Summary
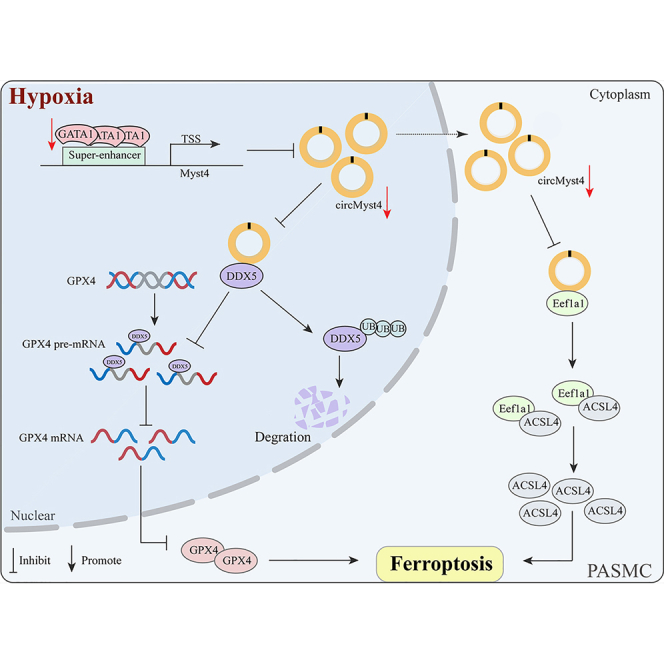
The abnormal expression of circular RNAs (circRNAs) is emerging as a critical cause in regulation of pathological changes of hypoxic pulmonary hypertension (PH), in which ferroptosis is a new pathological change reported recently. However, how circRNAs regulate ferroptosis remains unclear. Here, we proved a significant decrease in circMyst4 expression in hypoxia. In vitro assays revealed that circMyst4 alleviated hypoxic pulmonary artery smooth muscle cell (PASMC) ferroptosis through directly combing with DDX5 in the nucleus to promote GPX4 mRNA processing and inhibiting the formation of the Eef1a1/ACSL4 complex in the cytoplasm. Additionally, superenhancer (SE) was verified to drive the generation of circMyst4. In vivo assays revealed that circMyst4 inhibited the progression of hypoxic PH. Overall, SE-driven circMyst4 may be a new potential therapeutic target for mediating PASMC ferroptosis through promoting DDX5-regulated GPX4 mRNA processing and inhibiting the binding between Eef1a1 and ACSL4.
Subject areas: Nucleic acids, Molecular interaction, Cell biology
Graphical abstract
Highlights
-
•
CircMyst4 is significantly downregulated in hypoxic PH
-
•
CircMyst4 is involved in PASMC ferroptosis and acts as a key regulator
-
•
SE-driven circMyst4 promotes DDX5-regulated GPX4 mRNA processing in the nucleus
-
•
SE-driven circMyst4 inhibits the binding between Eef1a1 and ACSL4 in the cytoplasm
Nucleic acids; Molecular interaction; Cell biology
Introduction
Pulmonary hypertension (PH) is a high morbidity and mortality cardiopulmonary disease associated with a progressive increase in mean pulmonary artery pressure and right ventricular dysfunction, and is characterized by abnormal vasoconstriction and structural vascular remodeling.1,2 Pathological changes in the pulmonary artery smooth muscle cell (PASMC) phenotype are critical factors in pulmonary vascular remodeling. Abnormal cell phenotypes include apoptosis, proliferation, autophagy, pyroptosis, and fibrosis.2,3,4,5 A recent study reported that ferroptosis contributes to the progression of MCT-induced PH.6 However, the role and precise molecular mechanisms of ferroptosis in the pathogenesis of PH are still poorly understood.
Ferroptosis is a form of regulated cell death characterized by the iron-dependent accumulation of lipid hydroperoxides to lethal levels.7 As a key regulatory factor of ferroptosis, when the expression of glutathione peroxidase 4 (GPX4) becomes disturbed, free toxic lipid hydroperoxides cannot be effectively eliminated, causing lipid peroxidation, compromising the integrity of the cell membrane, and finally resulting in ferroptosis.7,8,9 In addition, acyl-CoA synthase, a long-chain family member 4 (ACSL4), as an enzyme involved in the activation of unsaturated fatty acids (PUFAs), can also lead to lipid peroxidation in cells and induce ferroptosis.10 It has been reported that ferroptosis is involved in the pathogenesis of several diseases. For example, nuclear receptor coactivator 4 (NCOA4) is upregulated, and markedly promotes ferritinophagy caused by ischemia/reperfusion injury to accelerate ferroptosis and participate in ischemic stroke.11 YL-939 (a new ferroptosis inhibitor) substantially ameliorated liver damage in a ferroptosis-related acute liver injury model by targeting the prohibitin 2/ferritin/iron axis.12 As many factors and complex mechanisms are involved in ferroptosis, the key factor mediating the process of ferroptosis urgently needs to be discovered.
Circular RNAs (circRNAs) are a unique class of RNAs with covalently closed loop structures that are produced by back-splicing of protein-coding gene exons or introns in eukaryotic cells with a cell- or tissue-specific expression pattern as well as high stability.13,14 CircRNAs are essential for diverse regulatory mechanisms, including competing endogenous RNAs (ceRNAs), transcriptional regulation, protein interactions, and translational regulation.15,16,17,18 Initially, circRNAs are widely investigated for their role as miRNA sponges. Subsequently, they also are proven to affect the structure and function of proteins. Additionally, it has also been found that EIciRNAs enhance RNA polymerase II-mediated transcription by binding to U1 small nuclear ribonucleoproteins, whereas circURI1 modulates alternative splicing of migration-related genes through its interaction with hnRNPM. Recently, a small fraction of endogenous circRNAs containing open reading frames have been also demonstrated to directly undergo translation into peptides or proteins.19 Mounting evidence indicates that circRNAs participate in various biological functions related to different diseases, such as immunity, metabolism, and cell proliferation.20 Indeed, our previous reports,21,22 as well as those of others,23,24 have demonstrated that circRNAs have act as miRNA molecular sponges to regulate PASMC proliferation and pyroptosis. These studies provide compelling evidence for the critical functionality of circRNAs in pulmonary vascular remodeling. However, the role of circRNAs in the context of PH is yet to be fully examined.
In this study, we aimed to screen to a key circRNA capable of serving as a diagnostic and therapeutic target for PH and to clarify its regulatory mechanism in PASMC ferroptosis. A novel circRNA, mmu_circ_0000505 (named circMyst4), was screened by bioinformatics analysis, and was demonstrated that it was significantly downregulated and caused ferroptosis in hypoxic PASMCs. The mechanism of action of circMyst4 was further analyzed, and it was found that in the nucleus, circMyst4 bound to the DEAD box helicase 5 (DDX5) protein and reduced splicing of GPX4 pre-mRNA; in the cytoplasm, circMyst4 acted as a protein sponge for Eef1a1 and inhibited the binding between it and its downstream target ACSL4, which enhanced the regulation of ferroptosis in PH. We also demonstrated that circMyst4 could alleviate the pathological changes resulting from hypoxia-induced PH in vivo. Overall, we identified circMyst4 as a suppressive circRNA in PH, offering a new therapeutic target for this disease.
Results
CircMyst4 is downregulated in hypoxia
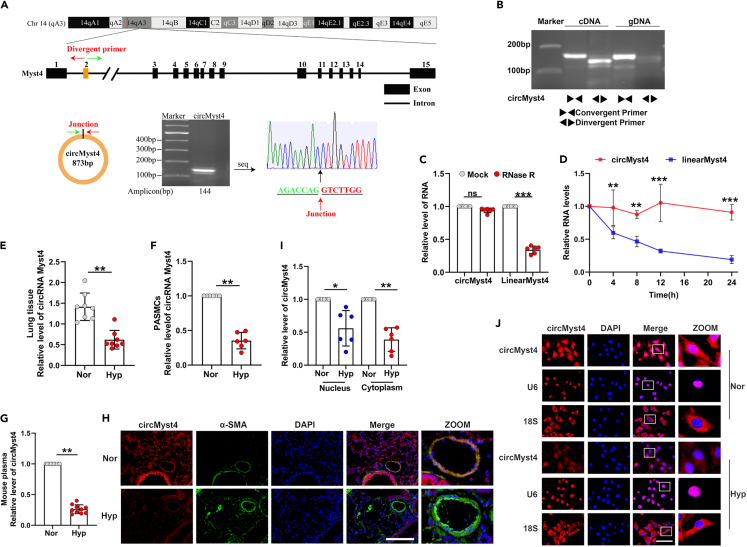
Mining of the circRNA databases including circBase (circBase: http://www.circbase.org/), CIRCpedia v2 (CIRCpedia v2: http://sea.edbc.org/) and circBank (circBank: http://www.circbank.cn/) screened 45 circRNAs having coding potential in lung tissues, with 9 found to be associated with superenhancer through database dbSUPER (dbSUPER: https://asntech.org/dbsuper/) and SEAv.3.0 (SEAv.3.0: http://sea.edbc.org) database (Figure S1A). Next, designed head-to-tail junction-specific primers to analyzed the expression of these circRNAs using quantitative real-time PCR (real-time qPCR) (Figure S1B). Considering the relative expression level and conservatism, we chose mmu_circ_0000505 (named as circMyst4) as the target for next study. Sanger sequencing results showed that circMyst4 was generated from exon 2 of the Myst4 gene with a length of 873 nt and located on mouse chromosome 14: 22335847–22336720 (Figure 1A). CircMyst4 was amplified by divergent primers in cDNA but not by divergent primers in genomic DNA (gDNA) (Figure 1B). Resistance to RNase R exonuclease digestion confirmed that circMyst4 harbors a closed loop structure (Figure 1C), and circMyst4 was more stable in PASMCs than linear Myst4 mRNA (Figure 1D).
Figure 1.
CircMyst4 is downregulated in PH
(A) Genomic location of circRNA Myst4 (circMyst4). CircMyst4 was formed by the back-splicing of exons 2 of Myst4. And the back-splicing junction site of circMyst4 was identified by Sanger sequencing.
(B) Real-time qPCR analysis of genomic DNA (gDNA) and cDNA using divergent and convergent primers of circMyst4 (n = 6).
(C) RNase R degradation analysis was performed to detect the stability of circMyst4 and linear Myst4 mRNA (n = 6).
(D) Real-time qPCR analysis was conducted to detect the amount of circMyst4 and linear Myst4 mRNA in PASMCs after actinomycin (D) treatment (n = 6).
(E and F) Real-time qPCR analysis of circMyst4 expression levels in lung tissues of hypoxic PH mouse (n = 8 mice/group) and hypoxic PASMCs (n = 6).
(G) Real-time qPCR analysis of circMyst4 expression levels in plasma of hypoxic PH mouse and normoxic mouse (n = 10).
(H) Fluorescence in situ hybridization analysis of circMyst4 location in lung tissues of mouse. CircMyst4 probes were labeled with Cy3 (red). Nucleus were stained with DAPI (blue), pulmonary smooth muscle stained with α-SMA (green). Scale bar, 100 μm (n = 6 mice/group).
(I) Real-time qPCR analysis was using to determine the circMyst4 expression in the nucleus and cytoplasm of PASMCs after exposure to hypoxia for 24 h (n = 6).
(J) RNA FISH analysis of the subcellular localization of circMyst4 in PASMCs. CircMyst4 probes were labeled with Cy3 (red). Nucleus were stained with DAPI (blue). U6 and 18S RNA, used as internal references, were labeled with Cy3 (red). Scale bar, 100 μm (n = 6). Data are shown as means ± SD. Statistical analysis was performed with Student’s t test. Hyp, hypoxia; Nor, normoxia; ns, not significantly different. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
We next examined the expression of circMyst4 in hypoxic PASMCs and lung tissues by real-time qPCR. The results showed that circMyst4 expression was decreased in hypoxia compared to normoxia, and circMyst4 was enriched in the lung tissue of mice compared to other tissues (Figures 1E, 1F, and S1C). The expression of circMyst4 was not significantly different in pulmonary artery endothelial cells (Figure S1D). Additionally, we also found that the content of circMyst4 in the plasma of PH mice was significantly lower than that of normal mice (Figure 1G). CircMyst4 colocalized with α-smooth muscle actin (α-SMA, a smooth muscle marker), suggesting that circMyst4 was expressed in pulmonary artery smooth muscle (Figure 1H). Furthermore, fluorescence in situ hybridization (FISH) and real-time qPCR analysis of nuclear and cytoplasmic RNA revealed that circMyst4 was localized in both the cytoplasm and the nucleus and was markedly downregulated under hypoxia (Figures 1I and 1J). Together, these data suggest that circMyst4 is a bona fide circRNA and is downregulated in PH.
