Abstract
Background
Limited epidemiological research has focused on translocations in soft tissue sarcomas, with no studies on bone sarcomas. This study aimed to clarify the epidemiology, prognosis, and genetic information of translocation-related sarcoma (TRS) and non-TRS patients.
Materials and methods
This retrospective cohort study used data from the Bone and Soft Tissue Tumor Registry in Japan (BSTTRJ) (2001-2019), the Kyushu University Hospital (KUH) repository (2001-2021), and a publicly available online dataset (MSK). The patients were categorized into TRS and non-TRS groups, and epidemiological, prognostic, and mutational diversity were compared.
Results
This study included 25 383 participants, of whom 4864 (19.2%) were TRS and 20 519 (80.8%) were non-TRS patients. TRS patients had significantly younger onset ages (median: 43 years, interquartile range: 29-59 years) than non-TRS patients (median: 63 years, interquartile range: 46-73 years). In the MSK cohort, microsatellite instability and tumor mutation burden scores in non-TRS were higher than in TRS, although they were rather low compared with the pan-cancer analysis. In the BSTTRJ cohort, survival analyses with the propensity score matching revealed that patients with TRS had better overall [hazard ratio (HR): 0.71, 95% confidence interval (CI) 0.63-0.81], metastasis-free (HR: 0.75, 95% CI 0.67-0.84), and recurrence-free (HR: 0.47, 95% CI 0.39-0.57) survival.
Conclusions
This study highlights differences in the epidemiology and genetic rearrangements of sarcoma.
Key words: sarcoma, bone sarcoma, soft tissue sarcoma, translocation-related sarcoma, epidemiology
Highlights
-
•
Patients with translocation-related sarcoma (TRS) exhibited a significantly younger onset age compared with non-TRS patients.
-
•
Non-TRS patients displayed higher levels of MSIsensor and TMB scores than those with TRS.
-
•
TRS patients exhibited better overall, metastasis-free, and recurrence-free survival rates than non-TRS patients.
Introduction
Sarcomas, which are notably less prevalent than carcinomas,1 are difficult to study; hence, research has progressed slowly. They are divided into two main categories based on their tumorigenic pathways: translocation-related sarcoma (TRS), characterized by tumor-specific fusion genes, and non-TRS, which lack fusion genes.2, 3, 4, 5, 6 Tumors with fusion genes constitute a greater percentage of sarcomas than carcinomas.5 TRS are commonly thought to manifest at a younger age and exhibit less genetic mutational diversity than non-TRS, as suggested by reports on individual tumors.7 Despite these observations, only a few comparative epidemiological studies have explicitly focused on classifying soft-tissue sarcomas based on the presence of fusion genes. To the best of our knowledge, no such studies have been conducted on bone tumors.
Recent large-scale and comprehensive genome projects have identified fusion genes involved in certain diseases that were previously considered non-TRS.8, 9, 10, 11 These findings raise questions as to whether the conventional concept of the TRS can uphold its traditional position.
The generalizability of the younger age of onset and the lack of genetic mutational diversity observed as distinctive features of TRS remains uncertain. Moreover, discussions regarding the tumorigenesis of fusion genes in non-TRS are only in the preliminary stages. In this study, we conducted a comparative analysis of the epidemiology of TRS and non-TRS patients using three distinct cohorts. The Bone and Soft Tissue Tumor Registry in Japan (BSTTRJ) was the largest cohort in our study but lacked genetic information. In contrast, the Kyushu University Hospital (KUH) repository cohort, which adhered to the same criteria as the BSTTRJ cohort, was a smaller local cohort that exclusively included patients with genetically confirmed translocations related to each TRS subtype. The medium-sized MSK cohort from the Memorial Sloan Kettering Cancer Center has publicly available online data on DNA mutations and translocations. Our study aimed to provide new epidemiological insights into TRS and non-TRS by combining cohorts of varying sizes and genetic information.
Materials and methods
Ethics
This study was conducted in accordance with the principles of the Declaration of Helsinki and was approved by the Musculoskeletal Tumor Committee of the Japanese Orthopaedic Association and the Institutional Review Board of Kyushu University, Japan (IRB number 21162-00).
Definition of TRS and non-TRS
TRS and non-TRS were defined based on previous reports and the latest World Health Organization classification,2, 3, 4, 5, 6 as shown in Supplementary Table S1, available at https://doi.org/10.1016/j.esmoop.2024.103726.
BSTTRJ cohort
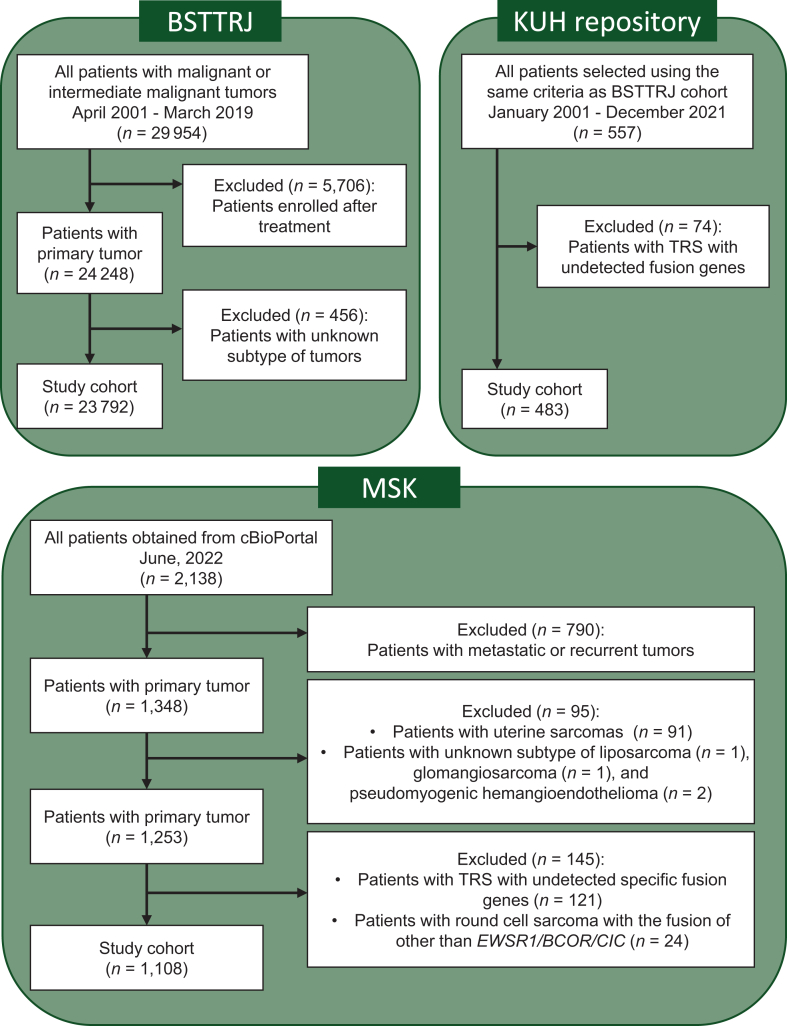
BSTTRJ is a nationwide, organ-specific cancer registry for bone and soft tissue tumors in Japan. The registry was launched in the 1950s, organized and funded by the Japanese Orthopaedic Association, and promoted by the National Cancer Centre in Japan.12,13 The Japanese Orthopaedic Association provides data on cases listed in this registry from 2001 to 2019. The extracted information included the patient’s age, sex, tumor size, location, depth, diagnosis, treatment details, and prognosis at the last follow-up. Note that the diagnosis of cases registered in this registry reflects the diagnosis made at each institution, and no central pathology review was made. In this cohort, no information on genetic alterations was not available. To prevent double entry, we obtained data from 29 954 patients, excluding those enrolled at KUH (Figure 1). Of these, only 24 248 primary patients were identified. Patients with liposarcoma or rhabdomyosarcoma of unknown subtype were excluded. Fibrosarcoma was reclassified as a plausible diagnosis if it could be determined from annotations. Patients with unknown details or those who could not be assigned to the present diagnostic criteria were excluded. Overall, 23 792 patients were included in the BSTTRJ cohort.
