Abstract
Plasmacytoid dendritic cells (pcDC) and myeloid dendritic cells (myDC) are shown to express CD4 and low levels of CCR5 and CXCR4, but only myDC express DC SIGN, a C-type lectin that binds human immunodeficiency virus but does not mediate virus entry. Both DC types were more susceptible to infection with a macrophage than a lymphotropic strain of human immunodeficiency virus type 1, but pcDC were more readily infected than myDC.
Plasmacytoid dendritic cells (pcDC) have only recently been recognized as a distinct population of blood dendritic cells (DC) (6, 8, 9). pcDC, like myeloid DC (myDC), stimulate strong T-cell-proliferative responses but differ in other aspects of their biology. Blood myDC take up residence in the peripheral tissues, where they intercept invading pathogens, and then migrate to secondary lymphoid tissue, secrete interleukin-12, and present processed antigens to T cells (1). By contrast, pcDC migrate directly from blood to lymphoid tissue, where on stimulation via CD40, they produce large amounts of α interferon and thus may play an important role in innate antiviral immunity (3, 10).
The question of the susceptibility of DC to infection by human immunodeficiency virus (HIV) has been controversial (2, 7). Some previous studies may have been unwittingly performed on mixed populations of myDC and pcDC, and this could explain conflicting findings. Here, we show that each DC population expresses the receptors and coreceptors necessary for HIV infection but that pcDC are more readily infected than myDC.
CD11c+ myDC and CD11c− pcDC were purified from buffy coats as previously described (9). Purified DC contained 2% or less contaminating T cells. Purified DC (105) were cultured in 96-well plates with recombinant granulocyte-macrophage colony-stimulating factor (100 ng/ml) for myDC or interleukin-3 (10 ng/ml) for pcDC (cytokines were from R&D Systems Europe Ltd). myDC and pcDC remained positive and negative, respectively, for CD11c expression throughout a 7- to 9-day culture period, and there was no evidence of expansion of a minor cell population.
CD4, CCR5, and CXCR4 expression by DC was analyzed in uncultured peripheral blood mononuclear cells (PBMC) by four-color fluorescence-activated cell sorter (FACS) analysis. DC were identified by the absence of labeling with phycoerythrin-conjugated antibodies against CD3, CD14, CD16, and CD19 and strong labeling with PerCp-conjugated anti HLA-DR (Becton Dickinson). myDC and pcDC were differentiated by staining them with fluorescein isothiocyanate-conjugated anti-CD11c (Dako). CD4, CXCR4, and CCR5 were detected by allophycocyanin-conjugated antibodies (Becton Dickinson). RNA message for CD4, CCR5, CXCR4, DC SIGN (a C-type lectin that binds HIV but does not mediate virus entry [5]), and GAPDH (glyceraldehyde-3-phosphate dehydrogenase) was detected in purified DC by reverse transcription (RT)-PCR on DNase-treated RNA. cDNA was amplified by PCR using the following primers: CD4 forward (ATAAAGATTCTGGGAAATCAGGGCTCC) and CD4 reverse (TGCAACTTTCCTGTTTTCGCTTCAAGG) (product, 751 bp); CXCR4 forward (TGACTCCATGAAGGAACCCTG) and CXCR4 reverse (CTTGGCCTCTGACTGTTGCTG) (product, 381 bp); CCR5 forward (CCAAAAGCACATTGCCAAACG) and CCR5 reverse (ACTTGAGTCCGTGTCACAAGCC) (product, 136 bp); DC SIGN forward (ACAGAGGAGAGCCCAACAACG) and DC SIGN reverse (AAGGGGGTGAAGTTCTGCTACG) (product, 202 bp); and GAPDH forward (ATGGAGAAGGCTGGGGCTC) and GAPDH reverse (AAGTTGTCATGGATGACCTTG) (product, 196 bp). Forty cycles of PCR were performed by denaturation for 30 s at 94°C, annealing for 1 min at 55°C, and then incubation for 2 min at 72°C.
Purified myDC and pcDC (105 cells) were infected with DNase-treated lymphotropic, CXCR4-utilizing IIIB or macrophage-tropic, CCR5-utilizing Ba-L (20 provirus-forming units assayed in PM1 cells) strains of HIV type 1 (HIV-1). After 18 h, they were washed three times and cultured for 7 to 9 days. DNA was extracted, and the HIV provirus load was estimated by limiting-dilution PCR using primers based on HIV-1 Pol, a technique shown to detect a single provirus copy (11). The first-round primers were forward, 5′ CATGGGTACCAGCACAAAGG 3′ and reverse, 5′ TCTACTTGTCCATGCATGGCTTC 3′. The second-round primers were forward, 5′ GGAGGAAATGAACAAGTAGATAAATTAGTCAG 3′, and reverse, 5′ TCACTAGCCATTGCTCTCCAATT 3′. To test for release of infectious virus, supernatants were cocultured with PM1 cells for 5 h, and newly synthesized proviral DNA was measured by limiting-dilution nested PCR.
