Abstract
The yeast retrotransposon Ty1 encodes a 7-nucleotide RNA sequence that directs a programmed, +1 ribosomal frameshifting event required for Gag-Pol translation and retrotransposition. We report mutations that block frameshifting, which can be suppressed in cis by “transplanting” the frameshift signal to a position upstream of its native location. These “frameshift transplant” mutants transpose with only a modest decrease in efficiency, suggesting that the location of the frameshift signal in a functional Ty1 element may vary. The genomic architecture of Ty1 is such that Gag, Ty1 PR (PR), and the Gag-derived p4 peptide share a common sequence. The functional independence of the movement of the frameshift signal to a new location within the Ty1 element is used to unambiguously attribute the effect of mutations deleterious to transposition in this region of overlapping coding sequences to effects on the Ty1 (PR). This work defines the amino terminus of the Ty1 PR and introduces a new technique for studying viral genome organization.
Ty1 is one of five endogenous retrotransposons of the yeast Saccharomyces cerevisiae. The life cycle of Ty1 begins with transcription of the genomic element and translation of the resulting mRNA. Like retroviruses, Ty1 encodes a protease (PR) that processes the element-encoded proteins (1, 18, 23). Ty1 Gag and Gag-Pol proteins, together with Ty1 mRNA, form virus-like particles (VLPs), which are essential transposition intermediates (8, 9). Reverse transcription of Ty1 mRNA takes place in VLPs. The newly synthesized cDNA enters the nucleus and then the yeast genome via an integration reaction (3, 11, 17).
Viruses and the closely related retrotransposons are under selective pressure to maintain a small genome. A number of unique strategies have evolved to ensure that all of the functions necessary for completion of their life cycle are carried out. Host protein functionalities may be usurped, obviating the need for a similar, element-encoded protein.
A genome streamlining strategy used by most long terminal repeat-containing retroelements is programmed ribosomal frameshifting. An mRNA that contains a frameshifting signal can encode multiple proteins, typically Gag and Gag-Pol, in a predetermined stoichiometric ratio that is determined by the efficiency of the frameshift signal. A frameshift-containing retroelement does not require separate promoters to drive the transcription of two messages and thereby increases the information content of its genome with minimal expense. In addition, the common sequences present in Gag and Gag-Pol direct the assembly of the Pol proteins into the virion or VLP.
A 7-nucleotide signal in Ty1 mRNA is necessary and sufficient for directing ribosomal frameshifting (2) and can function translationally in various heterologous contexts. This frameshift signal is required for retrotransposition and is thought to secure the appropriate Gag and Gag-Pol stoichiometry. The frameshifting signal is approximately 5 to 10% efficient and therefore results in a 10- to 20-fold excess of the structural protein Gag with respect to Gag-Pol (6, 7, 21). The Gag-Pol polyprotein contains the enzymatic components required for Ty1 replication.
Ty1 mRNA can have two translational fates. In the more common scenario, in which the frameshift signal is read through by the translating ribosome, a 49-kDa Gag species is made and processed in trans to a 45-kDa Gag species with concomitant liberation of its carboxy-terminal 40 amino acids. The 49-kDa precursor and 45-kDa (CA) processed forms migrate with apparent molecular masses of 58 and 54 kDa, respectively. For a complete description of our Ty1 protein nomenclature, see the study by Merkulov et al. (14). The liberated 40-mer, called the p4 peptide, was suspected to serve some role in particle formation and/or maturation, because mutations in this region result in smaller-than-normal VLPs and severe transposition defects (15). When frameshifting occurs, a Gag-Pol species is synthesized. Proteolytic processing of Gag-Pol liberates Ty1 PR, integrase (IN), and reverse transcriptase (RT) from the precursor polyprotein (10). A 45-kDa Gag species, CA, identical to that made from Gag is also produced, and we show here that it is through this processing event that the amino terminus of the PR is defined (Fig. 1). Most of the p4 peptide sequence is therefore common to the amino terminus of the PR and the carboxy terminus of 49-kDa Gag.
FIG. 1.
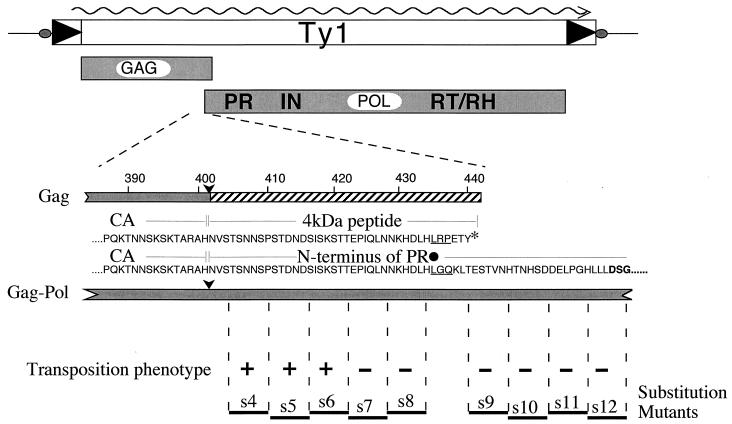
Schematic of the p4 region. The Gag and Gag-Pol polyproteins as well as the relevant amino acids are shown. The amino acids encoded fully or in part by the frameshift are underlined. The PR active site residues are in boldface. The PR cleavage sites in Gag and Gag-Pol are indicated by arrows. The p4 peptide is hatched. The large black dot represents the site of +1 translational frameshifting. The stop codon present in Gag is represented by an asterisk. Block substitution mutations s4 to s12 are shown as bars underneath the amino acids replaced by the peptide sequence AAGSAA. The transpositional competence of elements bearing these mutations is indicated.