CircMyst4 overexpression reversed hypoxia-induced PH in vivo
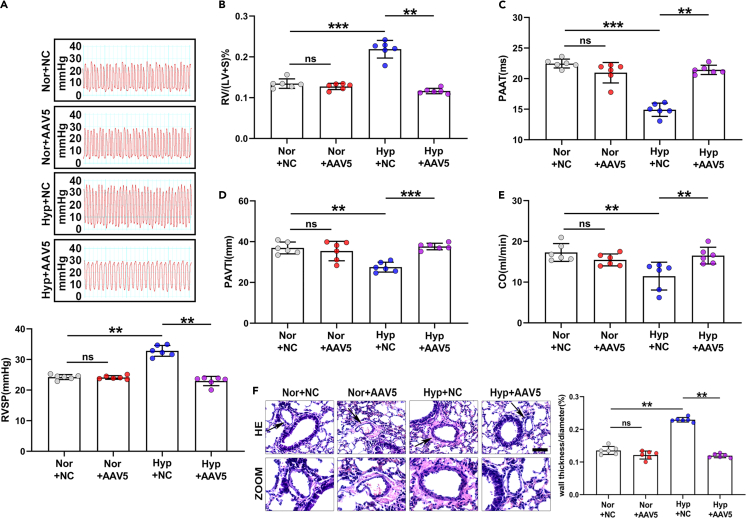
To investigate the function of circMyst4 in PH, mice with hypoxia were administered an adeno-associated virus vector 5 (AAV5) to overexpress circMyst4 (AAV5-circMyst4). The results showed that the abundance of circMyst4 was markedly increased by AAV5-circMyst4 in lung tissues (Figures S2A–S2C). Then we evaluated parameters related to PH, including right ventricular systolic pressure (RVSP), hemodynamics, cardiac function, and vascular remodeling. CircMyst4 overexpression alleviated the augmented RVSP and RV/(LV + S) induced by hypoxia (Figures 2A and 2B). Echocardiography showed that hypoxic mice exhibited significantly shortened pulmonary artery acceleration time (PAAT), lowered pulmonary artery velocity time integral (PAVTI), and reduced cardiac output (CO), which was reversed by AAV5-circMyst4, while did not affect the body weight of the model mice (Figures 2C–2E and S2D). We also confirmed that circMyst4 overexpression reduced hypoxia-induced pulmonary vascular wall thickening (Figure 2F). Additionally, circMyst4 overexpression significantly inhibited ferroptosis in vivo (Figure S2E–S2H). These results indicate that circMyst4 attenuates hypoxia-induced PH in hypoxic mouse models.
Figure 2.
Overexpression of circMyst4 in vivo alleviated the symptoms of hypoxia-mediated pulmonary hypertension
(A and B) Right ventricular systolic pressure (RVSP), and right ventricle (RV)/left ventricle (LV)+S weight ratio in the Nor+NC, Nor+AAV5, Hyp+NC, and Hyp+AAV5 groups are shown (n = 6 mice/group).
(C–E) Echocardiographic analysis of pulmonary artery acceleration time (PAAT), pulmonary artery velocity time integral (PAVTI) and cardiac output (CO) in the different groups (n = 6 mice/group).
(F) Morphological analysis of the pulmonary artery was performed using HE staining (n = 6 mice/group). Scale bar, 50 μm. Data are shown as means ± SD. Statistical analysis was performed with one-way ANOVA followed by Bonferroni correction. Hyp, hypoxia; Nor, normoxia; NC, negative control; AAV5, serotype 5 adenovirus-associated virus carrying circMyst4; ns, not significantly different. ∗∗p < 0.01, ∗∗∗p < 0.001.
CircMyst4 overexpression attenuates hypoxia-induced ferroptosis
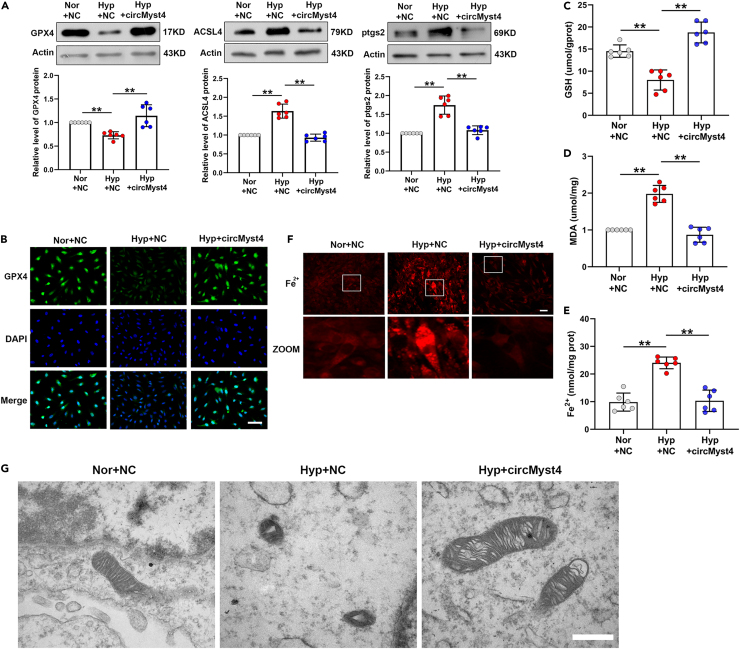
To further explore the biological roles of circMyst4, we predicted genes related to circMyst4 through the catRAPID database, performed KEGG pathway analysis, and found that these genes were concentrated in ferroptosis (Figure S3A). CircMyst4 can be overexpressed via a specific plasmid in cultured PASMCs (Figure S3B). CircMyst4 overexpression had no effect on the mRNA and protein expression of linear Myst4 (Figures S3C–S3E). The results of western blotting and immunofluorescence staining indicated that the protein levels of GPX4 were increased, while those of ptgs2 and ACSL4 were decreased in circMyst4-overexpressing PASMCs (Figures 3A and 3B). CircMyst4 overexpression reduced GSH depletion, MDA release and ferrous ion accumulation in hypoxic PASMCs (Figures 3C–3F). Moreover, increased mitochondrial membrane density and decreased mitochondrial cristae were observed under hypoxia, and circMyst4 overexpression minimized this effect (Figure 3G).
Figure 3.
CircMyst4 overexpression attenuates PASMC ferroptosis induced by hypoxia
(A) Western blotting analysis of the protein levels of GPX4, ACSL4 and ptgs2 in PASMCs (n = 6).
(B) Immunofluorescence analysis of GPX4 expression. Scale bars, 100 μm (n = 6). Green color denotes GPX4, stained with FITC, and blue color denotes nucleus, stained with DAPI.
(C–E) Detection of the GSH depletion, MDA release and ferrous ion accumulation in PASMCs (n = 6).
(F) Detection of the percentage of ferrous ion staining-positive cells by a specific probe of ferrous ion (red). Scale bars, 100 μm (n = 6).
(G) Representative electron micrograph of PASMCs in different groups. Asterisks represent mitochondrion. Scale bars, 0.5μm (n = 3). Data are shown as means ± SD. Statistical analysis of the graph (A) and (D) was performed with one-way ANOVA followed by Dunn’s post-test. Statistical analysis of the graph (C) and (E) was performed with one-way ANOVA followed by Bonferroni correction. Hyp, hypoxia; Nor, normoxia; NC, negative control. ∗∗p < 0.01.
Subsequently, we designed siRNA to knockdown the expression of circMyst4 in the cytoplasm and antisense oligonucleotide (ASO) to knockdown the expression of circMyst4 in the nucleus (Figures S4A–S4C). Western blotting results showed that circMyst4 knockdown markedly decreased the protein levels of GPX4 and increased the expression of ptgs2 and ACSL4 (Figures S4D and S4E). CircMyst4 knockdown promoted GSH depletion, MDA release and ferrous ion accumulation in PASMCs (Figures S4F–S4I). Meanwhile, we also found that the overexpression of circMyst4 inhibited ferroptosis in normoxic PASMCs (Figures S4J–S4M). In addition to this, we also found that deficiency of circMyst4 promoted the expression of inflammatory factors IL-18 and IL-1β, and did not affect PASMC proliferation (Figures S4N–S4P). Previous studies have confirmed that ferroptosis was closely related to inflammation. Taken together, these results indicate that circMyst4 plays suppressive roles in hypoxia-induced ferroptosis.
The association of circMyst4 with DDX5 in nucleus
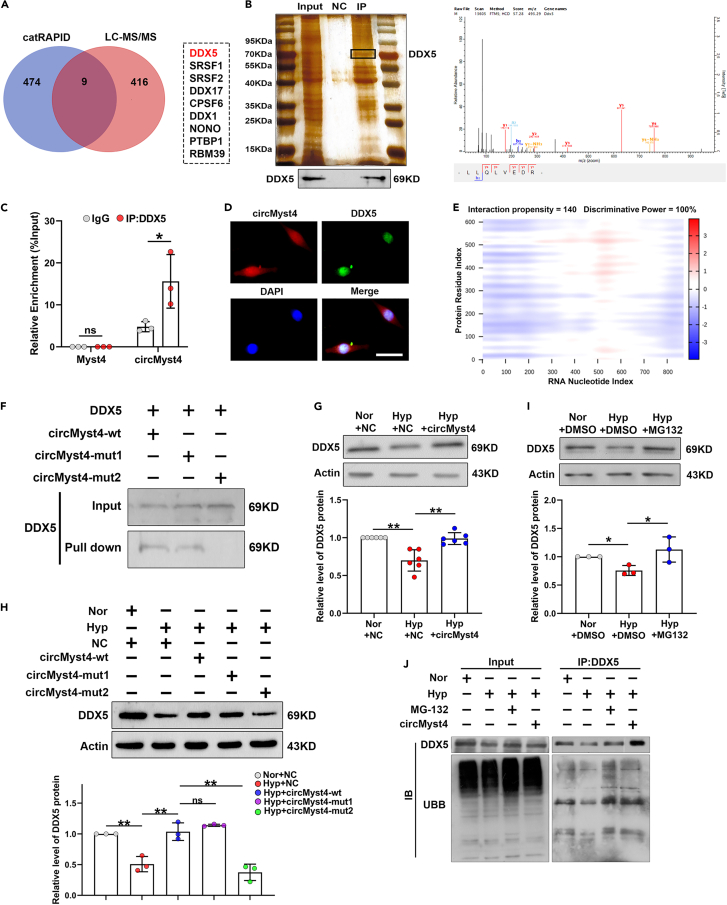
Our experiments confirmed that circMyst4 does not have the ability to encode peptides and proteins (Figure S5A). To test whether circMyst4 exerts its function by interacting with proteins, we conducted RNA pull-down assays and mass spectrometry analyses to detect the specific proteins bound with circMyst4. Nine potential circMyst4-interacting proteins were identified through comprehensive analysis (Figure 4A). Among them, we focused on DDX5 because it had the highest score in the mass spectrometry analyses. We then utilized RNA pull-down and western blotting to verify the interaction between circMyst4 and DDX5 protein (Figure 4B). Additionally, RIP assay results also indicated that circMyst4 was enriched in RNA coprecipitated by anti-DDX5 antibody in PASMCs, while linear Myst4 mRNA could not be enriched (Figure 4C). FISH fluorescence showed that circMyst4 and DDX5 colocalized in the nucleus (Figure 4D).
Figure 4.
The association of circMyst4 with DDX5
(A) catRAPID databases and Mass spectrometry analysis of the binding protein of circMyst4.
(B) RNA pull-down and western blotting analysis were used to identify proteins associated with circMyst4 (left), and mass spectrometry of specific segments of DDX5 (right).
(C) RIP-PCR analysis was performed to prove the association of circMyst4 to DDX5 protein (n = 3).
(D) The colocalization of circMyst4 and DDX5 in PASMCs. Scale bars, 100 μm (n = 3). Green color denotes DDX5, stained with FITC, and blue color denotes nucleus, stained with DAPI, whereas circMyst4 probes were labeled with Cy3 (red).
(E) Predicted interaction of circMyst4 (nucleotide positions 326-377nt and 701-752nt) and DDX5 protein.
(F) RNA pull-down and western blotting analysis were used to identify the binding site of DDX5 to circMyst4 (mut1, circMyst4 nucleotide positions 326-377nt; mut2, circMyst4 nucleotide positions 701-752nt).
(G and H) Detection of the level of DDX5 protein in PASMCs treated with overexpressing circMyst4 wild type (circMyst4-wt) or circMyst4 mutant type (circMyst4-mut) (n = 6, 3).
(I) Western blotting analysis of DDX5 in PASMCs treated with MG-132 (an inhibitor of ubiquitin protease system) (n = 3).
(J) Co-immunoprecipitation (CoIP) analysis was performed using anti-DDX5, followed by probing with anti-UBB in PASMCs transfected with circMyst4 overexpressing plasmid or treated with MG-132 (n = 3). Data are shown as means ± SD. Statistical analysis of the graph (C) was performed with Student’s t test. Statistical analysis of the graph (G, H, and I) was performed with one-way ANOVA followed by Dunn’s post-test. Hyp, hypoxia; Nor, normoxia; ns, not significantly different. ∗p < 0.05, ∗∗p < 0.01.
