Abstract
Monitoring the evolution of human immunodeficiency virus type 1 (HIV-1) drug resistance requires measuring the frequency of closely related genetic variants making up the complex viral quasispecies found in vivo. In order to resolve both major and minor (≥2%) protease gene variants differing by one or more nucleotide substitutions, we analyzed PCR products derived from plasma viral quasispecies by using a combination of denaturing gradient gel electrophoresis and DNA heteroduplex tracking assays. Correct population sampling of the high level of genetic diversity present within viral quasispecies could be documented by parallel analysis of duplicate, independently generated PCR products. The composition of genetically complex protease gene quasispecies remained constant over short periods of time in the absence of treatment and while plasma viremia fell >100-fold following the initiation of protease inhibitor ritonavir monotherapy. Within a month of initiating therapy, a strong reduction in the genetic diversity of plasma viral populations at the selected protease locus was associated with rising plasma viremia and the emergence of drug resistance. The high levels of protease genetic diversity seen before treatment reemerged only months later. In one patient, reduction in genetic diversity at the protease gene was observed concomitantly with an increase in diversity at the envelope gene (E. L. Delwart, P. Heng, A. Neumann, and M. Markowitz, J. Virol. 72:2416-2421, 1998), indicating that opposite population genetic changes can take place in different HIV-1 loci. The rapid emergence of drug-resistant HIV-1 was therefore associated with a strong, although only transient, reduction in genetic diversity at the selected locus. The denaturing gradient-heteroduplex tracking assay is a simple method for the separation and quantitation of very closely related, low-frequency, genetic variants within complex viral populations.
The short generation time, high mutation rate, and large population size of human immunodeficiency virus type 1 (HIV-1) make it one of the fastest evolving viruses known (2, 3). Genetically complex HIV-1 quasispecies rapidly evolve following generally clonal primary infection in men (7, 25, 46, 49, 50). In women, heterosexually acquired HIV-1 appears more genetically diverse than in men (23). In the absence of selection, drug-resistant mutants are expected to be of lower replicative fitness than wild-type viruses and therefore are only expected and actually detected as minority variants (2, 3, 19, 31). Antiviral drug selection can then rapidly drive such mutants into the majority. The analysis of HIV-1 quasispecies in vivo is needed to improve our understanding of the complex viral population changes associated with such rapid evolution. Detailed analysis of differentiated viral populations is complicated by several factors. Low-frequency variants are difficult to detect by direct population sequencing of PCR products (45), while a subcloning and sequencing approach will focus sequencing on the most frequent variants and may result in artifactual variant frequencies due to improper population sampling (4, 5, 22). Alternative methods used to detect low-frequency mutants involve designing mutant-specific probes or primers (1, 40). These methods require knowledge of the specific mutation sought and invariable flanking nucleotides and are limited to the analysis of single nucleotide positions. Recently a multiple-site-specific heteroduplex tracking assay (HTA) based on the universal heteroduplex generator concept (48) was developed and shown to detect HIV-1 protease variants at pretargeted codons (36).
Denaturing gradient gel electrophoresis (DGGE) was initially described by Myers et al. (28–30) and was shown to be able to detect most single base pair substitutions. Minute differences in the melting properties and resulting electrophoretic mobilities of DNA fragments differing by a single base pair or by the presence of a single mismatched nucleotide pair could be detected using denaturing gradient polyacrylamide gels. DNA HTAs allow the enumeration of multiple coamplified sequence variants and the rapid determination of their frequency in the viral population (5, 7, 8). Using a combination of both methods we measured population genetic changes at the protease loci of plasma viruses during the emergence of protease inhibitor resistance.
Requirement for denaturing gradient conditions to resolve intrapatient protease gene DNA heteroduplexes.
A minimum of 1 to 2% nucleotide mismatches are required for DNA heteroduplexes to exhibit mobility retardation in nondenaturing polyacrylamide gels (8, 9, 32, 44). As expected, protease gene DNA heteroduplexes containing only 1 to 3 mismatched nucleotides could not be distinguished from their homoduplexes, while DNA heteroduplexes of protease genes from different HIV-1 group M subtypes (differing by >5% substitution) could be readily resolved in nondenaturing polyacrylamide gels (data not shown). Heteroduplex mobility or tracking assays of the protease locus in nondenaturing polyacrylamide gels may therefore be used alongside env and gag heteroduplex mobility assays (9, 14, 43) for subtyping different regions of HIV-1 group M viruses but are unsuitable assays for intrapatient protease quasispecies analysis.
DGGE.