Certain mutations in the p4 region common to Gag, p4, and PR block transposition. However, interpretation of this mutant phenotype is complicated by the fact that these mutations simultaneously alter Gag, PR, and p4 sequences. The transposition defect observed in these mutants could be secondary to a loss or gain of function in any or all of these proteins. The functional independence of the frameshift signal suggests a unique manner by which the critical Ty1 protein(s) affected by these mutations can be identified.
In this study, we describe the creation of frameshift transplant mutants of Ty1. These mutants enable us to specifically and unambiguously attribute the deleterious effects of mutations in the p4 region to the Ty1 PR. The p4 peptide, previously thought to be required for transposition, is shown here to be dispensable. We also physically define the amino terminus of the PR by microsequencing and thereby confirm the location of the Gag-PR cleavage site. We confirm that the mutations in this region affect the N terminus of the Ty1 PR; frameshift transplant mutants combined in cis with P4 region mutations are as defective as the same mutants in an otherwise wild-type Ty1 element. Frameshift transplantation is a novel approach that permits the isolated study of one or more proteins encoded by a single mRNA. This approach should prove useful in the study of viruses that are under selective pressure to maintain small, information-dense genomes.
MATERIALS AND METHODS
Strains and plasmids.
All experiments were performed with S. cerevisiae strain YH51 (MATa ura3–52 his4–539 lys2–801 spt3–202). All of the yeast Ty1 expression plasmids were constructed in the context of plasmid pJEF1105, in which the Ty1 element is marked with the Tn903 neomycin phosphotransferase (neo) gene and carried on a 2μm, URA3-marked plasmid (5).
pD109, containing the fs− mutation (in which the native Ty1 Gag-Pol frameshift is erased via a 1-bp deletion), was constructed by bridge mutagenesis (12). The sequences of the oligonucleotides used in this study are given in Table 1. The template was plasmid pJEF1081 previously linearized with MstII. The bridging oligonucleotide, JB30, deletes 1 bp from the GAG-POL overlap region, merging them into a single open reading frame (ORF) in plasmid pJEF1946. Plasmid pJEF1081 was constructed by substituting the internal HpaI-HindIII fragment from Ty1–912, for the equivalent fragment in plasmid pJEF724, an unmarked GAL-Ty1 plasmid (4). By this maneuver, we introduced the frameshift site of Ty1–912, which, unlike that of Ty1-H3, contains a unique MstII site near the frameshift site. The BstEII-BamHI fragment containing the 3′ end of Ty1-neo was subcloned into pJEF1946, generating pD109. A similarly constructed wild-type Ty1 H3/Ty1–912-neo element was transpositionally competent (not shown). Also, whereas pD109 itself was not competent for Ty1 transposition, it was competent for transposition in the presence of a plasmid producing only Gag (D. Moore and J.D.B., unpublished data).
TABLE 1.
Oligonucleotides used in this study
| Oligonucleo- tide | Sequencea |
|---|---|
| JB30 | CGACCTTCACTTGGGCCAGGAAC |
| JB1329 | CTTGTATGCATATGCTAATTGATGGTGGGTACGGAGA |
| JB1378 | AGCCTTTACATATGGAATCCCAACAATTATCTCAAC |
| JB1553 | CTCCTTCTCGAGGCAGCAGCATCACGA |
| JB1890 | GCGGATCCGAAGAACAACAGGGATCGAGAAAC |
| JB1891 | GCGGATCCTTAATGATGATGATGATGATGATTGATGGTGGGTACGGAGATATT |
| JB1992 | GCGGATCCGCGTCTGATGATGAACTCCCTGGA |
| JB1993 | ATAATGTCAGACCACCACCAAT |
| JB1994 | TGTCCTGGAAGTGAAATTGTA |
| JB2016 | GCGGATCCCATCTTAGGCCATAGCACGGACAACGATTCCATC |
| JB2017 | GCGGATCCGGATACATTGTGAGCCCTGGC |
| JB2018 | TTCCGGATCCCATCTTAGGCCATGAACCGATTCAATTGAACAATAAG |
| JB2019 | TATGGGATCCGGAATCGTTGTCCGTG |
| JB2531 | GCGGATCCGCTGCTCTTAGGCCAGAAACTTAC |
| JB2537 | AGCAGCCGATCCAGCAGCAGTAGTTGATTTACTGATGG |
| JB2538 | GCTGCTGGATCGGCTGCTCTGGGCCAGAAACTTACTG |
| JB2539 | AGCAGCCGATCCAGCAGCGTTCAATTGAATCGGTTC |
| JB2626 | CCATCAGTAAATCAACTACTGCAGCCGGCTCCGCAGCAAATAAGCACGACCTTCAT |
| JB2628 | TGAACCGATTCAATTGAACGCAGCCGGCTCCGCAGCACTGGGCCAGAAACTTACT |
Restriction sites are underlined. Nucleotides that introduce mutations are in boldface.