We further predicted the binding ability between circMyst4 and DDX5 through the catRAPID database, and the prediction results showed that nucleotides 326 to 377 and 701 to 752 of circMyst4 bound to DDX5 with an interaction propensity of 140 (Figures 4E and S5B). Subsequently, two mutant vector plasmids were constructed based on the previous binding sites, and we found that circMyst4-mut2 (701–752) remarkably reduced interactions between DDX5 and circMyst4 (Figure 4F). In addition, we found that DDX5 was significantly reduced under hypoxia and was mainly localized in the nucleus (Figures S5C–S5E). Western blotting analysis indicated that circMyst4 overexpression markedly increased the protein level of DDX5 in hypoxic PASMCs, and then circMyst4-mut2 (701–752) had no effect on DDX5 expression (Figures 4G and 4H). Correlation analysis showed that the expression of DDX5 was positively correlated with circMyst4 levels (Figure S5F). It has been reported that the ubiquitin proteasome pathway participates in most protein degradation.25 We predicted that DDX5 had multiple ubiquitin modifying sites through Protein Lysine Modifications Database (Figure S5G). Western blotting analysis showed that the reduction in DDX5 induced by hypoxia was rescued after treatment with MG-132 (a ubiquitin protease system inhibitor) (Figure 4I). Additionally, ubiquitinated DDX5 was detected in PASMCs, while MG-132 treatment and circMyst4 overexpression resulted in the accumulation of ubiquitin protein (Figure 4J). Together, these results demonstrate a specific association between the nucleotides 701–752 of circMyst4 and DDX5.
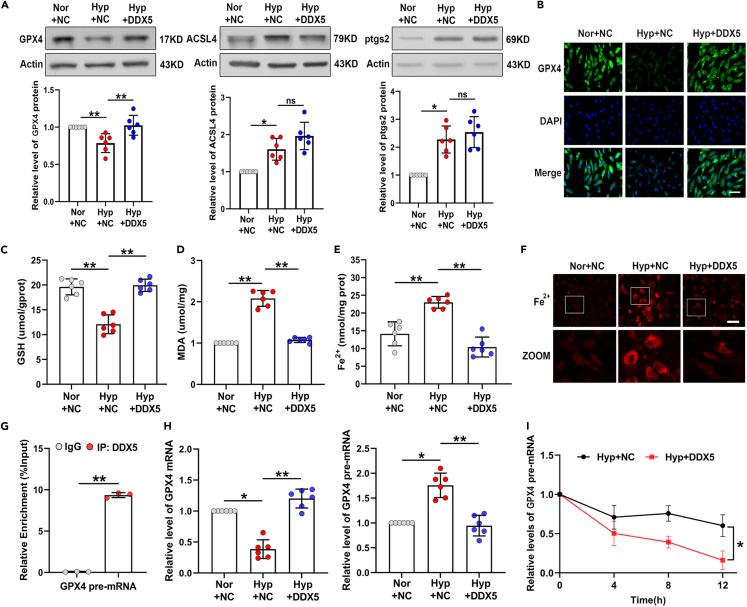
CircMyst4/DDX5 regulates ferroptosis by promoting GPX4 mRNA processing in the nucleus of PASMCs
To clarify whether DDX5 plays a role in hypoxia-induced ferroptosis, PASMCs were transfected with a DDX5 overexpression plasmid, and the efficiency of overexpression DDX5 in nucleus was verified (Figure S6A). DDX5 overexpression increased the expression of GPX4 upon hypoxic stimulation. However, DDX5 overexpression had no effect on the levels of ptgs2 and ACSL4 (Figures 5A and 5B). We further observed that overexpression of DDX5 rescued GSH depletion, and reduced MDA release and ferrous ion accumulation than hypoxic group (Figures 5C–5F). The results showed that DDX5 regulated hypoxia-induced ferroptosis in PASMCs.
Figure 5.
DDX5 attenuates hypoxia-induced ferroptosis through promoting GPX4 mRNA processing
(A) Western blotting analysis of the protein levels of GPX4, ACSL4, and ptgs2 in PASMCs transfected with DDX5 plasmid (n = 6).
(B) Immunofluorescence analysis of GPX4 expression. Scale bars, 100 μm (n = 6). Green color denotes GPX4, stained with FITC, and blue color denotes nucleus, stained with DAPI.
(C–E) Detection of the GSH depletion, MDA release and ferrous ion accumulation in PASMCs overexpressed DDX5 (n = 6).
(F) Detection of the percentage of ferrous ion staining-positive cells by a specific probe of ferrous ion (red). Scale bars, 100 μm (n = 6).
(G) RIP-PCR analysis of the association of GPX4 pre-mRNA to DDX5 protein (n = 3).
(H) After transfection with DDX5 plasmid, real-time qPCR analysis of the level of GPX4 mRNA and GPX4 pre-mRNA (n = 6).
(I) After Actinomycin D treatment, real-time qPCR analysis of the impact of DDX5 overexpression on the stability of GPX4 pre-mRNA (n = 3). Data are shown as means ± SD. Statistical analysis of the graph (A, D, and H) was performed with one-way ANOVA followed by Dunn’s post-test. Statistical analysis of the graph (C) and (E) was performed with one-way ANOVA followed by Bonferroni correction. Statistical analysis of the graph (G) and (I) was performed with Student’s t test. Hyp, hypoxia; Nor, normoxia; NC, negative control, ns, not significantly different. ∗p < 0.05, ∗∗p < 0.01.
Studies have shown that nucleus-retained circRNAs can perform their functions in regulating splicing by interacting with proteins.20,26 DDX5 has been reported to be involved in precursor messenger RNA processing or alternative splicing.27,28 Therefore, we speculated that DDX5 might mediate the splicing process of GPX4 pre-mRNA. We performed correlation analysis and found that DDX5 was positively correlated with GPX4 mRNA, and negatively correlated with GPX4 pre-mRNA (Figure S6B). RIP experiments demonstrated that DDX5 binds to GPX4 pre-mRNA (Figure 5G). Subsequently, we found that DDX5 overexpression increased GPX4 mRNA levels and decreased GPX4 pre-mRNA levels compared with the hypoxia group (Figure 5H). Moreover, after treating PASMCs with actinomycin D, the real-time qPCR results showed that DDX5 overexpression significantly reduced the stability of the GPX4 pre-mRNA (Figure 5I). Consistent with these results, circMyst4 overexpression increased GPX4 mRNA levels and decreased GPX4 pre-mRNA levels (Figure S6C). After treating PASMCs with actinomycin D, circMyst4 overexpression significantly reduced the stability of the GPX4 pre-mRNA (Figure S6D). But circMyst4-mut2 overexpression significantly increased the stability of the GPX4 pre-mRNA (Figure S6E). Then we also performed rescue experiments by overexpressing circMyst4 while disrupt DDX5, after treating PASMCs with actinomycin D, the results showed that increased the stability of the GPX4 pre-mRNA after interference with DDX5 relative to the circMyst4 overexpression group (Figure S6F). Additionally, we also measure GPX4 pre-mRNA splicing activity in the presence or absence of circMyst4. The results found that knocking down circMyst4 increased the stability of the GPX4 pre-mRNA (Figure S6G). These results suggested that circMyst4 promoted GPX4 mRNA processing by combining with DDX5.
To further verify that circMyst4 regulates hypoxia-induced ferroptosis through DDX5, we constructed DDX5 interfering RNA. The interference efficiency of DDX5 was detected (Figures S7A–S7C). Next, western blotting and immunofluorescence staining results showed that compared with the circMyst4 overexpression group, knockdown of DDX5 significantly reduced GPX4 expression, but had no significant effect on ACSL4 and ptgs2 expression (Figures S7D and S7E). DDX5 knockdown resulted in GSH depletion, MDA release and ferrous ion accumulation (Figures S7F–S7I). These results suggest that circMyst4 regulates ferroptosis via the DDX5/GPX4 axis under hypoxic conditions.
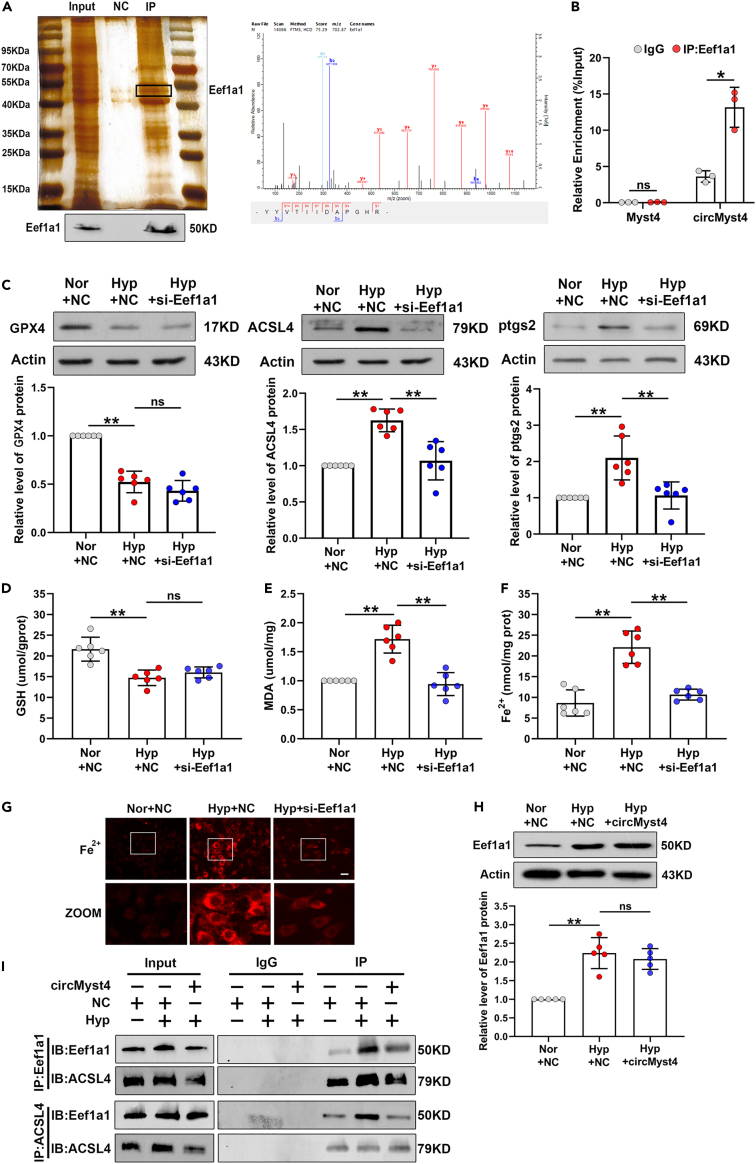
CircMyst4 regulates ferroptosis by suppressing the formation of the Eef1a1/ACSL4 complex in the cytoplasm of PASMCs
We then explored the potential mechanism of circMyst4 in the cytoplasm. Based on the mass spectrometry results, we screened for Eef1a1. We utilized RNA pull-down and western blotting to verify the interaction between circMyst4 and the Eef1a1 protein (Figure 6A). Additionally, RIP assay results indicated that circMyst4 was enriched in RNA coprecipitated by anti-Eef1a1 antibody in PASMCs, while linear Myst4 mRNA could not be enriched (Figure 6B). Subsequently, we found that Eef1a1 expression was significantly increased in hypoxia and mainly localized in the cytoplasm (Figures S8A and S8B). We constructed small interfering RNA to knockdown Eef1a1, and the transfection specificity and efficiency were evaluated (Figures S8C–S8E). Eef1a1 knockdown decreased the expression levels of ACSL4 and ptgs2 upon hypoxia but did not significantly affect the expression of GPX4 (Figure 6C). We further observed that knockdown of Eef1a1 reduced MDA release but did not rescue GSH depletion (Figures 6D and 6E). Silencing Eef1a1 diminished the accumulation of ferrous ion induced by hypoxia (Figures 6F and 6G). Moreover, western blotting indicated that circMyst4 overexpression had no effect on the protein level of Eef1a1 in hypoxic PAMSCs (Figure 6H). However, we observed that circMyst4 overexpression could inhibit the binding between Eef1a1 and ACSL4 (Figure 6I). The results show that circMyst4 may regulate hypoxia-induced ferroptosis by suppressing the interaction between Eef1a1 and ACSL4 in PASMCs.
Figure 6.
Silencing Eef1a1 attenuates hypoxia-induced ferroptosis
(A) RNA pull-down and western blotting analysis were used to identify proteins associated with circMyst4 (left), and mass spectrometry of specific segments of Eef1a1 (right).
(B) RIP-PCR analysis of the association of circMyst4 to Eef1a1 protein (n = 3).
(C) Western blotting analysis of effects of silencing Eef1a1 on the protein levels of GPX4, ACSL4 and ptgs2 in hypoxic PASMCs (n = 6).
(D–F) The GSH, MDA and ferrous ion analysis of the effects of Eef1a1 on cell ferroptosis (n = 6).
(G) The percentage of ferrous ion-positive cells were detected after transfection with Eef1a1 siRNA in hypoxia-treated PASMCs. Scale bars, 100 μm. Images of fluorescence staining with a ferrous ion probe (red).
(H) Western blotting analysis of effects of circMyst4 overexpression on the protein levels of Eef1a1 in hypoxic PASMCs (n = 5).
(I) CoIP analysis of the binding of Eef1a1 with ACSL4 after circMyst4 overexpression in hypoxic PASMCs. Data are shown as means ± SD. Statistical analysis of the graph B was performed with Student’s t test. Statistical analysis of the graph (C, E, and H) was performed with one-way ANOVA followed by Dunn’s post-test. Statistical analysis of the graph (D) and (F) was performed with one-way ANOVA followed by Bonferroni correction. Hyp, hypoxia; Nor, normoxia; NC, negative control, ns, not significantly different. ∗p < 0.05, ∗∗p < 0.01.
