Abstract
Rare actinomycete genera are highly recognized as a promising source of structurally diverse and bioactive natural products. Among these genera, Allokutzneria and Kibdelosporangium are two phylogenetically closely related and have been reported to encode some valuable biosynthetic enzymes and secondary metabolites. However, there is currently no relevant systematic research available to outline the linkage of genomic and metabolomics for specific secondary metabolites in these two promising genera. In this study, we first investigated the genus-specific secondary metabolic potential in Allokutzneria and Kibdelosporangium by comparing the diversity and novelty of their secondary metabolite biosynthetic gene clusters (BGCs). The specific secondary metabolites produced by two representative strains of these genera were comprehensively investigated using untargeted metabolomics techniques. The findings unveiled that the majority (95.4%) of the gene cluster families (GCFs) encoded by Allokutzneria and Kibdelosporangium were genus-specific, including NRPS GCFs encoding siderophores. The untargeted metabolomics analysis revealed that the metabolic profiles of two representative strains exhibit extensive specificity, with the culture medium having a big impact on the metabolic profiles. Besides, an MS-cluster featuring a series of hydroxamate-type siderophores was identified from Allokutzneria albata JCM 9917, with two of them, including a novel one (N-deoxy arthrobactin A), being experimentally validated. The present study offers valuable insights for the targeted discovery of genus-specific natural products from microorganisms.
Keywords: Genus-specific secondary metabolism, Allokutzneria, Kibdelosporangium, Genome mining, Untargeted metabolomics, Siderophore
Graphical abstract
1. Introduction
Over the past few decades, actinobacteria have consistently been one of the major sources of microbial secondary metabolites [1]. Among them, Streptomyces is the most prominent genus in Actinobacteria, known for its remarkable ability to produce valuable secondary metabolites such as antibiotics, anticancer drugs, immunosuppressants, and enzyme inhibitors [2]. Unfortunately, due to the growing number of active compounds isolated from Streptomyces, it has become increasingly challenging to discover novel natural products from this genus. Currently, a rapid surge of new compounds with broad biological activity and pharmacological properties are being discovered from non-streptomyces actinobacteria, that is, rare actinobacteria [3]. Rare actinobacteria encompass over 200 genera, including Actinomadura, Allokutzneria, Amycolatopsis, Catenuloplanes, Cystobacter, Kibdelosporangium, Kutzneria, Pseudonocardia, Saccharomonospora, Thermomonospora, Virgosporangium and so on [4].
The genus Kibdelosporangium was first proposed by Shearer et al., in 1986 [5] and comprised ten recognized species and subspecies. Kibdelosporangium strains are renowned for their production of novel glycopeptide antibiotics, such as aridicins and kibdelins [6,7]. Based on the phylogenetic and chemotaxonomic characteristics, K. albatum has been reclassified as the “type species” of the new genus Allokutzneria by Labeda and Kroppenstedt in 2008 [8]. The genus of Allokutzneria comprised three species, namely A. albata, A. multivorans, and A. oryzae [9]. Although the number of species within these two genera is relatively small, many intriguing natural products and biosynthetic enzymes have been characterized from these strains. For instance, two novel N-phenylacetylated heptapeptides have been isolated from Kibdelosporangium sp. AK-AA56 [10]. Similarly, two diterpene synthases and their catalytic products spiroalbatene and cembrene A have been identified from A. albata [11].
Genome mining studies have suggested the presence of a substantial number of secondary metabolite biosynthetic gene clusters (BGCs) in these rare actinomycetes [12]. Our recent findings have uncovered that Kibdelosporangium possesses the highest secondary metabolic potential among bacterial genera, with an average of 40 BGCs per genome [13]. Allokutzneria is phylogenetic closely related to Kutzneria and Kibdelosporangium, our recent study has demonstrated that Kutzneria exhibited great potential to synthesize novel secondary metabolites and encode a variety of valuable biosynthetic enzymes [14]. These findings lead us to contemplate whether the secondary metabolic potential of these phylogenetic closely related genera is comparable to each other and explore the specific secondary metabolites from these strains.
In this study, we first compared the specific secondary biosynthetic potential of Allokutzneria and Kibdelosporangium by examining the distribution of BGCs in these two genera. Then, the secondary metabolites produced by two representative strains of these genera were investigated under different culture conditions using untargeted metabolomics. Furthermore, an MS-cluster featuring a series of specific secondary metabolites was preliminarily identified using UPLC-QTOF/MS, and two of these metabolites were extensively characterized by HR-MS/MS, 1D, and 2D NMR, and their biological activity was determined.
2. Materials and methods
2.1. Genome dataset and bacterial strains
All available genome sequences from Allokutzneria and Kibdelosporangium were manually downloaded from the NCBI database. The GenBank accession number, genome size, and taxonomic information of all the genomes were collected and summarized in Supplementary Table 1. Genome assembly quality was assessed by CheckM version 1.2.2 [15]. Allokutzneria albata JCM 9917 and Kibdelosporangium phytohabitans KLBMP1111 were obtained from the German Collection of Microorganisms and Cell Cultures GmbH (DSMZ) and Ningbo testobio. Co., Ltd, Zhejiang, China, respectively.
2.2. Biosynthetic gene cluster identification and networking
All genome sequences were processed using the online version of antiSMASH 7.0 with the bacterial setting and relaxed strictness [16]. The number of each class of BGCs in these genomes was extracted from the HTML files (antiSMASH outputs) using a custom Python toolkit, as described in our previous report [17]. The genomic features and numbers of BGCs in each genome were also summarized inSupplementary Table 1. All predicted BGCs were assigned to different biosynthetic gene cluster families (GCFs) using BiG-SLiCE at a distance cutoff of 0.5 [18]. The specificity of GCF depends on whether the BGCs in it originate from a single bacterial genus or not. The origin, class, GCF classification result, and specificity of each BGC were summarized in Supplementary Table 2.
2.3. High-Throughput elicitor screening (HiTES) for A. albata JCM 9917 and K. phytohabitans KLBMP1111
A. albata JCM 9917 and K. phytohabitans KLBMP1111 were cultured under nine different liquid media (M1: GYE; M2: DSMZ 554; M3: JCMM 58; M4: JCMM 44; M5: JCMM 42; M6: GYM; M7: R5M; M8: TSB; M9: NL148) for three days, then six previously reported chemical elicitors, namely LaCl3·7H2O [31], ScCl3·6H2O [32], N-acetylglucosamine (NAG) [33], sodium butyrate [34], streptomycin in combination with trimethoprim (TMP) [35], and triclosan [36], were individually supplemented to the cultures and allowed to incubate for an additional seven days. Simultaneously, the blank and control groups were set for subsequent analysis. All HiTES screens were carried out in a single replicate. The culture broth was extracted by an equal volume of ethyl acetate and analyzed using a UPLC-QTOF/MS system (SCIEX X500B Q-TOF spectrometer coupled to an ExionLC AC system) according to our recent report [17].
