Abstract
This report examines the role of African swine fever virus (ASFV) structural protein pE120R in virus replication. Immunoelectron microscopy revealed that protein pE120R localizes at the surface of the intracellular virions. Consistent with this, coimmunoprecipitation assays showed that protein pE120R binds to the major capsid protein p72. Moreover, it was found that, in cells infected with an ASFV recombinant that inducibly expresses protein p72, the incorporation of pE120R into the virus particle is dependent on p72 expression. Protein pE120R was also studied using an ASFV recombinant in which E120R gene expression is regulated by the Escherichia coli lac repressor-operator system. In the absence of inducer, pE120R expression was reduced about 100-fold compared to that obtained with the parental virus or the recombinant virus grown under permissive conditions. One-step virus growth curves showed that, under conditions that repress pE120R expression, the titer of intracellular progeny was similar to the total virus yield obtained under permissive conditions, whereas the extracellular virus yield was about 100-fold lower than in control infections. Immunofluorescence and electron microscopy demonstrated that, under restrictive conditions, intracellular mature virions are properly assembled but remain confined to the replication areas. Altogether, these results indicate that pE120R is necessary for virus dissemination but not for virus infectivity. The data also suggest that protein pE120R might be involved in the microtubule-mediated transport of ASFV particles from the viral factories to the plasma membrane.
African swine fever virus (ASFV), the only member of the new family Asfarviridae, is a complex enveloped deoxyvirus responsible for a severe disease of domestic pigs (19, 23, 39, 49). ASFV infects soft ticks of the Ornithodoros genus and different species of suids, being the only known arbovirus that contains DNA, ASFV is unique among DNA viruses in that it resembles the poxviruses in its genome structure and gene expression strategy but morphologically is similar to the iridoviruses (39). The viral genome is a double-stranded DNA molecule of 170 to 190 kbp with terminal inverted repetitions and terminal cross-links (29, 47). The genome of the ASFV strain BA71V encodes more than 150 polypeptides, including structural proteins; a variety of enzymes involved in DNA replication and repair, gene transcription, and protein modification, and proteins potentially involved in the modulation of the virus-host interaction (39, 50).
The virus particle possesses a complex structure composed by several concentric domains with an overall icosahedral shape and an average diameter of 200 nm (4, 5, 14). The viral core consists of a DNA-containing nucleoid covered by a thick protein coat, the core shell. The core is surrounded by an inner lipid envelope and an icosahedral protein capsid. Extracellular particles possess an additional envelope derived from the plasma membrane (9). ASFV particles assemble within cytoplasmic viral factories (4, 9, 10, 31) from precursor membranous structures that probably represent collapsed cisternae derived from the endoplasmic reticulum (5, 18, 38). These membranes give rise to the inner viral envelope, which becomes an icosahedral structure by the progressive assembly of the capsid layer (5, 27). Envelopment and capsid formation depend on calcium gradients and ATP (18). The core is formed beneath the inner envelope through the consecutive assembly of the core shell domain and the electron-dense DNA-containing nucleoid (4, 11). The intracellular particles associate with microtubules to reach the plasma membrane (3, 16) and are finally released from the cell by budding (9).
The mature virion contains about 50 proteins (25), some of which are produced by the proteolytic processing of two viral polyproteins by the viral cysteine proteinase pS273R (6). The core shell proteins p150, p37, p34, and p14, which represent about 25% of the total protein mass of the virus particle, are derived from polyprotein pp220 (4, 43). Similarly, the structural proteins p35 and p15 are derived from polyprotein pp62 (44). At least three major structural proteins have DNA-binding properties (8, 30, 32); one of them (protein 5AR) has been located within the nucleoid and is similar to bacterial histone-like proteins (8). Among the 26 putative membrane proteins encoded by the ASFV genome (36), the structural proteins p12 and pE183L have been involved in the virus attachment to the host cell (1, 13, 28, 34). The icosahedral capsid is mainly composed by protein p72, which represents about one-third of the virus protein mass and probably forms the hexagonal capsomers (4, 27).
Despite the emerging information about the ASFV structural components, little is known on their particular role in virus morphogenesis. To facilitate this study, our laboratory recently adapted to ASFV an inducible expression system based on the Escherichia coli lac operon (27). By using an ASFV recombinant with an inducible copy of protein p72 gene, the role of the major capsid protein in virus assembly and also the origin of the inner viral envelope were analyzed (5, 27). In this report, we have employed the same strategy to investigate the role of protein pE120R in ASFV replication. Protein pE120R, also called p14.5, has been previously characterized as a structural protein expressed as different molecular weight forms late after infection (30). In vitro assays revealed that protein pE120R has DNA-binding properties and that it interacts with a late virus-induced protein of 72 kDa.
Data presented here show that protein pE120R is a capsid component which associates with the major capsid protein p72. Furthermore, protein pE120R was found to be essential for virus dissemination but not for infectivity.
MATERIALS AND METHODS
Cells and viruses.
Vero cells (ATCC CCL81) were grown in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal calf serum (FCS), which was reduced to 2% during viral infection. The ASFV strain BA71V, adapted to grow in Vero cells, and the recombinant viruses vGUSREP and vA72 have been already described (24, 27). Highly purified extracellular BA71V was obtained by Percoll equilibrium centrifugation (12).
Antibodies.
The monospecific rabbit polyclonal serum against protein pE120R and the mouse monoclonal antibody (MAb) 17L.D3 against the major capsid protein p72 have been described previously (27, 30, 41). The rabbit polyclonal serum against protein p72 was raised by immunization with protein p72 obtained from polyacrylamide gels after electrophoresis of highly purified ASFV.
Plasmid construction.
(i) pIND1 and pIND2. The transfer vectors pIND1 and pIND2 for inducible ASFV gene expression were constructed as follows. A 3.4-kb DNA fragment, containing the lacZ gene under the control of the strong late ASFV promoter p72, was purified from SmaI/SalI-digested plasmid p72GAL10T (26). After treatment with Klenow enzyme, the fragment was cloned into the SmaI-digested plasmid p72.I, immediately upstream of a viral inducible promoter (p72.I) consisting of the strong late promoter p72.4 separated by 6 bp from the core sequence of the E. coli lac operator O1 (27). The resulting plasmids were called p72.I.GAL(r) and p72.I.GAL(l). A NotI restriction site was added to plasmid pUC119 by inserting the self-hybridized oligonucleotide 5′-AATTGCGGCCGC into the unique EcoRI site. The resulting plasmid was called pUC119-NotI. Finally, pIND1 and pIND2 plasmids were obtained by inserting the 3.5-kb KpnI/HindIII fragments from p72.I.GAL(r) and p72.I.GAL(l) into pUC119-NotI-linearized with KpnI and HindIII. These vectors are designed to allow the inducible expression of a target gene after homologous recombination with the virus vGUSREP, which constitutively expresses the lacI repressor (27). They contain a cassette formed by the viral inducible promoter p72.I and the lacZ gene under the control of the strong late promoter p72 (27) that allows the color identification of the recombinant viruses. This cassette is flanked by a multiple cloning site formed by XbaI, SalI, PstI, and HindIII sites, immediately downstream of the inducible promoter to allow the cloning of the target gene and the necessary downstream-flanking sequences. A second multiple cloning site formed by SmaI, KpnI, and NotI sites is located upstream of the lacZ gene to allow the cloning of the upstream-flanking sequences. The transfer vectors pIND1 and pIND2 differ in the direction of transcription of the lacZ gene with respect to the inducible promoter.
(ii) pIND1.E120R and pIND2.E120R.
A synthetic DNA fragment of 1,456 bp, which contains the nucleotide sequence from positions −1417 to +19 relative to the translation initiation codon of the ASFV E120R gene, was obtained by PCR, using the primers 5′-CCTGCGGCCGCAGCTCGGAAATCGAAGGG and 5′-ACTGGTACCGAGAATTAAAATCTGCCATC, which contain NotI and KpnI restriction sites (underlined) at their respective 5′ ends. Plasmids pIND1.E120R.Fl and pIND2.E120R.Fl were generated by inserting the KpnI/NotI-digested PCR fragment into KpnI/NotI-digested pIND1 and pIND2, respectively. These plasmids contained the upstream flanking sequences of the E120R gene. The oligonucleotides 5′-GCGCCCGGGGATCCTCTAGAGTCGACATGGCAGATTTTAATTCTCC and 5′-TAACTGCAGGACATTCGCTAAAACTCATCC were used to obtain a 1,341-bp PCR DNA fragment containing the complete E120R open reading frame sequence (the primers include, respectively, SmaI and PstI restriction sites at their 5′ ends). The PCR fragment was digested with SmaI and PstI and inserted into the plasmids pIND1.E120R.Fl and pIND2.E120R.Fl, previously linearized with XbaI, treated with mung bean nuclease and digested with PstI, producing the final transfer plasmids pIND1.E120R and pIND2.E120R, respectively.
