape current understandings in the field. In the 1960s and 1970s, investigations into potential genetic markers for vitiligo such as ABO, haptoglobin, erythrocyte enzymes, and various blood proteins were commonplace, yet the outcomes yielded no significant correlations. The 1970s saw a surge in studies exploring the role of HLA in vitiligo, resulting in a body of literature that presented mixed and often perplexing findings, adding layers of complexity to the genetic landscape of the condition. The evolving nature of genetic research in vitiligo underscores the intricate web of factors influencing disease development and progression, highlighting the need for further exploration and clarification in this intricate field.7 After this, numerous additional potential genes were scrutinized and associated with vitiligo; nevertheless, currently, only PTPN22 and possibly CTLA4, both of which encode immune modulators, are deemed highly reliable. The discovery of SLEV1 on chromosome 17p13, a genetic marker in families with systemic lupus erythematosus that also included at least one individual with vitiligo, indicated a causal genetic connection between these two conditions and marked the inception of the contemporary epoch of genetic investigations on widespread vitiligo.8 Subsequent investigations unveiled that SLEV1 was actually NLRP1 (NALP1), a critical regulator of the innate immune system.9
Aim of the Study
Our research endeavor was undertaken with the overarching objective of identifying the pivotal genes and upstream regulators implicated in the pathogenesis of vitiligo. To accomplish this goal, we conducted a rigorous screening process to pinpoint the pathogenic genes associated with vitiligo through an integrated analysis of microarray datasets sourced from the Gene Expression Omnibus (GEO) database. This meticulous approach involved leveraging bioinformatics tools and methodologies to discern genes that exhibit a specific and significant association with the development and progression of vitiligo. Moreover, our investigation is particularly focused on elucidating the crucial role played by microRNAs (miRNAs) in regulating the molecular pathways involved in the pathophysiology of vitiligo. Additionally, we are dedicated to identifying the most pertinent transcription factors (TFs) that exert control over the expression of the identified genes, thereby shedding light on the intricate regulatory networks underpinning vitiligo. Through the employment of this comprehensive and systematic strategy, we envision the discovery of innovative therapeutic targets and the exploration of state-of-The-art investigative approaches aimed at revolutionizing the management and treatment of vitiligo.
Material and Methods
Microarray Expression Profiling in GEO
The study received ethical approval from ethical committee of Shaqra University (CMD/DWD/SU/2023/03/043). The National Centre for Biotechnology Information is responsible for the creation and management of the largest database of high-throughput gene expression data known as the GEO dataset. To identify gene expression profiling studies related to vitiligo, a thorough search was conducted within the GEO dataset available at http://www.ncbi.nlm.nih.gov/geo/. The search strategy included specific keywords such as “gse” [Filter], “Homo sapiens” [porgn], AND “vitiligo” [MeSH Terms] OR “vitiligo” [All Fields]. Various selection criteria were applied to ensure the relevance and quality of the retrieved data. Following a meticulous screening process, a single vitiligo mRNA data set meeting the established criteria was successfully identified. The essential characteristics of the chosen dataset included the presence of genome-wide mRNA transcriptome data, the origin of data from skin tissue samples obtained from individuals diagnosed with vitiligo, and the availability of both normalized and raw datasets for analysis. Subsequently, the differentially expressed genes (DEGs) within the dataset were singled out for further investigation. By comparing the gene expression profiles in lesional tissues with those in healthy tissues, the DEGs specific to vitiligo-affected skin were successfully pinpointed and characterized.
According to the predetermined inclusion criteria, we acquired a singular microarray study sourced from the NCBI GEO database, specifically identified with the GEO access number GSE65127 https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE65127 and utilizing the Platform GPL570 (HG-U133_Plus_2) Affymetrix Human Genome U133 Plus 2.0 Array, constituting a total of 10 healthy and 10 lesional samples.10
Analysis of Microarray Set GSE65127
The GSE65127 set analyzed by GEO2R tool available in NCBI https://www.ncbi.nlm.nih.gov/geo/geo2r/?acc=GSE65127. We define groups into healthy and lesional. The data analyzed was classified into upregulated and downstream genes, and we selected upregulation in the analysis.
Protein-Protein Interactions Network and Module Analysis
The Search Tool for the Retrieval of Interacting Genes STRING, available at http://string-db.org and specifically version 10.5, was harnessed in this study to forecast interactions among DEGs. This online database was effectively utilized to identify potential interactions between genes.11 Following this, a network of Protein-Protein Interactions (PPI) was meticulously crafted using Cytoscape, which was at version 3.7.1 during the implementation of this research project. Furthermore, within Cytoscape, the CytoHubba tool was applied to ascertain and rank the top ten proteins along with DEGs, providing crucial insights into the network topology and potential key players within the system.
Biological, Cellular, Molecular Process, and KEGG Pathway Analysis
The examination of the biological, cellular, and molecular process pathways through enrichment analyses was conducted by utilizing the Shiny GO 0.80 software available at http://bioinformatics.sdstate.edu/go/ with a stringent criterion of FDR < 0.05.12 Simultaneously, the investigation involved the utilization of the Amigo gene ontology database found at https://amigo.geneontology.org/amigo. Moreover, the KEGG pathway was scrutinized using the Shiny GO 0.80 software, maintaining the criteria of FDR < 0.05, in conjunction with https://www.genome.jp/kegg/pathway.html. This comprehensive approach facilitated a detailed exploration of the intricate biological, cellular, and molecular mechanisms underlying the pathways of interest, enhancing the depth of understanding in this research endeavor. The utilization of these sophisticated tools and databases exemplifies a rigorous methodology aimed at uncovering valuable insights into the complex interplay of biological processes at a fundamental level.
Prediction and Selection of Suitable TFs
TFs were anticipated through the utilization of the online TFEA tool ChEA3 https://maayanlab.cloud/chea3/#top. The submission of the top ten gene lists to the website led to the acquisition of ten potential TFs based on their top rank as determined by the amalgamation of various databases. The construction of molecular networks is facilitated and analyzed through Cytoscape software.
Prediction and Analysis of Suitable miRNAs for Genes
We used various tools such as the TargetScan https://www.targetscan.org/vert_80/, miRDB https://mirdb.org/, MiRTarBase 9.0 https://mirtarbase.cuhk.edu.cn/, and miRWalk http://mirwalk.umm.uni-heidelberg.de/databases to anticipate their potential target genes with a confidence interval (CIs) of 95%. The construction of molecular networks is facilitated and analyzed through Cytoscape software.
Results
DEGs in the Integrated Analysis of Microarray Datasets
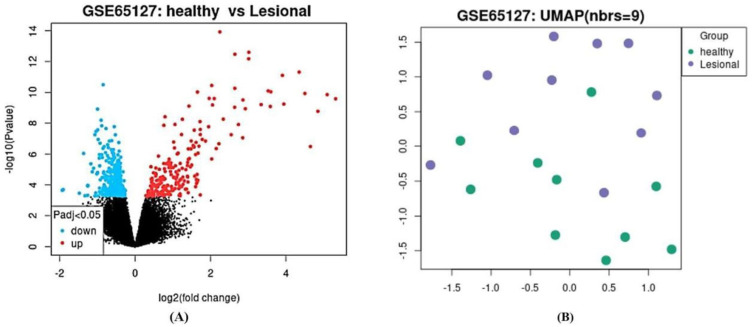
According to the predetermined inclusion criteria, we acquired a singular microarray study sourced from the NCBI GEO database, specifically identified with the GEO access number GSE65127 and utilizing the Platform GPL570 (HG-U133_Plus_2) Affymetrix Human Genome U133 Plus 2.0 Array, constituting a total of 10 healthy and 10 lesional samples. Through a comprehensive integrated analysis approach, a substantial total of 54,093 genes were scrutinized, leading to the identification of 582 DEGs, 367 downregulated DEGs, and 216 upregulated DEGs, all meeting the strict statistical criteria (FDR < 0.05, |Combined ES| > 1). The detailed outcomes of this analytical investigation are meticulously depicted in [Figure 1A and B], offering a visual representation of the identified DEGs and their respective expression patterns for further scientific scrutiny and interpretation.