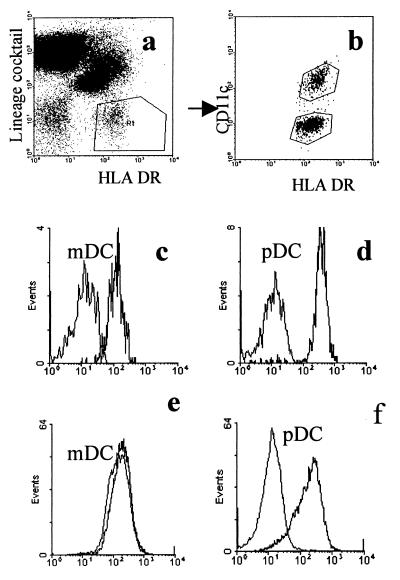
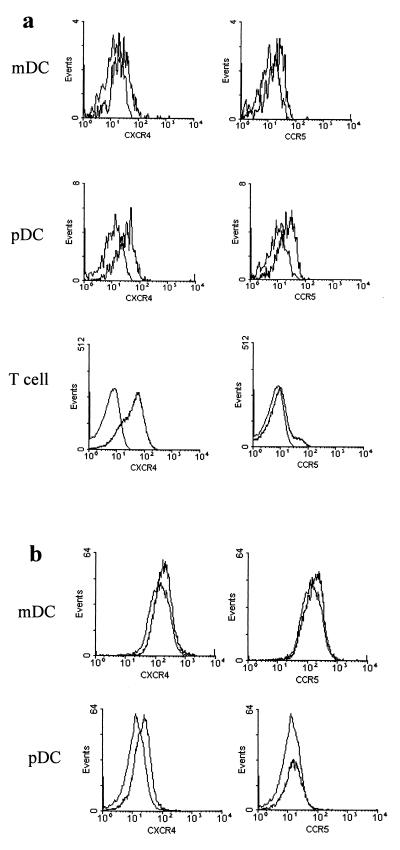
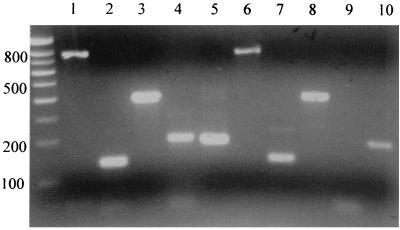
myDC and pcDC were identified by FACS analysis (Fig. 1a and b). CD4 was detected on both populations but was more highly expressed on pcDC (Fig. 1c and d). After 48 h of culture, surface expression of CD4 was downregulated on myDC but not on pcDC (Fig. 1e and f). Data from one of five experiments to detect chemokine expression on DC are shown, and for comparison, we also show expression on T cells. Labeling for CXCR4 was very low on myDC but higher on pcDC (Fig. 2a). There were low levels of CCR5 on myDC and slightly higher levels on pcDC. CCR5 became undetectable on the surfaces of both types of DC cultured for 48 h whereas CXCR4 expression was little changed (Fig. 2b). Surprisingly, mRNA for all three receptors was detectable in cultured as well as fresh myDC and pcDC (Fig. 3). DC-SIGN mRNA was detected in cultured and freshly isolated myDC but not in pcDC (Fig. 3).
FIG. 1.
FACS analysis of DC for expression of CD4. (a) Identification of DC in peripheral blood (R1); (b) myDC and pcDC are differentiated by expression of CD11c; (c and d) expression of CD4 on myDC (c) and pcDC (d) in freshly isolated PBMC. (e and f) CD4 expression on purified myDC (e) and pcDC (f) cultured for 2 days. On all histograms, the left-hand curve represents staining with an isotype control antibody.
FIG. 2.
Expression of CXCR4 and CCR5. (a) FACS labeling for CXCR4 and CCR5 on myDC, pcDC, and T cells in freshly isolated PBMC. (b) Expression of CXCR4 and CCR5 on purified myDC and pcDC cultured for 48 h. The left-hand curve in each histogram represents staining with the isotype control antibody.
FIG. 3.
Detection of mRNA for CD4, CXCR4, CCR5, DC SIGN, and GAPDH on cultured myDC and pcDC. Gel analysis of an RT-PCR to detect mRNAs in purified DC cultured for 48 h. Lanes 1 to 5, myDC; lanes 6 to 10, pcDC; lanes 1 and 6, CD4; lanes 2 and 7, CCR5; lanes 3 and 8, CXCR4; lanes 4 and 9, DC SIGN; lanes 5 and 10, GAPDH.