The fs− mutant, from which the frameshift signal was removed, was constructed by being subcloned into pJEF1105 as an HpaI-BstEII fragment from pD109. To make the fst1 and fst2 mutants, two PCR products were synthesized and combined in a three-piece ligation with HpaI-BstEII-digested pJEF1105. PCR product 1 was amplified with primers JB1993 and JB2017 and digested with HpaI and BamHI. PCR product 2 was synthesized with primers JB2016 and JB1994 and digested with BamHI and BstEII. For fst2, PCR product 1 was synthesized with JB1992 and JB1993. Product 2 was synthesized with JB2018 and JB2019. pD109 served as the template in all of these reactions. fst1-s7 and fst1-s8 were constructed via PCR-based site-directed mutagenesis of an XhoI-KpnI fragment of the fst1 mutant with oligonucleotides JB2626 and JB2628, respectively (20). The mutagenized fragments were subcloned into pJEF1105.
Prokaryotic expression construct pET3a GagPRHis6 was prepared via PCR amplification of pD109 with primers JB1378 and JB1329. The amplified products were cloned into pET3a via the NdeI site. pD109 was used as the template. To construct pET3a GagPR−His6, site-directed mutagenesis was performed on an XhoI-SalI fragment of pD109 in pBluescript with oligonucleotide JB1553. This changes the PR active site residues DSG to EAA and incorporates a unique XbaI site.
pET12a PPRHis6 was made by PCR amplification of pD109 with oligonucleotides JB1890 and JB1891. This fragment was cloned into the BamHI site of pET12a. pET12a PPRHis6 was prepared via site-directed mutagenesis with oligonucleotide JB1553.
PPRHis6-s7 and PPRHis6-s8 were synthesized in two stages via chimeric PCR. For PPrHis6-s7, oligonucleotides JB1890 and JB2537 were used to synthesize product 1 and oligonucleotides JB2536 and JB1891 were used to synthesize product 2. Similarly for PPRHis6-s8, oligonucleotide pairs JB1890 and JB2539 and JB2531 and JB1891 were used to synthesize products 1 and 2, respectively. The products of the first reactions were combined as a template in a second reaction with primers JB1890 and JB1891. The resulting products were cloned into the BamHI site of pET12a.
Expression and purification of Ty1 PR.
Escherichia coli strain BL21(DE3), containing various pET vector derivatives, was inoculated and grown to an optical density at 600 nm (OD600) of 0.6 in Luria-Bertani medium containing 100 μg of ampicillin per ml and 1 mM isopropyl-β-d-thiogalactopyranoside. Protein expression was induced for 4 h at 26°C. Following induction, the cells were pelleted and lysed via sonication in binding buffer (0.5 M NaCl, 20 mM Tris-HCl [pH 7.9], 5 mM imidazole, 10% glycerol). The lysate was cleared by centrifugation in an SS-34 rotor at 19,000 rpm for 30 min. The His6-tagged protein was purified under native conditions over Ni-nitrilotriacetic acid resin (Qiagen). The 0.75-ml column was washed with 10 volumes of binding buffer. Ty1PRHis6 fusion protein was eluted in binding buffer containing 300 mM imidazole.
cDNA synthesis assays.
Yeast cells harboring the indicated plasmid were grown for 24 h at 24°C in uracil-negative SC medium containing 2% raffinose (SC −ura, 2% raffinose). The cells were then diluted to an OD600 of 0.6 and induced with the addition of galactose to a final concentration of 2%. Five milliliters of cells at an OD600 of 2.0 was then pelleted and washed once in water. Nucleic acids were extracted from the cell pellet by bead beating with glass beads in 400 μl of DNA extraction buffer (0.5 M NaCl, 20 mM Tris-HCl [pH 7.5], 1 mM EDTA, 1% sodium dodecyl sulfate), 200 μl of buffer-saturated phenol, and 200 μl of chloroform. The lysate was centrifuged for 10 min in an Eppendorf microcentrifuge at 14,000 rpm. The supernatant was extracted with phenol chloroform and then with chloroform before the nucleic acids were ethanol precipitated. Approximately 10 μg of nucleic acid was digested with 20 U of EcoRI and 2 μg of RNase A in a 15-μl reaction mixture.
Immunoblotting.
Ty1 expression was induced for 24 h at 24°C as described above. The cell pellets from 2 ml of culture with an OD600 of 1.0 to 1.5 were lysed in 40 μl of buffer B-EDTA (20 mM Tris [pH 7.9], 15 mM KCl, 0.1 mM EDTA) with PR inhibitors and phenylmethylsulfonyl fluoride (1 mM) with a Mini-Bead Beater (Biospec Products) at a high setting for 1 min. The lysate was cleared of cell debris by centrifugation in an Eppendorf centrifuge at 14,000 rpm. The protein concentration was determined by the method of Bradford. Samples (5 μg) were then used for immunoblotting. After electrophoresis, proteins were transferred to Immobilon membranes at 300 mA for 2 h. Membranes were blocked in 3% milk–0.1% Tween 20 in phosphate-buffered saline (PBS). Primary antibody R2-F was applied in blocking solution for 1 h at 1:10,000 dilution. The blots were washed three times for 10 min in PBS–0.1% Tween 20. Secondary antibody (antirabbit-horseradish peroxidase conjugate [Amersham]) was applied at a 1:7,500 dilution for 40 min. The membranes were washed as described above and developed with the Amersham ECL enhanced chemiluminescent system as per the manufacturer's instructions.
Transposition assays.