Figure 1.
Patient selection and cohort composition. In the BSTTRJ and KUH repository cohorts, patients with liposarcoma and rhabdomyosarcoma of unknown subtypes were excluded. Fibrosarcoma was reclassified based on annotations. TRS with undetectable specific fusion genes were excluded from the KUH repository. The analyzed cohort comprised 23 792 and 483 patients from the BSTTRJ and KUH repository cohorts, respectively. In the MSK cohort, 1348 primary patients were initially selected from a pool of 2138 patients. The exclusion criteria were uterine tumors, liposarcoma of unknown subtype, glomangiosarcoma, and pseudomyogenic hemangioendothelioma. The round cell sarcoma, other’ (excluding EWSR1/BCOR/CIC-related sarcoma), and TRS patients with undetected or uncertain specific fusion genes were also excluded. The resulting MSK cohort comprised 1108 patients. BSTTRJ, Bone and Soft Tissue Tumor Registry in Japan; KUH, Kyushu University Hospital; MSK, Memorial Sloan Kettering; TRS, translocation-related sarcoma.
KUH repository cohort
The data of patients diagnosed with sarcoma from 2001 to 2021 were retrieved from the archive of the Department of Orthopaedic Surgery, Kyushu University, Japan. We selected 557 patients based on the same inclusion and exclusion criteria as used in the BSTTRJ cohort (Figure 1). Additionally, only patients with specific gene rearrangements confirmed by reverse transcription polymerase chain reaction and direct sequencing or fluorescence in situ hybridization were included as having TRS. Thus, in this cohort, in addition to the same clinical information as in the BSTTRJ cohort, data on fusion genes were appended, but information on other genetic alterations was not available. A total of 483 patients were included in the KUH repository cohort.
MSK cohort
The MSK cohort data were obtained using cBioPortal.14 Data extracted from the MSK cohort included clinical information, follow-up data [overall survival (OS)], and genetic information from the targeted sequence using MSK-IMPACT (Memorial Sloan Kettering Cancer Centre – Integrated Mutation Profiling of Actionable Cancer Targets),15 such as data on fusion genes and somatic mutations (primarily analyzed), microsatellite instability scores (calculated as MSIsensor16), and tumor mutation burden (TMB)17 scores. Further cohort details have been described previously.11 We selected 1348 primary cases from 2138 cases for which clinical data were available (Figure 1). Then, we extracted 587 cases from the fusion gene data, excluding those with intragenic rearrangements. The clinical and fusion gene data were integrated. Patients with uterine tumors, liposarcoma of unknown subtype, glomangiosarcoma, or pseudomyogenic hemangioendothelioma, which lacked consensus on their TRS classification, were excluded. Patients with ‘round cell sarcoma, other’ other than EWSR1/BCOR/CIC-related sarcoma were excluded. Moreover, patients with undetectable specific fusion genes or uncertain fusion genes were excluded. Ultimately, 1108 patients were included in the MSK cohort.
The percentages of TRS patients with somatic DNA mutations were also calculated. Among the detected mutations, we excluded ‘benign’ or ‘likely benign’ pathogenicity, as classified by ClinVar (a public archive on human genetic variations and phenotypes with free access).18 Then, we identified pathogenic mutations with a cut-off of 0.75 in the functional analysis through hidden Markov models (version 2.3).19,20 Mutations not listed were considered nonpathogenic.
Statistical analysis
All the statistical analyses were conducted using R software (version 4.1.2, R Foundation for Statistical Computing, Vienna, Austria). Significance was defined as a P < 0.01. The sexes were identified and evaluated according to their biological characteristics. Patients with missing values for the analysis items were excluded from each analysis. The two groups were compared using the t-test or Mann–Whitney U test for quantitative variables and the chi-square test for qualitative variables. Survival curves for OS, metastasis-free survival (MFS), and recurrence-free survival (RFS) were estimated using the Kaplan–Meier method and compared using the log-rank test in the survival package. The P values were corrected using the Bonferroni method, which is used to compare variables among three or more groups. Hazard ratios (HRs) and 95% confidence intervals (CIs) were estimated using Cox univariate regression models. Furthermore, among the variables, age (<65 or ≥65 years), distant metastasis at diagnosis, chemotherapy, radiotherapy, and resection margin (defined as complete resection: R0 or incomplete resection: non-R0) were adjusted by propensity score matching (1 : 1 matching) using logistic regression in the MatchIt package.21
Results
Cohort characteristics
The clinical characteristics of each cohort are presented in Table 1. The BSTTRJ cohort had 23 792 patients and included 4627 (19.4%) TRS patients and 19 165 (80.6%) non-TRS patients (Supplementary Table S2, available at https://doi.org/10.1016/j.esmoop.2024.103726). Information on the missing values in the BSTTRJ cohort is shown in Supplementary Figure S1, available at https://doi.org/10.1016/j.esmoop.2024.103726. In the KUH repository cohort, 66 (13.7%) patients with TRS and 417 (86.3%) patients without TRS were included (Supplementary Table S3, available at https://doi.org/10.1016/j.esmoop.2024.103726). In the MSK cohort, 171 (15.4%) TRS patients and 937 (84.6%) non-TRS patients were enrolled in this study (Supplementary Table S4, available at https://doi.org/10.1016/j.esmoop.2024.103726).