Figure 1.
(A) Volcano plots showing the upregulated and downregulated genes in GSE65127 (The volcano plots shown identify genes that are p < 0.05 and are used as criteria for significance). (B) Principal component analysis (PCA) map of DEGs. The data was analyzed by GEO2R on NCBI website.
PPI Network Construction for Upregulate Genes
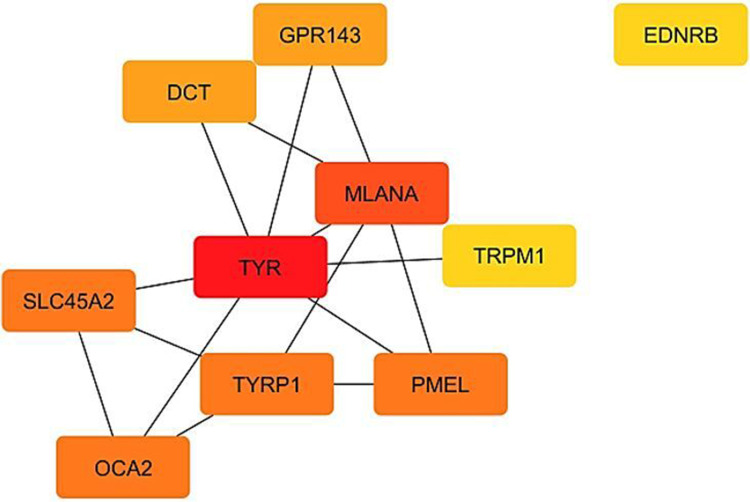
We have successfully identified the interaction network of the top 106 upregulated DEGs using the String tool, followed by calculation and filtering through the Cytoscape software to focus on the top 10 genes for further analysis. The graphical representation of this analysis can be observed in [Figure 2]. It is important to note that the network comprises nodes and edges, with nodes symbolizing the proteins and edges denoting the interactions between them. The top 10 networks are specifically comprised of 10 nodes and 15 edges, exhibiting an average number of neighbors amounting to 3.33. Additionally, the network diameter is measured at 3, while the network radius stands at 2. In terms of network characteristics, the average pathway length is determined to be 1.61, accompanied by a clustering coefficient of 0.54. Furthermore, the network density is calculated to be 0.417, with a network heterogeneity of 0.51 and a network centralization value of 0.589. Among the genes analyzed, those with higher degrees were identified as TYR with a degree of 9, MLANA with a degree of 8, TYRP1 with a degree of 4, PMEL with a degree of 4, OCA2 with a degree of 4, SLC45A2 with a degree of 4, GPR143 with a degree of 2, DCT with a degree of 2, TRPM1 with a degree of 1, and EDNRB with a degree of 1.
Figure 2.
The PPI network of the top 10 DEGs in vitiligo. The red color indicates the highest degree, and yellow indicates the lowest degree. The analysis was done by Cytoscape software and CytoHubba tool.
Functional Enrichment Analysis for Top Ten Genes
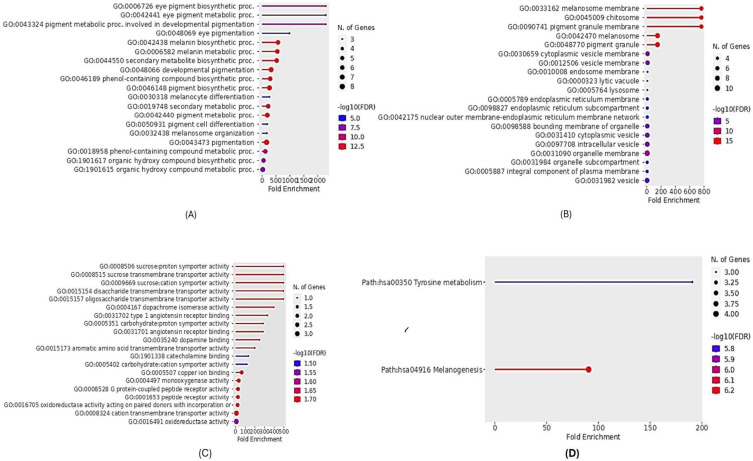
We carried out an analysis of GO and KEGG categories to explore the biological functions of the identified DEGs by utilizing the web-based tool Shiny GO 0.80. Our investigation revealed that the GO term enriched for Biological Process was developmental pigmentation (GO:0048066) with an FDR of 4.15E-15 and a Fold Enrichment of 326.8. This pathway was linked to 7 genes including DCT, OCA2, TYRP1, EDNRB, SLC45A2, TYR, and GPR143. Additionally, we observed enrichment in the pigment metabolic process (GO:0042440) with an FDR of 6.78E-14 and a Fold Enrichment of 188.4, involving 7 genes such as TYR, DCT, OCA2, TYRP1, SLC45A2, PMEL, and GPR143. Furthermore, the melanin biosynthetic process (GO:0042438) showed significant enrichment (FDR = 1.865E-14, Fold Enrichment = 572.025) with 6 genes participating, including TYR, DCT, OCA2, TYRP1, SLC45A2, and PMEL. Likewise, the melanin metabolic process (GO:0006582) exhibited enrichment (FDR = 1.93E-14, Fold Enrichment = 549.1) with 6 shared genes like TYR, DCT, OCA2, TYRP1, SLC45A2, and PMEL, and the secondary metabolite biosynthetic process (GO:0044550) also displayed enrichment (FDR = 2.1E-14, Fold Enrichment = 528.0) with 6 involved genes consisting of TYR, DCT, OCA2, TYRP1, SLC45A2, and PMEL as depicted in [Figure 3A].
Figure 3.
Gene Ontology functional enrichment and KEGG pathways of DEGs (FDR < 0.05). (A) Biological process. (B) Cellular components. (C) Molecular functions. (D) KEGG pathway. FDR, false discovery rate. The charts are designed by Shiny GO 0.80 tool.
On the other hand, the GO terms related to Cellular Processes included melanosome (GO:0042470) with an FDR of 2.69E-16 and a Fold Enrichment of 122, where 8 out of the top 10 genes were involved such as DCT, GPR143, OCA2, TYRP1, MLANA, PMEL, TYR, and SLC45A2. Additionally, the pigment granule pathway (GO:0048770) exhibited enrichment (FDR = 2.679E-16, Fold Enrichment = 122) with 8 involved genes like DCT, GPR143, OCA2, TYRP1, MLANA, PMEL, TYR, and SLC45A2. Moreover, the melanosome membrane (GO:0033162) showed enrichment (FDR = 2.679E-16, Fold Enrichment = 762.7) with 6 genes involved, including GPR143, OCA2, TYR, DCT, TYRP1, and SLC45A2, and the chitosome (GO:0045009) displayed enrichment (FDR = 2.679E-16, Fold Enrichment = 762.7) with 6 genes participating like GPR143, OCA2, TYR, DCT, TYRP1, and SLC45A2 as shown in [Figure 3B].