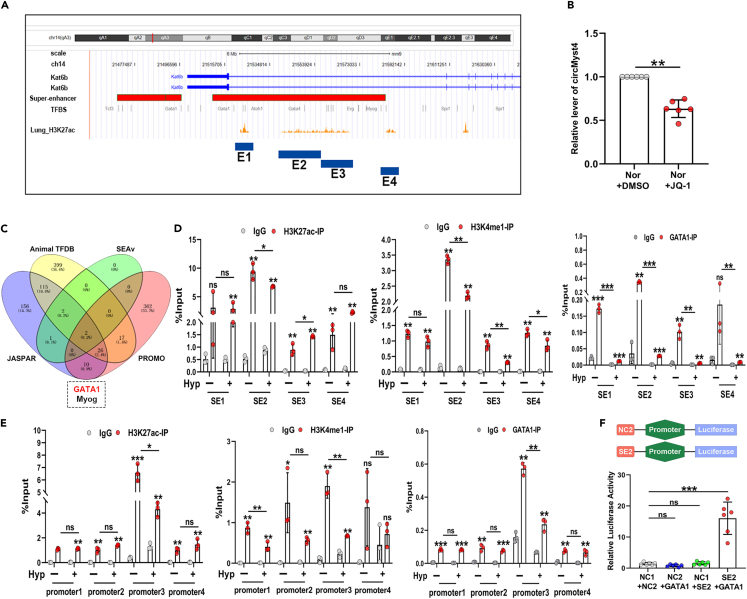
GATA1 controls circMyst4 expression by binding to the superenhancer at the Myst4 locus
Superenhancers (SEs) consist of a cluster of many enhancers bound to a great number of transcription factors and have previously been reported to regulate the expression of circRNAs.29,30 Therefore, we predicted whether SE exists at the Myst4 (also called kat6b) locus via the SEAv.3.0 database and found that the Myst4 locus was enriched with acetyl H3K27 (H3K27ac) signal peaks (a superenhancer mark) (Figure 7A). We also confirmed that JQ-1 (a superenhancer inhibitor) could inhibit the expression of circMyst4 (Figure 7B). We therefore screened two transcription factors that could bind to both the SE and promoter regions of Myst4 via superenhancer databases (SEAv) and promoter databases (JASPAR, AnimalTFDB, and PROMO) (Figure 7C). Only GATA1 was downregulated under hypoxic conditions (Figures S9A and S9B). Then, we performed a coimmunoprecipitation (CoIP) assay and found that GATA1 could interact with both H3K27ac and H3K4me1 (Figure S9C). SEs often form long-range chromatin loops with the promoters of target genes to control gene expression.31 To investigate the specific formation of chromatin loops, we next divided the SE region of Myst4 into four constituents (E1-E4) and confirmed that GATA1, H3K27ac, and H3K4me1 bound to the E2 segment by ChIP-qPCR in PASMCs (Figure 7D). Subsequently, we divided the Myst4 promoter equally into four segments (−100–2000, P1-P4) according to the distribution of binding sites between GATA1 and the Myst4 promoter (Figure S9D). Using ChIP-qPCR assays, we confirmed that GATA1, H3K27ac, and H3K4me1 bound to the P3 segment (Figure 7E). Moreover, to further validate expression of circMyst4 was mediated by GATA1 and superenhancer (SE2), we constructed a dual-luciferase reporter gene plasmid containing the SE2 and promoter regions of Myst4 and a GATA1 plasmid, and confirmed by dual-luciferase assays that GATA1 promoted circMyst4 expression by targeting the SE region (Figure 7F). Meanwhile, we also found that the expression of circMyst4 was increased by overexpression of GATA1 in hypoxic PASMCs (Figure S9F). Taken together, these data demonstrate that H3K27ac, H3K4me1, and GATA1 interact with both the E2 superenhancer region and P3 promoter regions of Myst4 to regulate the expression of circMyst4 in PASMCs.
Figure 7.
Superenhancers drive the generation of circMyst4
(A) Gene tracks depicted the superenhancer region of Myst4 in lung tissue with measured H3K27ac marks. According to H3K27ac signal peak, the superenhancer region of Myst4 was divided into four constituents (E1, chr14: 21523088–21524806; E2, chr14: 21546613–21561236; E3, chr14: 21585003–21587120; E4, chr14: 21617293–21618401).
(B) After JQ-1 (a kind of inhibitor of superenhancer) treatment, real-time qPCR analysis of the expression of circMyst4 (n = 6) in PASMCs.
(C) Venn diagram shown that candidate transcription factors bound to the promoter and superenhancer of Myst4.
(D) ChIP-qPCR analysis showed GATA1, H3K27ac and H3K4me1 bound to the second constituent (E2) of superenhancer region of Myst4 (n = 3).
(E) ChIP-qPCR analysis showed GATA1, H3K27ac and H3K4me1 bound to the third segment of promoter region (P3) of Myst4 (n = 3).
(F) The luciferase activities of superenhancer element 2 were measured through dual-luciferase reporter assay in PASMCs (n = 6). Data are shown as means ± SD. Statistical analysis was performed with one-way ANOVA followed by Bonferroni correction and Student’s t test for 2 means. Hyp, hypoxia; Nor, normoxia; ns, not significantly different. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
To provide more concrete evidence for the SE’s role in circMyst4 regulation, we used JQ-1 to disruption of the SE for exploring the role of superenhancers in circMyst4-mediated ferroptosis. The results showed that JQ-1 significantly decreased circMyst4 expression and promoted PASMC ferroptosis under normoxic conditions. But overexpression of circMyst4 significantly improved this phenomenon (Figures S9G–S9J). These suggested that SE was involved in the occurrence of PASMC ferroptosis by regulating circMyst4 expression.
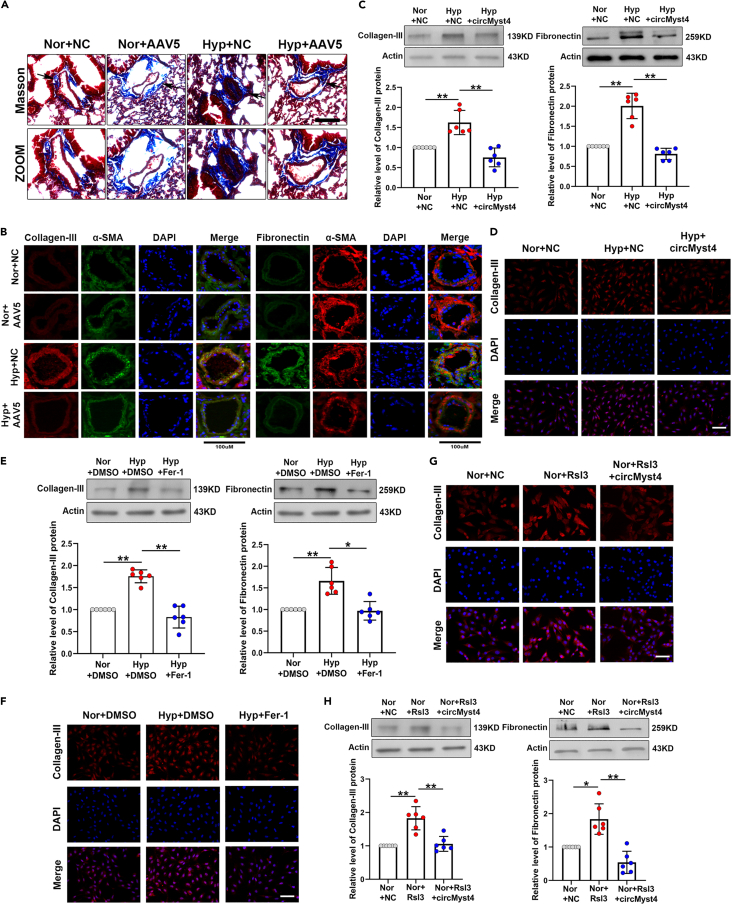
CircMyst4 decreased pulmonary fibrosis levels by regulating ferroptosis
The development of pulmonary fibrosis is an established biomarker of PH.4 Masson staining demonstrated that circMyst4 overexpression sharply decreased pulmonary fibrosis induced by hypoxia (Figure 8A). Immunofluorescence analyses also showed that the expression levels of collagen-III and fibronectin were reduced by overexpressing circMyst4 compared to the hypoxia groups (Figure 8B). These results of circMyst4-regulated fibrosis were further demonstrated by in vitro. We found that circMyst4 overexpression significantly depressed the high expression of collagen-III and fibronectin induced by hypoxia in cultured mouse PASMCs, indicating that fibrosis is regulated by circMyst4 both in vivo and in vitro (Figures 8C and 8D). Moreover, we found ferrostatin-1 (Fer-1, a ferroptosis inhibitor) significantly depressed the high expression of collagen-III and fibronectin induced by hypoxia (Figures 8E and 8F). To further investigate whether circMyst4 regulates hypoxia-induced fibrosis through ferroptosis, we cultured PASMCs with a ferroptosis inducer (Rsl3) and found that the expression levels of fibrosis-related proteins were significantly increased, while overexpression of circMyst4 reversed this phenomenon (Figures 8G and 8H). These data suggest that circMyst4 is involved in the pathological process of PH by ferroptosis mediated fibrosis.
Figure 8.
CircMyst4 decreases pulmonary fibrosis levels by regulating ferroptosis
(A and B) Masson staining and immunofluorescence analysis of mouse pulmonary fibrosis of in different groups. Scale bar, 100μm (n = 6). Red color denotes collagen-III or α-SMA, stained with Cy3, green denotes fibronectin or α-SMA, stained with FITC and blue color denotes nucleus, stained with DAPI.
(C and D) After overexpression with circMyst4, western blotting and immunofluorescence analysis of the levels of collagen III and fibronectin. Scale bar, 100μm (n = 6). Red color denotes collagen-III, stained with Cy3, and blue color denotes nucleus, stained with DAPI.
(E and F) After Ferrostatin-1 (an inhibitor of ferroptosis, Fer-1) treatment, western blotting and immunofluorescence analysis of the levels of collagen-III and fibronectin. Scale bar, 100μm (n = 6). Red color denotes collagen-III, stained with Cy3, and blue color denotes nucleus, stained with DAPI.
(G and H) After circMyst4 overexpression and Rsl3 (an inducer of ferroptosis) treatment, western blotting and immunofluorescence analysis of the levels of collagen-III and fibronectin. Scale bar, 100μm (n = 6). Red color denotes collagen-III, stained with Cy3, and blue color denotes nucleus, stained with DAPI. Data are shown as means ± SD. Statistical analysis of the graph (C, E, and H) was performed with one-way ANOVA followed by Dunn’s post-test. Hyp, hypoxia; Nor, normoxia; NC, negative control; AAV5, serotype 5 adenovirus-associated virus carrying circMyst4. ∗p < 0.05, ∗∗p < 0.01.
Discussion
Due to the complex pathogenesis of pulmonary vascular remodeling, there is still a lack of effective therapeutic targets for PH. With the maturity and application of high-throughput technology, increasing evidence suggests that circRNAs are key regulators of genes encoded at transcriptional or post-transcriptional levels in PH.21,22,32,33 In this study, by analyzing the datasets in the database, it was revealed that superenhancer-driven circMyst4, a novel circRNA, was significantly downregulated in hypoxic PH, played a role in regulation of PASMC ferroptosis and affected pulmonary vascular remodeling. Mechanistically, circMyst4 promoted GPX4 pre-mRNA splicing by regulating DDX5 in the nucleus, and inhibited the combination of Eef1a1 and ACSL4 in the cytoplasm, which suppressed the occurrence of hypoxia-induced ferroptosis. Our study demonstrates the meaningful role and mechanism of circMyst4 in ferroptosis and clarifies the therapeutic significance of circMyst4 in hypoxic PH.
CircRNAs were initially considered as by-products of mRNA splicing within cells and had no function.34 Nowadays, there are growing evidences that functional circRNAs play prominent roles a variety of diseases in different ways, such as adsorbing miRNAs, acting as protein scaffolds, competitively binding proteins, and even encoding peptides.35 Among them, circRNAs with coding potential have attracted attention. Zeng’s study found that m6A-modified circYAP promoted YAP dephosphorylation and nuclear translocation by encoding a novel 220aa peptide, resulting in colorectal cancer cell migration and invasion.36 CircFAM53B, a tumor-specific circRNA, elicit infiltration of T cells and enhanced anti-tumor immunity by encoding cryptic peptides, thereby controlling of tumor development.37 In the initial stage of our study, circMyst4 was found to have coding potential. Subsequently, we constructed FLAG-tagged circMyst4 overexpression plasmids, but unfortunately did not detect FLAG tag protein, suggesting that circMyst4 may be unable to encode peptides. We speculate that there may be several reasons why the database predictions do not match the detection results. First, database prediction only uses algorithms to analyze the protein-coding conserved regions present in circRNA sequences or search for homologous protein sequences for comparison, while the actual translation process in vivo is complex and variable. Second, circRNAs may be due to the special three-dimensional structures and the multi-gene interactions in vivo. Third, the actual open reading frame (ORF) of circRNA also affects how it is translated.38,39,40 However, further research is needed to redefine the database screening criteria by including more predictive elements to exclude circRNAs with ORF but no coding ability.