Table 1.
The patients’ characteristics in each cohort. Information on missing values is not presented in the table
| Factor | Group | BSTTRJ | KUH repository | MSK |
|---|---|---|---|---|
| N | 23792 | 483 | 1108 | |
| Age, median (IQR) | 60 (41-72) | 58 (37-71) | 54 (32-66) | |
| Sex (%) | Male | 13 073 (54.9) | 264 (54.7) | 603 (54.4) |
| Female | 10 719 (45.1) | 219 (45.3) | 505 (45.6) | |
| Site (%) | Bone | 5677 (23.9) | 145 (30.0) | 184 (16.6) |
| Soft tissue | 18 115 (76.1) | 338 (70.0) | 924 (83.4) | |
| Site, detail (%) | Extremity | 14713 (61.9) | 308 (64.3) | 208 (22.2) |
| Trunk | 8431 (35.4) | 165 (34.4) | 56 (6.0) | |
| Head and neck | 648 (2.7) | 6 (1.3) | 66 (7.0) | |
| Other | 0 (0.0) | 0 (0.0) | 608 (64.8) | |
| Last follow-up data (%) | AWD | 4749 (20.0) | 113 (23.4) | |
| NED | 14 507 (61.0) | 300 (62.1) | ||
| DOD | 3255 (13.7) | 63 (13.0) | ||
| DOC | 385 (1.6) | 7 (1.4) | ||
| Alive | 791 (71.7) | |||
| Dead | 312 (28.3) | |||
| Diagnosis (%) | Atypical lipomatous tumor/well-differentiated liposarcoma | 4131 (17.4) | 52 (10.8) | 23 (2.1) |
| Undifferentiated pleomorphic sarcoma/malignant fibrous histiocytoma | 3138 (13.2) | 35 (7.4) | 93 (8.4) | |
| Osteosarcoma | 2290 (9.6) | 59 (12.2) | 77 (6.9) | |
| Myxofibrosarcoma | 1886 (7.9) | 57 (11.8) | 25 (2.3) | |
| Chondrosarcoma | 1603 (6.7) | 33 (6.8) | 25 (2.3) | |
| Myxoid liposarcoma | 1584 (6.7) | 33 (6.8) | 7 (0.6) | |
| Desmoid fibromatosis | 1375 (5.8) | 21 (4.3) | 19 (1.7) | |
| Leiomyosarcoma | 1133 (4.8) | 42 (8.7) | 54 (4.9) | |
| Dedifferentiated liposarcoma | 1037 (4.4) | 49 (10.1) | 109 (9.8) | |
| Synovial sarcoma | 783 (3.3) | 12 (2.5) | 5 (0.5) | |
| Malignant peripheral nerve sheath tumor | 706 (3.0) | 16 (3.3) | 38 (3.4) | |
| Ewing sarcoma | 616 (2.6) | 5 (1.0) | 66 (6.0) | |
| Chordoma | 454 (1.9) | 14 (2.9) | 0 (0.0) | |
| Solitary fibrous tumor | 347 (1.5) | 1 (0.2) | 21 (1.9) | |
| Pleomorphic liposarcoma | 309 (1.3) | 5 (1.0) | 14 (1.3) | |
| Dermatofibrosarcoma protuberans | 265 (1.1) | 1 (0.2) | 0 (0.0) | |
| Low-grade fibromyxoid sarcoma | 233 (1.0) | 1 (0.2) | 4 (0.4) | |
| Extraskeletal myxoid chondrosarcoma | 197 (0.8) | 1 (0.2) | 3 (0.3) | |
| Angiosarcoma | 194 (0.8) | 2 (0.4) | 77 (6.9) | |
| Epithelioid sarcoma | 179 (0.8) | 2 (0.4) | 20 (1.8) | |
| Alveolar soft part sarcoma | 169 (0.7) | 5 (1.0) | 2 (0.2) | |
| Alveolar rhabdomyosarcoma | 158 (0.7) | 2 (0.4) | 3 (0.3) | |
| Clear cell sarcoma | 136 (0.6) | 3 (0.6) | 7 (0.6) | |
| Pleomorphic rhabdomyosarcoma | 144 (0.6) | 2 (0.4) | 0 (0.0) | |
| Parosteal osteosarcoma | 130 (0.5) | 3 (0.6) | 1 (0.1) | |
| Dedifferentiated chondrosarcoma | 117 (0.5) | 4 (0.8) | 7 (0.6) | |
| Embryonal rhabdomyosarcoma | 72 (0.3) | 1 (0.2) | 36 (3.2) | |
| Mesenchymal chondrosarcoma | 54 (0.2) | 2 (0.4) | 0 (0.0) | |
| Inflammatory myofibroblastic tumor | 53 (0.2) | 0 (0.0) | 6 (0.5) | |
| Clear cell chondrosarcoma | 51 (0.2) | 1 (0.2) | 0 (0.0) | |
| Low-grade central osteosarcoma | 45 (0.2) | 3 (0.6) | 0 (0.0) | |
| Malignant giant cell tumor | 35 (0.1) | 7 (1.4) | 0 (0.0) | |
| Adamantinoma | 34 (0.1) | 3 (0.6) | 0 (0.0) | |
| Periosteal osteosarcoma | 27 (0.1) | 1 (0.2) | 0 (0.0) | |
| Malignant granular cell tumor | 23 (0.1) | 0 (0.0) | 0 (0.0) | |
| Periosteal chondrosarcoma | 20 (0.1) | 1 (0.2) | 0 (0.0) | |
| Round cell sarcoma (with EWSR1, BCOR, CIC rearrangement) | 18 (0.1) | 0 (0.0) | 8 (0.7) | |
| Malignant rhabdoid tumor | 12 (0.1) | 0 (0.0) | 0 (0.0) | |
| Spindle cell/sclerosing rhabdomyosarcoma | 12 (0.1) | 0 (0.0) | 10 (0.9) | |
| Infantile fibrosarcoma | 12 (0.1) | 0 (0.0) | 1 (0.1) | |
| Extraskeletal osteosarcoma | 6 (0.0) | 3 (0.6) | 4 (0.4) | |
| Intimal sarcoma | 2 (0.0) | 0 (0.0) | 15 (1.4) | |
| Sclerosing epithelioid fibrosarcoma | 1 (0.0) | 0 (0.0) | 4 (0.4) | |
| Malignant epithelioid hemangioendothelioma | 1 (0.0) | 0 (0.0) | 0 (0.0) | |
| Gastrointestinal stromal tumor | 0 (0.0) | 0 (0.0) | 272 (24.5) | |
| Desmoplastic small round cell tumor | 0 (0.0) | 0 (0.0) | 27 (2.4) | |
| Perivascular epithelioid cell tumor | 0 (0.0) | 0 (0.0) | 15 (1.4) | |
| Epithelioid hemangioendothelioma | 0 (0.0) | 0 (0.0) | 7 (0.6) | |
| Myxoinflammatory fibroblastic sarcoma | 0 (0.0) | 0 (0.0) | 2 (0.2) | |
| Myxoid chondrosarcoma | 0 (0.0) | 0 (0.0) | 1 (0.1) |
AWD, alive with disease; BSTTRJ, Bone and Soft Tissue Tumor Registry in Japan; DOC, died of other causes; DOD, died of disease; IQR, interquartile range; KUH, Kyushu University Hospital; MSK, Memorial Sloan Kettering; NA, not available; NED, no evidence of disease.