Different electrophoretic conditions were tested to determine if DGGE could be used to separate the closely related protease variants found within an individual. DNA fragments spanning the 3′ half of the protease gene (protease amino acids 52 to 99) were PCR amplified from pNL4-3-derived plasmids containing single or multiple mutations at sites associated with resistance to HIV-1 protease inhibitors using primers EDPR5 (AATGATAGGGGGAATTGGAG [HXB2 positions 2386 to 2406]) and EDPR4GC (GCCCGCCGCGCCCCGCGCCCGGCCCGCCGCCCCCGCTGGTACAGTTTCAATAGGACTAATGG [HXB2 positions 2550 to 2577]). Primer EDPR4GC contained a 35-nucleotide 5′ GC clamp to provide a high melting domain and to prevent total strand separation during the DGGE. DNA homoduplexes and heteroduplexes with 1 to 3 mismatched nucleotides could be electrophoretically resolved (Fig. 1A) in a 7.5% acrylamide (37.5 acrylamide:1 Bis) 1× TAE denaturing gradient gel (top to bottom, 8% formamide and 8.4% [wt/vol] urea to 28% formamide and 29.4% [wt/vol] urea) held at a constant temperature of 53°C at 250 V for 3 h, 15 min in a D-Gene electrophoretic system (Bio-Rad). In all cases tested, DNA heteroduplexes showed reductions in mobilities relative to those of homoduplexes (Fig. 1A). DNA heteroduplexes containing an identical number of mismatched nucleotides at different locations on the PCR fragment exhibited distinct mobilities due to their different melting characteristics. As for heteroduplex mobility assays in nondenaturing conditions, there was a general correlation between the number of mismatched nucleotides and the degree of mobility retardation (8, 9, 14, 32, 44).
FIG. 1.
(A) Ethidium bromide-stained DGGE gel of protease gene PCR products showing the different electrophoretic mobilities of DNA homoduplexes and heteroduplexes. Ho, position of DNA homoduplexes; He, position of DNA heteroduplexes. The numbers of mismatches in the DNA heteroduplexes are indicated above the gel. Changes in PCR products relative to that of pNL4–3 and the pairs of PCR fragments reannealed to form DNA heteroduplexes are indicated below the gel. (B) DG-HTA of plasma quasispecies collected over 7 to 14 days in three untreated subjects showing the resolved protease sequence variants and correct (i.e., reproducible) population sampling in independent nPCRs. Range of plasma viral RNA per milliliter: A lanes, 1.5 × 105 to 3.5 × 105; B lanes, 2.5 × 105 to 3 × 104; C lanes, 2 × 105 to 8 × 105.
DG-HTA.
We next analyzed the reverse transcription-nested PCR (RT-nPCR) products derived from complex quasispecies in clinical samples. Viral RNA was purified from plasma and reverse transcribed as described previously (5). PCR primers used for the first round of nPCR protease gel amplification were EDPR1 (GAGCAGACCAGAGCCAACAGCCCCA [HXB2 positions 2139 to 2163]) and EDPR2 (TTGTTTAACTTTTGGGCCATCC [HXB2 positions 2597 to 2618]). Second-round primers were EDPR5 and EDPR4GC. Direct analysis of the RT-nPCR products from clinical samples by DGGE followed by ethidium bromide staining yielded DNA smears with only faintly discernible bands (data not shown). When multiple sequence variants (number of variants = x) are coamplified and their complementary strands are randomly reannealed, the number of different DNA heteroduplexes formed is equal to x2 − x. In order to reduce the number of detected DNA heteroduplexes to equal the actual number of sequence variants, we combined DGGE with HTA (DG-HTA). During HTA a single-stranded, clonal, radiolabeled probe is reannealed with an excess of unlabeled PCR product amplified from the viral population under study. The labeled HTA probe therefore hybridizes with each of the different variants in the target population forming labeled DNA heteroduplexes. Following electrophoresis only the labeled heteroduplexes, equivalent in number to the number of coamplified sequence variants, are detected by autoradiography or the use of a phosphorimager (4, 5, 7). Because different DNA heteroduplexes may comigrate, this method provides a minimum estimate of the number of variants in the quasispecies. Each labeled HTA band therefore reflects the presence of at least one distinct protease variant, and the intensity of the radiolabeled heteroduplex band provides an estimate of the frequency of that variant within the population (4, 51). In Fig. 1B, RT-nPCR products derived from the cryopreserved plasma samples of three untreated HIV-1-infected subjects were analyzed by DG-HTA. Clonal probes for the DG-HTA were derived from each patient by serially diluting the viral cDNA (5× dilutions) from their latest plasma sample prior to nPCR in order to derive a PCR product from a single cDNA molecule (i.e., an end-point nPCR) (6, 42). The clonal nature of the end-point nPCR product was confirmed by DGGE of the nPCR products followed by ethidium bromide staining, as done in Fig. 1A, to confirm the presence of only a single DNA homoduplex band (data not shown). The second-round PCR was then repeated using a 5′ biotin-tagged EDPR5 primer and a 5′ 32P-labeled EDPR4GC in order to purify single-stranded, clonal, and labeled HTA probes as previously described (4, 5, 7). The HTA probes from each of the three untreated subjects were then reannealed with a 50- to 100-fold excess of unlabeled nPCR products amplified from the undiluted cDNA, and the resulting radiolabeled DNA heteroduplexes were resolved by DG-HTA. The gels were dried and the radioactive signal was detected with a PhosphorImager (Molecular Dynamics) (Fig. 1B). Distinct radiolabeled DG-HTA bands could now be seen in all samples tested (Fig. 1B). The stability of the DG-HTA patterns in these untreated subjects over the course of 7 to 14 days indicated that no quasispecies changes took place at the pro locus over these short time intervals.
Documenting sufficient population sampling.