Furthermore, the GO terms associated with molecular processes included copper ion binding (GO:0005507) with an FDR of 0.015 and a Fold Enrichment of 72.6, where 2 genes were involved, namely TYR and DCT. Additionally, the monooxygenase activity (GO:0004497) demonstrated enrichment (FDR = 0.015, Fold Enrichment = 37.8) with 2 genes participating such as TYR and TYRP1. Moreover, the oxidoreductase activity acting on paired donors with incorporation (GO:0016705) displayed enrichment (FDR = 0.015, Fold Enrichment = 23.4) with 2 involved genes including TYR and TYRP1, and the cation transmembrane transporter activity (GO:0008324) showed enrichment (FDR = 0.015, Fold Enrichment = 9.8) with 3 genes participating like TRPM1, SLC45A2, and OCA2 as depicted in [Figure 3C].
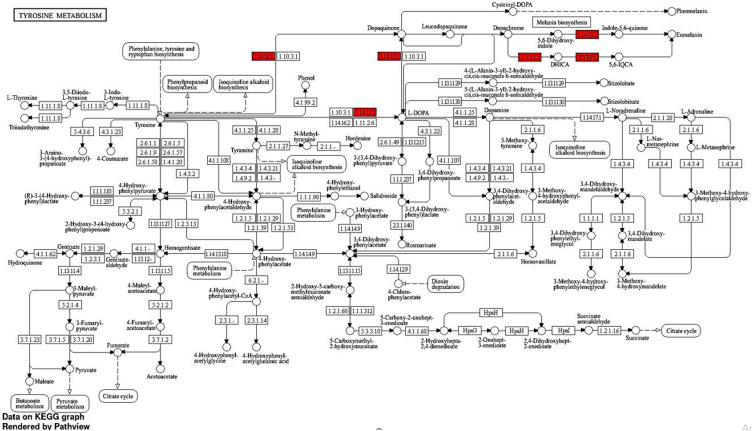
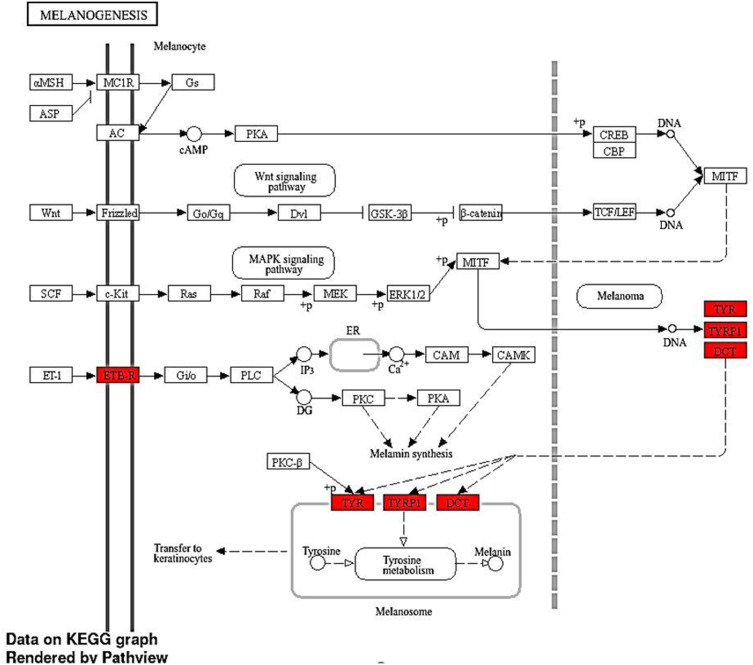
KEGG analysis revealed a significant enrichment of DEGs. The analysis indicated a notable enrichment of DEGs in Tyrosine metabolism (Path: hsa00350) with an FDR value of 1.703E-06 and a Fold Enrichment of 190.6. Among the top 10 genes, three genes, namely DCT, TYR, and TYRP1, were found to be involved in this pathway. Furthermore, the analysis also showed enrichment in Melanogenesis (Path: hsa04916) with an FDR of 5.84E-07 and Fold Enrichment of 90.6. Out of the top 10 genes in this pathway, four genes (DCT, EDNRB, TYR, and TYRP1) were identified. These findings are visually represented in [Figures 3D, 4 and 5].
Figure 4.
Tyrosine metabolism KEGG pathway. The red color indicates the gene’s effect in the pathway from this study (1.14.18.1 is TYR gene, and 533.12 is DCT gene). The diagram is designed by Shiny GO 0.80 tool and KEGG pathway website.
Figure 5.
Melanogenesis KEGG pathway the red color indicates gene effects in the pathway from the top ten genes in this study. The diagram was designed by the Shiny GO 0.80 tool and the KEGG pathway website.
The genes that have been identified and predicted within the context of this particular study are expected to provide significant insights that will greatly facilitate a more comprehensive understanding of the intricate molecular mechanisms underlying the condition known as vitiligo. Particularly in relation to their influence on T-cells and the subsequent ramifications that these interactions may have on the overall functioning of the immune system, especially considering.
TFs Prediction and Analysis for the Top Ten Selected Genes
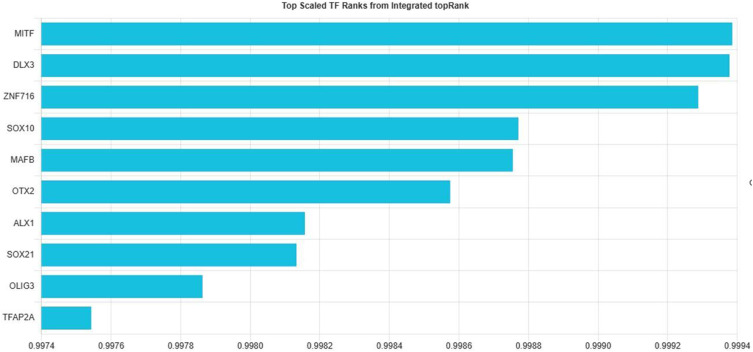
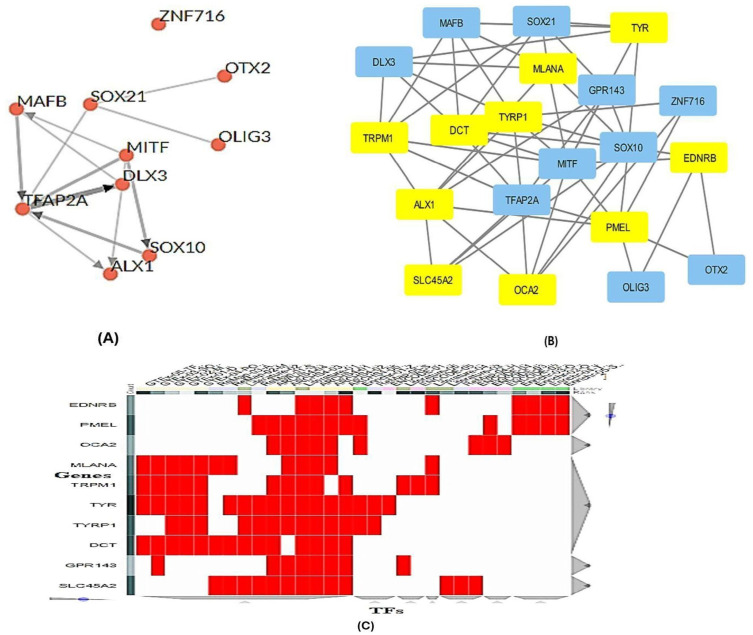
TFs were predicted using the online TFEA tool ChEA3, which allowed for the identification of potential TFs based on the submission of ten gene lists to the website. The top ten TFs were determined through the amalgamation of data from various databases in accordance with their rank as illustrated in [Figure 6]. It is crucial to note that the level of certainty in these predictions increases as the mean rank decreases. The regulatory network involving these TFs and the GO analysis linked to them are depicted in [Figure 7A and B]. Additionally, visual representations of the potential interactions among these TFs and the local network are presented in [Figure 8A]. The analysis of the network, as shown in [Figure 8A], consisted of 20 nodes, 55 edges, an average number of neighbors of 5.5, a network diameter of 1, a network radius of 1, a characteristic path length of 1, a clustering coefficient of 0.0001, and a network density of 0.145. Furthermore, the cluster heatmap analysis is detailed in [Figure 8C], where the red color signifies multiple TFs sharing a gene. The correlation between the TFs and the ten genes is outlined in [Table 1], revealing that MITF controlled 10 genes, followed by SOX10 controlling 9 genes, and ALX1 and TFAP2A controlling 7 genes each.