Age distribution in the TRS and non-TRS groups
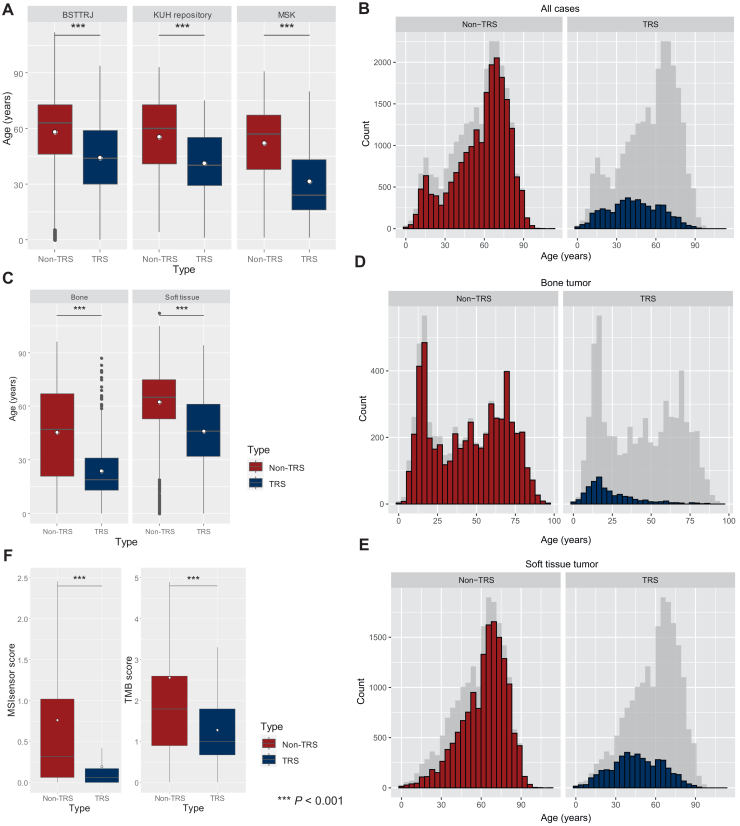
The age distribution at onset is shown in Figure 2. All three cohorts included any age, i.e. pediatric patients. In the BSTTRJ cohort, the median age of the participants was 44 years (interquartile range [IQR]: 30-59 years, mean: 44 years) for the TRS group and 63 years (IQR: 46-73 years, mean: 58 years) for the non-TRS group (P < 0.001 by Mann–Whitney U test) (Figure 2A and Supplementary Table S2, available at https://doi.org/10.1016/j.esmoop.2024.103726). In the KUH cohort, the median age was 40 years (IQR: 29-55 years, mean: 41 years) for the TRS group and 60 years (IQR: 41-73 years, mean: 56 years) for the non-TRS group (P < 0.001) (Figure 2A and Supplementary Table S3, available at https://doi.org/10.1016/j.esmoop.2024.103726). In the MSK cohort, the median age was 24 years (IQR: 16-43 years, mean: 20 years) for patients with TRS and 57 years (IQR: 38-67 years, mean: 52 years) for non-TRS patients (P < 0.001) (Figure 2A and Supplementary Table S4, available at https://doi.org/10.1016/j.esmoop.2024.103726).
Figure 2.
Distribution of onset age and MSIsensor and TMB scores between TRSs and non-TRSs. Bar plots of the three cohorts show a significantly younger age of TRS onset in each cohort (A). In the combined cohort, TRS had a single peak at ∼40 years, while non-TRS showed a bimodal peak at the teenage years and ∼70 years (B). Age distribution of bone and soft tissue (C-E). Bone non-TRS shows marked bimodality (D) and a broader spectrum (C and D), whereas soft tissue non-TRS shows unimodality (E). The MSIsensor scores were significantly lower in the TRS group (median: 0.06, IQR: 0.00-0.17) than in the non-TRS group (median: 0.32, IQR: 0.06-1.02). Similarly, TMB scores were significantly lower in the TRS group (median: 1.00 mut/Mb, IQR: 0.68-7.80 mut/Mb) and in the non-TRS group (median: 1.80 mut/Mb, IQR: 0.90-2.60 mut/Mb) (F). The outliers were considered significant but are not shown in Figures (F). White dots indicate mean values (A, C, and F). BSTTRJ, Bone and Soft Tissue Tumor Registry in Japan; IQR, interquartile range; KUH, Kyushu University Hospital; MSIsensor, microsatellite instability sensor score; MSK, Memorial Sloan Kettering; TMB, tumor mutation burden; TRS, translocation-related sarcoma. ∗∗∗P < 0.001.