2.4. Molecular networking and annotation of MS/MS data
The high-resolution liquid chromatography-mass spectrometry (LC-HRMS) data was converted using MSConvert, and processed using MZmine2 software [19]. Feature detection, grouping, alignment, and molecular networking were performed following the feature-based molecular networking (FBMN) and the Dereplicator + workflow in GNPS (http://gnps.ucsd.edu) [20,21]. Molecular networks were generated with a minimum of four matching peaks, a cosine score of 0.7, and a maximum of 250 connected component sizes. The SIRIUS 4 was also employed for feature annotation with the default settings [22]. The molecular network generated from FBMN was visualized by using Cytoscape 3.6.1. The mirror match of spectra was constructed using a Python package spectrum_utils using a custom Python toolkit [23]. All MS/MS peaks are listed in Supplementary Table 3.
2.5. Fermentation, isolation, and structure elucidation
JCM medium 42 and elicitor E (streptomycin in combination with TMP) were selected for large-scale fermentation of A. albata JCM 9917. For 15 L fermentation, seed inoculum was cultured in 500 mL Erlenmeyer flasks containing 200 mL medium at 30 °C for 72 h on a shaker incubator at 180 r.p.m. Aliquot (5–6 mL) of the seed culture was inoculated into each of 2000 mL Erlenmeyer flasks containing 1000 mL of the JCM medium 42. The microorganism was cultured at 30 °C for 14 days in a shaker incubator at 180 r.p.m. Then, the 15 L fermentation broth collected from cultures grown in 2 L Erlenmeyer flasks was centrifuged to remove cells, and the resulting supernatant was extracted with n-butanol and evaporated for concentration. The crude extract was eluted by MCI gel with gradient elution using MeOH/H2O (from 10:90 to 100:0). The targeted fraction was further purified by semi-prep-HPLC with gradient elution using MeOH/H2O (from 40:60 to 90:10 over 25 min) at a flow rate of 3 mL/min to yield two pure compounds 1 (20 mg) and 2 (5 mg).
2.6. Chrome azurol S (CAS) assay
The iron-chelating activity of compounds 1 and 2 was assessed using a modified CAS assay as described in our recent study [24]. The cetyltrimethylammonium bromide (CTAB)-CAS-Fe(III) solution was prepared by mixing 25 mL of CTAB solution (2 mM), 1.5 mL of FeCl3 solution (1 mM), and 7.5 mL of CAS solution (2 mM). The CAS assay solution was obtained by adding the CAS-Fe(III) solution into the MES buffer. The test compounds were mixed with CAS solution and then incubated at 30 °C for 3 h, the resulting color changes were recorded by measuring the absorption changes at 630 nm. DFOB (Desferrioxamine B) is used as a positive control, and MeOH (methanol) is used as a negative control.
3. Results
3.1. Genomic features and the overall distribution of BGCs in Allokutzneria and Kibdelosporangium
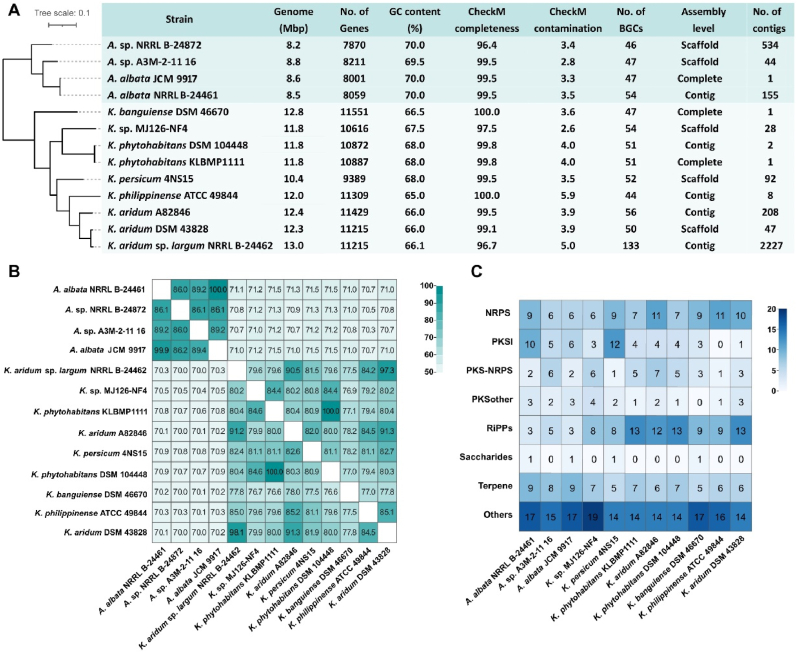
To assess the secondary metabolite potential of Allokutzneria and Kibdelosporangium, we collected all the available genome sequences for both genera and conducted an in-depth bioinformatic analysis. Our findings suggest that the strains of these two genera exhibited significant differences in genomic characteristics. In comparison to Kibdelosporangium genomes, the genomes of Allokutzneria exhibit slightly smaller sizes and lower number of genes, while their GC content is higher than that of Kibdelosporangium (Fig. 1A). It is worth noting that there exists a significant inter-generic difference in the similarity between these two genera. Genomes within the Allokutzneria genus share a sequence similarity above 86%, while in the Kibdelosporangium genus, the similarity is above 76% (Fig. 1B). The similarity between genomes from these two different genera ranges from 70% to 71.5% (Fig. 1B). These findings further confirm that Allokutzneria is a distinct genus separate from Kibdelosporangium. The total numbers of predicted BGCs in these genomes range from 46 to 56, except for Kibdelosporangium aridum subsp. largum NRRL B-24462, showed no significant difference between these two genera (Fig. 1A). The genome of K. aridum subsp. largum NRRL B-24462 showed a completeness of 96.7% and a contamination rate of up to 5%. It contained 2227 contigs, and the fragmented genome resulted in an overestimated number of BGCs (Fig. 1A). Therefore, we selected 11 high-quality genomes from the two genera and counted the number of each class of BGCs in these genomes (Table S1). The results show that NRPS, PKSI, and Terpene are the top three dominant classes of BGCs in the genomes of the Allokutzneria, while RiPPs, NRPS, and Terpene are the top three dominant in the Kibdelosporangium genomes (Fig. 1C). For instance, the number of RiPPs BGCs in Kibdelosporangium is approximately three times higher compared to Allokutzneria, with an average of 10.63 versus 3.67. Besides, both genera possess a wide variety of BGC types, and many of them are classified as “Others”. Notably, the genomes of two strains of the same species (Allokutzneria albata NRRL B-24461 and Allokutzneria albata JCM 9917) have extremely high genomic similarity (Fig. 1B). Nevertheless, they showed some difference in the numbers of NRPS and PKSI BGCs (Fig. 1C), which was mainly due to some larger NRPS and PKSI BGCs being located at the edges of contigs, resulting in the overestimation.