Figure 6.
Bar charts corresponding to the top TFs of the selected library display the -log10 (p-value) for ChEA TF target library results and an integrated rank score for the integrated library results. The top 10 TFs are on the y-axis.
Figure 7.
Global network analysis representation of top-ranked TFs displaying putative interactions according to tissue-specific, based on averaged integrated ranks across multiple libraries and databases (ChEA3 libraries). (A) Based on TCGFA TF network (The blue color indicates the top ten TFs). (B) Based on GTEx network.
Figure 8.
(A) Local network analysis representation of top-ranked TFs displaying putative interactions, based on averaged integrated ranks across multiple libraries and databases (ChEA3 libraries). (B) TFs and genes regulatory network for ten genes using Cytoscape software. The yellow color indicates genes, and the blue color indicates TFs. (C) Heat map cluster analysis showing relationships between genes and TFs the red color indicates several TFs share in one gene. The analysis was done by ChEA3 libraries.
Table 1.
TFs and Overlapping Genes
| TF | Overlapping Genes | Integrated Scall Ranking | Library |
|---|---|---|---|
| MITF (Microphthalmia-associated transcription factor) | TRPM1, PMEL, OCA2, SLC45A2, GPR143, EDNRB, DCT, MLANA, TYRP1, TYR | 6.143E-4* | ARCHS4 Coexpression |
| DLX3 (Distal-less homeobox 3) | TRPM1, DCT, MLANA, TYRP1, TYR | 6.223E-4* | GTEx Coexpression |
| ZNF716(Zinc finger protein 716) | PMEL, OCA2, TYRP1 | 7.123E-4* | Enrichr Queries |
| SOX10 (SRY (sex determining region Y)-box 10) | PMEL, OCA2, SLC45A2, GPR143, EDNRB, DCT, MLANA, TYRP1, TYR | 0.001229* | ARCHS4 Coexpression |
| MAFB (V-maf avian musculoaponeurotic fibrosarcoma oncogene homolog B) | TRPM1, DCT, MLANA, TYRP1, TYR | 0.001245* | GTEx Coexpression |
| OTX2 (Orthodenticle homeobox 2) | PMEL, EDNRB | 0.001425* | Enrichr Queries |
| ALX1 (ALX homeobox 1) | TRPM1, PMEL, OCA2, SLC45A2, GPR143, MLANA, TYRP1 | 0.001843* | ARCHS4 Coexpression |
| SOX21 (SRY (sex determining region Y)-box 21) | TRPM1, GPR143, DCT, MLANA, TYR | 0.001867* | GTEx Coexpression |
| OLIG3(Oligodendrocyte transcription factor 3) | PMEL, EDNRB | 0.002137* | Enrichr Queries |
| TFAP2A (Transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha) | TRPM1, PMEL, OCA2, SLC45A2, GPR143, DCT, TYRP1 | 0.002457* | ARCHS4 Coexpression |
Abbreviations: TRPM1, transient receptor potential cation channel subfamily M member 1; PMEL, premelanosome protein; OCA2, melanosomal transmembrane protein; SLC45A2, solute carrier family 45 members 2; GPR143 G protein-coupled receptor 143; EDNRB, endothelin receptor type B; DCT, dopachrome tautomerase; MLANA, melan-A; TYRP1, tyrosinase-related protein 1; TYR, tyrosinase. * Highly significant differences, p ≤ 0.05.
miRNAs Prediction and Analysis for the Top Ten Selected Genes
To investigate thoroughly the potential target miRNAs of the differentially expressed top ten genes, an analysis was carried out using the esteemed miRWalk tool, renowned for its reliability in such studies. This extensive analysis involved the application of numerous target prediction algorithms, such as TargetScan, mirDB, and MirTarBase, among others. The central aim of this analysis was to categorize and provide annotations for the miRNAs located within the 3’UTR. The selection process for these target miRNAs was based on a precise set of criteria, which included factors like high binding probability, low binding energy, and the presence of Adenylate-uridylate (AU)-rich elements (AREs) known for their significance. The data retrieved from the miRWalk tool revealed information about approximately 2793 miRNAs. This dataset was then filtered based on specific conditions, including a binding energy equal to, or exceeding −15, an AU content equal to or greater than 0.412, and a binding probability of 1. These stringent criteria allowed for the identification of suitable miRNAs for nine out of the ten selected genes. We identified a total of 73 miRNAs that are considered appropriate for the regulation of nine specific genes associated with vitiligo. These genes are further categorized as 15 for EDNRB, 1 for OCA2, 2 for TYR, 1 for TYRP1, 13 for MLANA, 9 for TRPM1, 13 for DCT, 18 for SLC45AC, and 1 for PMEL, showcasing the intricate regulatory network involved in the pathogenesis of this skin disorder., as outlined in [Table 2].
Table 2.
Predicted Target miRNAs for Genes Selected in This Study by Bioinformatics Tools
| Gene (Ensemble ID) | Reference Sequence ID | Binding Probability | Binding Energy | AU Rich Region Fraction | Position |
|---|---|---|---|---|---|
| EDNRB (ENSG00000136160) | hsa-miR-200c-3p | 1 | −16.6 | 0.559 | 3’UTR |
| EDNRB (ENSG00000136160) | hsa-miR-640 | 1 | −16.96 | 0.5 | 3’UTR |
| EDNRB (ENSG00000136160) | hsa-miR-885-5p | 1 | −18.9 | 0.5 | 3’UTR |
| EDNRB (ENSG00000136160) | hsa-miR-941 | 1 | −17.0 | 0.676 | 3’UTR |
| EDNRB (ENSG00000136160) | hsa-miR-942-5p | 1 | −15.5 | 0.647 | 3’UTR |
| EDNRB (ENSG00000136160) | hsa-miR-297 | 1 | −16.8 | 0.779 | 3’UTR |
| EDNRB (ENSG00000136160) | hsa-miR-378c | 1 | −15.5 | 0.731 | 3’UTR |
| EDNRB (ENSG00000136160) | hsa-miR-6855-3p | 1 | −17.1 | 0.5 | 3’UTR |