In the integrated cohort, TRS patients had significantly younger onset ages (median: 43 years, IQR: 29-59 years, mean: 44 years) than non-TRS patients (median: 63 years, IQR: 46-73 years, mean: 58 years) (P < 0.001). The histogram of non-TRS patients in the three cohorts showed clear bimodality, with peaks at ages 15 and 70 years (Figure 2B). Furthermore, we analyzed the bone and soft tissue tumors separately (Figure 2C-E). The non-TRS bone tumor group showed marked bimodality and a broader spectrum (Figure 2D), but non-TRS in the other tumor groups, except for the osteosarcoma group, indicated unimodality (Supplementary Figure S2, available at https://doi.org/10.1016/j.esmoop.2024.103726), similar to that in the soft tissue non-TRS group (Figure 2E).
MSI and TMB between the TRS and non-TRS groups
MSIsensor and TMB scores were compared between the TRS and non-TRS groups in the MSK cohort (Figure 2F). The MSIsensor scores were significantly greater in the non-TRS group (median: 0.32, IQR: 0.06-1.02) than in the TRS group (median: 0.06, IQR: 0.00-0.17) (P < 0.001). TMB scores were also significantly greater in the non-TRS group (median: 1.80 mut/Mb, IQR: 0.90-2.60) than in the TRS group (median: 1.00 mut/Mb, IQR: 0.68-1.80, P < 0.001).
DNA mutations in the TRS group
One hundred and sixty-three different somatic mutations were detected in the TRS group in the MSK cohort (Supplementary Figure S3, available at https://doi.org/10.1016/j.esmoop.2024.103726). Of these, 30 were pathogenic. Among the 171 TRS patients, 87 (50.9%) had mutations, regardless of their functions; however, only 26 patients (15.2%) harbored at least one known pathogenic mutation.
Treatments and survival analyses between the TRS and non-TRS groups
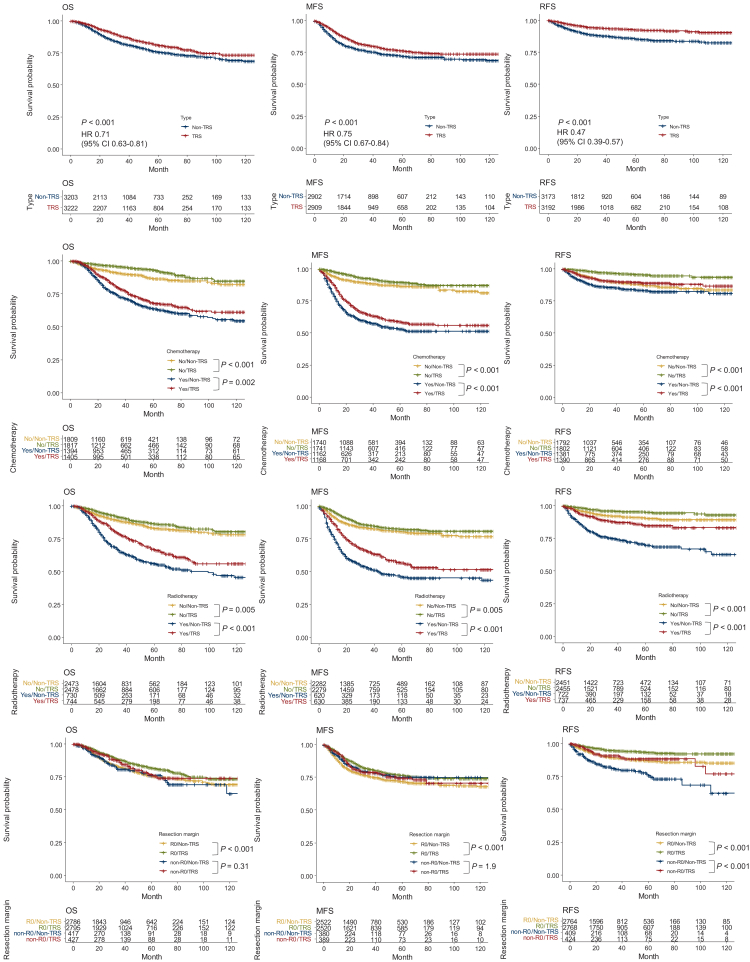
In the BSTTRJ cohort, the OS of the TRS group did not differ significantly from that of the non-TRS group (HR: 0.96, 95% CI: 0.89-1.04, P = 0.36) (Supplementary Figure S4A, available at https://doi.org/10.1016/j.esmoop.2024.103726). Significantly more patients with TRS, however, already had multiple lesions at diagnosis in the BSTTRJ cohort (P < 0.001) (Supplementary Table S2, available at https://doi.org/10.1016/j.esmoop.2024.103726). MFS in patients with local disease at the entry point did not differ between the TRS and non-TRS groups (HR: 0.96, 95% CI 0.88-1.05, P = 0.96) (Supplementary Figure S4B, available at https://doi.org/10.1016/j.esmoop.2024.103726). Furthermore, RFS in patients who had undergone surgery was significantly longer in the TRS group (HR: 0.50, 95% CI 0.43-0.59, P < 0.001) (Supplementary Figure S4C, available at https://doi.org/10.1016/j.esmoop.2024.103726). The MSK cohort also showed no significant differences in OS (HR: 1.03; 95% CI 0.76-1.39; P = 0.86) (Supplementary Figure S4D, available at https://doi.org/10.1016/j.esmoop.2024.103726).
Alternatively, survival analyses were carried out in the BSTTRJ cohort dataset and adjusted for potential confounding factors (Figure 3, top row). Details of the propensity matching procedure are presented in Supplementary Figure S5, available at https://doi.org/10.1016/j.esmoop.2024.103726. The results showed that OS (HR: 0.71, 95% CI 0.63-0.81, P < 0.001), MFS (HR: 0.75, 95% CI 0.67-0.84, P < 0.001), and RFS (HR: 0.47, 95% CI 0.39-0.57, P < 0.001) were better in the TRS group than in the non-TRS group. We also carried out survival analyses stratified by treatment histories. Patients with TRS had better prognoses regardless of whether they were treated with chemotherapy or radiotherapy (Figure 3, middle two rows); however, in cases of incomplete resection, there was no significant difference in OS or MFS between the two groups (Figure 3, bottom row).
Figure 3.