Fig. 1.
(A) Genomic features of four Allokutzneria genomes and nine Kibdelosporangium genomes. (B) Genomic similarities between thirteen genomes were measured by the pattern similarity score. (C) The numbers of each class of BGCs in the 11 high-quality genomes of Allokutzneria and Kibdelosporangium.
3.2. Genus-specific secondary metabolic potential in Allokutzneria and Kibdelosporangium
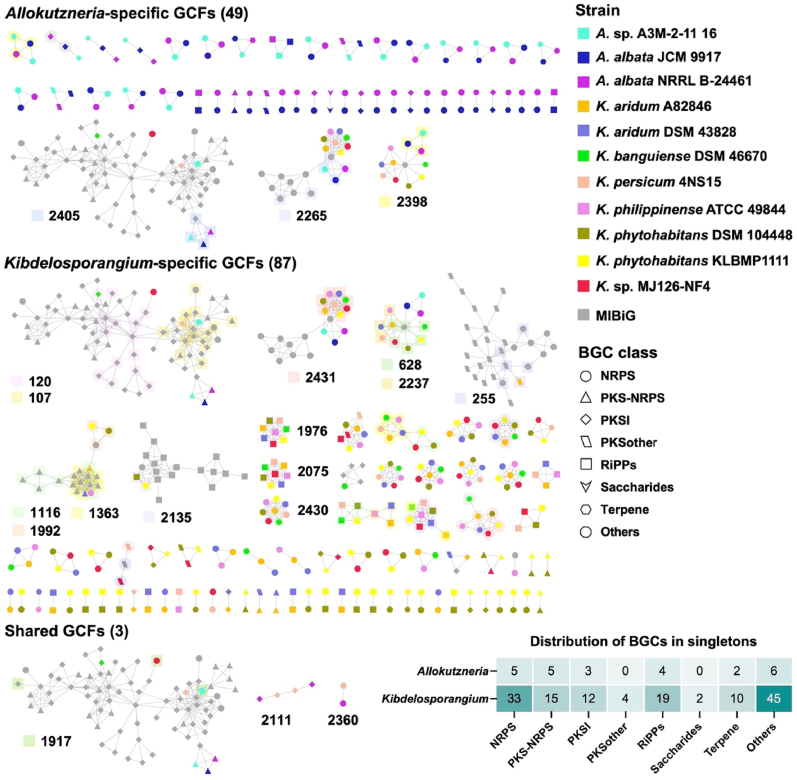
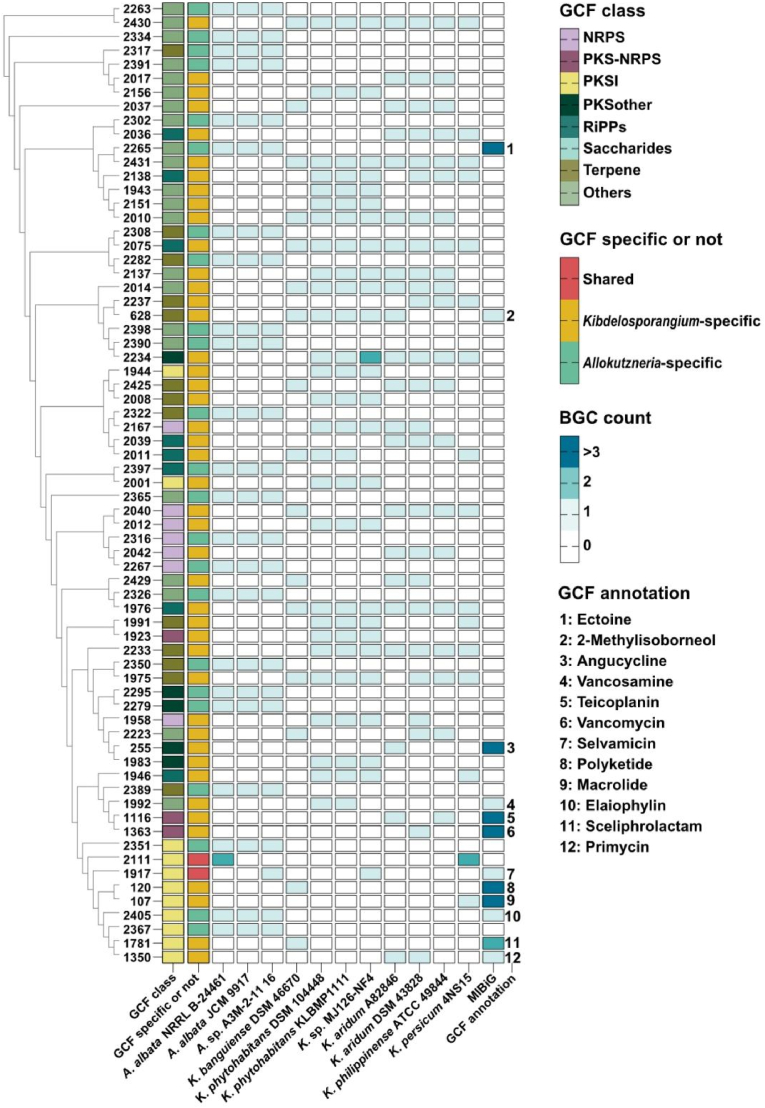
The diversity and novelty of all 553 BGCs predicted from the 11 high-quality genomes were investigated using BiG-SCAPE. The results showed that these BGCs were divided into 304 gene cluster families (GCFs) at a threshold of 0.5. Among them, 139 GCFs contained at least two BGCs, while the remaining 165 GCFs were singletons. Out of the 304 GCFs obtained, only 14 GCFs contained characterized BGCs from the MIBiG database, implying that a significant majority (95.4%) of these GCFs remain unexploited. Besides, 74 GCFs (including 49 GCFs containing more than two BGCs and 25 singletons) are specific to the Allokutzneria, 227 GCFs (including 87 GCF containing more than two BGCs and 140 singletons) are unique to the Kibdelosporangium, and only 3 GCFs are shared between them, demonstrating a remarkable specificity in the secondary metabolic potential between Allokutzneria and Kibdelosporangium. Three genomes of Allokutzneria encoded 52 GCFs with more than two BGCs and 25 singletons. Allokutzneria albata shared 23 GCFs with Allokutzneria sp. A3M-2–11, including two GCFs encoding elaiophylin (no. 2405) and ectoine (no. 2265) analogous, respectively. Notably, A. albata also contained 23 species-specific GCFs that were not present in Allokutzneria sp. A3M-2–11 16, and all of them did not contain known BGCs from the MIBiG database. Eight genomes of Kibdelosporangium encoded 90 GCFs with more than two BGCs and 140 singletons. Only four GCFs were found to contain BGCs from all eight strains of Kibdelosporangium, suggesting that the remaining GCFs exhibited a certain degree of species-specificity (Fig. 2). Majority of the BGCs in the singletons of Kibdelosporangium were originated from others, NRPS, and RiPPs, suggesting that different species of Kibdelosporangium have the potential to produce species-specific and biosynthetically complex natural products, particularly peptidic natural products.