| EDNRB (ENSG00000136160) | hsa-miR-7159-5p | 1 | −15.5 | 0.647 | 3’UTR |
| EDNRB (ENSG00000136160) | hsa-miR-9903 | 1 | −18.9 | 0.731 | 3’UTR |
| EDNRB (ENSG00000136160) | hsa-miR-629-3p | 1 | −17.1 | 0.662 | 3’UTR |
| EDNRB (ENSG00000136160) | hsa-miR-1183 | 1 | −15.4 | 0.544 | 3’UTR |
| EDNRB (ENSG00000136160) | hsa-miR-664a-3p | 1 | −17.2 | 0.662 | 3’UTR |
| EDNRB (ENSG00000136160) | hsa-miR-6879-5p | 1 | −16.2 | 0.544 | 3’UTR |
| EDNRB (ENSG00000136160) | hsa-miR-562 | 1 | −15.56 | 0.632 | 3’UTR |
| OCA2 (ENSG00000104044) | hsa-miR-361-5p | 1 | −15.9 | 0.544 | 3’UTR |
| TYR (ENSG00000077498) | hsa-miR-3163 | 1 | −17.1 | 0.559 | 3’UTR |
| TYR (ENSG00000077498) | hsa-miR-3614-5p | 1 | −16.9 | 0.618 | 3’UTR |
| TYRP1 (ENSG00000107165) | hsa-miR-1247-3p | 1 | −15.09 | 0.676 | 3’UTR |
| MLANA (ENSG00000120215) | hsa-miR-135a-3p | 1 | −15.71 | 0.515 | 3’UTR |
| MLANA (ENSG00000120215) | hsa-miR-34c-3p | 1 | −21.4 | 0.574 | 3’UTR |
| MLANA (ENSG00000120215) | hsa-miR-520d-3p | 1 | −17.1 | 0.706 | 3’UTR |
| MLANA (ENSG00000120215) | hsa-miR-718 | 1 | −18.3 | 0.5 | 3’UTR |
| MLANA (ENSG00000120215) | hsa-miR-3121-3p | 1 | −15.7 | 0.721 | 3’UTR |
| MLANA (ENSG00000120215) | hsa-miR-4268 | 1 | −15.7 | 0.632 | 3’UTR |
| MLANA (ENSG00000120215) | hsa-miR-3911 | 1 | −15.74 | 0.456 | 3’UTR |
| MLANA (ENSG00000120215) | hsa-miR-5705 | 1 | −15.7 | 0.471 | 3’UTR |
| MLANA (ENSG00000120215) | hsa-miR-6074 | 1 | −19.4 | 0.426 | 3’UTR |
| MLANA (ENSG00000120215) | hsa-miR-6771-3p | 1 | −16.7 | 0.618 | 3’UTR |
| MLANA (ENSG00000120215) | hsa-miR-6887-5p | 1 | −16.2 | 0.419 | 3’UTR |
| MLANA (ENSG00000120215) | hsa-miR-6892-5p | 1 | −16.80 | 0.574 | 3’UTR |
| MLANA (ENSG00000120215) | hsa-miR-7112-3p | 1 | −21.56 | 0.618 | 3’UTR |
| TRPM1 (ENSG00000134160) | hsa-miR-105-5p | 1 | −17.1 | 0.485 | 3’UTR |
| TRPM1 (ENSG00000134160) | hsa-miR-371b-5p | 1 | −17.1 | 0.632 | 3’UTR |
| TRPM1 (ENSG00000134160) | hsa-miR-320d | 1 | −15.7 | 0.618 | 3’UTR |
| TRPM1 (ENSG00000134160) | hsa-miR-3925-5p | 1 | −16.8 | 0.618 | 3’UTR |
| TRPM1 (ENSG00000134160) | hsa-miR-4539 | 1 | −15.2 | 0.412 | 3’UTR |
| TRPM1 (ENSG00000134160) | hsa-miR-4749-3p | 1 | −17.2 | 0.412 | 3’UTR |
| TRPM1 (ENSG00000134160) | hsa-miR-6512-5p | 1 | −16.4 | 0.485 | 3’UTR |
| TRPM1 (ENSG00000134160) | hsa-miR-516a-5p | 1 | −15.2 | 0.5 | 3’UTR |
| TRPM1 (ENSG00000134160) | hsa-miR-5681b | 1 | −19.4 | 0.647 | 3’UTR |
| DCT(ENSG00000080166) | hsa-miR-17-3p | 1 | −15.70 | 0.529 | 3’UTR |
| DCT(ENSG00000080166) | hsa-miR-217-5p | 1 | −19.42 | 0.603 | 3’UTR |
| DCT(ENSG00000080166) | hsa-miR-381-3p | 1 | −18.30 | 0.559 | 3’UTR |
| DCT(ENSG00000080166) | hsa-miR-450a-5p | 1 | −15.56 | 0.662 | 3’UTR |
| DCT(ENSG00000080166) | hsa-miR-585-5p | 1 | −16.2 | 0.544 | 3’UTR |
| DCT(ENSG00000080166) | hsa-miR-1185-2-3p | 1 | −15.5 | 0.544 | 3’UTR |
| DCT(ENSG00000080166) | hsa-miR-1185-1-3p | 1 | −15.5 | 0.544 | 3’UTR |
| DCT(ENSG00000080166) | hsa-miR-873-5p | 1 | −18.81 | 0.618 | 3’UTR |
| DCT(ENSG00000080166) | hsa-miR-4259 | 1 | −16.4 | 0.529 | 3’UTR |
| DCT(ENSG00000080166) | hsa-miR-3692-3p | 1 | −18.3 | 0.721 | 3’UTR |
| DCT(ENSG00000080166) | hsa-miR-4799-5p | 1 | −17.96 | 0.544 | 3’UTR |
| DCT(ENSG00000080166) | hsa-miR-10523-5p | 1 | −16.8 | 0.588 | 3’UTR |
| DCT (ENSG00000080166) | hsa-miR-520d-3p | 1 | −15.2 | 0.588 | 3’UTR |
| SLC45A2 (ENSG00000164175 | hsa-miR-4269 | 1 | −16.9 | 0.529 | 3’UTR |
| SLC45A2 (ENSG00000164175 | hsa-miR-4524b-5p | 1 | −15.5 | 0.529 | 3’UTR |
| SLC45A2 (ENSG00000164175 | hsa-miR-200c-3p | 1 | −17.1 | 0.515 | 3’UTR |
| SLC45A2 (ENSG00000164175 | hsa-miR-371a-5p | 1 | −17.1 | 0.485 | 3’UTR |
| SLC45A2 (ENSG00000164175 | hsa-miR-584-5p | 1 | −17.1 | 0.544 | 3’UTR |
| SLC45A2 (ENSG00000164175 | hsa-miR-621 | 1 | −15.7 | 0.588 | 3’UTR |
| SLC45A2 (ENSG00000164175 | hsa-miR-668-5p | 1 | −17.7 | 0.426 | 3’UTR |
| SLC45A2 (ENSG00000164175 | hsa-miR-1185-1-3p | 1 | −19.42 | 0.544 | 3’UTR |
| SLC45A2 (ENSG00000164175 | hsa-miR-3163 | 1 | −18.16 | 0.471 | 3’UTR |
| SLC45A2 (ENSG00000164175 | hsa-miR-3184-3p | 1 | −19.42 | 0.559 | 3’UTR |
| SLC45A2 (ENSG00000164175 | hsa-miR-3150b-5p | 1 | −19.83 | 0.544 | 3’UTR |
| SLC45A2 (ENSG00000164175 | hsa-miR-4640-5p | 1 | −15.6 | 0.426 | 3’UTR |
| SLC45A2 (ENSG00000164175 | hsa-miR-4731-3p | 1 | −17.1 | 0.529 | 3’UTR |
| SLC45A2 (ENSG00000164175 | hsa-miR-5003-5p | 1 | −16.1 | 0.485 | 3’UTR |
| SLC45A2 (ENSG00000164175 | hsa-miR-6730-3p | 1 | −15.47 | 0.515 | 3’UTR |
| SLC45A2 (ENSG00000164175 | hsa-miR-6776-3p | 1 | −18.46 | 0.544 | 3’UTR |
| SLC45A2 (ENSG00000164175 | hsa-miR-6788-3p | 1 | −19.15 | 0.574 | 3’UTR |
| SLC45A2 (ENSG00000164175 | hsa-miR-6865-3p | 1 | −15.5 | 0.5 | 3’UTR |
| PMEL(ENSG00000185664) | hsa-miR-1247-3p | 1 | −17.5 | 0.603 | 3’UTR |
Abbreviations: TRPM1, transient receptor potential cation channel subfamily M member 1; PMEL, premelanosome protein; OCA2, melanosomal transmembrane protein; SLC45A2, solute carrier family 45 members 2; EDNRB, endothelin receptor type B; DCT, dopachrome tautomerase; MLANA, melan-A; TYRP1, tyrosinase-related protein 1; TYR, tyrosinase, 3’UTR; 3′ untranslated region; AU rich region fraction, adenylate uridylate rich region fraction.