Survival curves between TRS patients and non-TRS patients in the propensity score-matched BSTTRJ cohort. The Kaplan–Meier curves for OS, MFS, and RFS were adjusted using propensity score matching for age (<65 or ≥65 years), distant metastasis at diagnosis, chemotherapy, radiotherapy, and resection margin (R0 or others). Patients with TRS exhibited a better prognosis than non-TRS patients in terms of OS, MFS, and RFS (top row). Survival analyses stratified by treatment history showed that patients with TRS tended to have a better prognosis, regardless of whether they were treated with chemotherapy or radiotherapy (2nd and 3rd row from the top). In patients who underwent incomplete resection, however, there was no significant difference in OS or MFS between the TRS and non-TRS groups (bottom row). BSTTRJ, Bone and Soft Tissue Tumor Registry in Japan; CI, confidence interval; HR, hazard ratio; MFS, metastasis-free survival; OS, overall survival; RFS, recurrence-free survival; TRS, translocation-related sarcoma.
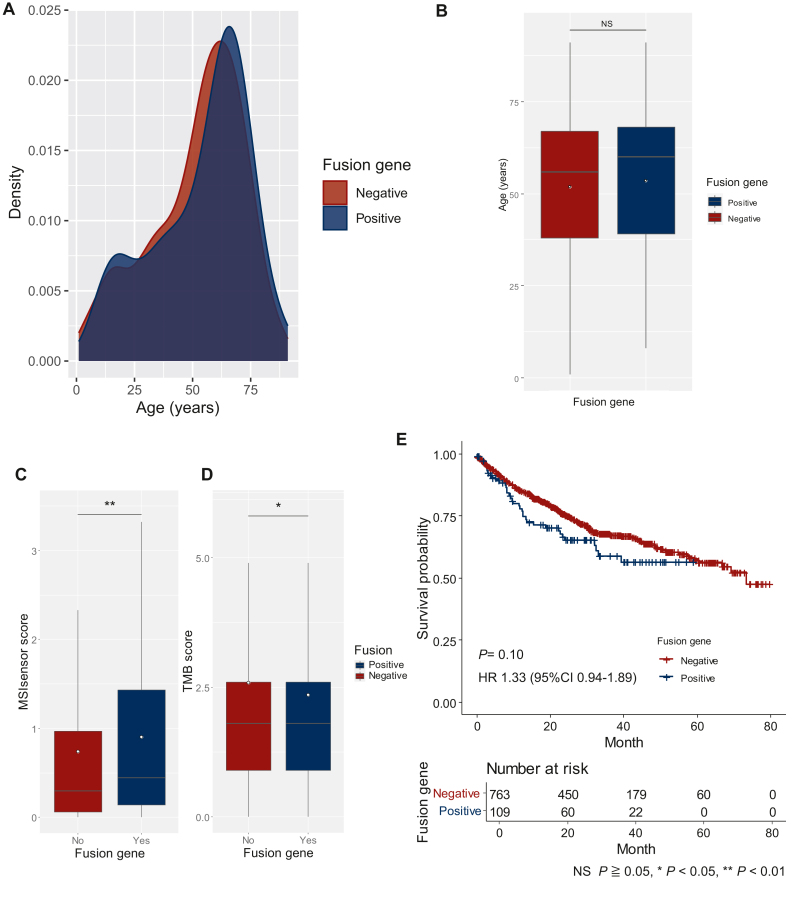
Non-TRS with uncertain fusion genes
The epidemiology and clinical impact of fusion genes in non-TRS patients were estimated using the MSK cohort. Fusion genes were detected in 13.0% of non-TRS patients (Supplementary Table S5, available at https://doi.org/10.1016/j.esmoop.2024.103726). Each non-TRS patient had unique fusion genes. In the non-TRS group, age was compared separately based on the presence or absence of fusion genes (Figure 4A and B). The ages of patients with fusion genes (median age: 60 years, IQR: 39-68 years, mean age: 54 years), however, did not differ significantly from those without fusion genes (median age: 56 years, IQR: 38-67 years, mean age: 52 years) (P = 0.22 by Mann–Whitney U test). Additionally, MSIsensor and the TMB scores were compared (Figure 4C and D). The MSIsensor scores differed significantly (P = 0.006), but the difference was much smaller than that between the TRS and non-TRS groups (median score: 0.45, IQR: 0.14-1.43 in the fusion gene-positive group versus median score: 0.30, IQR: 0.06-0.97 in the fusion gene-negative group) (Figure 4C). The TMB did not differ between the two groups (median score: 1.80 mut/Mb, IQR: 0.90-2.60 mut/Mb in both groups, P = 0.049) (Figure 4D). There was also no difference in OS (HR: 1.33, 95% CI 0.94-1.89, P = 0.10) (Figure 4E).
Figure 4.
The variables of the fusion gene-detected and non-fusion groups of non-TRS patients in the MSK cohort. The probability distributions of the onset age of fusion in the detected (blue) and non-detected (red) groups were almost identical (A). No difference in onset age was observed between non-TRS patients with fusion genes (mean: 54 years; SD: 20.6) compared with those without fusion genes (mean: 52 years; SD: 20.2) (B). The MSIsensor score was significantly lower in non-TRS patients without fusion genes (median: 0.30, IQR: 0.06-0.97) than among those with fusion genes (median: 0.44, IQR: 0.14-1.43) (C). The TMB did not differ between the two groups (median: 1.80 mut/Mb, IQR: 0.90-2.60 mut/Mb in both groups) (D). The outliers were considered significant but are not shown in Figures (C and D). White dots indicate mean values (B-D). IQR, interquartile range; MSIsensor, microsatellite instability sensor score; NS, non-significant; SD, standard deviation; TMB, tumor mutation burden; TRS, translocation-related sarcoma. NS P ≥ 0.05. ∗P < 0.05. ∗∗P < 0.01.
Discussion
We conducted a comparative epidemiological analysis focusing on the classification of TRS and non-TRS patients using three independent cohorts from different databases. For two cohorts (the KUH repository and MSK), only genetically confirmed TRS were included, and specific gene rearrangement details were collected. Conversely, the BSTTRJ cohort lacked fusion gene information; however, the distribution of the clinical information was similar to that of the KUH repository cohort, supporting the validity of the diagnosis in the BSTTRJ cohort. The two cohorts were from the same country, and the same criteria were applied. For the same reason, survival analysis was carried out for the larger cohort (the BSTTRJ cohort) but not for the KUH repository cohort.