Insufficient quasispecies samplings were initially seen when duplicate nPCRs (each initiated with different aliquots of undiluted cDNA) resulted in different DG-HTA patterns (data not shown). Insufficient sampling was expected to occur if the number of protease cDNA molecules amplified by nPCR was so low as to result in the stochastic amplification of only a few of the quasispecies' variants. Since an apparent reduction in quasispecies diversity or quasispecies changes could therefore be the artifactual consequence of amplifying a nonrepresentative sampling of a viral population, care was taken to document sufficient sampling throughout this study. Sufficient sampling was achieved following the optimization of the RNA extraction and RT and nPCR procedures and was documented by the generation of identical DG-HTA patterns in independently generated duplicate nPCRs analyzed in parallel (Fig. 1B, 2, and 3). Because the relative intensity of each distinct DG-HTA band (i.e., the relative frequency of distinct sequence variants) was demonstrably reproducible, correct population sampling of these quasispecies was ensured.
FIG. 2.
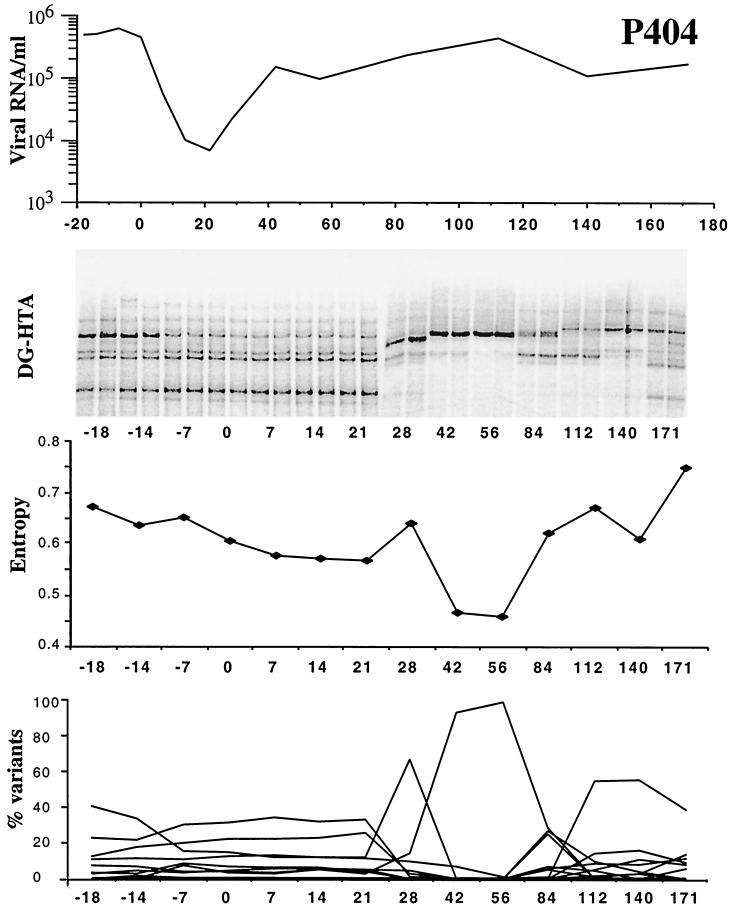
Longitudinally collected plasma viral quasispecies from P404 before and after initiation of ritonavir treatment at day 0, showing DG-HTA patterns together with Shannon entropy and the percent variant values. Viral loads were determined by branched DNA.
FIG. 3.
Longitudinally collected plasma viral quasispecies from P105 before and after initiation of ritonavir treatment at day 0, showing DG-HTA patterns together with Shannon entropy and the percent variant values. Viral loads were determined by branched DNA.
The ability of this method to detect low-frequency variants was determined by scanning the radioactive signal distribution along DG-HTA lanes using the Molecular Dynamics Imagequant 1.2 program to measure the area under the curve of each distinct DG-HTA band. DG-HTA bands consisting of as little as 2% of the total signal of all distinguishable bands could be reproducibly generated in independent duplicate nPCRs (Fig. 1B, 2, and 3). Since PCR amplifies variant templates in direct proportion to their fractional representation (38), DG-HTA could detect variants found at frequencies of ≥2% in the quasispecies, a degree of sensitivity consistent with previous reconstitution experiments using single-stranded HTA probes (4, 51).
Protease quasispecies in the absence of antiviral drug selection.
Two patients enrolled in an early study of the protease inhibitor ritonavir were selected because of their rapid viral load rebound to pretreatment levels following initially successful ritonavir treatment. CD4 counts for P404 and P105 have been described previously (5). The ritonavir drug regimen was 600 mg twice a day and was continued uninterrupted throughout the time of study. Adherence was monitored by interrogation and counting remaining pills at each visit. Eleven and fourteen plasma samples were longitudinally collected over periods of 70 to 189 days, including 3 to 4 samples collected prior to the initiation of ritonavir treatment. Again, each time point was analyzed in duplicate nPCRs to ensure proper sampling (a single instance of insufficient sampling was seen in one of the two duplicate nPCRs of P105 at day 14 [see Fig. 3, DG-HTA]). The clonal DG-HTA probes were derived from the last time point by end-point dilution. Six to seven protease variants in the form of distinct DG-HTA bands ranging in frequency from 2 to 40% could be detected in both subjects prior to therapy (Fig. 2 and 3, DG-HTA and % variants). As for the untreated subjects shown in Fig. 1B, the quasispecies composition (i.e., the relative frequency of distinct variants) remained largely stable during a sampling periods of 14 to 18 days prior to therapy. Change was observed for only one P404 variant, whose frequency fell from 40 to 20% during the pretherapy period (Fig. 3, DG-HTA and % variants).