Yeast cells, grown as patches, were replica plated from SC −ura, 2% glucose to SC −ura, 2% galactose and grown at 26°C for 4 days. The patches were then replica plated to SC −ura, 2% glucose and grown overnight at 30°C. The patches were replica plated to yeast-peptone-dextrose (YPD) and, after overnight growth at 30°C, replica plated to SC plus 0.1% 5-fluoroorotic acid. A portion of the patch was then diluted and plated on both YPD and YPD plus 75 μg of G418 per ml. The transposition frequency of the wild-type element is defined as 100%.
RESULTS
Figure 1 gives a schematic representation of the p4 region of Ty1. This region is defined at its amino terminus by the Gag-p4 cleavage site and at its carboxy terminus by the Ty1 frameshift signal. The p4 peptide shares sequences with Gag and PR. Block substitution mutations in the region have been described previously: two of them, s7 and s8, block transposition (15).
The sequence of the carboxy terminus of processed Ty1 CA (capsid) protein has previously been determined from yeast VLP preparations to be TARAH (15). This places the CA-p4 cleavage site between amino acids 401 and 402. The p4 peptide liberated by the proteolytic maturation of Gag to CA has not been detected in yeast lysates or VLPs, however, which raises the possibility that additional proteolytic events may be responsible for shaping the amino terminus of the PR. If this is the case, linker insertion mutants in the p4 region that alter the primary structure of the protein might block such further processing and thereby compromise transposition.
Definition of the Ty1 PR N terminus.
To determine the identity of the amino terminus of Ty1 PR, as defined by autoproteolytic processing, we expressed it in E. coli. Two series of constructs were prepared (Fig. 2A). The first contained the entire Gag ORF in frame with the PR ORF, and the endogenous frameshift signal was “erased,” via the fs− mutation, to permit expression in E. coli, in which the frameshift signal is nonfunctional. A carboxy-terminal hexahistidine tag was affixed to facilitate purification. Control constructs in which an active site mutation (DSG to EAA) was introduced in the PR were also prepared.
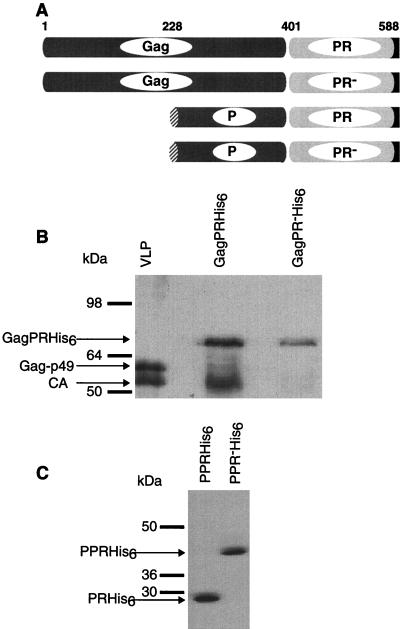
FIG. 2.
Expression of Gag-PR in E. coli and demonstration of autoprocessing. (A) Schematic representation of the expression constructs used to generate the underlying blots. The carboxy-terminal hexahistidine tag is shown in black. Relevant amino acid positions are shown. (B) Immunoblot of bacterial lysates in which the Gag-PR constructs were expressed. VLPs from yeast were separated in lane 1. (C) Appearance of the PR following expression of the PPR precursor protein. Polyclonal antibody R2-F, used in the immunoblot shown in panel B, recognizes Ty1 Gag. Antipeptide antibody JH695 (15), directed against an epitope in the p4 region, was used in the immunoblot shown in panel C.
The GagPRHis6 fusion is expressed as an 85-kDa precursor polypeptide in E. coli and is incompletely processed into two products. The intact fusion as well processed product can be detected with anti-Gag antibody; their electrophoretic mobilities correspond to those of the full-length and processed products, respectively (Fig. 2B). The latter product comigrates with authentic Ty1 CA from VLPs. Antibodies raised against a synthetic peptide corresponding to amino acids 418 to 440 of Ty1 Gag detect an immunoreactive band at approximately 29 kDa that corresponds to the Ty1 PR (Fig. 2C). The GagPRHis6 precursor and the Ty1 PR bands could also be detected by antihexahistidine antibodies (data not shown). No processing is observed in lysates prepared from GagPRHis6-expressing bacteria, as evidenced by a single 85-kDa band on the immunoblot detected with anti-Gag antibody (Fig. 2B). Taken together, these findings strongly suggest that the processing observed is autocatalytic and not the result of an adventitious E. coli PR.
A second series of constructs were prepared in which the first 300 amino acids of Gag were replaced with a 45-residue periplasmic targeting signal. These constructs, denoted PPR, are efficiently expressed and are capable of more complete autocatalytic processing. The processed PPRHis6 fusion proteins were purified from cells expressing the GagPRHis6 construct and the PPRHis6 construct and subjected to N-terminal microsequencing. The first five residues of PRHis6 from the GagPRHis6 construct were determined to be NVSTS. The first seven residues of PRHis6 from the PPRHis6 construct were determined to be NVSTSNN (Table 2). These sequences precisely match the predicted amino terminus of the PR, assuming it is produced by cleavage similar to that producing CA and p4. The identity of the carboxy-terminal residue of the CA protein was independently determined to be NNSKSKTARAH via mass spectroscopic analysis of Ty1 VLPs (J.F.L., R. Newitt, R. Aebersold, and J.D.B., unpublished results). We conclude that the carboxy terminus of Gag and the amino terminus of the PR are defined by a single autocatalytic processing event. For reasons that are unclear, we have been unable to detect the Ty1 PR by immunoblotting to confirm that it comigrates with the heterologously produced PR. However, in the mass spectroscopic analysis, we also found the PR-derived peptide IRSAHHIHSASSNPDINVVDAQKR in VLP preparations, directly confirming the presence of PR in VLPs (10).