Generation of recombinant virus vE120Ri.
Recombinant viruses were generated essentially as previously described (35) with minor modifications. Briefly, Vero cells were infected with virus vGUSREP (27) and transfected with plasmids pIND1.E120R or pIND2.E120R in the presence of 1 mM IPTG. At 48 h postinfection (hpi), the cells were harvested and the recombinant viruses were isolated by sequential rounds of plaque purification in the presence of 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside). Similar results were obtained with the two plasmids used and, after three rounds, one virus clone coming from the pIND1.E120R-transfected cells was selected for further characterization. The structure of this recombinant virus, named vE120Ri, was confirmed by DNA hybridization analysis.
Plaque assays.
Preconfluent monolayers of Vero cells seeded in six-well plates were infected with 600 PFU of recombinat vE120Ri or parental BA71V. After 1 h, the inoculum was removed and the cells were overlaid with DMEM containing 0.6% Noble agar and 2% FCS in the presence or absence of 1 mM IPTG. Five days later, the medium was removed and the monolayers were stained with 1% crystal violet.
One-step virus growth curves.
Preconfluent monolayers of Vero cells were infected with recombinant vE120Ri or parental BA71V at a multiplicity of infection of 5 PFU per cell. After 1 h, the inoculum was removed and the cells were washed with fresh DMEM and overlaid with DMEM supplemented with 2% FCS. IPTG (1 mM) was added immediately after the adsorption period or at 12, 16, or 20 hpi. Infected cells with their culture supernatants were harvested at different times postinfection and centrifuged at 1,000 × g for 5 min. The cell sediment was resuspended in DMEM supplemented with 2% FCS. Both the cellular fraction and the culture supernatant were sonicated and separately titrated by plaque assay on monolayers of Vero cells in the presence of 1 mM IPTG.
Metabolic labeling, immunoprecipitation, and Western immunoblotting.
Preconfluent monolayers of mock- and BA71V-infected Vero cells were pulse-labeled from 16 to 18 hpi with 500 μCi of [35S]methionine-[35S]cysteine (Promix In Vitro Cell Labeling Mix; Amersham Pharmacia Biotech) per ml. The cells were lysed at 4°C with immunoprecipitation buffer (0.01 M Tris-HCl, pH 7.5; 0.15 M NaCl; 1% sodium deoxycholate; 1% IGEPAL CA-630; 0.1% sodium dodecyl sulfate [SDS]) supplemented with protease inhibitors (Complete EDTA-free Cocktail; Roche). Extracts were immunoprecipitated with anti-pE120R antibodies immobilized on protein A-Sepharose (Sigma). Proteins were resolved by SDS–12% polyacrylamide gel electrophoresis and detected by autoradiography.
For the pulse-chase experiment, preconfluent monolayers of Vero cells, cultured in 60-mm plates, were infected with ASFV at 20 PFU/cell. At 11 hpi, cells were pulse-labeled for 1 h with 1 mCi of [35S]methionine-[35S]cysteine in methionine- and cysteine-free DMEM per ml. Before the pulse, the medium was replaced with methionine- and cysteine-free DMEM for 15 min to remove any residual methionine and cysteine. At the end of the labeling period, the medium was removed and the cells were incubated with fresh DMEM for different chase periods. The soluble cytoplasm and the membrane-particulate fractions were obtained as described below. The extracellular virus fraction was collected from the medium by centrifugation in a Beckman Airfuge at 133,000 × g for 20 min. Equivalent amounts of the different fractions were immunoprecitated with anti-pE120R antibodies.
For immunoblotting, samples were electrophoresed in SDS–12% polyacrylamide gels and transferred to nitrocellulose as described elsewhere (4). Protein detection was carried out with peroxidase-conjugated antibodies and the ECL System (Amersham Pharmacia Biotech) according to the manufacturer's indications. Quantitation of protein bands was performed with a Bio-Rad GS710 densitometer and Quantity One software (Bio-Rad).
Subcellular fractionation.
Vero cells were mock infected or were infected with 5 PFU of the BA71V strain per ml or with the recombinant virus vA72 in the presence or absence of 1.25 mM IPTG. At 24 hpi, cells were resuspended at 107 cells/ml in homogenization buffer containing 20 mM Tris-HCl (pH 7.5)–0.25 M sucrose–1 mM EDTA and passed through a 25-gauge syringe 20 times. Cell breakage was monitored by phase-contrast microscopy. The homogenate was centrifuged at 500 × g for 5 min to sediment the nuclei and unbroken cells, and the supernatant was subsequently centrifuged at 100,000 × g for 15 min to separate the soluble cytoplasm from the membrane-particulate fraction. Analysis of equivalent amounts of both fractions was performed by Western immunoblotting.
Immunofluorescence microscopy.
Preconfluent Vero cells grown on coverslips were infected with recombinant vE120Ri at 1 PFU per cell in the presence or absence of 1 mM IPTG. At 18 hpi, infected cells were fixed with methanol at −20°C for 5 min. After being washed with phosphate-buffered saline (PBS), cells were blocked for 30 min with 1% cold fish skin gelatin in PBS. Cells were then incubated for 1 h with rabbit serum anti-pE120R and mouse MAb (17LD3) anti-p72 diluted 1/200 and 1/20, respectively, in blocking solution. After an extensive washing with PBS, cells were incubated for 1 h with Alexa 488 goat anti-rabbit immunoglobulin G (IgG) and Alexa 594 goat anti-mouse IgG (Molecular Probes) diluted 1/1,000 in blocking solution. Finally, coverslips were washed with PBS and mounted with Mowiol/Dabco on glass slides. Preparations were examined with a Bio-Rad Radiance 2000 confocal laser scanning microscope. Images were processed using Adobe Photoshop software.
Electron microscopy.
For conventional Epon section analysis, Vero cells were infected with 10 PFU per cell and fixed at the indicated times with 2% glutaraldehyde in 200 mM cacodylate buffer (pH 7.4) for 1 h at room temperature. Postfixation was carried out with 1% OsO4 and 1.5% K3Fe(CN)6 in cacodylate buffer at 4°C for 30 min. After an extensive washing with distilled water, the samples were dehydrated and embedded in Epon.
For freeze substitution, the infected cells were fixed for 1 h with 4% formaldehyde and 0.1% glutaraldehyde in 200 mM HEPES (pH 7.2) on ice and cryoprotected with 30% glycerol for 30 min. Specimens were rapidly frozen in liquid propane (−180°C) and stored in liquid nitrogen until use. Freeze substitution was carried out as described earlier (5).
For pre-embedding immunolabeling, ASFV-infected Vero cells were permeabilized at 24 hpi essentially as described previously (45). Permeabilization of the plasma membrane was carried out with 20 U of streptolysin O (Sigma) per ml in SLO buffer (0.25 M sucrose, 50 mM potassium acetate, 5 mM magnesium acetate, 1 mM dithiothreitol, 25 mM HEPES; pH 7.4). After 15 min at 4°C, the cells were incubated in SLO buffer for 15 min at 37°C, extensively washed with 0.2 M HEPES (pH 7.2), and lightly fixed with 4% paraformaldehyde in the same buffer for 10 min at 4°C. Subsequently, the cells were washed with ice-cold PBS, blocked with 0.5% cold fish skin gelatin in PBS for 1 h, and labeled with a 1/25 dilution of anti-pE120R antibody in PBS with 1% egg albumin. After being washed for 2 h with PBS, the cells were incubated with protein A-gold (10 nm) for 1 h, washed again for 2 h with PBS, fixed with 2% glutaraldehyde for 1 h, and processed for conventional Epon embedding.
Postembedding labeling of ultrathin Lowicryl K4M sections was performed as described elsewhere (4). The rabbit polyclonal antibodies against pE120R and p72 were used at a 1/25 and 1/100 dilutions, respectively, in PBS with 1% egg albumin.