Recently, some studies have shown that circRNAs are easily detected in plasma or serum due to their strong stability.41,42 Therefore, the detection of circRNA levels in plasma or serum can be used as a potential diagnostic marker for diseases. Zhang et al. have found circCDK17 was downregulated in the serum of PH patients.43 Additionally, circRNAs are abnormally expressed in plasma of pediatric patients with PH due to congenital heart disease.44 In our study, we detected the content of circMyst4 in the plasma of hypoxic PH mouse models found that circMyst4 was significantly lower than that of normal mice. This implied that circMyst4 may serve as a plasma biomarker for the diagnosis of PH. Moreover, circRNA SCMH1 has been shown to achieve targeted delivery as nucleic acid drugs for the treatment of stroke.45 Currently, in vivo overexpression of circMyst4 has been manifested to alleviate hypoxia-induced vascular pathologic alterations in PH. Implying, circMyst4 could be developed to become potential nucleic acid-based drugs by targeting lung tissue in future, thereby attenuating the mortality of PH patients and provide theoretical basis for the development of nucleic acid-based drugs for the treatment of PH. Nevertheless, more research is needed in the future to apply it to the treatment and diagnosis of PH.
Ferroptosis, a novel nonapoptotic iron-dependent death form, is morphologically distinct from the general features of apoptosis and necrosis.46,47 Ferroptosis is considered a key pathological change in a highly lethal disease. However, its exact regulatory mechanisms in diseases have not yet been fully elucidated. Previous reports have demonstrated that several ferroptosis-related genes are abnormally expressed in the lung tissue of patients with pulmonary arterial hypertension and that the ferroptosis inhibitor ferrostatin-1 can inhibit endothelial cell inflammation to reverse pulmonary vascular remodeling in MCT-induced rats.6,48 However, little is known about whether ferroptosis participates in hypoxia-induced PH. In our study, we proved that hypoxia induced PASMC ferroptosis. While the overexpression of circMyst4 significantly inhibited the occurrence of ferroptosis in hypoxic PASMCs. Ferroptosis been proven to exert pro-inflammatory effects in different diseases.49 In MCT-induced rats, ferroptosis triggered inflammatory responses through the HMGB1/TLR4/NLRP3 inflammatory signaling pathways to resulting in pulmonary vascular remodeling.6 Interestingly, we also found that circMyst4 mediated the expression of IL-18 and IL-1β in PASMCs. This implies that circMyst4 may be involved in PH through ferroptosis-mediated inflammatory response, but more functions of circMyst4 still require further clarification.
CircRNAs have different roles in different localization. In the nucleus, they may regulate gene transcription and splicing, while in the cytoplasm, they may regulate mRNA stability and translation, protein stability and activity.20,50 We detected the subcellular localization of circMyst4 by FISH assays and found that it was distributed in both the nucleus and cytoplasm. In the nucleus, we validated that circMyst4 directly bound to and colocalized with DDX5. We further confirmed that circMyst4 overexpression upregulated the hypoxia-induced decrease in DDX5 by inhibiting its ubiquitination. Additionally, we demonstrated that circMyst4 regulated ferroptosis by binding to DDX5 to promote GPX4 pre-mRNA splicing in PASMCs. In addition, we found that circMyst4 regulates ferroptosis by suppressing the formation of the Eef1a1/ACSL4 complex in the cytoplasm. Our results demonstrated that circMyst4 regulated ferroptosis through different mechanisms in the nucleus and cytoplasm, which provided a new perspective to gain insight into the mechanism of ferroptosis occurrence.
DEAD box helicase 5 (DDX5), also called RNA helicase p68, is a member of the ATP-dependent RNA helicase family.51 It was previously reported that DDX5 acts as an important human splicing factor that acts on U1 snRNA and 5′ splice site (5′ss) duplexes during pre-mRNA splicing, which regulates the processing of mRNA.52,53 A recent study demonstrated that DDX5 and the human RNA chaperone protein hnRNPA1 coregulate alternative pre-mRNA splicing events.27 Our study confirmed that DDX5 could bind to GPX4 pre-mRNA. We also found that circMyst4 or DDX5 overexpression decreased GPX4 pre-mRNA levels and increased GPX4 mRNA levels. These results showed that circMyst4 in the nucleus directly bound to DDX5 and promoted GPX4 expression by facilitating splicing of the GPX4 pre-mRNA, thereby participating in PASMCs ferroptosis.
Eukaryotic translation elongation factor 1 alpha 1 (Eef1a1) was originally thought to be a translation factor that is ubiquitous in a variety of cells and plays a role in peptide elongation during protein translation.54 However, increasing evidence suggests that Eef1a1 is also involved in many other biological processes, including tumorigenesis, transcriptional regulation, and apoptosis.55,56 Previous reports have suggested that aberrant expression of Eef1a1 is closely associated with prognosis in several solid tumors.57,58,59 A new study shows that Eef1a1 can be used as a novel covalent target for pancreatic cancer treatment, covalently combined with the natural product BE-43547A to exert anticancer effects.60 However, whether Eef1a1 is involved in ferroptosis has not been reported. In our study, we found that Eef1a1 knockdown inhibited the occurrence of hypoxia-induced ferroptosis, especially ACSL4 expression. CoIP experiments confirmed that Eef1a1 could bind to ACSL4, but the binding of both was inhibited after overexpression of circMyst4. These findings suggest that circMyst4 in the cytoplasm regulates ferroptosis by suppressing the formation of the Eef1a1/ACSL4 complex.
Superenhancers are a class of large clusters composed of typical enhancers, which enrich key transcription factors, cofactors, and histone 3 lysine 27 acetylation (H3K27ac), have superior transcriptional activation activity and can regulate the expression of protein-coding genes important for the control of cell identity and differentiation.61,62,63 Recent studies have shown that SEs can also regulate the expression of key noncoding RNAs in various diseases. For example, the liver-specific SE cooperates with the transcription factor HNF4G to drive the production of lncRNA-DAW, which promotes the proliferation of hepatoma cells by mediating EZH2 degradation to activate the Wnt/β-catenin pathway.64 Loss of superenhancer-regulated circNfix promotes cardiac regenerative repair and improves cardiac function after myocardial infarction by inhibiting Ybx1 ubiquitin-dependent degradation and increasing miR-214 activity.30 In our studies, we found that JQ-1, a superenhancer inhibitor, may inhibit the expression of circMyst4. Bioinformatic analysis showed that SE region was present in the Myst4 locus and was enriched in H3K27ac signal peaks. Next, we screened for GATA1, as the key transcription factor. ChIP-qPCR analysis showed that GATA1 can bind to circMyst4-related superenhancers, and mainly binds to the E2 segment. At the same time, we also confirmed by ChIP-qPCR that H3K27ac, H3K4me1, and GATA1 mainly bind to the promoter P3 segment. These findings suggest that SE drives the expression of circMyst4.
Conclusions
In our study, we identified a novel superenhancer-mediated circRNA, circMyst4. CircMyst4 expression was downregulated in hypoxia, and circMyst4 overexpression inhibited PASMC ferroptosis and fibrosis in response to hypoxia by binding DDX5 to promote GPX4 pre-mRNA processing and by competitively antagonizing the binding of Eef1a1 to ACSL4 to regulate ACSL4 expression. Additionally, circMyst4 overexpression improved the symptoms of hypoxia-induced PH. Therefore, our findings provide a potential target for PH therapy.
Limitations of the study
First, due to circMyst4 also contains multiple miRNA binding sites, circMyst4 as a ceRNA mechanism requires to be further explored for more entirely understand the role of circMyst4. Second, as an exon-type circRNAs, the mechanisms of circMyst4 translocation between the nucleus and cytoplasm are not well understood, and require to be further characterized in future studies. Third, that is also urgently needed to detect the expression and function of circMyst4 in PASMCs of PH patients to better acquiring clinical data of circMyst4 on human PH.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact Daling Zhu (zhudaling@hrbmu.edu.cn).
Materials availability
This study did not generate new unique reagents.
Data and code availability
-
•
This paper does not report any original code.
-
•
All data reported in this paper will be shared by the lead contact upon request.
-
•
Any additional information required to reanalyze the data reported in this paper is available form the lead contact upon request.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (31820103007, 31971057, 31771276 to D.Z., 82170059 to C.M.), Natural Science Foundation of Heilongjiang Province (ZD2023H003 to C.M.), Heilongjiang Touyan Innovation Team Program to D.Z., Natural Science Foundation of Heilongjiang Province (LH2020H028 to L.Z.).
Author contributions
All authors contributed to the study conception and design. D.Z. designed the experiments. Data collection and analysis were performed by S.H., J.B., L.Z., and H.Y. One part data collection was performed by X.W., J.M., and X.Z. Material preparation was performed by X.G. and W.X. The first draft of the manuscript was written by S.H. and J.B., and the critical revision of the article written by D.Z. and C.M. All authors read and approved the final manuscript.
Declaration of interests
The authors declare no conflict of interest.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-GPX4 Antibody | Boster | Cat# BM5231; RRID: AB_3661639 |
| Recombinant Anti-DDX5 antibody | abcam | Cat# ab126730; RRID: AB_11130291 |
| Recombinant Anti-eEF1A1/EF-Tu+eEF1A1+eEF1AL3 antibody | abcam | Cat# ab157455; RRID: AB_3094654 |
| Recombinant Anti-Collagen III antibody | abcam | Cat# ab184993; RRID: AB_2895112 |
| Rabbit Anti-Fibronectin/Ugl-Y3 antibody | Bioss | Cat# bs-13455R; RRID: AB_3661640 |
| smooth muscle actin specific Monoclonal antibody | Proteintech | Cat# 67735-1-Ig; RRID: AB_2918504 |
| ACSL4 Rabbit mAb | ABclonal | Cat# A20414; RRID: AB_2909505 |
| Anti-Cox2 PTGS2 Monoclonal Antibody | Boster | Cat# M00084-1; RRID: AB_3661638 |
| KAT6B Rabbit pAb | ABclonal | Cat# A17116; RRID: AB_2770037 |
| GATA1 Polyclonal antibody | Proteintech | Cat# 10917-2-AP; RRID: AB_2108279 |
| Chemicals, peptides, and recombinant proteins | ||
| actinomycin D | MedChem Express | Cat # HY-17559 |
| JQ-1 | Selleck | Cat #S7110 |
| MG-132 | Selleck | Cat #S2619 |
| Ferrostatin-1 | Selleck | Cat #S7243 |
| RSL3 | Selleck | Cat #S8155 |
| Critical commercial assays | ||
| Fluorescence In Situ Hybridization Kit | GenePharma | N/A |
| Cytoplasmic & Nuclear RNA Purification kit | NORGEN Biotrk Corp | Cat # 21000 |
| RNA Immunoprecipitation (RIP) Kit | BersinBio | Cat # Bes5101 |
| RNA pull-down kit | BersinBio | Cat # Bes5102 |
| ChIP Assay Kit | Beyotime | Cat #P2078 |
| Lipid Peroxidation MDA Assay Kit | Beyotime | Cat #S0131S |
| Glutathione Assay Kit | Sigma | Cat # CS0260 |
| Ferrous Ion Content Assay Kit | Solarbio | Cat # BC5415 |
| Experimental models: cell lines | ||
| Mouse pulmonary artery smooth muscle cells | Procell Life Science & Technology | N/A |
| Experimental models: organisms/strains | ||
| Male C57BL/6 mice | Changchun Yisi Experimental Animal Technology Limited | N/A |
| Oligonucleotides | ||
| Mus-circMyst4: Forward (5′–3′): CCCAGTGCTTTTCCGTCCTC |
This paper | N/A |
| Mus-circMyst4: Reverse (5′–3′): CCAGGCAACGTGGAGAGTTG |
This paper | N/A |
| Mus-Myst4: Forward (5′–3′): TAGCCTCTCGTGTCCTGTCA |
This paper | N/A |
| Mus-Myst4: Reverse (5′–3′): CCCCTCCCCCAAAGTCAAAA |
This paper | N/A |
| Mus-GPX4: Forward (5′–3′): GCCAAAGTCCTAGGAAACGC |
This paper | N/A |
| Mus-GPX4: Reverse (5′–3′): CCGGGTTGAAAGGTTCAGGA |
This paper | N/A |
| Mus-GPX4 pre-mRNA: Forward (5′–3′): GGCACGTCTGAACGAGTTATC |
This paper | N/A |
| Mus-GPX4 pre-mRNA: Reverse (5′–3′): TAGGGGCACACACTTGTAGG |
This paper | N/A |
| Mus-DDX5: Forward (5′–3′): CATTTCCATCGGTCCTTGC |
This paper | N/A |
| Mus-DDX5: Reverse (5′–3′): AAGTAGAATCCTGCGGCGAC |
This paper | N/A |
| Mus-Eef1a1: Forward (5′–3′): CGCAGGTGTTGTGAAAACCA |
This paper | N/A |
| Mus-Eef1a1: Reverse (5′–3′): ACGTGTCCGATTACGACGAT |
This paper | N/A |
| Mus-β-Actin: Forward (5′–3′): TCAGGTCATCACTATCGGCAAT |
This paper | N/A |
| Mus-β-Actin: Reverse (5′–3′): AAAGAAAGGGTGTAAAACGCA |
This paper | N/A |
| Software and algorithms | ||
| ImageJ | National Institutes of Health | N/A |
| GraphPad Prism 9 | GraphPad Software | N/A |
Experimental model and study participant details
Animals
Male C57BL/6 mice (25-30g, six-week-old) were obtained from the Changchun Yisi Experimental Animal Technology Limited. All mouse care and use conformed to the Guide for the Care and Use of Laboratory Animals (National Institutes of Health Publication 85-23, revised 1996), and were approved by the Institutional Animal Care and Use Committee of Harbin Medical University (HMUDQ20221111001). All mice were randomly divided into different groups. After isoflurane anesthesia, mice were infected with the negative control vector (NC) or the serotype 5 adenovirus-associated virus particles carrying circMyst4 (AAV5-circMyst4) via dropwise nasal instillation. The virus particles were constructed by GENECHEM (Shanghai, China), and the virus titer is 1011 genome equivalents. Next, mice were treated in normoxic and hypoxic environments for 28 days with inspired oxygen (FiO2) fractions of 0.21 and 0.10 respectively, as previously described.65
Method details
Echocardiography
At the end of hypoxic treatment, mice were anesthetized with avertin (200 mg/kg, intraperitoneal injection), and detected echocardiography using a VisualSonics Vevo2100 imaging system (VisualSonicse Inc., Toronto, Ontario, Canada) with a 40 mHZ probe. Stable images were obtained in the M, B and Doppler modes, and the pulmonary artery acceleration time (PAAT), pulmonary artery velocity time integral (PAVTI) and cardiac output (CO) were measured.