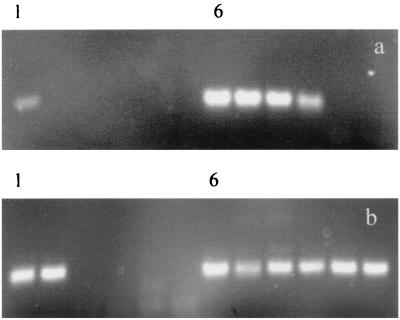
HIV proviral DNA was detected in six of seven in vitro infection experiments (Table 1). Experiment 5 shows a higher proviral load in cultures infected with macrophage-tropic Ba-L and also shows that virus grows more efficiently in pcDC than in myDC (Fig. 4). This pattern was seen in all experiments, although the differences were not always as great as those observed in experiment 5. Nevertheless, in no experiment using DC derived from the same donor was IIIB found to replicate to higher levels than Ba-L nor were provirus loads ever observed to be higher in myDC than in pcDC. Infectious virus, demonstrated by the ability of DC supernatant to form provirus in PM1 cells, was detected in six of eight DC cultures tested (experiments 2, 3, and 5), but the largest amounts of virus were found in the pcDC cultures infected with Ba-L (Table 1).
TABLE 1.
Summary of seven in vitro infection experiments
| Experiment | DC | Virus | Provirus load/105 cells | Provirus-forming units/ml in DC supernatant |
|---|---|---|---|---|
| 1aa | mDC | Ba-L | <50 | Not tested |
| 1ba | pDC | Ba-L | <50 | Not tested |
| 2aa | mDC | Ba-L | 50 | 1 × 103 |
| 2ba | pDC | Ba-L | 12,500 | 2.5 × 103 |
| 3ab | mDC | IIIB | 50 | None detected |
| 3bb | pDC | IIIB | 50 | None detected |
| 4ab | mDC | Ba-L | 50 | Not tested |
| 4bb | pDC | Ba-L | 31,250 | Not tested |
| 5ab | mDC | IIIB | 50 | 2 × 102 |
| 5bb | mDC | Ba-L | 6,250 | 2.5 × 103 |
| 5cb | pDC | IIIB | 250 | 2 × 102 |
| 5db | pDC | Ba-L | 156,000 | 12.4 × 103 |
| 6aa | mDC | IIIB | <50 | Not tested |
| 6ba | mDC | Ba-L | <50 | Not tested |
| 6ca | pDC | IIIB | 50 | Not tested |
| 6da | pDC | Ba-L | 250 | Not tested |
| 7aa | pDC | IIIB | 50 | Not tested |
| 7ba | pDC | Ba-L | 31,250 | Not tested |
Virus load and infectious virus measured at 7 days.
Virus load and infectious virus measured at 9 days.
FIG. 4.
Estimation of HIV-1 proviral DNA copy number on myDC and pcDC infected in vitro with lymphotropic and macrophage-tropic strains of HIV-1. Limiting-dilution nested PCR to detect HIV-1 provirus in in vitro-infected myDC (a) and pcDC (b) (experiment 5; 9-day infection [Table 1]). PCR was performed on five fivefold dilutions of the DNA template. Lanes 1 to 5 and 6 to 10 on each gel represent assays on IIIB-and Ba-L-infected cultures, respectively.
We show by FACS, analysis and RT-PCR that pcDC express CD4, CCR5, and CXCR4, the receptors required for infection by macrophage and lymphotropic strains of HIV-1. These receptors were also detected on myDC, but FACS analysis showed lower levels of expression, particularly for CD4 and CXCR4. On culture, surface expression of CXCR4 was little changed, whereas there was a loss of membrane CCR5 on both cell types and downregulation of CD4 on myDC. However, a positive signal for all three receptors on both populations was detected by RT-PCR on fresh and cultured cells. Whether these findings reflect inefficient transport to the cell surface, a high rate of membrane receptor turnover, or other possible explanations is not yet clear.
In vitro infection experiments with HIV-1 Ba-L confirmed that both cell types could be infected by HIV-1; however, the virus replicated more efficiently in pcDC. This may reflect lower CD4 expression by myDC. Alternatively, cellular entry into myDC may be blocked by attachment to DC SIGN. Lower provirus loads were detected in DC infected with the IIIB lymphotropic strain than in those infected with the Ba-L strain of HIV-1. For the myDC, this may be explained by poor expression of CXCR4; however, in pcDC, CCR5 and CXCR4 were expressed at similar levels. A further inconsistency of the data was that for both DC populations, in vitro culture resulted in downregulation of CCR5 but not of CXCR4. The findings may partly reflect a defective vpr gene, thought to facilitate infection of nondividing cells, in the IIIB strain of virus (4).