Specimens were examined at 80 kV in a Jeol 1010 electron microscope and photographed with a Bioscan 792 charge-coupled device camera (1,024 × 1,024 pixels; Gatan). Digital images were processed with DigitalMicrograph (Gatan) and Adobe Photoshop software (Adobe).
RESULTS
Protein pE120R localizes at the surface of the intracellular virions.
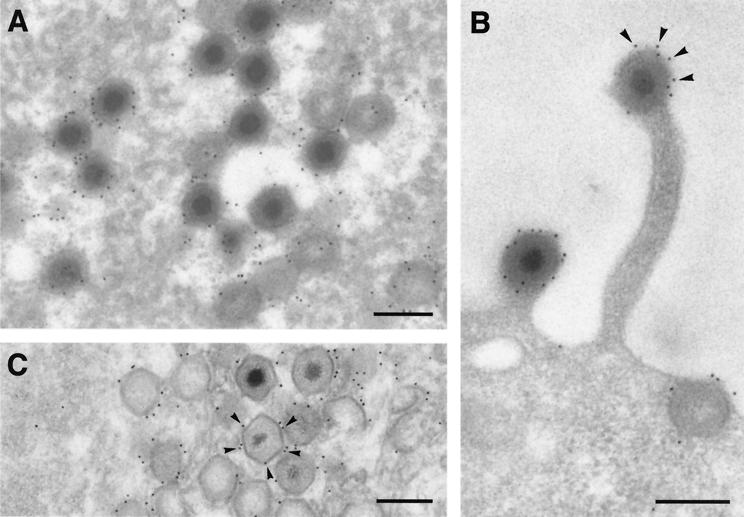
We first analyzed the localization of protein pE120R in ASFV-infected cells by immunoelectron microscopy. Ultrathin Lowicryl sections of infected cells processed at 24 hpi by freeze substitution were incubated with a monospecific rabbit anti-pE120R antibody (30) prior to incubation with protein A-gold complexes. The signal was essentially localized in the virus factories (Fig. 1A), as well as on virus particles spreading throughout the cytoplasm (not shown) or budding at the plasma membrane (Fig. 1B). Within the assembly sites, gold particles were found mainly associated with both immature and mature icosahedral virus particles (Fig. 1A). In contrast, nonpolyhedral membranous structures, which are thought to be precursor forms of the ASFV particles (4), were poorly labeled. No significant pE120R labeling was associated with the endoplasmic reticulum or other cellular membranes (not shown). Within the virus particles, most of the labeling was associated with the periphery of the virions, which was particularly evident when individual cross-sectioned budding particles were visualized (Fig. 1B).
FIG. 1.
Immunoelectron microscopy of protein pE120R in BA71V-infected cells. (A and B) Ultrathin Lowicryl K4M sections of infected Vero cells fixed at 24 hpi and processed by freeze substitution were incubated with anti-pE120R antibodies followed by protein A-gold (10 nm). (A) Within the virus factory, the labeling was mainly associated with mature particles and icosahedral immature virions lacking an electrondense nucleoid. In contrast, the precursor membranous structures surrounding the virus particles were weakly labeled. (B) A strong and peripheral labeling was also detected on ASFV particles budding through the plasma membrane. (C) Infected Vero cells were permeabilized with streptolysin O at 24 hpi and labeled with anti-pE120R antibody and protein A-gold (10 nm) before conventional Epon embedding. Gold particles (arrowheads) decorated the surface of ASFV particles within the virus factories. Bars, 200 nm.
The localization of protein pE120R in the virus particle was also examined by pre-embedding immunolabeling on permeabilized ASFV-infected cells. At 24 hpi, infected cells were permeabilized with streptolysin O and, after a brief fixation, were incubated with anti-pE120R antibodies followed by protein A-gold. As shown in Fig. 1C, gold particles decorated the external layer, i.e., the capsid, of the icosahedral particles present at the assembly sites. Altogether, immunoelectron microscopy studies indicate that protein pE120R is a capsid component exposed on the surface of the intracellular virions.
Protein pE120R interacts with the major capsid protein p72.
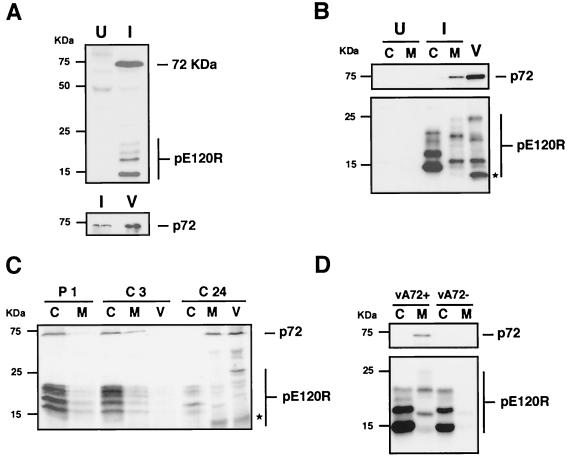
Based on in vitro binding assays with purified pE120R protein and on coimmunoprecipitation experiments with anti-pE120R antibodies, Martínez-Pomares et al. (30) found that protein pE120R binds to a late virus-induced polypeptide of 72 kDa. To ascertain whether this polypeptide is the major capsid protein p72, uninfected and ASFV-infected cells were labeled with [35S]methionine-[35S]cysteine from 16 to 18 hpi and then immunoprecipitated with anti-pE120R antibody. As previously described (30) and as shown in Fig. 2A (upper panel), the anti-pE120R antibodies immunoprecipitated several low-molecular-mass proteins ranging from 14.5 to 22 kDa, which correspond to the different pE120R forms, as well as an additional 72-kDa polypeptide that was not detected in uninfected cells. The identity of this protein was ascertained by Western immunoblotting with MAb 17L.D3 against p72 (27, 41) of the material previously immunoprecipitated with anti-pE120R antibody. As shown in Fig. 2A (lower panel), a 72-kDa protein band comigrating with the major capsid protein from highly purified virus was detected.
FIG. 2.
Protein pE120R interacts with the major capsid protein p72. (A) Uninfected (U) Vero cells or cells infected with BA71V virus (I) were pulse-labeled with [35S]methionine-[35S]cysteine from 16 to 18 hpi. The cell extracts were immunoprecipitated with anti-pE120R serum and analyzed by SDS-polyacrylamide gel electrophoresis and autoradiography (upper panel). The immunoprecipitated material from infected cell extracts was further analyzed, together with highly purified extracellular ASFV (V), by Western immunoblotting with anti-p72 MAb 17L.D3 (lower panel). (B) Western immunoblotting with MAb anti-p72 (upper panel) and anti-pE120R (lower panel) antibodies of cytosolic (C) and membrane-particulate (M) fractions from uninfected cells (U) or cells infected with parental BA71V (I). Analysis of highly purified extracellular virions (V) is also shown. The asterisk indicates the position of the major structural pE120R form of 12 kDa. (C) Infected Vero cells were pulse-labeled with [35S]methionine-[35S]cysteine from 11 to 12 hpi (P1), chased for 3 h (C3) and 24 h (C24), and then fractionated into soluble cytosolic (C), membrane-particulate (M), and extracellular virus (V) fractions. Equivalent amounts of the fractions were immunoprecipitated with anti-pE120R antibodies and analyzed by SDS-polyacrylamide gel electrophoresis. The asterisk indicates the position of the major structural pE120R form of 12 kDa. (D) Immunoblotting with anti-p72 (upper panel) and anti-pE120R (lower panel) antibodies of cytosolic (C) and membrane-particulate (M) fractions from extracts of vA72-infected cells grown in the presence (vA72+) or in the absence (vA72−) of IPTG. The migration position of molecular mass markers is indicated on the left. The bands corresponding to protein p72 and the different forms of protein pE120R are indicated on the right.
Incorporation of protein pE120R into the virus particle depends on expression of the major capsid protein p72.