TABLE 2.
Amino-terminal sequences of PRHis6 proteins in this study
| Sequencea | Construct |
|---|---|
| H2-NVSTS | GagPRHis6 |
| NH2-NVSTSNN | PPRHis6 |
| NH2-NVSTS | PPRHis6-s7 |
| NH2-NVSTS | PPRHis6-s8 |
The sequences were determined by amino-terminal microsequencing of affinity-purified fusion protein.
The possibility that mutations in the p4 region alter the specificity of autoprocessing and thereby compromise transposition was addressed by expressing PPR constructs bearing the s7 or s8 mutation and microsequencing their cleavage site. However, the autoprocessing sites in s7 and s8 mutants are identical to those in the wild type (Table 2). These results suggest that the transposition defects observed in s7 and s8 Ty1 elements do not result from aberrant processing at the Gag-PR junction.
Frameshift signal transplantation.
The endogenous +1 frameshift signal is essential for Ty1 transposition and is believed to secure the appropriate stoichiometry of Gag and the Gag-Pol polyprotein. It is located in a region of the Ty1 genome that encodes the carboxy terminus of Gag and the amino terminus of the PR (Fig. 1). The minimal frameshift signal is only 7 nucleotides long and can function independently of the surrounding context.
We reasoned that erasing the endogenous frameshift signal and relocating it to a region permissive for amino acid substitution mutants immediately adjacent to the Gag-PR cleavage site would result in an element incapable of producing the p4 peptide as a part of the Ty1 Gag protein (15). In these constructs, Gag should be synthesized exclusively as a CA protein with four extra C-terminal amino acids. To avoid complementation of the mutant elements in trans, a host strain (YH51) in which endogenous Ty1 elements are not expressed was used. This strain lacks the SPT3 gene, which is required for genomic Ty1 element transcription (22).
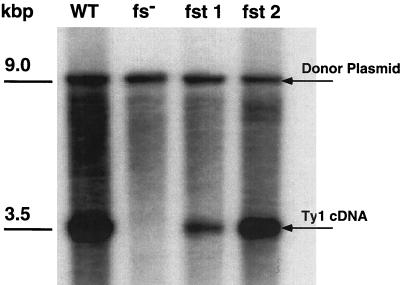
To erase the endogenous signal, its sequence, CTTAGGC, was replaced with CTGGGC, which deletes one nucleotide, substitutes a second (T →G), and preserves the native amino acid sequence of Gag-Pol. Ty1 elements in which the frameshift signal has been erased show a 100-fold decrease in transposition frequency (Fig. 3). The CA and IN proteins are detected in cell lysates, however, suggesting that proteolytic processing is not impaired (Fig. 4). These frameshift-null mutants do form VLPs, but do not synthesize cDNA (Fig. 5). No Gag immunoreactivity is observed in the control lysates (Fig. 4, lane 1), independently confirming that the host strain does not express endogenous Ty1 elements.
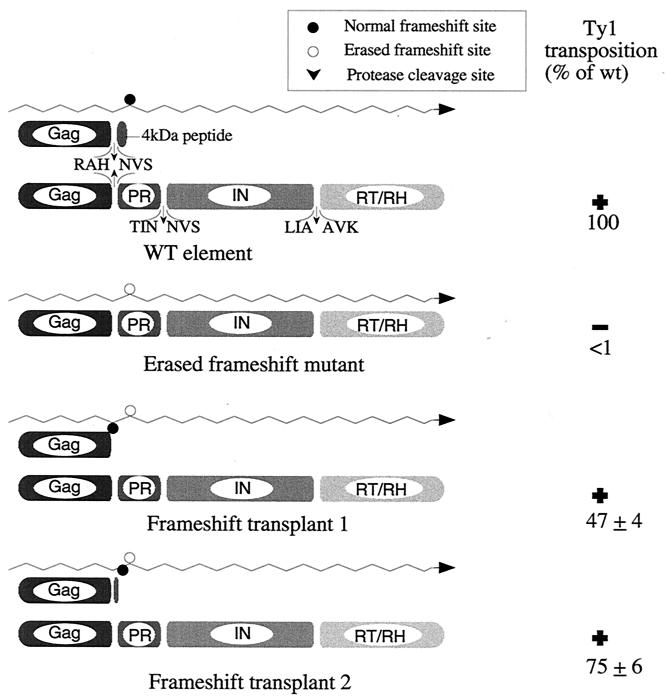
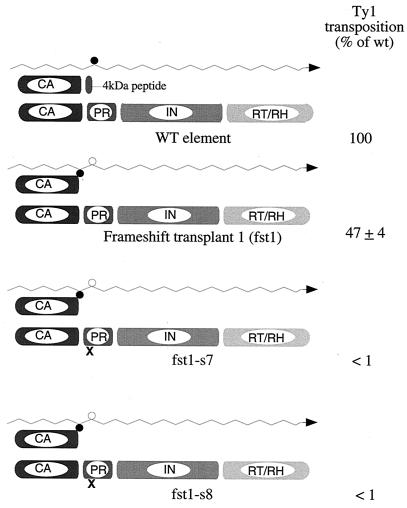
FIG. 3.