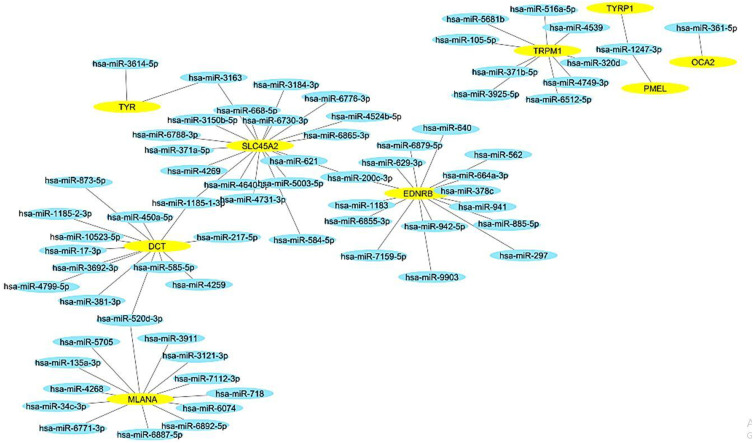
The network linking the miRNAs to the nine genes was constructed utilizing the Cytoscape software, a widely used tool for visualizing biological networks. The resulting network comprised 77 nodes and 73 edges, with an average number of neighbors calculated at 1.8. Furthermore, the network exhibited a diameter of 1 and a radius of 1, indicating the compactness of the network structure. The characteristic path length of the network was determined to be 1, underscoring the efficiency of information transfer within the network. The clustering coefficient, a measure of the network’s tendency to form clusters, was found to be 0.0001, suggesting a low level of clustering. Lastly, the network density, which signifies the number of possible connections in relation to the total number of connections, was calculated at 0.012, indicating a sparse network connectivity [Figure 9].
Figure 9.
miRNAs and genes regulatory network for nine genes using Cytoscape software. The yellow color indicates genes, and the blue color indicates miRNAs.
Discussion
Genome-wide association studies have unveiled numerous susceptibility genes associated with the complex genetic disease known as vitiligo, demonstrating a connection between immunomodulatory genes and specific melanocyte-specific genes that contribute to the gradual loss of pigmentation.7,13,14 The research conducted in this study involves the utilization of microarray data analysis techniques and bioinformatic methodologies to pinpoint the top 10 genes responsible for regulating vitiligo progression. These identified genes include TYR, MLANA, TYRP1, PMEL, OCA2, SLC45A2, GPR143, DCT, TRPM1, and EDNRB, all of which play crucial roles in the development and manifestation of vitiligo. By employing advanced computational tools and genetic analysis, this study sheds light on the intricate molecular mechanisms underlying vitiligo pathogenesis, offering valuable insights for further research and potential therapeutic interventions.
Our discoveries are consistent with the research conducted by Jin et al, who also observed noteworthy correlations between generalized vitiligo and genetic variations in the TYR gene.15 Additionally, the study by Awad et al provided evidence that the levels of the three proteins Tyr, Tyrp1, and Tyrp2 were reduced, indicating similar changes in the progression of vitiligo.16 Another investigation, which delved into the rs1847134 and rs1393350 variants of the TYR gene in non-segmental vitiligo patients, uncovered significant variations between the different groups of subjects. In particular, the rs1393350 polymorphism of the TYR gene displayed considerable differences among the subject groups, with a statistically significant p-value of less than 0.001. This suggests a potential role of this genetic variant in the development or manifestation of vitiligo. The findings from these studies contribute to a better understanding of the genetic factors involved in vitiligo pathogenesis. Furthermore, they underscore the importance of further research to elucidate the mechanisms underlying these associations and their implications for potential therapeutic interventions in vitiligo patients. In conclusion, the identification of genetic variations such as the rs1393350 polymorphism in the TYR gene highlights the complexity of vitiligo etiology and the need for personalized approaches to treatment based on individual genetic profiles.17
Our findings align closely with the research conducted by Yuan and his team, who also identified six DEGs (PMEL, MLANA, DCT, SOX10, TYRP1, and MC1R) that play a role in the development of vitiligo.18 Furthermore, Zhao et al conducted a study that suggested DCT and KIF1A as potential biomarkers associated with vitiligo.19 In contrast, another research study proposed TYR, TYRP1, DCT, and LARP7 as potential biomarkers related to vitiligo.20 These studies collectively contribute to the growing body of knowledge surrounding the molecular mechanisms underlying vitiligo and the potential biomarkers that could aid in its diagnosis and treatment.
In the current investigation, we have delineated various sets of biological, cellular, molecular, and KEGG pathways in our analysis. The most notable biological processes identified include developmental pigmentation (GO:0048066) and pigment metabolic processes (GO:0042440), characterized by a substantial overlap in gene participation within the pathway, as evidenced by significant False Discovery Rate (FDR) and Fold Enrichment values. These findings underscore the pivotal role of these pathways in regulating pigmentation and skin coloration. Moreover, the predominant cellular pathway identified is the melanosome pathway (GO:0042470) along with the pigment granule pathway, elucidating their importance in cellular processes related to pigmentation. Conversely, the KEGG pathways that exhibited significance in our analysis are Tyrosine metabolism (Path: hsa00350) and Melanogenesis (Path: hsa04916), further emphasizing the intricate network of pathways governing pigmentation. It is noteworthy that these pathways collectively contribute to the diverse array of biological mechanisms underlying pigmentation and skin color regulation. Pigmentation, which refers to the distribution of pigment in skin, hair, and eyes for color expression, serves as a distinguishing feature due to its association with genetic diversity in the pathways controlling melanocytes, the pigment-producing cells.21
Numerous studies have highlighted the involvement of genes associated with human hypo- and hyperpigmentation disorders in normal phenotypic variations in basal skin color, including TYR, TYRP1, SLC45A2, SLC24A5, SLC24A4, OCA2, ASIP, IRF4, TPCN2, and MC1R.21–24
Mutations and variants in these genes play a direct role in the pigmentation process, underscoring their significance in regulating skin pigmentation.21,25 This wealth of information corroborates our identification of the top ten genes outlined in this study and their predicted involvement in the pathways associated with pigmentation. Thus, these findings provide valuable insights into the complex interplay of genetic factors and pathways influencing pigmentation and skin coloration.
In the present investigation, the top-ranking TFs that coincide with the predicted genes were identified, and through a concentrated examination of TFs, it was discerned that MITF was found to overlap with 10 genes, while SOX10 exhibited overlap with 9 genes. Additionally, it was observed that ALX1 and TFAP2A demonstrated overlap with 7 genes each, shedding light on their potential regulatory significance. These findings underscore the pivotal regulatory function of TFs such as MITF, SOX10, ALX1, and TFAP2A in the pathogenesis of vitiligo, thereby highlighting their crucial involvement in the development and progression of this skin disorder. We can explain the role of TFs recommended in this study by their effect on the up or down-regulation of key regulatory genes and this mechanism is very important in developing vitiligo.