To further explore the relationship between the capsid proteins pE120R and p72, we analyzed their intracellular distribution. Mock- and BA71V-infected cells were fractionated at 24 hpi into a soluble and a membrane-particulate cytoplasmic fraction as described in Materials and Methods. A Western blot analysis showed that protein p72, a peripheral membrane protein (17), was essentially present in the membrane-particulate fraction (Fig. 2B, upper panel). In contrast, protein pE120R was predominantly cytosolic, although a significant proportion was also detected in the membrane-particulate cytoplasm (Fig. 2B, lower panel). Interestingly, some pE120R forms, ranging from 14.5 to 22 kDa, were mainly found in the cytosol, while others, ranging from 12 to 25 kDa, were present almost exclusively in the cytoplasmic sediment. Moreover, the analysis of protein pE120R in highly purified extracellular ASFV revealed a multiple band profile resembling that observed in the membrane-particulate cell fraction, although with some significant differences. As shown in Fig. 2B (lower panel), the main difference was the detection of a prominent 12-kDa structural form which was almost undetectable in the cytoplasmic sediment. This striking mass heterogeneity of protein pE120R suggests that the protein undergoes posttranslational modifications before and during its incorporation to the intracellular virus particle, as well as during or after virus release. To evaluate whether the species found in the membrane-particulate and extracellular virus fractions are derived from the cytosolic pE120R forms, we analyzed the subcellular distribution of pE120R in a pulse-chase experiment. ASFV-infected cells were labeled with [35S]methionine-[35S]cysteine from 11 to 12 hpi and then chased in the presence of cold methionine-cysteine for different periods. After the pulse and chase periods, the infected cells were fractionated into a soluble and a membrane-particulate fraction and the extracellular medium was centrifuged to collect extracellular released virions. Equivalent amounts of the fractions were then analyzed by immunoprecipitation with anti-pE120R antibodies. As shown in Fig. 2C, protein pE120R (as 14.5- to 22-kDa isoforms) and coprecipitated protein p72 were essentially detected in the soluble cytoplasmic fraction after the 1-h pulse-labeling. Following a 3-h chase, pE120R and p72 levels slightly decreased in the cytosol and, concomitantly, slightly increased in the cytoplasmic sediment. Finally, after a 24-h chase, proteins pE120R (as 12- to 25-kDa isoforms) and p72 were mostly detected in the membrane-particulate and extracellular virus fractions. These results indicate that protein pE120R is initially expressed in the cytosol and subsequently, a fraction of it is slowly incorporated into the assembling virus particles, with the appearance of some new species. On the other hand, since anti-pE120R antibodies coimmunoprecipitated both proteins pE120R and p72 from the pulse-labeled cytosolic extract, it seems that pE120R associates with cytosolic p72 before its incorporation into the assembling virions.
We next investigated if the intracellular distribution of protein pE120R is dependent on p72 expression using an ASFV recombinant (vA72) which inducibly expresses the major capsid protein (27). Vero cells were infected for 24 h with recombinant vA72 in the presence or absence of IPTG and subsequently fractionated as described above. As shown in Fig. 2D, under permissive conditions both protein p72 and pE120R fractionated as in control BA71V infections. In contrast, when p72 expression was abrogated, protein pE120R was detected almost exclusively in the soluble cytoplasmic fraction. The virtual absence of protein pE120R in the membrane-particulate sediment indicates that p72 expression influences the intracellular distribution of pE120R.
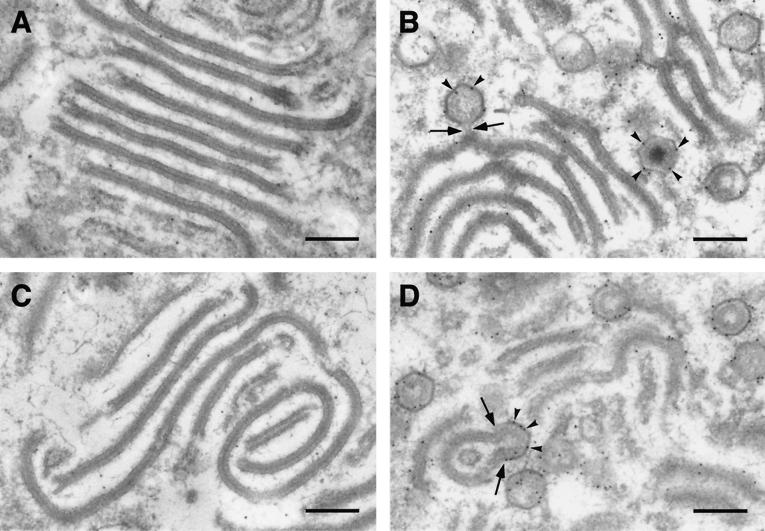
To further investigate this effect, immunoelectron microscopy with the anti-pE120R and anti-p72 antibodies was performed on sections of vA72-infected cells maintained for 16 h in the absence of IPTG (Fig. 3A and C) or treated with the inducer at 16 hpi during an additional 8-h period (Fig. 3B and D). As previously reported, the effect of p72 repression was the accumulation of aberrant virus forms, called zipper-like structures, at the virus factories (5, 27). These structures consist of an extended central domain, which is reminiscent of the core shell, flanked by inner viral envelopes. As shown in Fig. 3A, these zipper-like structures were poorly labeled with the anti-pE120R, which is consistent with the cytosolic distribution of protein pE120R observed under these conditions (Fig. 2C). As a negative control, the anti-p72 labeling was practically absent (Fig. 3C). After addition of the inducer, p72 expression led to the capsid assembly on the previously accumulated zipper-like structures, as well as to the de novo formation of normal virions (27). In these conditions, both anti-pE120R (Fig. 3B) and anti-p72 (Fig. 3D) antibodies strongly labeled the nascent icosahedral particles, as well as the zipper-like structures, especially in the areas where assembling capsids were evident.
FIG. 3.
Immunoelectron microscopy of protein pE120R on vA72-infected cells. Lowicryl sections of vA72-infected cells maintained 16 h in the absence of IPTG (A and C) or treated with the inducer at 16 hpi during an 8-h period (B and D) were incubated with anti-pE120R antibody (A and B) or anti-p72 antibody (C and D), followed by protein A-gold (10 nm). In the absence of IPTG the aberrant zipper-like structures were poorly labeled by both sera while, in the presence of the inducer, anti-pE120R and anti-p72 antibodies strongly labeled (arrowheads) icosahedral particles as well as polyhedral forms derived from previously assembled zipper-like structures. The arrows indicate icosahedral forms emerging from zipper-like structures. Bars, 200 nm.
In summary, these results indicate that the incorporation of protein pE120R to the virus particle is dependent on expression of the major capsid protein p72 and probably concomitant to capsid assembly.
Inducible expression of protein pE120R by recombinant virus vE120Ri.
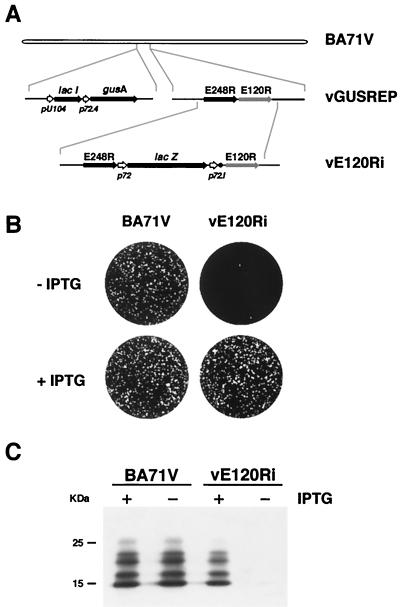
To further analyze the role of protein pE120R in virus replication, we constructed an ASFV recombinant (vE120Ri), derived from the parental BA71V strain, enabling the inducible expression of gene E120R (Fig. 4A). In this recombinant virus, the endogenous gene E120R was replaced by a copy under the transcriptional control of the inducible late promoter p72.1 consisting of the strong late promoter p72.4 (27) and the operator sequence O1 from the E. coli lac operon (Fig. 4A). Expression of gene E120R is regulated by the E. coli lac repressor encoded by gene lacI, which was inserted within the nonessential tk locus under the control of the early promoter pU104 (2).
FIG. 4.