Fig. 2.
Sequence similarity network of BGCs in 11 high-quality Allokutzneria and Kibdelosporangium genomes. Each node represents one BGC, and the gray symbols represent BGCs from the MIBiG database. BGCs distributed within the same cluster but belonging to different GCFs were differentiated using background colors. The heatmap displays the number of different classes of BGCs within the singletons of these two genera.
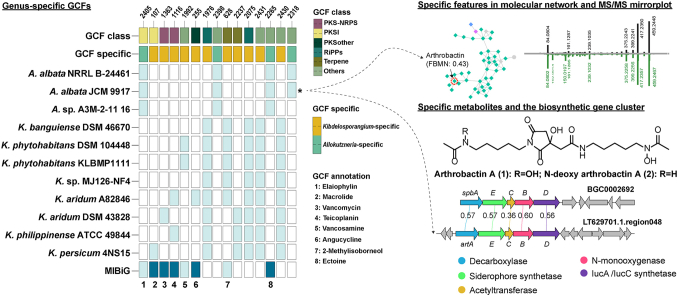
The distribution of BGCs in 69 predominant GCFs was presented in Fig. 3, further confirming the genus-specificity of Allokutzneria and Kibdelosporangium in secondary metabolic potential. Among these GCFs, 12 of them tentatively annotated according to the characterized metabolites encoded by BGCs from the MIBiG database, which mainly belonged to three major classes: PKSI [6], PKS-NRPS hybrid [2], and others [2]. For instance, Allokutzneria sp. A3M-2–11 16 and Kibdelosporangium sp. MJ126-NF4 were predicted to be capable of synthesizing selvamicin analogous (PKSI products). Kibdelosporangium aridum A82846, Kibdelosporangium aridum DSM 4328, and Kibdelosporangium philippinense ATCC 49844 encoded a PKS-NRPS BGC for the synthesis of glycopeptide antibiotics, but these three BGCs were divided into two GCFs (nos. 1116 and 1363), which are highly homologous to the BGCs encoding vancomycin and teicoplanin, respectively. Notably, all seven NRPS and eight RiPPs GCFs were genus-specific and showed high potential to produce novel peptidic natural products. The hierarchical clustering dendrogram of these GCFs was established using clinker, based on the sequence similarity matrix of similar genes in representative BGCs. Consequently, the GCFs that share biosynthetic similarities were gathered together. For instance, parts of the BGCs within two genetically closely related GCFs (nos. 2265 and 2431) were connected to each other in the network indicating a certain degree of similarity as well as noticeable differences between these BGCs. Comparative analysis of these BGCs revealed that they all contain ectoine synthase, parts of them harbored genes encoding aldo/keto reductases, and only BGCs in GCF 2431 contained genes encoding GDSL-like lipase/cylhydrolase and cyclic nucleotide-binding domain (Supplementary Fig. 1). Each of the three strains of Kibdelosporangium encoded a homologous gene cluster for either vancomycin or teicoplanin, which are distributed in two different GCFs (nos. 1116 and 1363). There are significant differences in the predicted substrates of the adenylation domains of their NRPS enzymes (Supplementary Fig. 2).
Fig. 3.
The hierarchical clustering dendrogram of 69 GCFs containing more than three BGCs and the distribution of these BGCs within Allokutzneria and Kibdelosporangium. The GCF class and its specificity were presented in the heatmap. GCFs were annotated based on the characterized metabolites encoded by BGCs from the MIBiG database.
3.3. Specific secondary metabolites in Allokutzneria and Kibdelosporangium
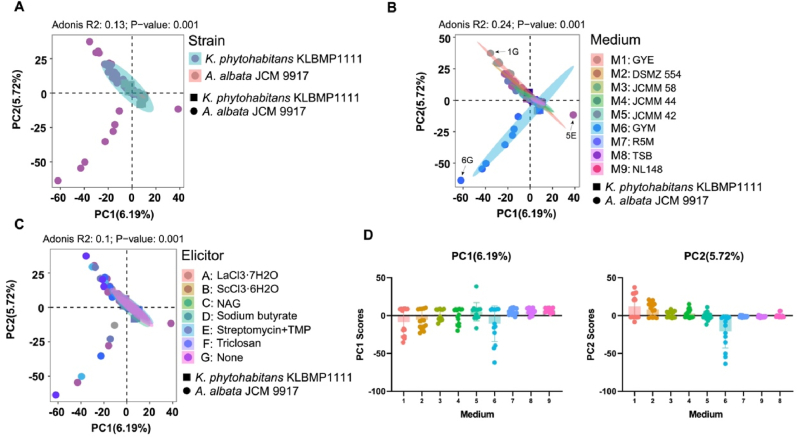
To investigate the specific secondary metabolites in Allokutzneria and Kibdelosporangium, A. albata JCM 9917 and K. phytohabitans KLBMP1111, which possess complete genomes and display a remarkable diversity and novelty of BGCs, were selected as the representative species for each genus. The secondary metabolites of these two strains were extensively studied using the HiTES approach. The HiTES experiment mainly investigated the impact of nine different culture media and six chemical elicitors on the specific secondary metabolites of these two strains by comparing the metabolic profiles with control samples that were prepared without elicitors or strains. The ethyl acetate extracts of these samples were analyzed using an untargeted metabolomics approach with LC-HRMS. The relative abundance of the detected metabolite features in these samples was subjected to principal component analysis (PCA). As shown in Fig. 4A–C, the secondary metabolites produced by A. albata JCM 9917 and K. phytohabitans KLBMP1111 exhibited significant differences from each other (Adonis R2 = 0.13, p < 0.001), and the metabolic profiles are also significantly affected by both culture media (Adonis R2 = 0.24, p < 0.001) and chemical elicitors (Adonis R2 = 0.1, p < 0.001). The principal component values indicated that culture media 6 (GYM), 5 (JCMM 42), and 1 (GYE) have a significant impact on the metabolic profiles of some samples of the two strains (Fig. 4D), particularly in the case of 6G (cultured in GYM without elicitor), 5E (cultured in JCMM 42 under the induction of streptomycin and trimethoprim), and 1G (cultured in GYE without elicitor).
Fig. 4.
Principal Component Analysis (PCA) of the metabolic profiles of A. albata JCM 9917 and K. phytohabitans KLBMP1111 under nine different culture media and six elicitors. Samples were colored according to the type of (A) strain, (B) culture media, and (C) elicitor. (D) The principal component values of samples when divided according to the culture media.