Effect of protease inhibitor resistance selection on genetic diversity.
After initiation of ritonavir monotherapy, plasma viral loads fell more than 100-fold. During this 2- to 3-week-long decrease in viral load the plasma viral quasispecies composition remained largely stable, with all variants maintaining the same relative frequency. Only one protease variant transiently increased in relative frequency at day 6 of therapy in P105. For both patients large changes in quasispecies composition then became noticeable during the earliest sign of plasma viral load rebound (day 28 for P404 and day 21 for P105). A large reduction in the number of variants was observed in P404 with only two, then one, major variant being detected at days 42 and 56, when the viral load had rebounded 1 to 1.5 logs over the nadir value. Over the next 3 months of continued monotherapy, the P404 quasispecies diversity gradually increased with the detection of multiple distinct protease variants (Fig. 2, DG-HTA and % variants). During that later phase of virus rebound the composition of the quasispecies (i.e., the DG-HTA pattern) continued to change and had not genetically stabilized by day 171.
In the case of P105, a reduction in quasispecies diversity was seen in the form of a new variant increasing in frequency to 70% of the total DG-HTA signal at day 28 during the early stage of plasma viremia rebound (Fig. 3, DG-HTA and % variants). During that time distinct minor variants were still detectable in the plasma virus population. P105 quasispecies complexity then increased with the appearance of new variants with more equally distributed frequencies. The similar DG-HTA patterns on days 42 and 56 indicated that evolutionary stasis had largely been reached by day 2.
The reduction and rapid rebound in quasispecies complexity in both patients was also quantified by measuring the Shannon entropy of their DG-HTA gels. Shannon entropy is a measure of signal distribution or spread in each gel lane and is used to derive a quantitative measure of genetic diversity (7, 10, 11, 33, 35). Signal distributions were measured using the Molecular Dynamics Imagequant program, and the signal background was set as the average intensity of the top 10 pixels in each lane. Shannon entropy was determined using the HDent program with the bin number set at 50 (the HDent program tool is available at the Los Alamos HIV sequence database web site, http://hiv-web.lanl.gov) (7). As could be seen from the DG-HTA gels and the percent frequency of distinct variants (Fig. 2 and 3), sharp reductions in signal entropy was also measured during early viral load rebounds which were rapidly followed by increases in entropy in both patients (Fig. 2 and 3, entropy).
Direct population DNA sequencing.
P404 nPCR products from before therapy (day −14), during the apparent genetic bottleneck (day 6), and after the return of a diverse pro quasispecies (day 71) were analyzed by big dye dideoxy terminator population sequencing (data not shown). The second-round PCR was repeated using EDPR3 (GAAGCAGGAGCCGATAGACAAGG [HXB2 positions 2210 to 2233]) and EDPR4 (CTGGTACAGTTTCAATAGGACTAATGG [HXB2 positions 2550 to 2577]). The PCR was purified using Qiagen PCR purification columns and was sequenced from the EDPR3 primer in a 3700 ABI automated capillary sequencer. Polymorphic bases were scored when the automated sequencer electrophoregram showed a second base peak with a height 20% or greater than that of the major peak. Approximately one third of the nucleotide positions in the protease gene of the day −14 quasispecies appeared polymorphic, including seven N base calls by the more stringent ABI base-calling algorithm. L63P, a common accessory drug resistance polymorphism, was present in the plasma quasispecies before and throughout therapy. Notable nucleotide changes between days −14 and 6 (when the population became homogeneous by DG-HTA) included every polymorphic base becoming homogeneous (according to the 20% peak height criterion) and four nucleotide base changes (including two nonsynonomous changes, I62V and V82F). V82F has been associated with ritonavir resistance (41). At day 71, seven bases were scored as N by the ABI algorithm (two being at identical positions as on day −14), and codon 82 was a mixture of GTC (wild-type Val) and GCC (drug-resistant Ala). Again, approximately a third of the protease nucleotide positions were polymorphic (according to the 20% peak height criterion). Direct population sequencing of nPCR products therefore corroborated the conclusion from DG-HTA that a highly diverse quasispecies in P404 underwent a severe reduction in genetic diversity associated with selection for ritonavir resistance which was rapidly followed by a return to pretreatment levels of protease genetic diversity.
Population genetic changes.
The population bottlenecks reported here were only transient, coinciding with the early phase of plasma viral load rebounds but occurring later than the nadir of plasma viremia. Reproducible population sampling ensured that these observations reflected genuine population genetic changes and were not the results of insufficient variant sampling (4, 5, 22). After several months of continued treatment a rapid return to pretreatment levels of protease gene diversity took place. A recent subcloning and sequencing study of indinavir resistance selection has also reported a reduction in protease gene diversity between the pretreatment and week 12 posttreatment quasispecies (16).