(A) Genomic structure of the recombinant ASFV virus vE120Ri. Recombinant virus vE120Ri was obtained from recombinant vGUSREP, which contains the lacI gene encoding the lac repressor inserted into the nonessential tk locus. In vE120Ri virus, the gene E120R is under the transcriptional control of an inducible promoter p72.1, which is composed by the strong late promoter p72.4 and the lac operator sequence (●). The reporter genes lacZ and gusA, used for selection and purification of the recombinants, are also represented. (B) Plaque phenotype of vE120Ri. Monolayers of Vero cells were infected in the absence or presence of 1 mM IPTG with parental BA71V or recombinant vE120Ri virus. Plaques were visualized with 1% crystal violet 5 days after infection. (C) Inducible expression of protein pE120R. Vero cells were infected with BA71V or recombinant vE120Ri in the presence (+) or absence (−) of 1 mM IPTG. At 24 hpi, samples were electrophoresed and analyzed by Western immunoblotting with a serum anti-pE120R. The electrophoretic mobility of molecular weight markers is indicated on the left.
To test the inducer dependence of recombinant vE120Ri, a plaque assay was performed at different concentrations of IPTG ranging from 0 to 2 mM (not shown). The plaque number was maximal at 1 mM IPTG. Figure 4B shows the plaque phenotype of parental BA71V and recombinant vE120Ri viruses in the presence or in the absence of 1 mM IPTG. In the presence of the inducer, both the number and the size of the plaques of recombinant vE120Ri were similar to those observed for the control virus. In contrast, in the absence of IPTG the plaque number of recombinant vE120Ri was reduced by about 2.5 orders of magnitude.
To verify that the plaque phenotype of vE120Ri virus correlates with the IPTG-dependent expression of protein pE120R, a Western blot analysis was performed with extracts of BA71V- and vE120Ri-infected cells maintained in the presence or in the absence of 1 mM IPTG for 24 hpi. As shown in Fig. 4C, under restrictive conditions the expression levels of protein pE120R were dramatically reduced with regard to those observed under permissive conditions or in BA71V infections. A densitometric quantitation showed that, in the absence of IPTG, expression of pE120R was 1 and 0.5% of that observed under permissive conditions and in control BA71V infections, respectively. These results indicate that plaque formation by recombinant vE120Ri is related to pE120R expression.
Protein pE120R is required for virus dissemination but not for infectivity.
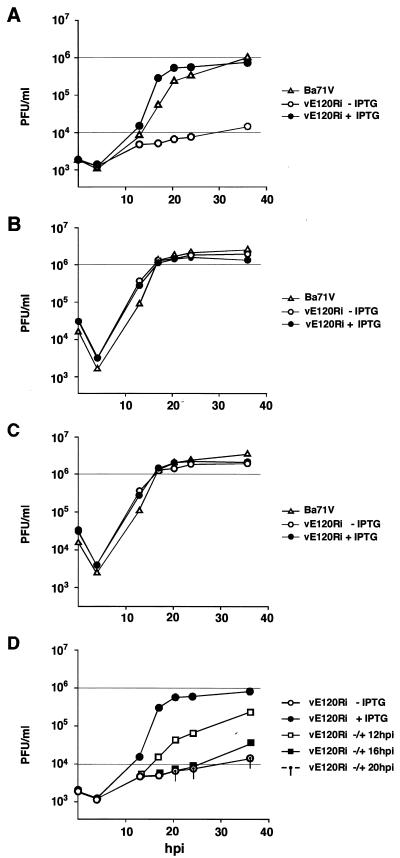
To study the effect of pE120R shutoff on viral replication, one-step growth curves were performed. Cells infected with recombinant vE120Ri in the presence or in the absence of inducer were harvested in their culture medium at different times postinfection and titrated by plaque assay under permissive conditions. Surprisingly, no significant difference was observed in the virus yield of recombinant vE120Ri grown under restrictive or permissive conditions, a yield which was, on the other hand, similar to that obtained for the control BA71V virus (not shown). To clarify the apparent contradiction between the plaque assays and the one-step growth curves, infected cells and culture supernatants were titrated separately after sonication. Figure 5 shows the titration curves of the extracellular (panel A) and intracellular (panel B) viruses, as well as the total virus yield (panel C) deduced from those curves. Under restrictive conditions, the titer of extracellular vE120Ri was, from 24 to 48 hpi, ca 2 log units lower than that obtained under permissive conditions or with the parental virus (Fig. 5A). In contrast, the yield of cell-associated recombinant virus grown under restrictive conditions (Fig. 5B) was similar to the total yield of the recombinant grown under permissive conditions or the parental virus (Fig. 5C). These results show, on the one hand, that mutant vE120Ri particles are infectious and, on the other, that protein pE120R is required for virus dissemination.
FIG. 5.
One-step growth curves of vE120Ri. Vero cells were infected with 10 PFU of BA71V or vE120Ri virus per cell in the presence or absence of 1 mM IPTG. Virus from the culture supernatants (A) and the infected cells (B) were collected at the indicated times of infection and titrated separately by plaque assay on fresh Vero cells in the presence of the inducer. (C) Curves of total virus yield were deduced from the intracellular and extracellular virus yields shown in panels A and B. (D) In the same experiment, recombinant virus vE120Ri was grown under nonpermissive conditions for the indicated times and then induced with IPTG. At different times postinfection, extracellular virus from the culture supernatant was titrated as described above in the presence of inducer. As a control, the extracellular virus yields of recombinant vE120Ri grown in the absence or presence of IPTG throughout the infection are shown.
In the same experiment, it was also tested the ability of vE120R-infected cells maintained for different times under nonpermissive conditions to produce extracellular infectious virus upon IPTG addition. As shown in Fig. 5D, the extracellular virus yield increased significantly when IPTG was added at 12 hpi, an early time for the virus assembly in normal infections (4, 10, 31). However, when pE120R expression was induced from 16 or 20 hpi onward, the extracellular virus titers did not increase significantly compared to a vE120Ri infection in the absence of IPTG. Since the intracellular virus yield is considerable at these late times of infection (see Fig. 5B), these results strongly suggest that the inhibitory effect of pE120R repression on virus egress is not reversible (see below).
Protein pE120R is required for the transport of virions from the assembly sites to the plasma membrane.
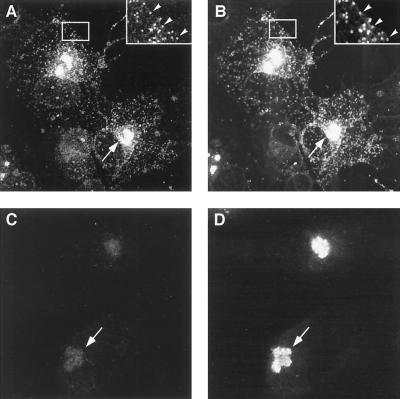
To characterize in more detail the effect of repression of pE120R on virus egress, immunofluorescence experiments were performed on vE120Ri-infected cells maintained for 18 h with or without 1 mM IPTG. Figure 6 shows a double labeling with the rabbit polyclonal serum against protein pE120R (panels A and C) and the mouse MAb against protein p72 (panels B and D). In the presence of the inducer (Fig. 6A and B), both anti-pE120R and anti-p72 antibodies strongly labeled the virus factories, as well as virus particles scattered throughout the cytoplasm (see also the inserts in these panels). In the absence of the inducer, labeling of protein pE120R (Fig. 6C) was drastically diminished, while an intense signal of p72 (Fig. 6D) was observed only in the assembly sites. These observations strongly suggest that, when expression of protein pE120R is inhibited, intracellular virus particles are retained in the virus factories and no transport to the cell periphery occurs.
FIG. 6.
Immunofluorescence microscopy of vE120Ri-infected cells. Vero cells infected with recombinant vE120Ri virus in the presence (A and B) or absence (C and D) of IPTG were fixed at 18 hpi and double-labeled with rabbit serum anti-pE120R (A and C) and mouse MAb 17L.D3 anti-p72 (B and D). Labeling was revealed with Alexa 488 goat anti-rabbit rabbit IgG and with Alexa 594 goat anti-mouse IgG. Insets in panels A and B show enlarged images of the delimited cytoplasmic areas. Viral factories and virions spread throughout the cytoplasm are indicated by arrows and arrowheads, respectively.