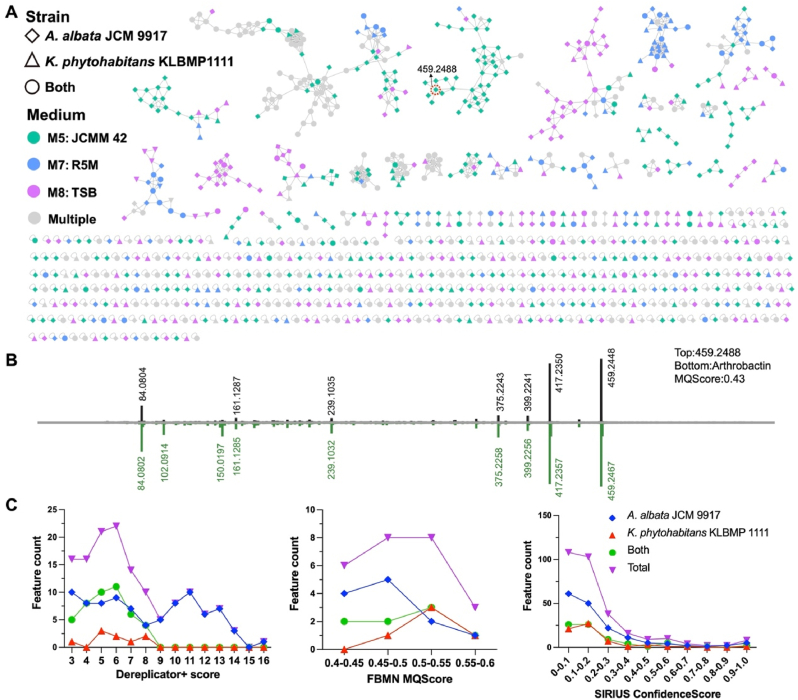
We further analyze the diversity and novelty of secondary metabolites produced by these two strains in the HiTES experiment using GNPS and related annotation platforms (FBMN, Dereplicator+, and SIRIUS). Among the 991 features detected under the culture media of M5, M7, and M8, 510 of them were unique to A. albata JCM 9917, 210 were unique to K. phytohabitans KLBMP1111, and the rest were detected in both strains (Supplementary Table 4), highlighting the former strain possessed a higher chemical diversity. These features were categorized into 69 molecular families (MFs) and 475 singletons at a cosine similarity cut-off of 0.6. It can be observed that features in some MFs are exclusively derived from a single culture medium, particularly the M5 (JCMM 42) and M8 (TSB) media (Fig. 5A). For example, nearly all features in the MF containing a feature annotated as arthrobactin (FBMN MQscore = 0.43) were only detected in samples cultured in JCMM 42. The Dereplicator + analysis identified 139 features with scores larger than 3. However, only 86 features had scores larger than 6 and p-values smaller than 0.001. The FBMN analysis yielded 25 features with MQScore greater than 0.4, with the majority (14/25) scoring between 0.4 and 0.5. The SIRIUS analysis yielded 26 features with Confidence Score greater than 0.5, and 51 features with Confidence Score greater than 0.3 (Fig. 5B). These findings indicated that the majority of these features were potential undescribed metabolites. Besides, these annotated features, under most annotation thresholds, mainly originated from samples of A. albata JCM 9917.
Fig. 5.
(A) Molecular network of secondary metabolites produced by A. albata JCM 9917 and K. phytohabitans KLBMP1111 in the HiTES experiment. Features were colored according to the culture media and shaped according to the presence or absence of these two strains. Features were annotated by three methods (Dereplicator+, FBMN, and SIRIUS), and a unique compound is annotated in the figure. (B) The mirror MS/MS plot of the selected feature and Arthrobactin. (C) The number and composition of features annotated by Dereplicator+, FBMN, and SIRIUS at various thresholds.
3.4. Characterization of specific secondary metabolites in Allokutzneria
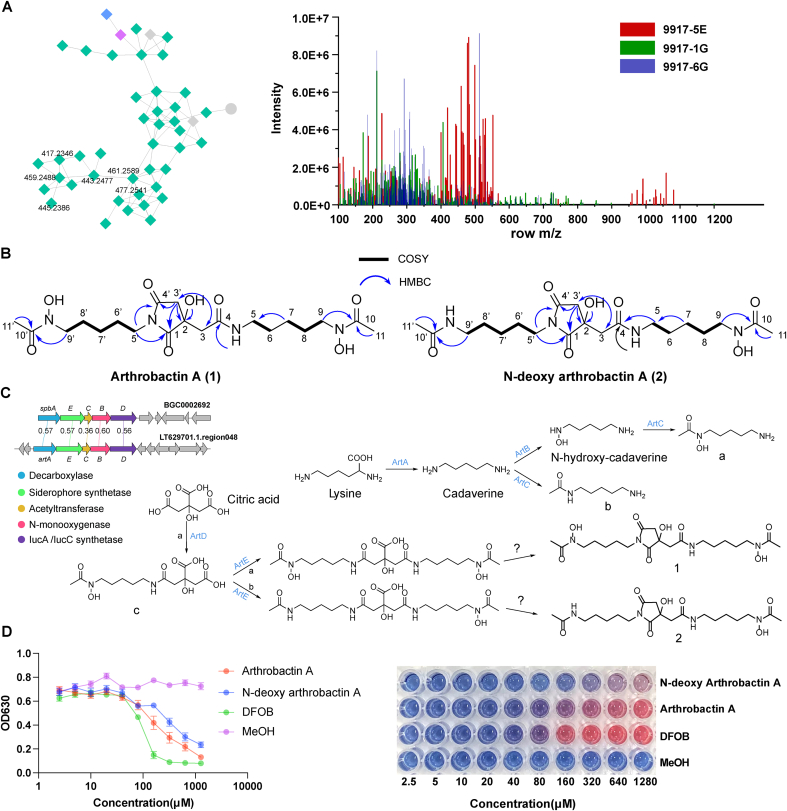
Among the specific secondary metabolites produced by A. albata JCM 9917, two of them (1 and 2) showed high abundance and were assigned as arthrobactin analogs (Fig. 6A). Consequently, they were purified and structurally elucidated. The structures of Arthrobactin A (1) and N-deoxy arthrobactin A (2) were characterized by extensive analysis of 1D and 2D NMR spectroscopic data.
Fig. 6.
(A) A group of specific secondary metabolites produced by A. albata JCM 9917. (B) Chemical structures of Arthrobactin A (1) and N-deoxy arthrobactin A (2), key COSY (bold lines), and HMBC (single-headed arrows) correlations for them. (C) Comparison of the spb and art gene clusters. The putative biosynthetic pathways for compounds 1 and 2. (D) Fe(III)-binding properties of compounds 1 and 2 were assessed using CAS assay. The OD630 values were obtained after mixing the compound with CAS solution at different concentrations. DFOB (Desferrioxamine B) was used as a positive control, while MeOH served as a negative control.