Right ventricular systolic pressure (RVSP) and RV hypertrophy measurements
Using the PowerLab monitoring equipment (AD Instruments, Colorado Springs, Colo) measured RVSP. In a nutshell, a 1.2 French Pressure Catheter (Scisense Inc.) was connected to the Scisense FA-404 recorder. Exposing the right jugular vein of mouse, the catheter was inserted into the superior vena cava, and finally into the right ventricular vein. RVSP was continuously recorded for 40 min. Subsequently, heart was dissected and weighed to calculate the right ventricular hypertrophy index (ratio of right ventricular free wall weight over sum of septum plus left ventricular free wall weight: RV/LV + S).
Histology and morphologic analysis
The fresh lung tissues were extracted and fixed in 4% paraformaldehyde for 48 h. Then tissues were dehydrated and embedded in paraffin wax, and cut into 5μm thick sections to stain with hematoxylin and eosin (H&E) or Masson’s trichrome stain. The changes of lung tissue morphology were observed via a fluorescence microscope (Nikon, Japan).
Cell lines and cell culture
Mouse pulmonary artery smooth muscle cells (PASMCs) were obtained from Procell Life Science & Technology (Wuhan, China), and were cultured in Dulbecco’s Modified Eagle Medium (DMEM) containing 15% fetal bovine serum (FBS) and 1% penicillin/streptomycin (PEST) and were placed in a 37°C humidified incubator containing 5% CO2. Then, PASMCs were cultured under hypoxia for 24 h in a TriGas incubator (Heal Force, Shanghai, China) comprising 3% O2, 5% CO2 and 92% N2. These cell lines are Mycoplasm free.
Immunofluorescence
PASMCs were fixed with 4% paraformaldehyde for 30 min at 4°C and permeabilized with 0.3% Triton X-100 for 30 min at room temperature. Next, using 5% Bovine Serum Albumin blocked the cell at room temperature for 30 min. And the cells were incubated with antibodies against GPX4 (1:30, BM5231, Boster, Wuhan, China), DDX5 (1:100, ab126730, abcam, Cambridge, UK), Eef1a1 (1:50, ab157455, abcam, Cambridge, UK), Collagen-III (1:100, ab184993, abcam, Cambridge, UK), Fibronectin (1:100, bs-13455R, BIOSS, Beijing, China) and α-SMA (1:50) at 4°C overnight. The cells were washed three times with PBS, and incubated with Cy3/FITC-conjugated secondary antibody (1:100, A0516/A0568, Beyotime, Shanghai, China) at 37°C for 2h. DAPI, used to stain nuclei, away from light for 10 min. The images of immunofluorescence staining were evaluated by a living cell workstation (AF6000; Leica, Germany).
Transmission electron microscopy (TEM)
TEM was preformed to analysis ferroptosis in the ultra-microstructure. Briefly, PASMCs were cultured and collected, then were fixed with 2.5% glutaraldehyde for 48-72 h at 4°C. After washing the fixed samples, the samples were gradually dehydrated in ethanol (30–100%), embedded in epoxy resin to slice for preparing ultrathin sections (0.1 mm). Samples were examined using a Hitachi-7650 TEM (Hitachi, Japan).
RNA fluorescence in situ hybridization
Fluorescence in situ hybridization (FISH) assay was performed according to the manufacturer’s instructions (Fluorescence in Situ Hybridization Kit, GenePharma, Suzhou, China). Briefly, PASMCs were exposure to hypoxia for 24h, then were fixed with 4% paraformaldehyde. Using pre-hybridization solution dealt with cell at 37°C for 30 min, and the cell were incubated with Cy3-labeled circMyst4 probes, Cy3-labeled U6 probes (the internal reference of the nuclei) or Cy3-labeled 18S probes (the internal reference of the cytoplasm) for overnight at 37°C. Next, PBS washed with cell three times, then the nuclei were stained with DAPI. The fluorescence quantitative analysis was evaluated by a living cell workstation (AF6000; Leica, Germany).
RNase R treatment
Total RNA (2μg) was incubated with or without 3 U/μg of RNase R (Epicenter Biotechnologies, Madison, WI, USA) for 15 min at 37°C, then was used for detecting the expression of circMyst4 and linearMyst4.
Actinomycin D assay
PASMCs were dealt with actinomycin D (2 μg/mL, HY-17559, MedChem Express, NJ, USA) for 0h, 4h, 8h and 12h respectively. Then, total RNA was extracted with Trizol reagent (Invitrogen, Carlsbad, CA) according to manufacturer’s instructions, and used for subsequent experiments.
Isolation of cytoplasmic and nuclear RNAs
Cytoplasmic and nuclear RNAs were extracted using a Cytoplasmic & Nuclear RNA Purification kit (NORGEN Biotrk Corp, Canada) according to the manufacturer’s protocol. Briefly, up to 3 × 106 PASMCs were collected. The cells were then resuspended in 200 μL ice-cold Lysis Buffer J and vortexed for 15 s. Centrifuge and take the supernatant. The supernatant and remaining precipitate were then resuspended in 200–400 μL ice-cold Buffer SK, respectively, vortexed for 10s, and added 200 μL 100% ethanol. Finally, centrifugation and elution were performed using an adsorption column. The supernatant was eluted to obtain cytoplasmic RNA. Similarly, the precipitate elutes to obtain nuclear RNA.
Plasmid construction, siRNA and ASO design and transfection
The overexpression plasmid of circMyst4 and DDX5 was constructed with the GV486 vector by GeneChem (Shanghai, China), and using the vector alone served as a negative control (NC). The small interfering RNA against circMyst4, DDX5 and Eef1a1 was designed and synthesized by GenePharma (Suzhou, China), and ASO against circMyst4 was designed and synthesized by Ribobio (Guangzhou, China). Nontargeted control siRNA (siNC) was used as negative control. According to the manufacturer’s instructions for the Lipofectamine 2000 Reagent (Life Technologies, Carlsbad, USA), transfection was performed. PASMCs were cultured to 60–70% confuence, and added then the serum-free DMEM medium containing 2μg plasmids/NC or 2μg siRNA/ASO/NC. The cells were then incubated for 24–48h to be used as required. The sequences are listed below:
siNC:
sense, 5′-UUCUCCGAACGUGUCACGUTT-3′, antisense, 5′-ACGUGACACGUUCGGAGAATT-3′.
si-circMyst4:
sense, 5′-GAGAAAGACCAGGUCUUGGTT- 3′, antisense, 5′-AUCACUUCUAUGCCUGAGGTT-3′.
siDDX5:
sense, 5′-GCUCCUAUUCUGAUUGCUATT- 3′, antisense,5′-UAGCAAUCAGAAUAGGAGCTT-3′.
siEef1a1:
sense, 5′-GCUGGAGCCAAGUGCUAAUTT- 3′, antisense,5′-AUUAGCACUUGGCUCCAGCTT-3′.
ASO-circMyst4:
ATGAGAAAGACCAGGTCTTG.
Western blotting analysis
Lung tissues of mouse and cultured PASMCs were placed at lysis solution, and were then broken by sonication on ice, centrifuged at 13,500 rpm for 15 min at 4°C to collect supernatants and store at −80°C for using in t analysis. The samples of protein (approximately 25μg) were separated on 12% SDS-PAGE gel and were transferred onto the nitrocellulose membrane. Using 5% nonfat milk blocked the protein-adhered membranes for 1h and were incubated with specific antibodies against GPX4 (1:500, BM5231, Boster, Wuhan, China), ACSL4 (1:1000, A20414, ABclonal, Wuhan, China), ptgs2 (1:500, BM4419, Boster, Wuhan, China), Collagen-III (1:1000, b184993, abcam, Cambridge, UK), Fibronectin (1:500, bs-13455R, BIOSS, Beijing, China), DDX5 (1:1000, ab126730, abcam, Cambridge, UK), Eef1a1 (1:1000, ab157455, abcam, Cambridge, UK), GATA1 (1:1000, 10917-2-AP, Proteintech, USA) and Myst4 (Kat6b, 1:500, A17116, ABclonal, Wuhan, China). Then, the protein-adhered membranes were reacted with horseradish peroxidase-labeled secondary antibodies for 1h at room temperature, and boosted chemiluminescence reagents imaging to analyze the level of proteins.
Quantitative real-time PCR
In accordance with the manufacturer’s protocol, total RNA was extracted from mouse lung tissue and cultured PASMCs using TRIZOL reagent (Invitrogen, Carlsbad, USA). Subsequently, the extracted RNA was reverse transcribed into cDNA via using a reverse transcription kit according to the manufacturer’s protocol. Quantitative real-time PCR (real-time qPCR) was performed with SYBR Green (TOYOBO, Tokyo, Japan) via using a Roche Light Cycler 480II. β-Actin was used as the internal control. The sequences of real-time qPCR primers were as follows:
circMyst4 (mouse):
forward, 5′-CCCAGTGCTTTTCCGTCCTC-3′, reverse, 5′-CCAGGCAACGTGGAGAGTTG-3′.
Myst4 (mouse):
forward, 5′-TAGCCTCTCGTGTCCTGTCA-3′, reverse, 5′-CCCCTCCCCCAAAGTCAAAA-3′.
GPX4 (mouse):
forward, 5′-GCCAAAGTCCTAGGAAACGC-3′, reverse, 5′-CCGGGTTGAAAGGTTCAGGA-3′.
GPX4 pre-mRNA (mouse):
forward, 5′-GGCACGTCTGAACGAGTTATC-3′, reverse, 5′-TAGGGGCACACACTTGTAGG-3′.
DDX5 (mouse):
forward, 5′-CATTTCCATCGGTCCTTGCC-3′, reverse, 5′-AAGTAGAATCCTGCGGCGAC-3′.
Eef1a1 (mouse):
forward, 5′-CGCAGGTGTTGTGAAAACCA-3′, reverse, 5′-ACGTGTCCGATTACGACGAT-3′.
β-Actin (mouse):
forward, 5′-TCAGGTCATCACTATCGGCAAT-3′, reverse, 5′-AAAGAAAGGGTGTAAAACGCA-3′.
MDA assay of lipid peroxidation
The lipid oxidation levels were detected via using a lipid oxidation (MDA) assay kit (Beyotime, Shanghai, China) according to the manufacturer’s protocol. Briefly, lung tissues andcultured PASMCs placed at lysis solution, were broken by sonication on ice. Then, the supernatant was collected, and taken 100 μL to mix with MDA detection working solution. The mixture was boiled for 15 min, and centrifuged at 1000 g at room temperature for 10 min. Taking 200 μL supernatant were added to a 96-well plate, and then the absorbance was measured at 532 nm using an enzyme plate reader. The MDA content in the sample was calculated as μmol/mg.
GSH assay
Lung tissues and cultured PASMCs were collected to extract GSH according to the manufacturer’s instructions of the Glutathione Assay Kit (Sigma, CS0260). Briefly, lung tissues and at least 108 cells were collected, and resuspended in 5% 5-Sulfosalicylic Acid (SSA) Solution. Then, cells were frozen and thawed twice repeatedly, and centrifuged to collect the supernatant. 10 μL of supernatant were taken, added 150 μL of working solution, and incubated at room temperature for 5 min Finally, the mixture was added 50 μL of diluted NADPH solution and mixed, and the absorbance was determined at 412 nm using an enzyme plate reader (BioTek, Vermont, USA).
Ferrous ion content assay
According to the manufacturer’s instructions of Ferrous Ion Content Assay Kit (Solarbio, BC5415), lung tissue (100 mg) or PASMCs (5×106) were homogenized in 1mL reagent I, then was centrifuged at 10000×g for 10 min at 4°C. Then, the supernatant was collected, and taken 200 μL to mix with reagent II. The mixture was incubated for 10 min at 37°C and added 100 μL chloroform. Then, fully vortex oscillated for 5 min, and centrifuged at 12000×g for 10 min at room temperature. Taking 200 μL upper inorganic phase were added to a 96-well plate, and then the absorbance was measured at 593 nm using an enzyme plate reader. The ferrous ion content in the sample was calculated as μmol/mg prot.