A schematic representation of the frameshift transplant constructs. The wavy line represents Ty1 mRNA. The transposition efficiency in strain YH51 is shown. The sequences surrounding the PR cleavage sites are shown in the wild-type (WT or wt) construct.
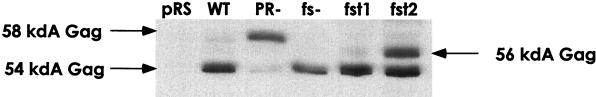
FIG. 4.
Frameshift transplant mutants produce the expected Gag proteins. A Western blot probed with polyclonal antibody R2-F, which recognizes Gag, is shown. Five micrograms of total yeast lysate from strain YH51 was loaded in each lane. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (9% polyacrylamide) was used to separate the Gag species. Lysates from a PR active site mutant (derived from pGM17) (16) are shown for comparison. pRS refers to plasmid pRS326, a 2μm, URA3-marked plasmid. The Gag immunoreactive band in the fst1 lane migrates as a tight doublet. The doublet cannot be resolved to a singlet with phosphatase treatment. The mobilities of the upper band in the fst1 doublet and of the 56-kDa fst2 species are consistent with the predicted mobilities of the unprocessed Gag species for these mutants. WT, wild type.
FIG. 5.
The frameshift transplant mutants reverse transcribe Ty1-neo mRNA. Total yeast nucleic acids from strain YH51 were digested with EcoRI, treated with RNase A, and separated on a 1% agarose gel. This gel was probed with 32P-labeled DNA generated from an XhoI-HindIII fragment of the Tn903 neo gene via random hexamer labeling. The lane background results from Ty1-neo insertions into yeast genomic DNA. WT, wild type.
Amino acid residues 405 to 422 of Gag (4 to 21 of PR) can be substituted for without adversely affecting transposition (15). Therefore, a new frameshift signal was inserted in this area (Fig. 3). Nucleotides 1512 through 1525 were substituted for with a 15-nucleotide cassette that contains a frameshift signal immediately followed by an ochre termination codon. This manipulation does not alter the length of the Ty1 transcript. The net effect is relocation of the frameshift signal immediately downstream of the Gag-PR cleavage site.
This frameshift transplant mutant (fst1) transposes at modestly reduced frequency (47% of that of the wild type) (Fig. 3), demonstrating that erasure of the endogenous frameshift can be suppressed in cis by introducing a wild-type copy of the frameshift signal upstream of it. As predicted, only a CA-sized species is observed (Fig. 4). The frameshift signal is therefore not limited to its native position in Ty1 mRNA. Interestingly, the extent of the transposition defect is closely paralleled by a decreased level of cDNA synthesis (Fig. 5).
A second frameshift transplant mutant was prepared in which the frameshift signal was relocated to a region approximately halfway into the p4 region. This construct (fst2) also transposes with only a modest reduction in efficacy (Fig. 3). In lysates from cells expressing this construct, Ty1 CA and a Gag species approximately 2 kDa larger than native CA can be detected if the relocated frameshift is functional, and the larger form of Gag presumably serves as a substrate for Ty1 PR (Fig. 4). The levels of cDNA synthesized by this mutant are also slightly reduced and consistent with the observed transposition defect (Fig. 5). Both fst1 and fst2 produced VLPs that behave indistinguishably from wild-type VLPs on sucrose density gradients.
We also tested the ability of these frameshift transplant mutants to transpose in a rad52 spt3 yeast strain (BY3227). RAD52 is required for multiple cellular functions, including homologous recombination and the repair of double-stranded breaks in cellular DNA. Newly synthesized Ty1 cDNA can enter the host genome via Ty1 integrase-mediated targeted integration or via homologous recombination with an existing element (13, 19). The latter pathway is RAD52 dependent. If the p4 region that is deleted as part of Gag in the frameshift transplant constructs is required for targeted recombination, then a transposition defect may not be unmasked in a RAD52 background. While the overall efficiency of transposition was decreased in a rad52 spt3 mutant strain, the relative efficiencies were preserved (data not shown). We therefore conclude that the p4 region is not required for targeted integration.
While the p4 region is dispensable for transposition, it remains formally possible that linker insertion mutations in the p4 region confer a gain-of-function phenotype upon Ty1 Gag. This gain of function could be responsible for the observed transposition defect. To address this possibility, we constructed linker insertion mutations s7 and s8, which are known to block transposition, in the context of frameshift transplant mutant no. 1 (fst1). The fst1-s7 and fst1-s8 double mutants were then tested for transpositional competence. As shown in Fig. 6, neither of the double mutants is capable of transposition. Linker insertion s7 and s8 mutants do not compromise Ty1 PR activity (15). This result effectively rules out the possibility that a gain of function in Ty1 Gag or p4 is responsible for the transposition defect observed in the s7 or s8 mutants.
FIG. 6.
Schematic of frameshift transplant s7 and s8 double mutants. Transposition frequencies in strain YH51 are indicated. The transposition frequencies reported are typical and did not vary by more than 5% in any independent experiment. WT or wt, wild type.