Prior studies have documented the appearance during the course of protease inhibitor monotherapies of viruses with progressively larger numbers of protease mutations relative to the baseline and increasingly drug-resistant phenotypes (24, 26). Mutations at V82 initially appeared which conferred minimal levels of in vitro drug resistance (24, 26). V82AF variants were later found in combination with other mutations that further increased levels of drug resistance. The transient population bottlenecks at the protease locus seen early in the viremia rebound of P404 were associated with the initial selection of the V82F variant. The association of the V82F mutation with an I62V change (which has not been reported in association with drug resistance in the Stanford HIV RT and protease sequence database [41]) may have been due to their fortuitous linkage on a selected V82F genome or may reflect an advantageous compensatory mutation specific to this particular genetic background. The reductions in population diversity of P404 and P105 are in accord with both stochastic (20, 21, 34) and selection-drift (37) models of HIV evolution whereby the pretreatment effective population size is low enough that selection of mutants theoretically expected at steady-state frequencies of 1/100 to 1/1,000 (3, 19, 31) would have the strong effect seen here on the genetic diversity of the quasispecies.
The rapid return of high levels of protease gene diversity has several possible explanations. The bottleneck variant seen in P404 may have evolved by continued cycles of replication, mutation, and selection into the multiple variants observed only weeks later. It may also be that later-emerging variants descended from pretreatment variants present at initially lower frequencies than the bottleneck variant. The surprising speed with which high levels of diversity returned and the partial reversion of the 62 and 82 codons provide some support for the later explanation. The bottleneck GTA (62V) codon partially changed back to the pretreatment codon (at day 71 codon 62 was a 50% mixture of the original ATA Ile and GTA Val). The bottleneck TTC (82F) codon also appeared to partially change back to the pretreatment codon (at day 71 codon 82 was a 50% mixture of the original GTC Val and GCC Ala). The mutational pathway for the later GCC 82A is shorter from the pretreatment GTC 82V than the bottleneck TTC 82F, indicating that the later variant is more likely to be derived from the earlier population than from the bottleneck variant. Furthermore, many of the numerous polymorphic nucleotides seen before treatment were seen again at the later time point (data not shown). Later-emerging variants therefore appeared to descend at least in part from the pretreatment population rather than evolve strictly from the bottleneck variant. Both scenarios may also be occurring simultaneously alongside recombination to rapidly increase plasma protease quasispecies diversity.
Discordant population genetic changes at different loci.
Using the same plasma samples, the evolution of HIV-1 was previously analyzed at the envelope locus, allowing a comparison of the viral population changes occurring at both loci (5). Similar to what happened at the protease gene during early viral load rebound, the P105 plasma quasispecies also underwent a transient reduction in envelope diversity. Contrary to changes seen at the pro locus, the P404 diversity at the env locus actually increased from a single to three major variants during the early rebound in viremia (at days 42 and 56) before rapidly returning to the single pretreatment variant (5). Comparison of the evolutionary changes seen at the pro and env loci therefore indicate that opposite population genetic changes took place in different regions of the HIV-1 genome. Linkage of the bottleneck protease variant to envelope sequences previously rare in the plasma population could reflect its origin from partially drug-privileged anatomical sites where HIV-1 envelope sequences may differ from those in the plasma (5). The well-documented compartmentalization of distinct env variants in different anatomical sites makes this scenario possible (6, 15, 17, 18, 47). The later reemergence of the pretreatment envelope variant while the quasispecies rapidly diversified at the pro locus could reflect selective pressure to restore the initial and presumably optimal env variant in the plasma quasispecies after drug resistance at the pro locus is fully established.
Overlapping immunological and pharmacological selective pressures changing in both time and space, compartmentalization of distinct variants, and potential recombination between loci make simple interpretations of the complex population genetic changes observed at both loci speculative, especially when based on analyses restricted to plasma populations. The severe but only transient reduction in pro quasispecies diversity does indicate that no long-term advantage in terms of lowered diversity at the selected locus was gained following the rapid loss of drug efficacy. This study also indicates that in a genome as recombination prone as that of HIV-1 (27), population genetic changes measured at one locus within a subject reflect the population history of that locus only and not necessarily that of the entire genome.
One of the limitations of present drug resistance assays is their inability to analyze minor variants (12). HIV-1 drug resistance genotyping methods directly sequence PCR products (population sequencing) or plasmid subclones, therefore deriving only the viral consensus sequence or that of its major variants. Population and oligonucleotide chip sequencing can detect, at best, variants present at >10% frequencies (13, 39, 45). Future adaptation of DG-HTA for the isolation of minor sequence variants followed by their direct sequencing may improve the detection of drug-resistant variants present at low frequencies.
Acknowledgments
We thank M. Markowitz and H. Mo for plasma samples and site-directed mutant plasmids.
This study was supported by a grant from the Blood Systems Foundation and NIH grants AI43224 and AI43895 to E.D.