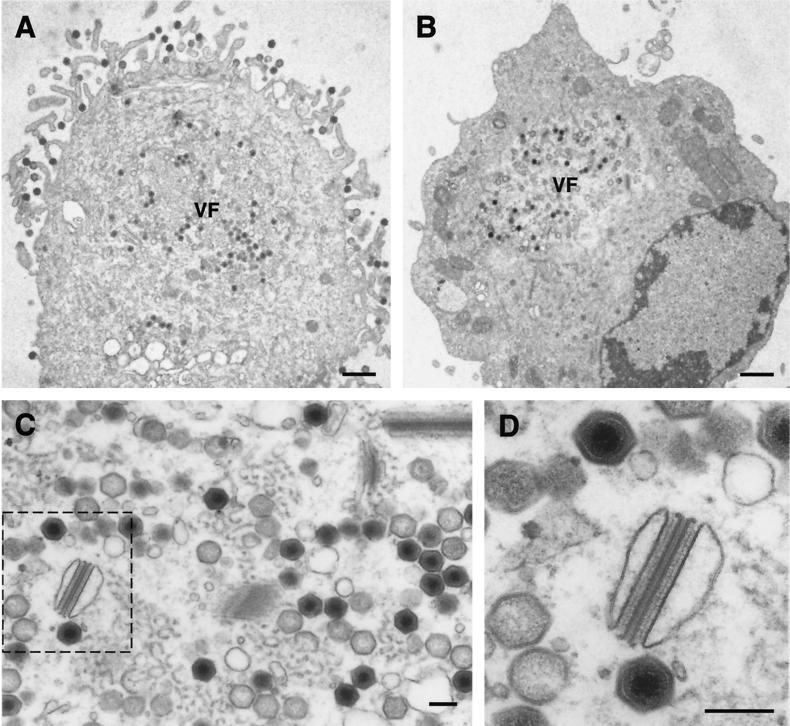
In another approach, electron microscopy (EM) studies were carried out on sections of vE120Ri-infected cells maintained for 18 h in the presence (Fig. 7A) or in the absence of 1 mM IPTG (Fig. 7B to D). When we compared both situations, it was evident that under restrictive conditions no budding occurred at the plasma membrane and essentially no virus particles were detected outside the virus factories (compare Fig. 7A and B). Within the assembly areas, all stages of virus morphogenesis, including large amounts of apparently mature virions, were observed (Fig. 7C). Close inspection of these mutant vE120Ri full particles (Fig. 7D) did not reveal any significant ultrastructural difference with normal mature virions. Interestingly, aberrant structures (Fig. 7D) reminiscent but more complex than the zipper-like structures found when p72 expression is abrogated (5, 27; see also Fig. 3), were observed at the assembly sites. As deduced from the one-step growth curves, the existence of such structures did not interfere significantly with the production of infectious virus particles. Finally, we also analyzed vE120Ri-infected cells maintained under restrictive conditions for 16 h and then induced with IPTG for 8 h. In agreement with the titration experiment shown in Fig. 5D, no significant transport or release of previously assembled mature particles could be detected (not shown).
FIG. 7.
Electron microscopy of vE120Ri-infected cells. Ultrathin Epon sections of vE120Ri-infected Vero cells incubated for 18 h in the presence (A) or in the absence (B, C, and D) of IPTG. While in the presence of inducer (A) ASFV particles move from the virus factories (VF) to the plasma membrane to be released by budding, in the absence of IPTG (B) virus particles are assembled within the virus factories but neither transport nor budding occur. Note also that, under restrictive conditions, the factories (panel C) contain all normal virus assembly stages, including large amounts of intracellular mature particles. Additionally, some aberrant virus structures can be detected. Panel D shows a higher magnification of the region delimited by the dashed line in panel C, which contains an aberrant structure and two apparently normal intracellular mature particles. Bars: A and B, 1 μm; C and D, 200 nm.
In conclusion, EM studies indicate that protein pE120R is essential for the transport of virus particles from the assembly sites to the plasma membrane but is not required for the assembly of morphologically mature intracellular particles.
DISCUSSION
Most of our knowledge on ASFV morphogenesis derives from the analysis at the electron microscope of the morphological stages of virus assembly (4, 5, 9, 10, 27, 31) and the immunolocalization of virus proteins on the virus precursors and mature virions (4, 6, 8, 15, 27). To facilitate the study of particular ASFV proteins on virus replication, our laboratory recently adapted to ASFV an inducible expression system based on the E. coli lac operon (27). In this report, we have used this strategy to investigate the role of ASFV protein pE120R.
Protein pE120R was described by Martínez-Pomares et al. (30) as a structural polypeptide expressed late after infection as different low-molecular -mass forms that, in our experiments, range from 12 to 25 kDa. Protein pE120R is 90.8% identical to its counterpart K3R in the pathogenic Malawi strain (22) and is expressed in cells infected by a variety of pathogenic and nonpathogenic virus strains (unpublished results). The protein lacks significant similarity with other viral and nonviral protein sequences available in the databases. Interestingly, Martínez-Pomares et al. (30) found that protein pE120R binds to a late-virus-induced protein of 72 kDa. By using coimmunoprecipitation assays, we show here that this polypeptide is the major capsid protein p72. This interaction is consistent with the external localization of protein pE120R in the virus particles. Immunoelectron microscopy showed that protein pE120R is mainly present in polyhedral virus particles, being exposed on the capsid of the intracellular mature virion. Using a recombinant virus that inducibly expresses protein p72 (27), we also found that expression of the major capsid protein is required for the association of protein pE120R with the virus. Taking into account that capsid protein p72 is a peripheral membrane protein initially expressed in the cytosol (17; this report) and that anti-pE120R antibodies coprecipitate p72 from cytosolic extracts (this report), it is likely that pE120R associates with assembling virions after being bound to cytosolic p72. The results also suggest that the incorporation of protein pE120R into nascent virus particles is concomitant with capsid formation.
Analysis of recombinant vE120Ri led to two important conclusions: (i) the protein pE120R is required for virus egress, and (ii) it is not essential for virus infectivity. Repression of pE120R synthesis drastically inhibited virus release from the host cell, as deduced from plaque assays and one-step virus growth curves. Immunofluorescence and immunoelectron microscopy revealed that this effect was a consequence of the retention of the intracellular mature particles within the viral factories. Collectively, these data indicate that pE120R expression is required for the transport of intracellular particles from the assembly sites to the plasma membrane. Interestingly, the induction of pE120R expression in vE120Ri-infected cells previously maintained for long periods under restrictive conditions did not significantly increase virus egress. The apparent irreversibility of the mutant phenotype suggests that newly synthesized protein pE120R is not able to associate with previously assembled mutant particles. This result supports the view that protein pE120R is recruited during capsid formation in normal infections, although it is not strictly essential for the assembly of the capsid layer nor for the production of intracellular infectious particles.
In general, virus movement during entry and exit from the host cell is dependent on the cytoskeleton (20, 40, 46, 48), which is commonly reorganized throughout the infection. With regard to the association of ASFV with the cytoskeleton, previous work showed a function of microtubules in the organization of the assembly sites (3, 16), the redistribution of mitochondria around the viral factories (37), and virus release (3, 16). Alves de Matos and Carvalho (3) found that incubation of ASFV-infected cells with drugs that depolymerize microtubules avoided the fusion of assembly sites into a unique viral factory that ocurrs in normal infections. Importantly, this treatment also inhibited strongly the migration of nascent virus particles to the cell surface, an effect that was reverted after drug removal. On the other hand, in vitro binding assays and EM studies with taxol-treated cells indicated that virions bind to microtubules (3). In this aspect, the involvement of microtubules in the transport of ASFV particles is analogous to that described for the intracellular movement of vaccinia virus. According to Sanderson et al. (40), the intracellular mature virus employs microtubules for efficient dispersion from the viral factories.
Since both protein pE120R and microtubules are required for the transport of virions to the cell periphery, it is tempting to speculate that protein pE120R could be involved in the virus-microtubule interaction. The external location of protein pE120R in the intracellular virion is compatible with such a role. On the other hand, the striking observation that protein pE120R exhibits different and complex band profiles in the cytosol, the membrane-particulate cytoplasm, and the extracellular virions might reflect a complex maturational process related to the regulation of the interaction with the microtubules and the subsequent transport of ASFV particles. The posttranslational modifications involved in the high mass heterogeneity of protein pE120R have not been elucidated at present, although glycosylation and disulfide-linked dimerization can be excluded from the sequence data (30). A more detailed study of protein pE120R will be necessary to determine the exact function of this protein in virus dissemination and to identify the basis of its size heterogeneity.