Our findings align with Listiawan and his research team, who conducted a study involving biopsies from vitiligo patients to assess MITF levels both before and after various therapies. They discovered a notable disparity in MITF quantities in vitiligo patients pre and post-NB-UVB treatment, with statistical significance indicated by a p-value of less than 0.001 (p=0.05). Their suggestion that MITF can serve as a valuable indicator for monitoring vitiligo treatment efficacy is noteworthy.26 Additionally, Papaccio et al reported an increase in MITF gene expression among individuals with vitiligo.27 Blokzijl et al shed light on the role of SOX10 in vitiligo by identifying heightened SOX10 levels in the serum of both Vitiligo and Melanoma patients.28 Furthermore, a study focusing on SOX10 and SOX19 proposed these TFs as novel autoantigens associated with vitiligo.29
The role of TFs highlighted in this research can be elucidated by their involvement in the regulation of key genes, either through upregulation or downregulation mechanisms. This intricate process plays a crucial role in the pathogenesis and progression of vitiligo, emphasizing the significance of understanding the molecular mechanisms underlying this skin condition.30 Further exploration of the interplay between TFs and regulatory genes could provide valuable insights into potential therapeutic targets for vitiligo management. Investigating the intricate network of transcriptional regulation in vitiligo may pave the way for personalized treatment approaches tailored to the specific molecular profiles of patients.31 Understanding the role of TFs in vitiligo pathophysiology is essential for advancing precision medicine strategies aimed at improving patient outcomes and quality of life.
In the present investigation, a total of 73 miRNAs have been identified as suitable candidates for the regulation of nine specific genes linked to vitiligo. Among these genes, there are 15 associated with EDNRB, 1 with OCA2, 2 with TYR, 1 with TYRP1, 13 with MLANA, 9 with TRPM1, 13 with DCT, 18 with SLC45AC, and 1 with PMEL. This comprehensive categorization highlights the complex and intricate regulatory mechanisms at play in the development of this skin disorder.
miRNAs, short for miRNAs, represent a category of minuscule noncoding RNA molecules that function by targeting mRNA to regulate gene expression With a typical length of approximately 22 nucleotides, these miRNAs play a crucial role in post-transcriptional gene regulation.32 Their mechanism of action involves binding in a partially complementary manner to specific sequences located in the 3′-UTR of the target mRNAs. By doing so, miRNAs effectively modulate the expression levels of the associated genes through this intricate regulatory process.33,34 It is widely recognized in the scientific community that both humoral and cellular immunity are essential components in the development of vitiligo.35 Recent studies have identified specific miRNAs that are associated with the immune response, shedding light on their potential role in this skin disorder. These small regulatory molecules, known as miRNAs, are responsible for modulating a diverse range of biological processes and have the capacity to influence the immune system’s functioning, particularly in relation to human cutaneous T cells. The intricate interplay between these miRNAs and the immunological imbalance observed in vitiligo underscores the complexity of the disease etiology.35–39
Conclusion
TYR, MLANA, TYRP1, PMEL, OCA2, SLC45A2, GPR143, DCT, TRPM1, and EDNRB are the top ten genes that are pivotal in the progression and exhibition of vitiligo. The biological, cellular, molecular, and KEGG pathways of those genes has an imperative role in the pathogenesis of vitiligo. TFs and miRNAs that interact with this gene are listed, shedding light on the regulatory mechanisms governing the expression of these key genes in vitiligo. The significance of these ten genes in vitiligo is not only noteworthy, but it also sheds light on the complicated interplay between genetic factors and regulatory components in the development of this dermatological disorder.
Future Recommendation
Future studies in vivo or in vitro focusing on the regulatory functions of TFs and miRNAs in vitiligo may uncover novel therapeutic avenues that target specific molecular pathways implicated in the disease progression. The identification of TFs and miRNAs as key players in vitiligo pathogenesis underscores the importance of utilizing targeted interventions that modulate transcriptional activity to restore skin pigmentation in affected individuals. Expanding our knowledge of the transcriptional landscape in vitiligo could lead to the development of innovative treatments that address the underlying molecular dysregulation driving this dermatological disorder.
Acknowledgments
The author would like to thank the Deanship of Scientific Research at Shaqra University for supporting this work. There are no sponsors or funds for the research; the author supported it.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Chen J, Li S, Li C. Mechanisms of melanocyte death in vitiligo. Med Res Rev. 2021;41(2):1138–1166. [PubMed PMID: 33200838. Pubmed Central PMCID: PMC7983894. Epub 2020/11/18]. doi: 10.1002/med.21754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kubelis-López DE, Zapata-Salazar NA, Said-Fernández SL, et al. Updates and new medical treatments for vitiligo (Review). Exp Ther Med. 2021;22(2):797. [PubMed PMID: 34093753. Pubmed Central PMCID: PMC8170669. Epub 2021/06/08]. doi: 10.3892/etm.2021.10229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Spritz RA, Andersen GH. Genetics of Vitiligo. Dermatol Clin. 2017;35(2):245–255. [PubMed PMID: 28317533. Pubmed Central PMCID: PMC5362127. Epub 2017/03/21]. doi: 10.1016/j.det.2016.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roberts GHL, Santorico SA, Spritz RA. The genetic architecture of vitiligo. Pigm Cell Melanoma Res. 2020;33(1):8–15. [PubMed PMID: 31743585. Pubmed Central PMCID: PMC6928395. Epub 2019/11/20]. doi: 10.1111/pcmr.12848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Spritz RA. Modern vitiligo genetics sheds new light on an ancient disease. J Dermatol. 2013;40(5):310–318. [PubMed PMID: 23668538. Pubmed Central PMCID: PMC3783942. Epub 2013/05/15]. doi: 10.1111/1346-8138.12147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Das S, Majumder PP, Majumdar T, Haldar B, Rao D. Studies on vitiligo. II. Familial aggregation and genetics. Gen Epidemiol. 1985;2(3):255–262. doi: 10.1002/gepi.1370020303 [DOI] [PubMed] [Google Scholar]
- 7.Spritz RA. The genetics of vitiligo. J Investig Dermatol. 2011;131(E1):E18–20. [PubMed PMID: 22094401. Pubmed Central PMCID: PMC3513341. Epub 2011/11/19]. doi: 10.1038/skinbio.2011.7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nath SK, Kelly JA, Namjou B, et al. Evidence for a susceptibility gene, SLEV1, on chromosome 17p13 in families with vitiligo-related systemic lupus erythematosus. Am J Hum Genet. 2001;69(6):1401–1406. doi: 10.1086/324470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jin Y, Mailloux CM, Gowan K, et al. NALP1 in vitiligo-associated multiple autoimmune disease. N Engl J Med. 2007;356(12):1216–1225. doi: 10.1056/NEJMoa061592 [DOI] [PubMed] [Google Scholar]
- 10.Regazzetti C, Joly F, Marty C, et al. Transcriptional Analysis of Vitiligo Skin Reveals the Alteration of WNT Pathway: a Promising Target for Repigmenting Vitiligo Patients. J Investig Dermatol. 2015;135(12):3105–3114. [PubMed PMID: 26322948. Epub 2015/09/01]. doi: 10.1038/jid.2015.335 [DOI] [PubMed] [Google Scholar]