REFERENCES
- 1.Boden D, Markowitz M. Resistance to human immunodeficiency virus type 1 protease inhibitors. Antimicrob Agents Chemother. 1998;42:2775–2783. doi: 10.1128/aac.42.11.2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coffin J M. Genetic diversity and evolution of retroviruses. Curr Top Microbiol Immunol. 1992;176:143–164. doi: 10.1007/978-3-642-77011-1_10. [DOI] [PubMed] [Google Scholar]
- 3.Coffin J M. HIV population dynamics in vivo: implications for genetic variation, pathogenesis, and therapy. Science. 1995;267:483–489. doi: 10.1126/science.7824947. [DOI] [PubMed] [Google Scholar]
- 4.Delwart E L, Gordon C J. Tracking changes in HIV-1 envelope quasispecies using DNA heteroduplex analysis. Methods Enzymol. 1997;12:348–354. doi: 10.1006/meth.1997.0489. [DOI] [PubMed] [Google Scholar]
- 5.Delwart E L, Heng P, Neumann A, Markowitz M. Rapid, transient changes at the env locus of plasma HIV-1 populations during the emergence of protease inhibitor resistance. J Virol. 1998;72:2416–2421. doi: 10.1128/jvi.72.3.2416-2421.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Delwart E L, Mullins J I, Gupta P, Learn G H J, Holodniy M, Katzenstein D, Walker B D, Singh M K. Human immunodeficiency virus type 1 populations in blood and semen. J Virol. 1998;72:617–623. doi: 10.1128/jvi.72.1.617-623.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Delwart E L, Pan H, Sheppard H W, Wolpert D, Neumann A U, Korber B, Mullins J I. Slower evolution of HIV-1 quasispecies during progression to AIDS. J Virol. 1997;71:7498–7508. doi: 10.1128/jvi.71.10.7498-7508.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Delwart E L, Sheppard H W, Walker B D, Goudsmit J, Mullins J I. Human immunodeficiency virus type 1 evolution in vivo tracked by DNA heteroduplex mobility assays. J Virol. 1994;68:6672–6683. doi: 10.1128/jvi.68.10.6672-6683.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Delwart E L, Shpaer E G, McCutchan F E, Louwagie J, Grez M, Rübsamen-Waigmann H, Mullins J I. Genetic relationships determined by a heteroduplex mobility assay: analysis of HIV env genes. Science. 1993;262:1257–1261. doi: 10.1126/science.8235655. [DOI] [PubMed] [Google Scholar]
- 10.Dickover R E, Garratty E M, Herman S A, Sim M S, Plaeger S, Boyer P J, Keller M, Deveikis A, Stiehm E R, Bryson Y J. Identification of levels of maternal HIV-1 RNA associated with risk of perinatal transmission. Effect of maternal zidovudine treatment on viral load. JAMA. 1996;275:599–605. [Google Scholar]
- 11.Dickover R E, Garratty E M, Plaeger S, Bryson Y J. Perinatal transmission of major, minor, and multiple maternal human immunodeficiency virus type 1 variants in utero and intrapartum. J Virol. 2001;75:2194–2203. doi: 10.1128/JVI.75.5.2194-2203.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Flexner C. HIV genotype and phenotype-arresting resistance. JAMA. 2000;283:2442–2444. doi: 10.1001/jama.283.18.2442. [DOI] [PubMed] [Google Scholar]
- 13.Gunthard H F, Wong J K, Ignacio C C, Havlir D V, Richman D D. Comparative performance of high-density oligonucleotide sequencing and dideoxynucleotide sequencing of HIV type 1 pol from clinical samples. AIDS Res Hum Retrovir. 1998;14:869–876. doi: 10.1089/aid.1998.14.869. [DOI] [PubMed] [Google Scholar]
- 14.Heyndrickx L, Janssens W, Zekeng L, Musonda R, Anagonou S, Van der Auwera G, Coppens S, Vereecken K, De Witte K, Van Rampelbergh R, Kahindo M, Morison L, McCutchan F E, Carr J K, Albert J, Essex M, Goudsmit J, Asjo B, Salminen M, Buve A, van Der Groen G. Simplified strategy for detection of recombinant human immunodeficiency virus type 1 group M isolates by gag/env heteroduplex mobility assay. Study Group on Heterogeneity of HIV Epidemics in African Cities. J Virol. 2000;74:363–370. doi: 10.1128/jvi.74.1.363-370.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hughes E S, Bell J E, Simmonds P. Investigation of the dynamics of the spread of human immunodeficiency virus to brain and other tissues by evolutionary analysis of sequences from the p17 gag and env genes. J Virol. 1997;71:1272–1280. doi: 10.1128/jvi.71.2.1272-1280.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ibanez A, Clotet B, Martinez M A. Human immunodeficiency virus type 1 population bottleneck during indinavir therapy causes a genetic drift in the env quasispecies. J Gen Virol. 2000;81:85–95. doi: 10.1099/0022-1317-81-1-85. [DOI] [PubMed] [Google Scholar]
- 17.Itescu S, Simonelli P F, Winchester R J, Ginsberg H S. Human immunodeficiency virus type 1 strains in the lungs of infected individuals evolve independently from those in peripheral blood and are highly conserved in the C-terminal region of the envelope V3 loop. Proc Natl Acad Sci USA. 1994;91:11378–11382. doi: 10.1073/pnas.91.24.11378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kroodsma K L, Kozal M J, Hamed K A, Winters M A, Merigan T C. Detection of drug resistance mutations in the human immunodeficiency virus type 1 (HIV-1) pol gene: differences in semen and blood HIV-1 RNA and proviral DNA. J Infect Dis. 1994;170:1292–1295. doi: 10.1093/infdis/170.5.1292. [DOI] [PubMed] [Google Scholar]