With regard to the infectivity of mutant vE120Ri, the results of this work support the idea that the intracellular mature form of ASFV is infectious. Under conditions that abrogate pE120R expression and virus egress, the intracellular virus yield was similar to the total yield obtained under permissive conditions or with parental BA71V virus. Consistent with this, EM studies showed that under restrictive conditions large amounts of intracellular mature particles, structurally indistinguishable from normal BA71V virions, are assembled at the virus factories. Previous work showed that protein pE120R has DNA-binding properties in vitro (30), which could suggest a potential role in DNA replication, DNA encapsidation, or proper assembly of the nucleoprotein core. Nevertheless, the localization of pE120R in the virus particle and the phenotype of recombinant vE120Ri do not support any essential function for pE120R related to its DNA-binding properties. Since protein pE120R binds to DNA with a low affinity constant (30), it cannot be excluded that this interaction merely reflects its ability to bind to negatively charged macromolecules.
The infectivity of intracellular vE120Ri particles is consistent with early work showing that preparations of partially purified cell-associated ASFV particles are infectious (42), that the outer viral envelope is not necessary for infectivity (42), and that antibodies against the capsid protein p72 can neutralize ASFV infection (7, 28). Unlike extracellular virions (12), intracellular mature particles have not been efficiently purified so far mainly due to their high contamination with cellular membranes as well as extracellular particles, which often lose the outer envelope (42). In relation to this, the recombinant vE120Ri can be a useful tool to purify intracellular ASFV mature particles and to establish the differences in composition and infectivity with the extracellular enveloped particles. Regarding this point, the existence of intracellular and extracellular infectious particles, which are structurally and antigenically distinct, is well established for other complex DNA viruses such as poxviruses (21, 33). Finally, it will be important to know if the infectious intracellular form of ASFV plays a direct role in virus dissemination in vivo. Since ASFV infection evolves finally toward cytolysis (9), the release of intracellular virions from lysed cells could constitute an alternative pathway to the budding at the plasma membrane. In such a case, the existence of two structurally and antigenically different infectious ASFV particles may have important implications in the host immune response against ASFV.
ACKNOWLEDGMENTS
We thank J. Salas and A. Alejo for critical reading of the manuscript. We also thank M. Guerra, M. Rejas, and M. J. Bustos for technical assistance.
This study was supported by grants from the Dirección General de Investigación Científica y Técnica (PB96-0902-C02 -01), the European Community (FAIR-CT97-3441), and the Ministerio de Educación y Cultura (AGF98-1352-CE) and by an institutional grant from Fundación Ramón Areces. G. Andrés was supported by a fellowship from Comunidad Autónoma de Madrid.
REFERENCES
- 1.Alcamí A, Angulo A, López-Otín C, Muñoz M, Freije J M P, Carrascosa A L, Viñuela E. Amino acid sequence and structural properties of protein p12, an African swine fever virus attachment protein. J Virol. 1992;66:3860–3868. doi: 10.1128/jvi.66.6.3860-3868.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Almazán F, Rodríguez J M, Andrés G, Pérez R, Viñuela E, Rodríguez J F. Transcriptional analysis of multigene family 110 of African swine fever virus. J Virol. 1992;66:6649–6654. doi: 10.1128/jvi.66.11.6655-6667.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alves de Matos A P, Carvalho Z G. African swine fever virus interaction with microtubules. Biol Cell. 1993;78:229–234. doi: 10.1016/0248-4900(93)90134-z. [DOI] [PubMed] [Google Scholar]
- 4.Andrés G, Simón-Mateo C, Viñuela E. Assembly of African swine fever virus: role of polyprotein pp220. J Virol. 1997;71:2331–2341. doi: 10.1128/jvi.71.3.2331-2341.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andrés G, García-Escudero R, Simón-Mateo C, Viñuela E. African swine fever virus is enveloped by a two-membraned collapsed cisterna derived from the endoplasmic reticulum. J Virol. 1998;72:8988–9001. doi: 10.1128/jvi.72.11.8988-9001.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Andrés G, Alejo A, Simón-Mateo C, Salas M L. African swine fever virus protease: a new viral member of the SUMO-1-specific protease family. J Biol Chem. 2001;276:780–787. doi: 10.1074/jbc.M006844200. [DOI] [PubMed] [Google Scholar]
- 7.Borca M V, Irusta P M, Carrillo C, Afonso C L, Burrage T, Rock D L. African swine fever virus structural protein p72 contains a conformational neutralizing epitope. Virology. 1994;201:413–418. doi: 10.1006/viro.1994.1311. [DOI] [PubMed] [Google Scholar]
- 8.Borca M V, Irusta P M, Kutish G F, Carrillo C, Afonso C L, Burrage T, Rock D L. A structural DNA-binding protein of African swine fever virus with similarity to bacterial histone-like proteins. Arch Virol. 1996;141:301–313. doi: 10.1007/BF01718401. [DOI] [PubMed] [Google Scholar]
- 9.Breese S S, Jr, DeBoer C J. Electron microscope observation of African swine fever virus in tissue culture cells. Virology. 1966;28:420–428. doi: 10.1016/0042-6822(66)90054-7. [DOI] [PubMed] [Google Scholar]
- 10.Brookes S M, Dixon L K, Parkhouse R M E. Assembly of African swine fever virus: quantitative ultrastructural analysis in vitro and in vivo. Virology. 1996;224:84–92. doi: 10.1006/viro.1996.0509. [DOI] [PubMed] [Google Scholar]
- 11.Brookes S M, Hyatt A D, Wise T, Parkhouse R M E. Intracellular virus DNA distribution and the acquisition of the nucleoprotein core during African swine fever virus particle assembly: ultrastructural in situ hybridisation and DNase-gold labelling. Virology. 1998;249:175–188. doi: 10.1006/viro.1998.9308. [DOI] [PubMed] [Google Scholar]
- 12.Carrascosa A L, del Val M, Santarén J F, Viñuela E. Purification and properties of African swine fever virus. J Virol. 1985;54:337–344. doi: 10.1128/jvi.54.2.337-344.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carrascosa A L, Sastre I, Viñuela E. African swine fever virus attachment protein. J Virol. 1991;65:2283–2289. doi: 10.1128/jvi.65.5.2283-2289.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carrascosa J L, Carazo J M, Carrascosa A L, García N, Santisteban A, Viñuela E. General morphology and capsid fine structure of African swine fever virus particles. Virology. 1984;132:160–172. doi: 10.1016/0042-6822(84)90100-4. [DOI] [PubMed] [Google Scholar]
- 15.Carrascosa J L, González P, Carrascosa A L, García-Barreno B, Enjuanes L, Viñuela E. Localization of structural proteins in African swine fever virus particles by immunoelectron microscopy. J Virol. 1986;58:377–384. doi: 10.1128/jvi.58.2.377-384.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carvalho Z G, Alves de Matos A P, Rodrigues-Pousada C. Association of African swine fever virus with the cytoskeleton. Virus Res. 1988;11:175–192. doi: 10.1016/0168-1702(88)90042-1. [DOI] [PubMed] [Google Scholar]
- 17.Cobbold C, Whittle J T, Wileman T. Involvement of the endoplasmic reticulum in the assembly and envelopment of African swine fever virus. J Virol. 1996;70:8382–8390. doi: 10.1128/jvi.70.12.8382-8390.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cobbold C, Brookes S M, Wileman T. Biochemical requirements of virus wrapping by the endoplasmic reticulum: involvement of ATP and endoplasmic reticulum calcium store during envelopment of African swine fever virus. J Virol. 2000;74:2151–2160. doi: 10.1128/jvi.74.5.2151-2160.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Costa J V. African swine fever virus. In: Darai G, editor. Molecular biology of iridoviruses. Boston, Mass: Kluwer Academic Publishers; 1990. pp. 247–270. [Google Scholar]