DISCUSSION
This study describes a novel technique called frameshift transplantation. This technique allows a previously unattainable insight into the relationship between the carboxy terminus of Ty1 CA and the amino terminus of the Ty1 PR and the p4 peptide. The amino-terminal sequence of the Ty1 PR is generated by the same cleavage that generates the carboxy terminus of Ty1 CA. Ty1 CA protein can be made in one of two ways: as the product of Gag-p49 processing or via processing of Gag-Pol-p199. We have shown that the cleavage site in both of these cases is the same. The p4 peptide liberated by the proteolysis of Gag-p49 to CA (Gag-p45) is shown to be dispensable for transposition. Mutations in the p4 region that block transposition are conclusively demonstrated to be uniquely attributable to an effect on the PR through the use of the frameshift transplant technique.
The exact positions of the Ty1 PR cleavage sites between PR and integrase (PR/IN) and integrase and reverse transcriptase (IN/RT) have previously been reported. The positions of these cleavage sites were recently confirmed by analysis of PR cleavage site mutants (14). The Gag-PR cleavage site was mapped based on the carboxy-terminal sequencing of endogenous Gag protein and confirmed by mass spectroscopic analysis (15). However, because we have never detected the p4 peptide in cell lysates or VLPs, it remained possible that additional events (such as multiple proteolytic cleavages) shaped the amino terminus of the PR. Heterologous expression of two different constructs confirms that the N terminus of the PR corresponds to its predicted position based on the C terminus of CA.
In this paper, the p4 region was shown to be dispensable for transposition both as part of the carboxy terminus of Gag and as an independent peptide. This was done by relocating the frameshift signal such that the p4 peptide sequence is expressed exclusively as part of the PR. The p4 peptide is similar in size to other retroviral nucleocapsid proteins, and it was thought that it might serve in a similar capacity. Recent evidence from our laboratory has shown the N-terminal segment of PR may facilitate at least one nucleocapsid-like function (J.F.L., G.M., and J.D.B., submitted for publication).
Ty1 cDNA can enter the host genome via targeted integration, an integrase-dependent event, or by homologous integration, a RAD52-dependent event (19). If the p4 region was necessary for targeted integration, it might have been possible for Ty1 cDNA synthesized by these mutants to enter the host genome via homologous recombination. However, we found that the frameshift transplant constructs were capable of transposition even in a rad52 spt3 strain background. This effectively rules out any role for the p4 region of Ty1 Gag and the p4 peptide in targeted integration.
Frameshift signals are found in a majority of retroviruses. In fact, the human T-cell leukemia virus type 1 and the mouse mammary tumor virus contain two independent frameshift signals. Many retroviruses position their frameshift signal between the structural and enzymatic components of the virus. The positioning typically ensures that structural components are synthesized in excess of the enzymatic components. The fact that we are able to relocate the frameshift signal and retain a functional retroelement raises interesting questions regarding how frameshifts arose in retroelement evolution. Both functional frameshift transplant constructs transpose at slightly reduced efficiencies, suggesting that the position of the native frameshift sequence may have evolved to maximize transposition frequency.
Among yeast retrotransposons, Ty1, the closely related Ty2 element, and the more distantly related Ty3 and Ty4 contain +1 frameshifting signals. Interestingly, only Ty1, Ty2, and Ty4 use the conserved 7-nucleotide motif CTT AGG C. The stop codon in Ty2 is predicted to produce a similar 4-kDa peptide by proteolysis. However, in Ty4, the Gag stop codon is located downstream of the PR active site residues. The peptide released from the Ty4 Gag C terminus is therefore considerably longer than its analogs in Ty1 and Ty2.
It has been shown that mutations in the C-terminal Gag-p6 peptide in human immunodeficiency virus might affect particle release from the cell, although little effort was made to distinguish between the effects of these mutations on the p6 and overlapping sequences (24). The generality of the frameshift transplant approach suggests that it can be used to genetically separate the effects of mutations that affect sequences present in multiple viral proteins.
ACKNOWLEDGMENTS
J. F. Lawler, Jr., and G. V. Merkulov contributed equally to this work.
We thank Daniel Moore for technical assistance and David Garfinkel for communicating unpublished data.
This work was supported by NIH grant GM 36481 to J.D.B. and Medical Scientist Training grant GM-07309 to J.F.L.