Arthrobactin A (1) was obtained as a yellow oil. The molecular formulas of compound 1 was established as C20H34N4O8 (m/z 459.2444 [M+H]+, calcd 476.2449; m/z 481.2283 [M+Na]+, calcd 481.2269) indicating six degree of unsaturation. The 1H, and 13C NMR (Table S5) of 1 revealed the presence of two methyl, twelve methylene, five carbonyls, and an oxygenated quaternary carbon. Comparison of 1H and 13C NMR data with HSQC correlations revealed that 1 has two amide carbonyl groups (δC 175.0 and 178.7). The 1H–1H COSY correlations of C5′ alkyl amine δH 3.32 (5′-H)/δH 1.48 (6′-H)/δH 1.20 (7′-H)/δH 1.48 (8′-H)/δH 3.44 (9′-H) and C5 alkyl amine δH 2.95 (5-H)/δH 1.35 (6-H)/δH 1.20 (7-H)/δH 1.48 (8-H)/δH 3.44 (9-H) allowed us to assemble two (CH2)5 moieties (Fig. 6B & Table S5). The long-range couplings observed in the HMBC experiments can be summarized (Fig. 6B) as follows: the cross-peaks from δH 3.32 (5′-H) to δC 178.7 (C-1) and δC 175.0 (C-4′), supported the presence of five-membered cyclic imide formed by condensation of citrate carboxyl with an amide of arthrobactin [25]. The cross-peaks from δH 1.96 (11-H/11′-H) to δC 170.2 (C-10/C-10′) supported the presence of two acetyl groups. The nitrogen-bearing methylene at δH 3.44 (9-H) showed a correlation with the carbonyl carbon at δC 170.2 (C-10), the same situation occurs in δH 3.44 (9′-H) and δC 170.2 (C-10′), thus establishing the connectivity of the C5 alkyl diamine to the N-acetyl group. Additionally, the correlation between δH 2.48 and 2.90 (3′-H), δH 2.61 and 2.75 (3-H), and δC 72.1 (C-2), led us to infer that the oxygenated carbon is linked to the two amide groups. Based on the molecular formula of 1, it is estimated that the remaining atoms are hydroxyl groups, one of which is attached to C-2, and the other two form N-hydroxyl groups.
The molecular formula of N-deoxy arthrobactin A (2) was established as C20H36N4O7 (m/z 443.2504 [M+H]+, calcd 443.2500; m/z 465.2330 [M+Na]+, calcd 465.2320). The 13C NMR and HSQC spectra confirmed the presence of 20 carbons in 2. Its NMR data were similar to those of 1 except, the protons on C-11’ (δH 1.92) were more shielded than those on the other side at C-11 (δH 2.09) (Table S5), suggesting a proton was substituted by a hydroxyl group on one hydroxamate of 2. Carefully examination of 13C and 2D NMR of 2 concluded the structure of N-deoxy of 1. We named this novel compound N-deoxy arthrobactin A.
The biosynthetic pathway for compounds 1 and 2 was proposed according to a recent report [26]. The art gene cluster showed a high sequence similarity to the previously putative spb gene cluster and contained all the essential genes encoding decarboxylase, acetyltransferase, l-lysine 6-monooxygenase, and siderophore synthetase (Fig. 6C). Meanwhile, the formation of a succinimide ring via the attack of the peptide bond nitrogen on the y-carboxyl carbonyl represents a non-enzymatic reaction [30]. The sequence variations between the decarboxylase-encoding genes in these BGCs may be related to the length of the carbon chain in the final natural products. The Fe(III)-binding ability of Arthrobactin A and N-deoxy arthrobactin A was evaluated through CAS assay, and both compounds exhibited remarkable affinity for Fe(III). However, the former demonstrated superior binding activity compared to the latter. Both compounds bound to Fe(III) in a concentration-dependent manner, but their binding capacity was weaker compared to the positive control, DFOB, at multiple concentrations (Fig. 6D).
4. Discussion
Rare actinomycetes are one of the most promising sources of natural products. The in-depth genome mining of the rare actinomycetes plays a crucial role in the efficient discovery of natural products. The present study compared the secondary biosynthetic potential and metabolic profiles of Allokutzneria and Kibdelosporangium through comparative genomics and metabolomics approaches. Our findings demonstrated that these two genera showed great specificity in the distribution of the GCFs. The untargeted metabolomics study revealed that a type strain, A. albata JCM 9917, produced multiple series of specific natural products, including a series of hydroxamate-type siderophores.
Allokutzneria and Kibdelosporangium are two phylogenetically related genera and share 89.0%–95.4% sequence identity in 16S rRNA gene sequences [8]. To date, only fifteen natural products have been reported from these two genera [27], which is highly incompatible with the reported number of BGCs in these genera. In particular, to explore secondary metabolites from strains of these two genera, it is crucial to conduct a comprehensive comparison of their specificity in secondary metabolic potential. It is worth noting that the genomic data of these two genera are still insufficient, and the genome quality of some strains is poor. These limitations will have a certain impact on the assignment of genus-specific GCFs. However, most of the identified Allokutzneria-specific GCFs are highly conserved within the genus, indicating that the current data is sufficient to demonstrate the significant genus-specificity in secondary biosynthetic potential between these two genera. On the other hand, because of the challenges in acquiring strains, we only conducted an in-depth analysis on the secondary metabolites of two strains. Further research can be considered on the secondary metabolites of strains that exhibit obvious species-specificity in the distribution of GCFs.
The distance cutoff value of 0.3 was used as the default parameter to define GCFs in BiG-SCAPE. In the present study, to eliminate the influence of the limited number of genome sequences and analyze the specificity of GCFs in these two genera more accurately, we chose a more relaxed threshold of 0.5 to define GCFs. Even at this threshold, we still observed a substantial number of genus-specific GCFs in both genera, and there were evident structural differences among BGCs within phylogenetically related GCFs. The targeted exploration of natural products encoded by BGCs within these genus-specific GCFs using strategies such as heterologous expression or synthetic-bioinformatics natural product (syn-BNP) holds tremendous potential [28,29]. In addition, the genus Kibdelosporangium harbors many species-specific BGCs, whose encoded natural products are also worthy of further exploration. It is important to note that although most of the genus-specific GCFs do not include known BGCs from the MIBiG database, they still share some degree of similarity, and the genus-specific BGCs may encode metabolites that are structurally similar to the known BGCs. For instance, two siderophore BGCs discovered in a specific GCF of Allokutzneria show a similarity score of 0.28 to the gene cluster BGC0002692 in MIBiG. Our research findings suggest that the products encoded by these two BGCs are quite similar.
The diversity and novelty of secondary metabolites produced by two representative strains, A. albata JCM 9917 and K. phytohabitans KLBMP1111were extensively investigated using HiTES experiments. The results showed that about 82% (annotation score less than 3, 0.4, or 0.5 for the Dereplicator+, FBMN, and SIRIUS analysis) of the features could not be successfully annotated, indicating their great potential to be novel natural products. Besides, the annotating results of some features with different tools were inconsistent, further emphasizing the novelty of these features. Through mass spectrometry-guided strategies, we successfully isolated and identified two siderophores, one of which is novel (N-deoxy arthrobactin A), while the remaining compounds were primarily derivatives of deoxygenation or demethylation. However, due to the low yield, we were unable to isolate the potential novel compounds identified by mass spectrometry analysis. In future studies, we will optimize the fermentation conditions to isolate these promising molecules.