Patients with TRS are generally presumed to have a younger age of onset, as indicated by studies on individual subtypes. Only one study, however, validated this assumption.3 In this study,3 the researchers did not confirm genetic translocations in TRS, consider the effects of confounding factors or include bone tumors. The present study confirms that TRS patients are significantly younger than non-TRS patients. Our findings suggest that the onset age may vary between Japanese and US cohorts; this discrepancy could be attributed to biases in included tumor subtypes. Notably, bone TRS peaked at a younger age than soft tissue TRS. The bimodal age distribution observed in non-TRS of the bone can be attributed to the fact that osteosarcoma (a representative of non-TRS) and Ewing sarcoma (a representative of TRS) have nearly identical onset ages.7
TRS patients are assumed to carry fewer somatic mutations than non-TRS patients. This notion has primarily been inferred from individual TRS reports,7 however, and its generalizability to the entire TRS population remains unclear. To address this uncertainty, we compared the MSIsensor and TMB scores between TRS and non-TRS groups. In general, an MSIsensor score ≥10 is presumed to be microsatellite instability-high (MSI-high),11,22 and TMB ≥10 or 20 mutations/megabase is often defined as TMB-high.23,24 In bone and soft tissue sarcomas, cases of high MSI or high TMB are considered rare; indeed, even without considering the TRS/non-TRS classification, the present results support this presumption. Similar to the previous study,24 both MSIsensor and TMB scores were significantly lower in the TRS group than for non-TRS in the selection criteria of this study, validating the prevailing theory and establishing the applicability of this concept to TRS. Some 15.2% of the TRS patients, however, exhibited at least one pathogenic mutation. A comprehensive mutation analysis may be needed, particularly for patients who are difficult to treat, as this may aid in identifying potential therapeutic targets.
Previous reports have indicated that TRS is associated with a greater rate of metastasis after initial treatment.3 Moreover, recurrence occurred less frequently in TRS patients than in non-TRS patients, according to crude analyses. Using adjustment for confounding factors, however, our findings revealed that patients with TRS had better MFS than non-TRS patients. Furthermore, patients with TRS consistently had better prognoses, including MFS, when stratified according to chemotherapy and radiation therapy. When resection margins were inadequate, however, no significant difference was observed between the TRS and non-TRS groups. These results suggest that TRS may cause more distant metastasis when the tumor is regionally present. This observation suggests that many patients with TRS present with multiple lesions at the time of diagnosis. Regarding local recurrence, patients with TRS exhibited a significantly longer RFS, which is consistent with findings from earlier reports. Therefore, resection with adequate surgical margins is important for the treatment of TRS.
Comprehensive sequencing analyses of specific non-TRS subtypes have recently revealed novel fusion genes in selected patients.8, 9, 10 This discovery sparked the intriguing possibility that translocations may act as drivers in some non-TRS patients. A thorough examination of the collection and exhaustive analysis of numerous rare sarcomas, however, are essential before validating this hypothesis. In the MSK cohort, fusion genes were identified in 13.0% of non-TRS patients, with a greater frequency in dedifferentiated liposarcoma, osteosarcoma, and undifferentiated pleomorphic sarcoma patients. These pleomorphic sarcomas are speculated to exhibit high genetic diversity. Notably, the detected fusion genes were unique to the individual patients and lacked reproducibility. To assess the clinical impact of these fusion genes, we categorized non-TRS patients into those with and without fusion genes. There was no significant difference in age at onset between the two groups, exhibiting nearly identical age distributions. Additionally, the TMB scores displayed no notable differences, while the MSIsensor score was slightly greater in the group with fusion genes. No discernible clinical differences were evident. Finally, OS did not vary between the two groups. These findings suggest that not all fusion genes are more likely to be expressed in younger individuals. Instead, a more accurate interpretation suggests that reproducible gene rearrangements that cause TRS are more prevalent in younger populations. It is also plausible that the mechanism of gene rearrangement in non-TRS differs from that in TRS. Furthermore, the fusion genes identified in non-TRS may not be etiologic agents but could merely be outcomes of a breakdown in the chromosome repair mechanism. Considering this perspective, trabectedin, which has demonstrated a beneficial effect on TRS,5,25, 26, 27, 28 may not necessarily exhibit the same effectiveness in non-TRS patients with fusion genes.
This study has several limitations. Most participants were Japanese, and we did not consider potential racial differences. The largest database (BSTTRJ) had inherent biases, as it compiles information based on independent evaluations from various institutions and has missing values. Moreover, translocation confirmation was lacking for all cases within this database despite the epidemiological distribution generally aligning with the smaller KUH repository cohort. Furthermore, genetic analyses were exclusively conducted in the USA-based cohort (MSK); this introduces a potential limitation due to differences in tumor subtype distributions between the Japan-based (KUH repository and BSTTRJ) and USA-based cohorts. Information on genetic alterations in the MSK cohort was based on annotated data, which is insufficient to address issues that depend on sequencing quality. More detailed molecular biology verification is needed before conclusions can be drawn regarding the pathogenicity of mutations and fusion genes.
Conclusion
This study encompasses multiple cohorts and comprehensively describes the epidemiology and genetic characteristics of TRS, highlighting differences in prognosis and mutational profiles. The epidemiological features of gene rearrangement in sarcoma, which have remained unclear due to a scarcity of data, have been successfully revealed.
Acknowledgements
The Centre for Clinical and Translational Research at Kyushu University was consulted regarding the statistical analyses without compensation. The authors would like to thank Editage for the English language review.
Funding
This work was supported by Grants-in-Aid for Scientific Research from the Japan Society for the Promotion of Science [grant numbers JP18K16627, JP23K08700] and research funds from the Graduate School of Medical Sciences, Kyushu University.
Disclosure
ME has received honoraria from Eisai, Novartis, and Taiho Pharma. All other authors have declared no conflicts of interest.
Data sharing
The data from the Bone and Soft Tissue Tumor Registry in Japan and the Kyushu University Hospital repository are not publicly accessible due to the privacy of individuals and are stored on the servers of the Musculoskeletal Tumor Committee of the Japanese Orthopaedic Association and Kyushu University Hospital, respectively. The remaining data, including genetic and clinical information, are available from the cBioPortal for Cancer Genomics (https://www.cbioportal.org/). The study protocol, statistical analysis plan, and analysis codes are available upon request to the corresponding author.