Ferrous ion staining
According to the manufacturer’s instructions (MX4558, MKBio, Shanghai, China), PASMCs were cultured in 12-well plates and washed with HBSS two times. Then, cells were added with 5uM FeRhoNox-1 working solution, and placed at a 37°C incubator containing 5% CO2 for 60 min. HBSS washed cell three times, and the results of Ferrous ion staining were observed by a living cell workstation (AF6000; Leica, Germany).
RNA immunoprecipitation (RIP)
Using an RNA Immunoprecipitation (RIP) Kit (Bes5101, BersinBio, Guangzhou, China) performed RNA immunoprecipitation assay following the manufacturer’s instructions. Briefly, PASMC precipitation was collected and lysed with RIP lysis buffer for 20 min. Then, samples were incubated with 20 μL protein A/G beads, which were conjugated anti-DDX5 (5 μL, ab126730, abcam, Cambridge, UK), anti-Eef1a1 (5 μL, ab157455, abcam, Cambridge, UK) and IgG, for 12-16h. After extracting and purifying RNA, the expression of immunoprecipitated RNA was detected by qRT-PCR.
RNA pull-down assay
According to the manufacturer’s instructions, the RNA pull-down kit (Bersinbio, Guangzhou, China) was used to perform pull down assays. Briefly, 2 × 107 PASMCs were collected and were cleaved by lysis buffer. Coupling the magnetic bead with the biotinylated circMyst4 probe or the negative control (NC) probe (GenePharma, Shanghai, China), the samples were then mixed with Composite of magnetic bead and probe, and incubated for 2h at 25°C. Magnetic beads were washed with wash buffer four times. Then, using a protein elution buffer washed magnetic beads for 2h at 37°C and collected the eluant, that is proteins pulled down by RNA probe. The western blotting and mass spectrometry analysis were used for detecting proteins. The mass spectrometry analysis was performed by Beijing Bio-Tech Pack Technology Company Ltd.
The circMyst4 probe sequence:
CUU UAGGAAA CCAAGACC UGGUCUUUC UCAUGGG.
The NC probe sequence:
CAGUACUUUUGUGUAGUACAA.
Co-immunoprecipitation
Cultured PASMCs were collected, and added 1 mL lysis solution containing 1% PMSF to lyse for 30 min on ice. Then, the supernatant was collected via centrifuging at 15,000 rpm for 30 min at 4°C, and added 5 μL target antibody (anti-H3K27ac, A7253, ABclonal; anti-H3K4me1, A2355, ABclonal; anti-GATA1, 10917-2-AP, Proteintech; anti-Eef1a1, ab157455, abcam; anti-ACSL4, A20414, ABclonal; anti-DDX5, ab126730, abcam; anti-UBB, 10201-2-AP, Proteintech, USA) or IgG for overnight in a 4°C shaker. Next day, Protein A + G agarose beads were added and incubated for 6h at 4°C, then beads were washed with cold PBS four times, then beads were collected and resuspended in protein loading buffer (2×). The use of western blotting detected the samples.
ChIP-qPCR
According to the manufacturer’s instructions of ChIP Assay Kits (P2078, Beyotime, Shanghai, China), PASMCs were cultured in 10 cm Petri dishes with 10 mL of culture medium containing 1% formaldehyde and incubated at 37°C for 10 min to cross-link. The cells were broken by sonication for making genomic DNA into 200-1000bp size, then the sonicated samples were incubated with Protein A + G Agarose/Salmon Sperm DNA combined with anti-H3K27ac (1:100, A7253, ABclonal), anti-H3K4me1 (1:100, A2355, ABclonal) and anti-GATA1 (1:50, 10917-2-AP, Proteintech) at 4°C overnight. The level of DNA was detected by qRT-PCR.
Dual-luciferase reporter assay
A dual-luciferase reporter assay was performed according to the protocol of the Dual-Lumi Luciferase Reporter Gene Assay Kit (RG088S, Beyotime, Shanghai, China). The Myst4 superenhancer segment (SE2) and promoter fragment containing the GATA1 binding site was cloned into the GV238 plasmid expressing luciferase (GenePharma, Suzhou, China). PASMCs were cotransfected with the GATA1 expression plasmid and a dual-luciferase reporter gene plasmid containing the SE2 and promoter regions of Myst4 with Lipofectamine 2000 for 48 h. Then, the luciferase activities were measured by the dual-luciferase reporter assay system (GloMax Multi Detection System, Promega, Madison).
Quantification and statistical analysis
Statistical analysis
Statistical analyses were performed using GraphPad Prism Software 9.0 (GraphPad Software Inc.). Data are expressed as mean ± SD. All expression values were checked for normal distribution before statistical testing. Statistical comparisons were performed with the two-tailed Student’s t test in 2 groups and one-way ANOVA with Bonferroni post hoc test in multiple groups. Nonparametric analyses were performed for non-normally distributed data, such as the Mann-Whitney U test for 1 or 2 groups or Kruskal-Wallis test followed by Dunn post-test for >2 groups. p < 0.05 were considered statistically significant. All experiments were performed in triplicate.
Published: September 12, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2024.110900.
Supplemental information
References
- 1.Maron B.A., Galiè N. Diagnosis, Treatment, and Clinical Management of Pulmonary Arterial Hypertension in the Contemporary Era: A Review. JAMA Cardiol. 2016;1:1056–1065. doi: 10.1001/jamacardio.2016.4471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dai J., Zhou Q., Chen J., Rexius-Hall M.L., Rehman J., Zhou G. Alpha-enolase regulates the malignant phenotype of pulmonary artery smooth muscle cells via the AMPK-Akt pathway. Nat. Commun. 2018;9:3850. doi: 10.1038/s41467-018-06376-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Long L., Yang X., Southwood M., Lu J., Marciniak S.J., Dunmore B.J., Morrell N.W. Chloroquine prevents progression of experimental pulmonary hypertension via inhibition of autophagy and lysosomal bone morphogenetic protein type II receptor degradation. Circ. Res. 2013;112:1159–1170. doi: 10.1161/CIRCRESAHA.111.300483. [DOI] [PubMed] [Google Scholar]
- 4.Zhang M., Xin W., Yu Y., Yang X., Ma C., Zhang H., Liu Y., Zhao X., Guan X., Wang X., Zhu D. Programmed death-ligand 1 triggers PASMCs pyroptosis and pulmonary vascular fibrosis in pulmonary hypertension. J. Mol. Cell. Cardiol. 2020;138:23–33. doi: 10.1016/j.yjmcc.2019.10.008. [DOI] [PubMed] [Google Scholar]
- 5.Samokhin A.O., Stephens T., Wertheim B.M., Wang R.S., Vargas S.O., Yung L.M., Cao M., Brown M., Arons E., Dieffenbach P.B., et al. NEDD9 targets COL3A1 to promote endothelial fibrosis and pulmonary arterial hypertension. Sci. Transl. Med. 2018;10 doi: 10.1126/scitranslmed.aap7294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xie S.S., Deng Y., Guo S.L., Li J.Q., Zhou Y.C., Liao J., Wu D.D., Lan W.F. Endothelial cell ferroptosis mediates monocrotaline-induced pulmonary hypertension in rats by modulating NLRP3 inflammasome activation. Sci. Rep. 2022;12:3056. doi: 10.1038/s41598-022-06848-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stockwell B.R., Friedmann Angeli J.P., Bayir H., Bush A.I., Conrad M., Dixon S.J., Fulda S., Gascón S., Hatzios S.K., Kagan V.E., et al. Ferroptosis: A Regulated Cell Death Nexus Linking Metabolism, Redox Biology, and Disease. Cell. 2017;171:273–285. doi: 10.1016/j.cell.2017.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee N., Carlisle A.E., Peppers A., Park S.J., Doshi M.B., Spears M.E., Kim D. xCT-Driven Expression of GPX4 Determines Sensitivity of Breast Cancer Cells to Ferroptosis Inducers. Antioxidants. 2021;10 doi: 10.3390/antiox10020317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stancic A., Saksida T., Markelic M., Vucetic M., Grigorov I., Martinovic V., Gajic D., Ivanovic A., Velickovic K., Savic N., Otasevic V. Ferroptosis as a Novel Determinant of beta-Cell Death in Diabetic Conditions. Oxid. Med. Cell. Longev. 2022;2022 doi: 10.1155/2022/3873420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Y., Feng D., Wang Z., Zhao Y., Sun R., Tian D., Liu D., Zhang F., Ning S., Yao J., Tian X. Ischemia-induced ACSL4 activation contributes to ferroptosis-mediated tissue injury in intestinal ischemia/reperfusion. Cell Death Differ. 2019;26:2284–2299. doi: 10.1038/s41418-019-0299-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li C., Sun G., Chen B., Xu L., Ye Y., He J., Bao Z., Zhao P., Miao Z., Zhao L., et al. Nuclear receptor coactivator 4-mediated ferritinophagy contributes to cerebral ischemia-induced ferroptosis in ischemic stroke. Pharmacol. Res. 2021;174 doi: 10.1016/j.phrs.2021.105933. [DOI] [PubMed] [Google Scholar]
- 12.Yang W., Mu B., You J., Tian C., Bin H., Xu Z., Zhang L., Ma R., Wu M., Zhang G., et al. Non-classical ferroptosis inhibition by a small molecule targeting PHB2. Nat. Commun. 2022;13:7473. doi: 10.1038/s41467-022-35294-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li X., Yang L., Chen L.L. The Biogenesis, Functions, and Challenges of Circular RNAs. Mol. Cell. 2018;71:428–442. doi: 10.1016/j.molcel.2018.06.034. [DOI] [PubMed] [Google Scholar]
- 14.Memczak S., Jens M., Elefsinioti A., Torti F., Krueger J., Rybak A., Maier L., Mackowiak S.D., Gregersen L.H., Munschauer M., et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 15.Shen S., Wu Y., Chen J., Xie Z., Huang K., Wang G., Yang Y., Ni W., Chen Z., Shi P., et al. CircSERPINE2 protects against osteoarthritis by targeting miR-1271 and ETS-related gene. Ann. Rheum. Dis. 2019;78:826–836. doi: 10.1136/annrheumdis-2018-214786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu X., Zhang J., Tian Y., Gao Y., Dong X., Chen W., Yuan X., Yin W., Xu J., Chen K., et al. CircRNA inhibits DNA damage repair by interacting with host gene. Mol. Cancer. 2020;19:128. doi: 10.1186/s12943-020-01246-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shen S., Yang Y., Shen P., Ma J., Fang B., Wang Q., Wang K., Shi P., Fan S., Fang X. circPDE4B prevents articular cartilage degeneration and promotes repair by acting as a scaffold for RIC8A and MID1. Ann. Rheum. Dis. 2021;80:1209–1219. doi: 10.1136/annrheumdis-2021-219969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang F., Hu A., Guo Y., Wang J., Li D., Wang X., Jin S., Yuan B., Cai S., Zhou Y., et al. p113 isoform encoded by CUX1 circular RNA drives tumor progression via facilitating ZRF1/BRD4 transactivation. Mol. Cancer. 2021;20:123. doi: 10.1186/s12943-021-01421-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu Y., Ding W., Wang J., Ao X., Xue J. Non-coding RNAs in lung cancer: molecular mechanisms and clinical applications. Front. Oncol. 2023;13 doi: 10.3389/fonc.2023.1256537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu C.X., Chen L.L. Circular RNAs: Characterization, cellular roles, and applications. Cell. 2022;185:2390. doi: 10.1016/j.cell.2022.06.001. [DOI] [PubMed] [Google Scholar]
- 21.Zhang J., Li Y., Qi J., Yu X., Ren H., Zhao X., Xin W., He S., Zheng X., Ma C., et al. Circ-calm4 Serves as an miR-337-3p Sponge to Regulate Myo10 (Myosin 10) and Promote Pulmonary Artery Smooth Muscle Proliferation. Hypertension. 2020;75:668–679. doi: 10.1161/HYPERTENSIONAHA.119.13715. [DOI] [PubMed] [Google Scholar]
- 22.Jiang Y., Liu H., Yu H., Zhou Y., Zhang J., Xin W., Li Y., He S., Ma C., Zheng X., et al. Circular RNA Calm4 Regulates Hypoxia-Induced Pulmonary Arterial Smooth Muscle Cells Pyroptosis via the Circ-Calm4/miR-124-3p/PDCD6 Axis. Arterioscler. Thromb. Vasc. Biol. 2021;41:1675–1693. doi: 10.1161/ATVBAHA.120.315525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou S., Jiang H., Li M., Wu P., Sun L., Liu Y., Zhu K., Zhang B., Sun G., Cao C., Wang R. Circular RNA hsa_circ_0016070 Is Associated with Pulmonary Arterial Hypertension by Promoting PASMC Proliferation. Mol. Ther. Nucleic Acids. 2019;18:275–284. doi: 10.1016/j.omtn.2019.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jing X., Wu S., Liu Y., Wang H., Huang Q. Circular RNA Sirtuin1 represses pulmonary artery smooth muscle cell proliferation, migration and autophagy to ameliorate pulmonary hypertension via targeting microRNA-145-5p/protein kinase-B3 axis. Bioengineered. 2022;13:8759–8771. doi: 10.1080/21655979.2022.2036302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meyer-Schwesinger C. The ubiquitin-proteasome system in kidney physiology and disease. Nat. Rev. Nephrol. 2019;15:393–411. doi: 10.1038/s41581-019-0148-1. [DOI] [PubMed] [Google Scholar]