In conclusion, the present study systematically compared the genus-specificity of Allokutzneria and Kibdelosporangium in secondary metabolism, highlighting their great variations in the distribution of the potential novel GCFs and the distinct metabolic profiles of two representative strains of these two genera. Two Allokutzneria-specific natural products, Arthrobactin A and N-deoxy arthrobactin A, were isolated and characterized for the first time from this rare genus. The present study offers valuable insights for the targeted discovery of genus-specific natural products from Allokutzneria and Kibdelosporangium.
CRediT authorship contribution statement
Man-Jing Ma: performed the experiment and prepared the original draft. Wen-Chao Yu: prepared the original draft and analyzed the data. Huai-Ying Sun: performed the experiment. Bing-Cheng Dong: performed the experiment. Gang-Ao Hu: provided technical help. Zhen-Yi Zhou: provided technical help. Yi Hua: provided helpful discussions. Buddha Bahadur Basnet: analyzed the NMR data. Yan-Lei Yu: provided helpful discussions. Hong Wang: conceived and designed the study. Bin Wei: conceived and designed the study, and revised the original draft. All authors have read and agreed to the published version of the manuscript.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was financially supported by the Fundamental Research Funds for the Provincial Universities of Zhejiang (No. RF-A2022013), the program of the National Natural Science Foundation of China (No. 42276137), the National Key Research and Development Programs (Nos. 2022YFC2804104, and 2022YFC2804700).
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.synbio.2024.03.015.
Contributor Information
Hong Wang, Email: hongw@zjut.edu.cn.
Bin Wei, Email: binwei@zjut.edu.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article.
References
- 1.Chater K.F. Streptomyces Inside-out: a new perspective on the bacteria that provide us with antibiotics. Philos Trans R Soc Lond B Biol Sci. 2006;361:761–768. doi: 10.1098/rstb.2005.1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hindra, Pak P., Elliot M.A. Regulation of a novel gene cluster involved in secondary metabolite production in Streptomyces coelicolor. J Bacteriol. 2010;192:4973–4982. doi: 10.1128/jb.00681-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tiwari K., Gupta R.K. Rare Actinomycetes: a potential storehouse for novel antibiotics. Crit Rev Biotechnol. 2012;32:108–132. doi: 10.3109/07388551.2011.562482. [DOI] [PubMed] [Google Scholar]
- 4.Bérdy J. Bioactive microbial metabolites. J Antibiot (Tokyo) 2005;58:1–26. doi: 10.1038/ja.2005.1. [DOI] [PubMed] [Google Scholar]
- 5.Shearer M.C., Colman P.M., Ferrin R.M., Nisbet L.J., Nash C.H. New genus of the actinomycetales: Kibdelosporangium aridum gen. nov., sp. nov. Int J Syst Bacteriol. 1986;36:47–54. doi: 10.1099/00207713-36-1-47. [DOI] [Google Scholar]
- 6.Shearer M.C., Actor P., Bowie B.A., Grappel S.F., Nash C.H., Newman D.J., Oh Y.K., Pan C.H., Nisbet L.J. Aridicins, novel glycopeptide antibiotics. I. Taxonomy, production and biological activity. J Antibiot. 1985;38:555–560. doi: 10.7164/antibiotics.38.555. [DOI] [PubMed] [Google Scholar]
- 7.Folena-Wasserman G., Poehland B.L., Yeung E.W., Staiger D., Killmer L.B., Snader K., Dingerdissen J.J., Jeffs P.W. Kibdelins (AAD-609), novel glycopeptide antibiotics. II. Isolation, purification and structure. J Antibiot (Tokyo) 1986;39:1395–1406. doi: 10.7164/antibiotics.39.1395. [DOI] [PubMed] [Google Scholar]
- 8.Labeda D.P., Kroppenstedt R.M. Proposal for the new genus Allokutzneria gen. nov. within the suborder Pseudonocardineae and transfer of Kibdelosporangium albatum Tomita et al. 1993 as Allokutzneria albata comb. Nov. Int J Syst Evol Micr. 2008;58:1472–1475. doi: 10.1099/ijs.0.65474-0. [DOI] [PubMed] [Google Scholar]
- 9.Duangmal K., Poomthongdee N., Pathom-aree W., Takè A., Thamchaipenet A., Matsumoto A., Takahashi Y. Allokutzneria oryzae sp. nov., isolated from rhizospheric soil of Oryza sativa L. Int J Syst Evol Micr. 2014;64:3559–3564. doi: 10.1099/ijs.0.065169-0. [DOI] [PubMed] [Google Scholar]
- 10.Izumikawa M., Takagi M., Shin-ya K.J.B.I.R.-78. JBIR-95: phenylacetylated peptides isolated from Kibdelosporangium sp. AK-AA56. J Nat Prod. 2012;75:280–284. doi: 10.1021/np2008279. [DOI] [PubMed] [Google Scholar]
- 11.Rinkel J., Lauterbach L., Rabe P., Dickschat J.S. Two diterpene synthases for spiroalbatene and cembrene A from Allokutzneria Albata. Angew Chem Int Ed Engl. 2018;57:3238–3241. doi: 10.1002/anie.201800385. [DOI] [PubMed] [Google Scholar]
- 12.Hoshino S., Okada M., Awakawa T., Asamizu S., Onaka H., Abe I. Mycolic acid containing bacterium stimulates tandem cyclization of polyene macrolactam in a lake sediment derived rare actinomycete. Org Lett. 2017;19:4992–4995. doi: 10.1002/anie.201800385. [DOI] [PubMed] [Google Scholar]
- 13.Wei B., Du A.-Q., Zhou Z.-Y., Lai C., Yu W.-C., Yu J.-B., Yu Y.-L., Chen J.-W., Zhang H.-W., Xu X.-W., Wang H. An atlas of bacterial secondary metabolite biosynthesis gene clusters. Environ Microbiol. 2021;23:6981–6992. doi: 10.1111/1462-2920.15761. [DOI] [PubMed] [Google Scholar]
- 14.Wei B., Du A.-Q., Ying T.-T., Hu G.-A., Zhou Z.-Y., Yu W.-C., He J., Yu Y.-L., Wang H., Xu X.-W. Secondary metabolic potential of Kutzneria. J Nat Prod. 2023;86:1120–1127. doi: 10.1021/acs.jnatprod.3c00007. [DOI] [PubMed] [Google Scholar]