DC infected with 20 infectious units of virus had a high provirus copy number after 7 to 9 days, suggesting productive virus infection. Release of infectious virus was confirmed by culturing DC supernatants with PM1 cells and analyzing them for provirus formation. Infectious virus was released by both pcDC and myDC cultures. Low levels of contaminating T cells, 2% or less CD3+ cells, were present in purified DC preparations. Although it cannot be ruled out that some of the provirus detected is derived from T cells, this is unlikely to significantly distort the in vitro infection data for the following reasons: (i) the number of provirus copies after 9 days greatly exceeded the maximum number of T cells, (ii) significant replication in T cells would mask the differences between myDC and pcDC cultures, and (iii) autologous T cells do not support productive infection unless activated by exogenous stimuli.
The finding that pcDC are more readily infected than myDC may explain previous inconsistencies and could reflect investigators working with different populations of cells. Infection and loss of pcDC in vivo may reduce levels of α interferon and result in higher virus loads and disease progression.
Acknowledgments
This work was supported by the United Kingdom Medical Research Council.
HIV-1 virus stocks and the PM-1 cell line (originally donated by M. Reitz) were provided by the MRC Centralised Facility for AIDS Reagents at NIBSC, South Mimms, Herts., United Kingdom.
REFERENCES
- 1.Banchereau J, Steinman R M. Dendritic cells and the control of immunity. Nature. 1998;392:245–251. doi: 10.1038/32588. [DOI] [PubMed] [Google Scholar]
- 2.Cameron P U, Freudenthal P S, Barker J M, Gezelter S, Inaba K, Steinman R M. Dendritic cells exposed to human immunodeficiency virus type-1 transmit a vigorous cytopathic infection to CD4+ T cells. Science. 1992;257:383–387. doi: 10.1126/science.1352913. [DOI] [PubMed] [Google Scholar]
- 3.Cella M, Jarrossay D, Facchetti F, Alebardi O, Nakajima H, Lanzavecchia A, Colonna M. Plasmacytoid monocytes migrate to inflamed lymph nodes and produce large amounts of type I interferon. Nat Med. 1999;5:919–923. doi: 10.1038/11360. [DOI] [PubMed] [Google Scholar]
- 4.Connor R I, Chen B K, Choe S, Landau N R. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology. 1995;206:935–944. doi: 10.1006/viro.1995.1016. [DOI] [PubMed] [Google Scholar]
- 5.Geijtenbeek T B, Kwon D S, Torensma R, van-Vliet S J, van-Duijnhoven G C, Middel J, Cornelissen I L, Nottet H S, Kewalramani V N, Littman D R, Figdor C G, van-Kooyk Y. DC-SIGN, a dendritic cell-specific HIV-1 binding protein that enhances trans-infection of T cells. Cell. 2000;100:587–597. doi: 10.1016/s0092-8674(00)80694-7. [DOI] [PubMed] [Google Scholar]
- 6.Grouard G, Rissoan M C, Filgueira L, Durand I, Banchereau J, Liu Y-J. The enigmatic plasmacytoid T cells develop into dendritic cells with interleukin (IL)-3 and CD40-ligand. J Exp Med. 1997;185:1101–1111. doi: 10.1084/jem.185.6.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Patterson S, Knight S C. Susceptibility of human peripheral blood dendritic cells to infection by human immunodeficiency virus. J Gen Virol. 1987;68:1177–1181. doi: 10.1099/0022-1317-68-4-1177. [DOI] [PubMed] [Google Scholar]
- 8.Rissoan M C, Soumelis V, Kadowaki N, Grouard G, Briere F, de-Waal-Malefyt R, Liu Y J. Reciprocal control of T helper cell and dendritic cell differentiation. Science. 1999;283:1183–1186. doi: 10.1126/science.283.5405.1183. [DOI] [PubMed] [Google Scholar]
- 9.Robinson S P, Patterson S, English N E, Davies D, Knight S C, Reid C D L. Human peripheral blood contains two distinct lineages of dendritic cells. Eur J Immunol. 1999;29:2769–2778. doi: 10.1002/(SICI)1521-4141(199909)29:09<2769::AID-IMMU2769>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 10.Siegal F P, Kadowaki N, Shodell M, Fitzgerald B P, Shah K, Ho S, Antonenko S, Liu Y J. The nature of the principal type 1 interferon-producing cells in human blood. Science. 1999;284:1835–1837. doi: 10.1126/science.284.5421.1835. [DOI] [PubMed] [Google Scholar]
- 11.Simmonds P, Balfe P, Peutherer J F, Ludlam C A, Bishop J O, Brown A J. Human immunodeficiency virus-infected individuals contain provirus in small numbers of peripheral mononuclear cells and at low copy numbers. J Virol. 1990;64:864–872. doi: 10.1128/jvi.64.2.864-872.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]