- 26.Wang X., Li J., Bian X., Wu C., Hua J., Chang S., Yu T., Li H., Li Y., Hu S., et al. CircURI1 interacts with hnRNPM to inhibit metastasis by modulating alternative splicing in gastric cancer. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2012881118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee Y.J., Wang Q., Rio D.C. Coordinate regulation of alternative pre-mRNA splicing events by the human RNA chaperone proteins hnRNPA1 and DDX5. Genes Dev. 2018;32:1060–1074. doi: 10.1101/gad.316034.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kar A., Fushimi K., Zhou X., Ray P., Shi C., Chen X., Liu Z., Chen S., Wu J.Y. RNA helicase p68 (DDX5) regulates tau exon 10 splicing by modulating a stem-loop structure at the 5' splice site. Mol. Cell Biol. 2011;31:1812–1821. doi: 10.1128/MCB.01149-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jia Q., Chen S., Tan Y., Li Y., Tang F. Oncogenic super-enhancer formation in tumorigenesis and its molecular mechanisms. Exp. Mol. Med. 2020;52:713–723. doi: 10.1038/s12276-020-0428-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang S., Li X., Zheng H., Si X., Li B., Wei G., Li C., Chen Y., Chen Y., Liao W., et al. Loss of Super-Enhancer-Regulated circRNA Nfix Induces Cardiac Regeneration After Myocardial Infarction in Adult Mice. Circulation. 2019;139:2857–2876. doi: 10.1161/CIRCULATIONAHA.118.038361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xiang J.F., Yin Q.F., Chen T., Zhang Y., Zhang X.O., Wu Z., Zhang S., Wang H.B., Ge J., Lu X., et al. Human colorectal cancer-specific CCAT1-L lncRNA regulates long-range chromatin interactions at the MYC locus. Cell Res. 2014;24:513–531. doi: 10.1038/cr.2014.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yuan P., Wu W.H., Gong S.G., Jiang R., Zhao Q.H., Pudasaini B., Sun Y.Y., Li J.L., Liu J.M., Wang L. Impact of circGSAP in Peripheral Blood Mononuclear Cells on Idiopathic Pulmonary Arterial Hypertension. Am. J. Respir. Crit. Care Med. 2021;203:1579–1583. doi: 10.1164/rccm.202005-2052LE. [DOI] [PubMed] [Google Scholar]
- 33.Ali M.K., Schimmel K., Zhao L., Chen C.K., Dua K., Nicolls M.R., Spiekerkoetter E. The role of circular RNAs in pulmonary hypertension. Eur. Respir. J. 2022;60:1. doi: 10.1183/13993003.00012-2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Misir S., Wu N., Yang B.B. Specific expression and functions of circular RNAs. Cell Death Differ. 2022;29:481–491. doi: 10.1038/s41418-022-00948-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen L.L. The expanding regulatory mechanisms and cellular functions of circular RNAs. Nat. Rev. Mol. Cell Biol. 2020;21:475–490. doi: 10.1038/s41580-020-0243-y. [DOI] [PubMed] [Google Scholar]
- 36.Zeng K., Peng J., Xing Y., Zhang L., Zeng P., Li W., Zhang W., Pan Z., Zhou C., Lin J. A positive feedback circuit driven by m(6)A-modified circular RNA facilitates colorectal cancer liver metastasis. Mol. Cancer. 2023;22:202. doi: 10.1186/s12943-023-01848-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang D., Zhu X., Ye S., Zhang J., Liao J., Zhang N., Zeng X., Wang J., Yang B., Zhang Y., et al. Tumour circular RNAs elicit anti-tumour immunity by encoding cryptic peptides. Nature. 2024;625:593–602. doi: 10.1038/s41586-023-06834-7. [DOI] [PubMed] [Google Scholar]
- 38.Wu P., Mo Y., Peng M., Tang T., Zhong Y., Deng X., Xiong F., Guo C., Wu X., Li Y., et al. Emerging role of tumor-related functional peptides encoded by lncRNA and circRNA. Mol. Cancer. 2020;19:22. doi: 10.1186/s12943-020-1147-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen Q., Shen H., Nie F., Sun M. A Whole New Comprehension about ncRNA-Encoded Peptides/Proteins in Cancers. Cancers. 2022;14 doi: 10.3390/cancers14215196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lei M., Zheng G., Ning Q., Zheng J., Dong D. Translation and functional roles of circular RNAs in human cancer. Mol. Cancer. 2020;19:30. doi: 10.1186/s12943-020-1135-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zang X., Jiang J., Gu J., Chen Y., Wang M., Zhang Y., Fu M., Shi H., Cai H., Qian H., et al. Circular RNA EIF4G3 suppresses gastric cancer progression through inhibition of β-catenin by promoting δ-catenin ubiquitin degradation and upregulating SIK1. Mol. Cancer. 2022;21:141. doi: 10.1186/s12943-022-01606-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hutchins E., Reiman R., Winarta J., Beecroft T., Richholt R., De Both M., Shahbander K., Carlson E., Janss A., Siniard A., et al. Extracellular circular RNA profiles in plasma and urine of healthy, male college athletes. Sci. Data. 2021;8:276. doi: 10.1038/s41597-021-01056-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang J., Li Y., Zhang J., Liu L., Chen Y., Yang X., Liao X., He M., Jia Z., Fan J., et al. ADAR1 regulates vascular remodeling in hypoxic pulmonary hypertension through N1-methyladenosine modification of circCDK17. Acta Pharm. Sin. B. 2023;13:4840–4855. doi: 10.1016/j.apsb.2023.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang Y., Chen Y., Yao H., Lie Z., Chen G., Tan H., Zhou Y. Elevated serum circ_0068481 levels as a potential diagnostic and prognostic indicator in idiopathic pulmonary arterial hypertension. Pulm. Circ. 2019;9 doi: 10.1177/2045894019888416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang L., Han B., Zhang Z., Wang S., Bai Y., Zhang Y., Tang Y., Du L., Xu L., Wu F., et al. Extracellular Vesicle-Mediated Delivery of Circular RNA SCMH1 Promotes Functional Recovery in Rodent and Nonhuman Primate Ischemic Stroke Models. Circulation. 2020;142:556–574. doi: 10.1161/CIRCULATIONAHA.120.045765. [DOI] [PubMed] [Google Scholar]
- 46.Dixon S.J., Lemberg K.M., Lamprecht M.R., Skouta R., Zaitsev E.M., Gleason C.E., Patel D.N., Bauer A.J., Cantley A.M., Yang W.S., et al. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell. 2012;149:1060–1072. doi: 10.1016/j.cell.2012.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yao X., Li W., Fang D., Xiao C., Wu X., Li M., Luo Z. Emerging Roles of Energy Metabolism in Ferroptosis Regulation of Tumor Cells. Adv. Sci. 2021;8 doi: 10.1002/advs.202100997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang F., Liu H. Identification of ferroptosis-associated genes exhibiting altered expression in pulmonary arterial hypertension. Math. Biosci. Eng. 2021;18:7619–7630. doi: 10.3934/mbe.2021377. [DOI] [PubMed] [Google Scholar]
- 49.Sun Y., Chen P., Zhai B., Zhang M., Xiang Y., Fang J., Xu S., Gao Y., Chen X., Sui X., Li G. The emerging role of ferroptosis in inflammation. Biomed. Pharmacother. 2020;127 doi: 10.1016/j.biopha.2020.110108. [DOI] [PubMed] [Google Scholar]
- 50.Kristensen L.S., Andersen M.S., Stagsted L.V.W., Ebbesen K.K., Hansen T.B., Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat. Rev. Genet. 2019;20:675–691. doi: 10.1038/s41576-019-0158-7. [DOI] [PubMed] [Google Scholar]
- 51.Legrand J.M.D., Chan A.L., La H.M., Rossello F.J., Änkö M.L., Fuller-Pace F.V., Hobbs R.M. DDX5 plays essential transcriptional and post-transcriptional roles in the maintenance and function of spermatogonia. Nat. Commun. 2019;10:2278. doi: 10.1038/s41467-019-09972-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu Z.R. p68 RNA helicase is an essential human splicing factor that acts at the U1 snRNA-5' splice site duplex. Mol. Cell Biol. 2002;22:5443–5450. doi: 10.1128/MCB.22.15.5443-5450.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lin C., Yang L., Yang J.J., Huang Y., Liu Z.R. ATPase/helicase activities of p68 RNA helicase are required for pre-mRNA splicing but not for assembly of the spliceosome. Mol. Cell Biol. 2005;25:7484–7493. doi: 10.1128/MCB.25.17.7484-7493.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Joung E.K., Kim J., Yoon N., Maeng L.S., Kim J.H., Park S., Kang K., Kim J.S., Ahn Y.H., Ko Y.H., et al. Expression of EEF1A1 Is Associated with Prognosis of Patients with Colon Adenocarcinoma. J. Clin. Med. 2019;8 doi: 10.3390/jcm8111903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Duman M., Vaquié A., Nocera G., Heller M., Stumpe M., Siva Sankar D., Dengjel J., Meijer D., Yamaguchi T., Matthias P., et al. EEF1A1 deacetylation enables transcriptional activation of remyelination. Nat. Commun. 2020;11:3420. doi: 10.1038/s41467-020-17243-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu Y., Jiang S., Yang P.Y., Zhang Y.F., Li T.J., Rui Y.C. EF1A1/HSC70 Cooperatively Suppress Brain Endothelial Cell Apoptosis via Regulating JNK Activity. CNS Neurosci. Ther. 2016;22:836–844. doi: 10.1111/cns.12581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu X., Chen L., Ge J., Yan C., Huang Z., Hu J., Wen C., Li M., Huang D., Qiu Y., et al. The Ubiquitin-like Protein FAT10 Stabilizes eEF1A1 Expression to Promote Tumor Proliferation in a Complex Manner. Cancer Res. 2016;76:4897–4907. doi: 10.1158/0008-5472.CAN-15-3118. [DOI] [PubMed] [Google Scholar]
- 58.Wu A., Tang J., Guo Z., Dai Y., Nie J., Hu W., Liu N., Ye C., Li S., Pei H., Zhou G. Long Non-Coding RNA CRYBG3 Promotes Lung Cancer Metastasis via Activating the eEF1A1/MDM2/MTBP Axis. Int. J. Mol. Sci. 2021;22 doi: 10.3390/ijms22063211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu S., Hausmann S., Carlson S.M., Fuentes M.E., Francis J.W., Pillai R., Lofgren S.M., Hulea L., Tandoc K., Lu J., et al. METTL13 Methylation of eEF1A Increases Translational Output to Promote Tumorigenesis. Cell. 2019;176:491–504.e21. doi: 10.1016/j.cell.2018.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu C., Wang L., Sun Y., Zhao X., Chen T., Su X., Guo H., Wang Q., Xi X., Ding Y., Chen Y. Probe Synthesis Reveals Eukaryotic Translation Elongation Factor 1 Alpha 1 as the Anti-Pancreatic Cancer Target of BE-43547A(2) Angew. Chem. Int. Ed. Engl. 2022;61 doi: 10.1002/anie.202206953. [DOI] [PubMed] [Google Scholar]
- 61.Pott S., Lieb J.D. What are super-enhancers? Nat. Genet. 2015;47:8–12. doi: 10.1038/ng.3167. [DOI] [PubMed] [Google Scholar]
- 62.Hnisz D., Abraham B.J., Lee T.I., Lau A., Saint-André V., Sigova A.A., Hoke H.A., Young R.A. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Peng L., Jiang B., Yuan X., Qiu Y., Peng J., Huang Y., Zhang C., Zhang Y., Lin Z., Li J., et al. Super-Enhancer-Associated Long Noncoding RNA HCCL5 Is Activated by ZEB1 and Promotes the Malignancy of Hepatocellular Carcinoma. Cancer Res. 2019;79:572–584. doi: 10.1158/0008-5472.CAN-18-0367. [DOI] [PubMed] [Google Scholar]
- 64.Liang W., Shi C., Hong W., Li P., Zhou X., Fu W., Lin L., Zhang J. Super-enhancer-driven lncRNA-DAW promotes liver cancer cell proliferation through activation of Wnt/beta-catenin pathway. Mol. Ther. Nucleic Acids. 2021;26:1351–1363. doi: 10.1016/j.omtn.2021.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhu D., Medhora M., Campbell W.B., Spitzbarth N., Baker J.E., Jacobs E.R. Chronic hypoxia activates lung 15-lipoxygenase, which catalyzes production of 15-HETE and enhances constriction in neonatal rabbit pulmonary arteries. Circ. Res. 2003;92:992–1000. doi: 10.1161/01.RES.0000070881.65194.8F. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
This paper does not report any original code.
-
•
All data reported in this paper will be shared by the lead contact upon request.
-
•
Any additional information required to reanalyze the data reported in this paper is available form the lead contact upon request.