- 19.Lech W J, Wang G, Yang Y L, Chee Y, Dorman K, McCrae D, Lazzeroni L C, Erickson J W, Sinsheimer J S, Kaplan A H. In vivo sequence diversity of the protease of human immunodeficiency virus type 1: presence of protease inhibitor-resistant variants in untreated subjects. J Virol. 1996;70:2038–2043. doi: 10.1128/jvi.70.3.2038-2043.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leigh-Brown A. Analysis of HIV-1 env gene sequences reveals evidence for a low effective number in the viral population. Proc Natl Acad Sci USA. 1997;94:1862–1865. doi: 10.1073/pnas.94.5.1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leigh-Brown A, Richman D D. HIV-1: gambling on the evolution of drug resistance. Nat Med. 1997;3:268–271. doi: 10.1038/nm0397-268. [DOI] [PubMed] [Google Scholar]
- 22.Liu S L, Rodrigo A G, Shankarappa R, Learn G H, Hsu L, Davidov O, Zhao L P, Mullins J I. HIV quasispecies and resampling. Science. 1996;273:415–416. doi: 10.1126/science.273.5274.415. [DOI] [PubMed] [Google Scholar]
- 23.Long E M, Martin H L, Jr, Kreiss J K, Rainwater S M, Lavreys L, Jackson D J, Rakwar J, Mandaliya K, Overbaugh J. Gender differences in HIV-1 diversity at time of infection. Nat Med. 2000;6:71–75. doi: 10.1038/71563. [DOI] [PubMed] [Google Scholar]
- 24.Markowitz M, Mo H, Kempf D J, Norbeck D W, Bhat T N, Erickson J W, Ho D D. Selection and analysis of human immunodeficiency virus type 1 variants with increased resistance to ABT-538, a novel protease inhibitor. J Virol. 1995;69:701–706. doi: 10.1128/jvi.69.2.701-706.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McNearney T, Hornickova Z, Markham R, Birdwell A, Arens M, Saah A, Ratner L. Relationship of human immunodeficiency virus type 1 sequence heterogeneity to stage of disease. Proc Natl Acad Sci USA. 1992;89:10247–10251. doi: 10.1073/pnas.89.21.10247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Molla A, Korneyeva M, Gao Q, Vasavanonda S, Schipper P J, Mo H M, Markowitz M, Chernyavskiy T, Niu P, Lyons N, Hsu A, Granneman G R, Ho D D, Boucher C A, Leonard J M, Norbeck D W, Kempf D J. Ordered accumulation of mutations in HIV protease confers resistance to ritonavir. Nat Med. 1996;2:760–766. doi: 10.1038/nm0796-760. [DOI] [PubMed] [Google Scholar]
- 27.Moutouh L, Corbeil J, Richman D D. Recombination leads to the rapid emergence of HIV-1 dually resistant mutants under selective drug pressure. Proc Natl Acad Sci USA. 1996;93:6106–6111. doi: 10.1073/pnas.93.12.6106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Myers R M, Fischer S G, Lerman L S, Maniatis T. Nearly all single base substitutions in DNA fragments joined to a GC-clamp can be detected by denaturing gradient gel electrophoresis. Nucleic Acids Res. 1985;13:3131–3145. doi: 10.1093/nar/13.9.3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Myers R M, Fischer S G, Maniatis T, Lerman L S. Modification of the melting properties of duplex DNA by attachment of a GC-rich DNA sequence as determined by denaturing gradient gel electrophoresis. Nucleic Acids Res. 1985;13:3111–3128. doi: 10.1093/nar/13.9.3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Myers R M, Maniatis T, Lerman L S. Detection and localization of single base changes by denaturing gradient gel electrophoresis. Methods Enzymol. 1987;155:501–527. doi: 10.1016/0076-6879(87)55033-9. [DOI] [PubMed] [Google Scholar]
- 31.Najera I, Holguin A, Quinones-Mateu M E, Munoz-Fernandez M A, Najera R, Lopez-Galindez C, Domingo E. Pol gene quasispecies of human immunodeficiency virus: mutations associated with drug resistance in virus from patients undergoing no drug therapy. J Virol. 1995;69:23–31. doi: 10.1128/jvi.69.1.23-31.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nelson J A, Fiscus S A, Swanstrom R. Evolutionary variants of the human immunodeficiency virus type 1 V3 region characterized by using a heteroduplex tracking assay. J Virol. 1997;71:8750–8758. doi: 10.1128/jvi.71.11.8750-8758.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nielsen K, Boyer P, Dillon M, Wafer D, Wei L S, Garratty E, Dickover R E, Bryson Y J. Presence of human immunodeficiency virus (HIV) type 1 and HIV-1-specific antibodies in cervicovaginal secretions of infected mothers and in the gastric aspirates of their infants. J Infect Dis. 1996;173:1001–1004. doi: 10.1093/infdis/173.4.1001. [DOI] [PubMed] [Google Scholar]