REFERENCES
- 1.Adams S E, Mellor J, Gull K, Sim R B, Tuite M F, Kingsman S M, Kingsman A J. The functions and relationships of Ty-VLP proteins in yeast reflect those of mammalian retroviral proteins. Cell. 1987;49:111–119. doi: 10.1016/0092-8674(87)90761-6. [DOI] [PubMed] [Google Scholar]
- 2.Belcourt M F, Farabaugh P J. Ribosomal frameshifting in the yeast retrotransposon Ty: tRNAs induce slippage on a 7 nucleotide minimal site. Cell. 1990;62:339–352. doi: 10.1016/0092-8674(90)90371-K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boeke J D, Devine S E. Yeast retrotransposons: finding a nice quiet neighborhood. Cell. 1998;93:1087–1089. doi: 10.1016/s0092-8674(00)81450-6. [DOI] [PubMed] [Google Scholar]
- 4.Boeke J D, Garfinkel D J, Styles C A, Fink G R. Ty elements transpose through an RNA intermediate. Cell. 1985;40:491–500. doi: 10.1016/0092-8674(85)90197-7. [DOI] [PubMed] [Google Scholar]
- 5.Boeke J D, Xu H, Fink G R. A general method for the chromosomal amplification of genes in yeast. Science. 1988;239:280–282. doi: 10.1126/science.2827308. [DOI] [PubMed] [Google Scholar]
- 6.Clare J J, Belcourt M, Farabaugh P J. Efficient translational frameshifting occurs within a conserved sequence of the overlap between the two genes of a yeast Ty1 transposon. Proc Natl Acad Sci USA. 1988;85:6816–6820. doi: 10.1073/pnas.85.18.6816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dinman J D, Wickner R B. Ribosomal frameshifting efficiency and gag/gag-pol ratio are critical for yeast M1 double-stranded RNA virus propagation. J Virol. 1992;66:3669–3676. doi: 10.1128/jvi.66.6.3669-3676.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eichinger D J, Boeke J D. The DNA intermediate in yeast Ty1 element transposition copurifies with virus-like particles: cell-free Ty1 transposition. Cell. 1988;54:955–966. doi: 10.1016/0092-8674(88)90110-9. [DOI] [PubMed] [Google Scholar]
- 9.Garfinkel D J, Boeke J D, Fink G R. Ty element transposition: reverse transcriptase and virus-like particles. Cell. 1985;42:507–517. doi: 10.1016/0092-8674(85)90108-4. [DOI] [PubMed] [Google Scholar]
- 10.Garfinkel D J, Hedge A-M, Youngren S D, Copeland T D. Proteolytic processing of pol-TYB proteins from the yeast retrotransposon Ty1. J Virol. 1991;65:4573–4581. doi: 10.1128/jvi.65.9.4573-4581.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kenna M A, Brachmann C B, Devine S E, Boeke J D. Invading the yeast nucleus: a nuclear localization signal at the C terminus of Ty1 integrase is required for transposition in vivo. Mol Cell Biol. 1998;18:1115–1124. doi: 10.1128/mcb.18.2.1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mandecki W, Caruthers M H. Mutants of the lac promoter with large insertions and deletions between the CAP binding site and the −35 region. Gene. 1984;31:263–267. doi: 10.1016/0378-1119(84)90219-1. [DOI] [PubMed] [Google Scholar]
- 13.Melamed C, Nevo Y, Kupiec M. Involvement of cDNA in homologous recombination between Ty elements in Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:1613–1620. doi: 10.1128/mcb.12.4.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Merkulov G V, Lawler J F, Jr, Eby Y, Boeke J D. Ty1 proteolytic cleavage sites are required for transposition: all sites are not created equal. J Virol. 2001;75:638–644. doi: 10.1128/JVI.75.2.638-644.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Merkulov G V, Swiderek K M, Brachmann C B, Boeke J D. A critical proteolytic cleavage site near the C terminus of the yeast retrotransposon Ty1 Gag protein. J Virol. 1996;70:5548–5556. doi: 10.1128/jvi.70.8.5548-5556.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Monokian G M, Braiterman L T, Boeke J D. In-frame linker insertion mutagenesis of yeast transposon Ty1: mutations, transposition and dominance. Gene. 1994;139:9–18. doi: 10.1016/0378-1119(94)90517-7. [DOI] [PubMed] [Google Scholar]
- 17.Moore S P, Rinckel L A, Garfinkel D J. A Ty1 integrase nuclear localization signal required for retrotransposition. Mol Cell Biol. 1998;18:1105–1114. doi: 10.1128/mcb.18.2.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Muller F, Bruhl K H, Freidel K, Kowallik K V, Ciriacy M. Processing of TY1 proteins and formation of Ty1 virus-like particles in Saccharomyces cerevisiae. Mol Gen Genet. 1987;207:421–429. doi: 10.1007/BF00331610. [DOI] [PubMed] [Google Scholar]
- 19.Sharon G, Burkett T J, Garfinkel D J. Efficient homologous recombination of Ty1 element cDNA when integration is blocked. Mol Cell Biol. 1994;14:6540–6551. doi: 10.1128/mcb.14.10.6540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Turchin A, Lawler J F., Jr The primer generator: a program that facilitates the selection of oligonucleotides for site-directed mutagenesis. BioTechniques. 1999;26:672–676. doi: 10.2144/99264st02. [DOI] [PubMed] [Google Scholar]
- 21.Wilson W, Malim M H, Mellor J, Kingsman A J, Kingsman S M. Expression strategies of the yeast retrotransposon Ty: a short sequence directs ribosomal frameshifting. Nucleic Acids Res. 1986;14:7001–7016. doi: 10.1093/nar/14.17.7001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Winston F, Durbin K J, Fink G R. The SPT3 gene is required for normal transcription of Ty elements in S. cerevisiae. Cell. 1984;39:675–682. doi: 10.1016/0092-8674(84)90474-4. [DOI] [PubMed] [Google Scholar]
- 23.Youngren S D, Boeke J D, Sanders N J, Garfinkel D J. Functional organization of the retrotransposon Ty from Saccharomyces cerevisiae: Ty protease is required for transposition. Mol Cell Biol. 1988;8:1421–1431. doi: 10.1128/mcb.8.4.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu S F, Edelmann K, Strong R K, Moebes A, Rethwilm A, Linial M L. The carboxyl terminus of the human foamy virus Gag protein contains separable nucleic acid binding and nuclear transport domains. J Virol. 1996;70:8255–8262. doi: 10.1128/jvi.70.12.8255-8262.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]