Supplementary data
References
- 1.Siegel R.L., Miller K.D., Fuchs H.E., Jemal A. Cancer statistics, 2021. CA Cancer J Clin. 2021;71:7–33. doi: 10.3322/caac.21654. [DOI] [PubMed] [Google Scholar]
- 2.Lucchesi C., Khalifa E., Laizet Y., et al. Targetable alterations in adult patients with soft-tissue sarcomas: insights for personalized therapy. JAMA Oncol. 2018;4:1398–1404. doi: 10.1001/jamaoncol.2018.0723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Penel N., Coindre J.M., Giraud A., et al. Presentation and outcome of frequent and rare sarcoma histologic subtypes: a study of 10,262 patients with localized visceral/soft tissue sarcoma managed in reference centers. Cancer. 2018;124:1179–1187. doi: 10.1002/cncr.31176. [DOI] [PubMed] [Google Scholar]
- 4.Schaefer I.M., Cote G.M., Hornick J.L. Contemporary sarcoma diagnosis, genetics, and genomics. J Clin Oncol. 2018;36:101–110. doi: 10.1200/JCO.2017.74.9374. [DOI] [PubMed] [Google Scholar]
- 5.Nakano K., Takahashi S. Translocation-related sarcomas. Int J Mol Sci. 2018;19:3784. doi: 10.3390/ijms19123784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mertens F., Antonescu C.R., Hohenberger P., et al. Translocation-related sarcomas. Semin Oncol. 2009;36:312–323. doi: 10.1053/j.seminoncol.2009.06.004. [DOI] [PubMed] [Google Scholar]
- 7.WHO Classification of Tumours Editorial Board . 5th ed. International Agency for Research on Cancer; Lyon, France: 2020. Soft Tissue and Bone Tumours. [Google Scholar]
- 8.Hirata M., Asano N., Katayama K., et al. Integrated exome and RNA sequencing of dedifferentiated liposarcoma. Nat Commun. 2019;10:5683. doi: 10.1038/s41467-019-13286-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Laetsch T.W., Roy A., Xu L., et al. Undifferentiated sarcomas in children harbor clinically relevant oncogenic fusions and gene copy-number alterations: a report from the Children's Oncology Group. Clin Cancer Res. 2018;24:3888–3897. doi: 10.1158/1078-0432.CCR-18-0672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chudasama P., Mughal S.S., Sanders M.A., et al. Integrative genomic and transcriptomic analysis of leiomyosarcoma. Nat Commun. 2018;9:144. doi: 10.1038/s41467-017-02602-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nacev B.A., Sanchez-Vega F., Smith S.A., et al. Clinical sequencing of soft tissue and bone sarcomas delineates diverse genomic landscapes and potential therapeutic targets. Nat Commun. 2022;13:3405. doi: 10.1038/s41467-022-30453-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ogura K., Higashi T., Kawai A. Statistics of bone sarcoma in Japan: report from the Bone and Soft Tissue Tumor Registry in Japan. J Orthop Sci. 2017;22:133–143. doi: 10.1016/j.jos.2016.10.006. [DOI] [PubMed] [Google Scholar]
- 13.Ogura K., Higashi T., Kawai A. Statistics of soft-tissue sarcoma in Japan: report from the Bone and Soft Tissue Tumor Registry in Japan. J Orthop Sci. 2017;22:755–764. doi: 10.1016/j.jos.2017.03.017. [DOI] [PubMed] [Google Scholar]
- 14.Center for Molecular Oncology at Memorial Sloan Kettering Cancer Center cBioPortal for Cancer Genomics. https://www.cbioportal.org/ Available at.
- 15.Cheng D.T., Mitchell T.N., Zehir A., et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): a hybridization capture-based next-generation sequencing clinical assay for solid tumor molecular oncology. J Mol Diagn. 2015;17:251–264. doi: 10.1016/j.jmoldx.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Niu B., Ye K., Zhang Q., et al. MSIsensor: microsatellite instability detection using paired tumor-normal sequence data. Bioinformatics. 2014;30:1015–1016. doi: 10.1093/bioinformatics/btt755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stenzinger A., Allen J.D., Maas J., et al. Tumor mutational burden standardization initiatives: recommendations for consistent tumor mutational burden assessment in clinical samples to guide immunotherapy treatment decisions. Genes Chromosomes Cancer. 2019;58:578–588. doi: 10.1002/gcc.22733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.National Center for Biotechnology Information ClinVar. https://www.ncbi.nlm.nih.gov/clinvar/ Available at.
- 19.Shihab H.A., Gough J., Cooper D.N., et al. Predicting the functional, molecular, and phenotypic consequences of amino acid substitutions using hidden Markov models. Hum Mutat. 2013;34:57–65. doi: 10.1002/humu.22225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shihab H.A. Functional analysis through hidden Markov models. http://fathmm.biocompute.org.uk/
- 21.Rosenbaum P.R., Rubin D.B. The central role of the propensity score in observational studies for causal effects. Biometrika. 1983;70:42–55. [Google Scholar]
- 22.Middha S., Zhang L., Nafa K., et al. Reliable pan-cancer microsatellite instability assessment by using targeted next-generation sequencing data. JCO Precis Oncol. 2017;2017 doi: 10.1200/PO.17.00084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chan T.A., Yarchoan M., Jaffee E., et al. Development of tumor mutation burden as an immunotherapy biomarker: utility for the oncology clinic. Ann Oncol. 2019;30:44–56. doi: 10.1093/annonc/mdy495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gounder M.M., Agaram N.P., Trabucco S.E., et al. Clinical genomic profiling in the management of patients with soft tissue and bone sarcoma. Nat Commun. 2022;13:3406. doi: 10.1038/s41467-022-30496-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kobayashi H., Iwata S., Wakamatsu T., et al. Efficacy and safety of trabectedin for patients with unresectable and relapsed soft-tissue sarcoma in Japan: a Japanese Musculoskeletal Oncology Group study. Cancer. 2020;126:1253–1263. doi: 10.1002/cncr.32661. [DOI] [PubMed] [Google Scholar]
- 26.Le Cesne A., Cresta S., Maki R.G., et al. A retrospective analysis of antitumour activity with trabectedin in translocation-related sarcomas. Eur J Cancer. 2012;48:3036–3044. doi: 10.1016/j.ejca.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 27.Blay J.Y., Leahy M.G., Nguyen B.B., et al. Randomised phase III trial of trabectedin versus doxorubicin-based chemotherapy as first-line therapy in translocation-related sarcomas. Eur J Cancer. 2014;50:1137–1147. doi: 10.1016/j.ejca.2014.01.012. [DOI] [PubMed] [Google Scholar]
- 28.Kawai A., Araki N., Sugiura H., et al. Trabectedin monotherapy after standard chemotherapy versus best supportive care in patients with advanced, translocation-related sarcoma: a randomised, open-label, phase 2 study. Lancet Oncol. 2015;16:406–416. doi: 10.1016/S1470-2045(15)70098-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.