- 20.Cudmore S, Cossart P, Griffiths G, Way M. Actin-based motility of vaccinia virus. Nature. 1995;378:636–638. doi: 10.1038/378636a0. [DOI] [PubMed] [Google Scholar]
- 21.Dales S, Pogo B G T. Biology of poxviruses. Virol Monogr. 1981;18:54–64. doi: 10.1007/978-3-7091-8625-1. [DOI] [PubMed] [Google Scholar]
- 22.Dixon L K, Twigg S R F, Baylis S A, Vydelingum S, Bristow C, Hammond J M, Smith G L. Nucleotide sequence and variability of a 55 kbp region from the right end of the genome of a pathogenic African swine fever virus isolate (Malawi LIL20/1) J Gen Virol. 1994;75:1655–1684. doi: 10.1099/0022-1317-75-7-1655. [DOI] [PubMed] [Google Scholar]
- 23.Dixon L K, Costa J V, Escribano J M, Rock D L, Viñuela E, Wilkinson P J. The Asfarviridae. In: Van Regenmortel M H V, Fauquet C M, Bishop D H L, Carsten E B, Estes M K, Lemon S M, Maniloff J, Mayo M A, McGeoch D J, Pringle C R, Wickner R B, editors. Virus taxonomy. Seventh Report of the International Committee for the Taxonomy of Viruses. New York, N.Y: Academic Press; 2000. pp. 159–165. [Google Scholar]
- 24.Enjuanes L, Carrascosa A L, Moreno M A, Viñuela E. Titration of African swine fever virus. J Gen Virol. 1976;32:471–477. doi: 10.1099/0022-1317-32-3-471. [DOI] [PubMed] [Google Scholar]
- 25.Esteves A, Marques M I, Costa J V. Two-dimensional analysis of African swine fever virus proteins and proteins induced in infected cells. Virology. 1986;152:192–206. doi: 10.1016/0042-6822(86)90384-3. [DOI] [PubMed] [Google Scholar]
- 26.García R, Almazán F, Rodríguez J M, Alonso M, Viñuela E, Rodríguez J F. Vectors for the genetic manipulation of African swine fever virus. J Biotechnol. 1993;40:121–131. doi: 10.1016/0168-1656(95)00037-q. [DOI] [PubMed] [Google Scholar]
- 27.García-Escudero R, Andrés G, Almazán F, Viñuela E. Inducible gene expression from African swine fever virus recombinants: analysis of the major capsid protein p72. J Virol. 1998;72:3185–3195. doi: 10.1128/jvi.72.4.3185-3195.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gomez-Puertas P, Rodríguez F, Oviedo J M, Ramiro-Ibañez F, Ruiz-Gonzalvo F, Alonso C, Escribano J M. Neutralizing antibodies to different proteins of African swine fever virus inhibit both virus attachment and internalization. J Virol. 1996;70:5689–5694. doi: 10.1128/jvi.70.8.5689-5694.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.González A, Talavera A, Almendral J M, Viñuela E. Hairpin loop structure of African swine fever virus DNA. Nucleic Acids Res. 1986;14:6835–6844. doi: 10.1093/nar/14.17.6835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martínez-Pomares L, Simón-Mateo C, López-Otín C, Viñuela E. Characterization of the African swine fever virus structural protein p14.5: a DNA binding protein. Virology. 1997;229:201–211. doi: 10.1006/viro.1996.8434. [DOI] [PubMed] [Google Scholar]
- 31.Moura Nunes J F, Vigario J D, Terrinha A M. Ultrastructural study of African swine fever virus replication in cultures of swine bone marrow cells. Arch Virol. 1975;49:59–66. doi: 10.1007/BF02175596. [DOI] [PubMed] [Google Scholar]
- 32.Muñoz M, Freije J M P, Salas M L, Viñuela E, López-Otín C. Structure and expression in Escherichia coli of the gene coding for protein p10 of African swine fever virus. Arch Virol. 1993;130:93–107. doi: 10.1007/BF01318999. [DOI] [PubMed] [Google Scholar]
- 33.Payne L G. Significance of extracellular enveloped virus in the in vitro and in vivo dissemination of vaccinia. J Gen Virol. 1980;50:89–100. doi: 10.1099/0022-1317-50-1-89. [DOI] [PubMed] [Google Scholar]
- 34.Rodríguez F, Alcaraz C, Eiras A, Yáñez R J, Rodríguez J M, Alonso C, Rodríguez J F, Escribano J M. Characterization and molecular basis of heterogeneity of the African swine fever virus protein p54. J Virol. 1994;68:7244–7252. doi: 10.1128/jvi.68.11.7244-7252.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rodríguez J M, Almazán F, Viñuela E, Rodríguez J F. Genetic manipulation of African swine fever virus: construction of recombinant viruses expressing the β-galactosidase gene. Virology. 1992;188:239–250. doi: 10.1016/0042-6822(92)90735-8. [DOI] [PubMed] [Google Scholar]
- 36.Rodríguez J M, Almazán F, Yáñez R J, García R, Viñuela E, Rodríguez J F. African swine fever virus membrane-associated and secreted proteins. In: McFadden G, editor. Viroceptors, virokines and related immune modulators encoded by DNA viruses. R. G. Austin, Tex: Landes Company; 1995. pp. 187–200. [Google Scholar]
- 37.Rojo G, Chamorro M, Salas M L, Viñuela E, Cuezva J M, Salas J. Migration of mitochondria to viral assembly sites in African swine fever virus-infected cells. J Virol. 1998;72:7583–7588. doi: 10.1128/jvi.72.9.7583-7588.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roullier I, Brookes S M, Hyatt A D, Windsor M, Wileman T. African swine fever virus is wrapped by the endoplasmic reticulum. J Virol. 1998;72:2373–2387. doi: 10.1128/jvi.72.3.2373-2387.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Salas J, Salas M L, Viñuela E. African swine fever virus: a missing link between poxviruses and iridoviruses? In: Domingo E, Webster R G, Holland J J, editors. Origin and evolution of viruses. London, England: Academic Press; 1999. pp. 467–480. [Google Scholar]
- 40.Sanderson C M, Hollinshead M, Smith G L. The vaccinia virus A27L protein is needed for the microtubule-dependent transport of intracellular mature particles. J Gen Virol. 2000;81:47–58. doi: 10.1099/0022-1317-81-1-47. [DOI] [PubMed] [Google Scholar]
- 41.Sanz A, García-Barreno B, Nogal M L, Viñuela E, Enjuanes L. Monoclonal antibodies specific for African swine fever virus proteins. J Virol. 1985;54:199–206. doi: 10.1128/jvi.54.1.199-206.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schloer G M. Polypeptides and structure of African swine fever virus. Virus Res. 1985;3:295–310. doi: 10.1016/0168-1702(85)90431-9. [DOI] [PubMed] [Google Scholar]
- 43.Simón-Mateo C, Andrés G, Viñuela E. Polyprotein processing in African swine fever virus: a novel gene expression strategy for a DNA virus. EMBO J. 1993;12:2977–2987. doi: 10.1002/j.1460-2075.1993.tb05960.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Simón-Mateo C, Andrés G, Almazán F, Viñuela E. Proteolytic processing in African swine fever virus: evidence for a new structural polyprotein, pp62. J Virol. 1997;71:5799–5804. doi: 10.1128/jvi.71.8.5799-5804.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sodeik B, Doms R W, Ericsson M, Hiller G, Machamer C E, van't Hof W, van Meer G, Moss B, Griffiths G. Assembly of vaccinia virus: role of the intermediate compartment between the endoplasmic reticulum and the Golgi stacks. J Cell Biol. 1993;121:521–541. doi: 10.1083/jcb.121.3.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sodeik B, Ebersold M W, Helenius A. Microtubule-mediated transport of incoming herpes simplex virus 1 capsids to the nucleus. J Cell Biol. 1997;136:1007–1021. doi: 10.1083/jcb.136.5.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sogo J M, Almendral J M, Talavera A, Viñuela E. Terminal and internal inverted repetitions in African swine fever virus DNA. Virology. 1984;133:271–275. doi: 10.1016/0042-6822(84)90394-5. [DOI] [PubMed] [Google Scholar]
- 48.Suomalainen M, Nakano M Y, Keller S, Boucke K, Stidwill R P, Greber U F. Microtubule-dependent plus- and minus end-directed motilities are competing processes for nuclear targeting of adenovirus. J Cell Biol. 1999;144:657–672. doi: 10.1083/jcb.144.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Viñuela E. Molecular biology of African swine fever virus. In: Becker Y, editor. African swine fever. Boston, Mass: Nijhoff; 1987. pp. 31–49. [Google Scholar]
- 50.Yáñez R J, Rodríguez J M, Nogal M L, Yuste L, Enriquez C, Rodríguez J F, Viñuela E. Analysis of the complete nucleotide sequence of African swine fever virus. Virology. 1995;208:249–278. doi: 10.1006/viro.1995.1149. [DOI] [PubMed] [Google Scholar]