- 11.Franceschini A, Szklarczyk D, Frankild S, et al. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013;41(Database issue):D808–15. [PubMed PMID: 23203871. Pubmed Central PMCID: PMC3531103. Epub 2012/12/04]. doi: 10.1093/nar/gks1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ge SX, Jung D, Yao R. ShinyGO: a graphical gene-set enrichment tool for animals and plants. Bioinform. 2020;36(8):2628–2629. [PubMed PMID: 31882993. Pubmed Central PMCID: PMC7178415. Epub 2019/12/29]. doi: 10.1093/bioinformatics/btz931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alikhan A, Felsten LM, Daly M, Petronic-Rosic V. Vitiligo: a comprehensive overview: part I. Introduction, epidemiology, quality of life, diagnosis, differential diagnosis, associations, histopathology, etiology, and work-up. J Am Acad Dermatol. 2011;65(3):473–491. doi: 10.1016/j.jaad.2010.11.061 [DOI] [PubMed] [Google Scholar]
- 14.Alkhateeb A, Fain PR, Thody A, Bennett DC, Spritz RA. Epidemiology of vitiligo and associated autoimmune diseases in Caucasian probands and their families. Pigment Cell Res. 2003;16(3):208–214. doi: 10.1034/j.1600-0749.2003.00032.x [DOI] [PubMed] [Google Scholar]
- 15.Jin Y, Birlea SA, Fain PR, et al. Variant of TYR and autoimmunity susceptibility loci in generalized vitiligo. N Engl J Med. 2010;362(18):1686–1697. doi: 10.1056/NEJMoa0908547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Awad SS, Moftah NH, Moustafa WM. Expression of Tyrosinase, Tyrp1 and Tyrp2 in vitiligo. Minia J Med Res. 2020;31(4):189–191. doi: 10.21608/mjmr.2022.217926 [DOI] [Google Scholar]
- 17.Męcińska-Jundziłł K, Tadrowski T, Jundziłł A, Witmanowski H, Czajkowki R. Evaluation of polymorphisms and expression of PTPN22, NLRP1 and TYR genes in vitiligo patients. Adv Dermatol Allergol. 2023;40(1):1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yuan X, Meng D, Cao P, et al. Identification of pathogenic genes and transcription factors in vitiligo. Dermatologic Therapy. 2019;32(5):e13025. doi: 10.1111/dth.13025 [DOI] [PubMed] [Google Scholar]
- 19.Zhao Y, Ge K, Zhang R. Identification of Dopachrome Tautomerase (DCT) and Kinesin Family Member 1A (KIF1A) as Related Biomarkers and Immune Infiltration Characteristics of Vitiligo Based on Lasso-SVM Algorithms. Clin Cosmet Invest Dermatol. 2023;Volume 16:3509–3520. doi: 10.2147/CCID.S443165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang J, Yu R, Guo X, et al. Identification of TYR, TYRP1, DCT and LARP7 as related biomarkers and immune infiltration characteristics of vitiligo via comprehensive strategies. Bioengineered. 2021;12(1):2214–2227. doi: 10.1080/21655979.2021.1933743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baxter LL, Pavan WJ. The etiology and molecular genetics of human pigmentation disorders. WIRES Dev Biol. 2013;2(3):379–392. doi: 10.1002/wdev.72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sturm RA. Molecular genetics of human pigmentation diversity. Human Mol Gen. 2009;18(R1):R9–R17. doi: 10.1093/hmg/ddp003 [DOI] [PubMed] [Google Scholar]
- 23.Valverde P, Healy E, Jackson I, Rees JL, Thody AJ. Variants of the melanocyte–stimulating hormone receptor gene are associated with red hair and fair skin in humans. Nature Genet. 1995;11(3):328–330. doi: 10.1038/ng1195-328 [DOI] [PubMed] [Google Scholar]
- 24.Beaumont KA, Shekar SN, Cook AL, Duffy DL, Sturm RA. Red hair is the null phenotype of MC1R. Human Mutation. 2008;29(8):E88–E94. doi: 10.1002/humu.20788 [DOI] [PubMed] [Google Scholar]
- 25.Bharti K, Nguyen MTT, Skuntz S, Bertuzzi S, Arnheiter H. The other pigment cell: specification and development of the pigmented epithelium of the vertebrate eye. Pigment Cell Res. 2006;19(5):380–394. doi: 10.1111/j.1600-0749.2006.00318.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Listiawan MY, Astari L, Wardhani PH. Comparison amount of microphthalmia-associated transcription factor in vitiligo before and after narrowband-ultraviolet B therapy. Dermatol Rep. 2019;11(s1):35–37. [Google Scholar]
- 27.Papaccio F, Bellei B, Ottaviani M, et al. A possible modulator of vitiligo metabolic impairment: rethinking a PPARγ agonist. Cells. 2022;11(22):3583. doi: 10.3390/cells11223583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blokzijl A, Chen LE, Gustafsdottir SM, et al. Elevated levels of SOX10 in serum from vitiligo and melanoma patients, analyzed by proximity ligation assay. PLoS One. 2016;11(4):e0154214. doi: 10.1371/journal.pone.0154214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hedstrand H, Ekwall O, Olsson MJ, et al. The transcription factors SOX9 and SOX10 are vitiligo autoantigens in autoimmune polyendocrine syndrome type I. J Biol Chem. 2001;276(38):35390–35395. [PubMed PMID: 11423552. Epub 2001/06/26. eng]. doi: 10.1074/jbc.M102391200 [DOI] [PubMed] [Google Scholar]
- 30.Oshchepkov D, Chadaeva I, Kozhemyakina R, et al. Transcription Factors as Important Regulators of Changes in Behavior through Domestication of Gray Rats: quantitative Data from RNA Sequencing. Int J Mol Sci. 2022;23(20):12269. [PubMed PMID: 36293128. Pubmed Central PMCID: PMC9603081. Epub 2022/10/28. eng]. doi: 10.3390/ijms232012269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lei Z, Yu S, Ding Y, et al. Identification of key genes and pathways involved in vitiligo development based on integrated analysis. Medicine. 2020;99(31):e21297. [PubMed PMID: 32756109. Pubmed Central PMCID: PMC7402735. Epub 2020/08/07. eng]. doi: 10.1097/MD.0000000000021297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ratti M, Lampis A, Ghidini M, et al. MicroRNAs (miRNAs) and Long Non-Coding RNAs (lncRNAs) as New Tools for Cancer Therapy: first Steps from Bench to Bedside. Targeted Oncol. 2020;15(3):261–278. [PubMed PMID: 32451752. Pubmed Central PMCID: PMC7283209 Mirchev and Jens C. Hahne have no conflicts of interest that are directly relevant to the content of this article. Nicola Valeri received speaker honorarium from Bayer, Eli-Lilly, Pfizer and Merck. Epub 2020/05/27. eng]. doi: 10.1007/s11523-020-00717-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ying SY, Chang DC, Lin SL. The microRNA (miRNA): overview of the RNA genes that modulate gene function. Mol Biotechnol. 2008;38(3):257–268. [PubMed PMID: 17999201. Pubmed Central PMCID: PMC7091389. Epub 2007/11/14. eng]. doi: 10.1007/s12033-007-9013-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O’Brien J, Hayder H, Zayed Y, Peng C. Overview of microRNA biogenesis, mechanisms of actions, and circulation. Front Endocrinol. 2018;9:388354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li L. The role of MicroRNAs in vitiligo: regulators and therapeutic targets. Ann Dermatol. 2020;32(6):441. doi: 10.5021/ad.2020.32.6.441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bronevetsky Y, Ansel KM. Regulation of mi RNA biogenesis and turnover in the immune system. Immunol Rev. 2013;253(1):304–316. doi: 10.1111/imr.12059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mansuri M, Singh M, Dwivedi M, Laddha N, Marfatia Y, Begum R. MicroRNA profiling reveals differentially expressed microRNA signatures from the skin of patients with nonsegmental vitiligo. Br J Dermatol. 2014;171(5):1263–1267. doi: 10.1111/bjd.13109 [DOI] [PubMed] [Google Scholar]
- 38.Wang P, Li Y, Hong W, et al. The changes of microRNA expression profiles and tyrosinase related proteins in MITF knocked down melanocytes. Mol Biosyst. 2012;8(11):2924–2931. doi: 10.1039/c2mb25228g [DOI] [PubMed] [Google Scholar]
- 39.Wu DT, Chen JS, Chang DC, Lin S-L. Mir-434-5p mediates skin whitening and lightening. Clin Cosmet Invest Dermatol. 2008;1:19–35. doi: 10.2147/ccid.s4181 [DOI] [PMC free article] [PubMed] [Google Scholar]