- 34.Nijhuis M, Boucher C A, Schipper P, Leitner T, Schuurman R, Albert J. Stochastic processes strongly influence HIV-1 evolution during suboptimal protease-inhibitor therapy. Proc Natl Acad Sci USA. 1998;95:14441–14446. doi: 10.1073/pnas.95.24.14441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ping L H, Cohen M S, Hoffman I, Vernazza P, Seillier-Moiseiwitsch F, Chakraborty H, Kazembe P, Zimba D, Maida M, Fiscus S A, Eron J J, Swanstrom R, Nelson J A. Effects of genital tract inflammation on human immunodeficiency virus type 1 V3 populations in blood and semen. J Virol. 2000;74:8946–8952. doi: 10.1128/jvi.74.19.8946-8952.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Resch W, Parkin N, Stuelke E L, Watkins T, Swanstrom R. A multiple-site-specific heteroduplex tracking assay as a tool for the study of viral population dynamics. Proc Natl Acad Sci USA. 2000;98:176–181. doi: 10.1073/pnas.011511298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rouzine I M, Coffin J M. Linkage disequilibrium test implies a large effective population number for HIV in vivo. Proc Natl Acad Sci USA. 1999;96:10758–10763. doi: 10.1073/pnas.96.19.10758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ruano G, Kidd K K. Modeling of heteroduplex formation during PCR from mixtures of DNA templates. PCR Methods Appl. 1992;2:112–116. doi: 10.1101/gr.2.2.112. [DOI] [PubMed] [Google Scholar]
- 39.Schuurman R, Demeter L, Reichelderfer P, Tijnagel J, de Groot T, Boucher C. Worldwide evaluation of DNA sequencing approaches for identification of drug resistance mutations in the human immunodeficiency virus type 1 reverse transcriptase. J Clin Microbiol. 1999;37:2291–2296. doi: 10.1128/jcm.37.7.2291-2296.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schuurman R, Nijhuis M, van Leeuwen R, Schipper P, de Jong D, Collis P, Danner S A, Mulder J, Loveday C C, Christopherson C, et al. Rapid changes in human immunodeficiency virus type 1 RNA load and appearance of drug-resistant virus populations in persons treated with lamivudine (3TC) J Infect Dis. 1995;171:1411–1419. doi: 10.1093/infdis/171.6.1411. [DOI] [PubMed] [Google Scholar]
- 41.Shafer R W, Stevenson D, Chan B. Human immunodeficiency virus reverse transcriptase and protease sequence database. Nucleic Acids Res. 1999;27:348–352. doi: 10.1093/nar/27.1.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Simmonds P, Balfe P, Peutherer J F, Ludlam C A, Bishop J O, Leigh Brown A J. Human immunodeficiency virus-infected individuals contain provirus in small numbers of peripheral mononuclear cells and at low copy numbers. J Virol. 1990;64:864–872. doi: 10.1128/jvi.64.2.864-872.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tatt I D, Barlow K L, Clewley J P. A gag gene heteroduplex mobility assay for subtyping HIV-1. J Virol Methods. 2000;87:41–51. doi: 10.1016/s0166-0934(00)00146-4. [DOI] [PubMed] [Google Scholar]
- 44.Upchurch D A, Shankarappa R, Mullins J I. Position and degree of mismatches and the mobility of DNA heteroduplexes. Nucleic Acids Res. 2000;28:E69. doi: 10.1093/nar/28.12.e69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wei X, Ghosh S K, Taylor M E, Johnson V A, Emini E A, Deutsch P, Lifson J D, Bonhoeffer S, Novak M A, Hahn B H, Saag M S, Shaw G M. Viral dynamics in human immunodeficiency virus type 1 infection. Nature. 1995;373:117–126. doi: 10.1038/373117a0. [DOI] [PubMed] [Google Scholar]
- 46.Wolfs T F, Zwart G, Bakker M, Goudsmit J. HIV-1 genomic RNA diversification following sexual and parenteral virus transmission. Virology. 1992;189:103–110. doi: 10.1016/0042-6822(92)90685-i. [DOI] [PubMed] [Google Scholar]
- 47.Wong J K, Ignacio C C, Torriani F, Havlir D, Fitch N J, Richman D D. In vivo compartmentalization of human immunodeficiency virus: evidence from the examination of pol sequences from autopsy tissues. J Virol. 1997;71:2059–2071. doi: 10.1128/jvi.71.3.2059-2071.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wood N, Tyfield L, Bidwell J. Rapid classification of phenylketonuria genotypes by analysis of heteroduplexes generated by PCR-amplifiable synthetic DNA. Hum Mutat. 1993;2:131–137. doi: 10.1002/humu.1380020213. [DOI] [PubMed] [Google Scholar]
- 49.Zhang L Q, MacKenzie P, Cleland A, Holmes E C, Leigh Brown A J, Simmonds P. Selection for specific sequences in the external envelope protein of human immunodeficiency virus type 1 upon primary infection. J Virol. 1993;67:3345–3356. doi: 10.1128/jvi.67.6.3345-3356.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhu T, Mo H, Wang N, Nam D S, Cao Y, Koup R A, Ho D D. Genotypic and phenotypic characterization of HIV-1 in patients with primary infection. Science. 1993;261:1179–1181. doi: 10.1126/science.8356453. [DOI] [PubMed] [Google Scholar]
- 51.Zhu T, Wang N, Carr A, Nam D S, Moor-Jankowski R, Cooper D A, Ho D D. Genetic characterization of human immunodeficiency virus type 1 in blood and genital secretions: evidence for viral compartmentalization and selection during sexual transmission. J Virol. 1996;70:3098–3107. doi: 10.1128/jvi.70.5.3098-3107.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]