- 15.Parks D.H., Imelfort M., Skennerton C.T., Hugenholtz P., Tyson G.W. CheckM: assessing the quality of microbial genomes recovered from isolates, single Cells, and metagenomes. Genome Res. 2015;25:1043–1055. doi: 10.1111/1462-2920.15761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blin K., Shaw S., Augustijn H.E., Reitz Z.L., Biermann F., Alanjary M., Fetter A., Terlouw B.R., Metcalf W.W., Helfrich E.J.N., van Wezel G.P., Medema M.H., Weber T. antiSMASH 7.0: new and improved predictions for detection, regulation, chemical structures and visualisation. Nucleic Acids Res. 2023;51:W46–W50. doi: 10.1093/nar/gkad344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wei B., Hu G.-A., Zhou Z.-Y., Yu W.-C., Du A.-Q., Yang C.-L., Yu Y.-L., Chen J.-W., Zhang H.-W., Wu Q., Xuan Q., Xu X.-W., Wang H. Global analysis of the biosynthetic chemical space of marine prokaryotes. Microbiome. 2023;11:144. doi: 10.1186/s40168-023-01573-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kautsar S.A., van der Hooft J.J.J., de Ridder D., Medema M.H. BiG-SLiCE: a highly scalable tool maps the diversity of 1.2 million biosynthetic gene clusters. GigaScience. 2021;10 doi: 10.1093/gigascience/giaa154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pluskal T., Castillo S., Villar-Briones A., Oresic M. MZmine 2: modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinf. 2010;11:395. doi: 10.1186/1471-2105-11-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nothias L.-F., Petras D., Schmid R., Dührkop K., Rainer J., Sarvepalli A., Protsyuk I., Ernst M., Tsugawa H., Fleischauer M., et al. Feature-based molecular networking in the GNPS analysis environment. Nat Methods. 2020;17:905–908. doi: 10.1038/s41592-020-0933-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mohimani H., Gurevich A., Shlemov A., Mikheenko A., Korobeynikov A., Cao L., Shcherbin E., Nothias L.-F., Dorrestein P.C., Pevzner P.A. Dereplication of microbial metabolites through database search of mass spectra. Nat Commun. 2018;9:4035. doi: 10.1038/s41467-018-06082-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dührkop K., Fleischauer M., Ludwig M., Aksenov A.A., Melnik A.V., Meusel M., Dorrestein P.C., Rousu J., Böcker S. Sirius 4: a rapid tool for turning tandem mass spectra into metabolite structure information. Nat Methods. 2019;16:299–302. doi: 10.1038/s41592-019-0344-8. [DOI] [PubMed] [Google Scholar]
- 23.Bittremieux W. Spectrum_utils : a python package for mass spectrometry data processing and visualization. Anal Chem. 2020;92:659–661. doi: 10.1021/acs.analchem.9b04884. [DOI] [PubMed] [Google Scholar]
- 24.Chen J., Guo Y., Wu Q., Wang W., Pan J., Chen M., Jiang H., Yin Q., Zhang G., Wei B., Zhang H., Wang H. Discovery of new siderophores from a marine Streptomycetes sp. via combined metabolomics and analysis of iron-chelating activity. J Agric Food Chem. 2023;71:6584–6593. doi: 10.1021/acs.jafc.3c00234. [DOI] [PubMed] [Google Scholar]
- 25.Linke W.D., Crueger A., Diekmann H. Metabolic products of microorganisms: 106. The structure of Terregens factor. Arch Mikrobiol. 1972;85:44–50. doi: 10.1007/BF00425143. [DOI] [PubMed] [Google Scholar]
- 26.Acquah K.S., Beukes D.R., Warner D.F., Meyers P.R., Sunassee S.N., Maglangit F., Deng H., Jaspars M., Gammon D.W. Novel south african rare actinomycete Kribbella speibonae strain SK5: a prolific producer of hydroxamate siderophores including new dehydroxylated congeners. Molecules. 2020;25:2979. doi: 10.3390/molecules25132979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Van Santen J.A., Poynton E.F., Iskakova D., McMann E., Alsup T.A., Clark T.N., Fergusson C.H., Fewer D.P., Hughes A.H., McCadden C.A., Parra J., Soldatou S., Rudolf J.D., Janssen E.M.-L., Duncan K.R., Linington R.G. The natural products atlas 2.0: a database of microbially-derived ntural products. Nucleic Acids Res. 2022;50:D1317–D1323. doi: 10.1093/nar/gkab941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luo Y., Enghiad B., Zhao H. New tools for reconstruction and heterologous expression of natural product biosynthetic gene clusters. Nat Prod Rep. 2016;33:174–182. doi: 10.1039/c5np00085h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Z., Koirala B., Hernandez Y., Zimmerman M., Park S., Perlin D.S., Brady S.F. A naturally inspired antibiotic to target multidrug-resistant pathogens. Nature. 2022;601:606–611. doi: 10.1038/s41586-021-04264-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stephenson R.C., Clarke S. Succinimide Formation from Aspartyl and Asparaginyl peptides as a model for the spontaneous degradation of proteins. J Biol Chem. 1989;264:6164–6170. doi: 10.1016/S0021-9258(18)83327-0. [DOI] [PubMed] [Google Scholar]
- 31.Xu D., Han L., Li C., Cao Q., Zhu D., Barrett N.H., Harmody D., Chen J., Zhu H., McCarthy P.J., Sun X., Wang G. Bioprospecting deep-sea actinobacteria for novel anti-infective natural products. Front Microbiol. 2018;9:787. doi: 10.3389/fmicb.2018.00787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kawai K., Wang G., Okamoto S., Ochi K. The rare earth, scandium, causes antibiotic overproduction in streptomyces spp. FEMS Microbiol Lett. 2007;274:311–315. doi: 10.1111/j.1574-6968.2007.00846.x. [DOI] [PubMed] [Google Scholar]
- 33.Pishchany G., Mevers E., Ndousse-Fetter S., Horvath D.J., Paludo C.R., Silva-Junior E.A., Koren S., Skaar E.P., Clardy J., Kolter R. Amycomicin is a potent and specific antibiotic discovered with a targeted interaction screen. Proc Natl Acad Sci U S A. 2018;115:10124–10129. doi: 10.1073/pnas.1807613115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zheng J., Li Y., Liu N., Zhang J., Liu S., Tan H. Multi-omics data reveal the effect of sodium butyrate on gene expression and protein modification in streptomyces. Genom Proteom Bioinform. 2022;S1672–0229(22) doi: 10.1016/j.gpb.2022.09.002. 00115-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seyedsayamdost M.R. High-throughput platform for the discovery of elicitors of silent bacterial gene clusters. Proc Natl Acad Sci U S A. 2014;111:7266–7271. doi: 10.1073/pnas.1400019111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mohammadipanah F., Kermani F., Salimi F. Awakening the secondary metabolite pathways of promicromonospora kermanensis using physicochemical and biological elicitors. Appl Biochem Biotechnol. 2020;192:1224–1237. doi: 10.1007/s12010-020-03361-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.