Abstract
Neuronal development is a highly regulated process that is dependent on the correct coordination of cellular responses to extracellular cues. In response to semaphorin axon guidance proteins, the MICAL1 protein is stimulated to produce reactive oxygen species that oxidize actin on specific methionine residues, leading to filamentous actin depolymerization and consequent changes in neuronal growth cone dynamics. Crossing genetically modified mice homozygous for floxed Mical1 (Mical1 fl/fl ) alleles with transgenic mice expressing Cre recombinase under the control of a tyrosinase gene enhancer/promoter (Tyr::Cre) enabled conditional Mical1 deletion. Immunohistochemical analysis showed Mical1 expression in the cerebellum, which plays a prominent role in the coordination of motor movements, with reduced Mical1 expression in Mical1 fl/fl mice co‐expressing Tyr::Cre. Analysis of the gaits of mice running on a treadmill showed that both male and female Mical1 fl/fl , Tyr::Cre mutant mice had significant alterations to their striding patterns relative to wild‐type mice, although the specific aspects of their altered gaits differed between the sexes. Additional motor tests that involved movement on a rotating rod, descending a vertical pole, or crossing a balance beam did not show significant differences between the genotypes, suggesting that the effect of the Mical1 fl/fl , Tyr::Cre genetic modifications was only manifested during specific highly coordinated movements that contribute to running. These findings indicate that there is a behavioral consequence in Mical1 fl/fl , Tyr::Cre mutant mice that affects motor control as manifested by alterations in their gait.
Keywords: actin, cytoskeleton, gait, genetic modification, Mical1, reactive oxygen species, running, stride, tyrosinase
Neuronal development relies on coordinated cellular responses, including actin depolymerization triggered by MICAL1 in response to semaphorin proteins. Conditional deletion of floxed Mical1 in mice expressing Cre recombinase from a tyrosinase gene enhancer/promoter led to reduced Mical1 expression in the cerebellum, with mutant mice showing altered gait patterns during treadmill running. However, no significant differences were observed in other motor tasks, suggesting that the Mical1 mutation affects highly coordinated movements contributing to running.

1. INTRODUCTION
The actin cytoskeleton plays a central role in neuronal development by influencing the complex architecture of neurons and facilitating their functionality. 1 , 2 During early stages of neurogenesis, actin cytoskeleton dynamics influence neural progenitor cell shape, division, and migration. 3 As neurons differentiate, the actin cytoskeleton is integrally involved with the formation of growth cones at the tips of developing neurites, which extend and retract as they navigate toward their synaptic targets. 4 The polymerization and depolymerization of actin filaments in growth cones controls their motility, which allows them to respond to guidance cues in the environment. 5 , 6 At latter stages, the actin cytoskeleton contributes to synapse formation and plasticity by providing the structural framework necessary for the formation and remodeling of the dendritic spines that are the primary sites of excitatory synapses. 7 As a consequence, the actin cytoskeleton is crucial for the proper development and function of the nervous system through its roles in cell shape, movement, and synapse formation.
One protein family that plays prominent roles in axon guidance during neural development is the semaphorins, which serve as chemorepulsive or chemoattractive signals that guide the growth cones of developing axons towards appropriate sites. 8 Semaphorins act by binding to specific plexin and neuropilin receptors on neuronal growth cones and initiating intracellular responses that lead to changes in the organization of the actin cytoskeleton. 9 , 10 The cytoskeleton rearrangements induced by semaphorins may result in changes in the direction and magnitude of growth cone extensions and retractions. Some regions of the brain, including the hippocampus and cerebellum, continue to undergo post‐natal remodeling of neuronal connections to enable structural plasticity, which is accompanied by high expression of semaphorins and their receptors 11 Aberrant regulation of semaphorin signaling can lead to improper formation of neural circuits that could contribute to neurological disorders and diseases.
To identify proteins mediating the chemorepulsive effects of Drosophila melanogaster semaphorin‐1A (Sema1A) via its cognate receptor plexin A (PlexA), Terman et al. performed a yeast 2‐hybrid screen and discovered MICAL as a protein interacting with the PlexA cytoplasmic domain. 12 Deletion of mical resulted in defects in motor axon pathfinding, similar to effects observed for Sema1A and PlexA mutants, indicating that it plays an essential role in Drosophila neural development. Furthermore, numerous mutant mical alleles were identified in a Drosophila screen for abnormally structured neuromuscular junctions that connect the terminal end of motor nerves and muscles. 13 In vertebrates, there are three MICAL homologs (MICAL1, MICAL2, and MICAL3) that are expressed in overlapping but distinct patterns in the rat developing and adult nervous systems, with broad MICAL1 and MICAL3 expression and restriction of MICAL‐2 expression largely to the peripheral nervous system. 14 Deletion of Mical1 in mice led to defects in mossy fiber projections by dentate gyrus neurons in the hippocampus, 15 indicating that there is a conserved role for MICAL proteins in neuronal development in vertebrates and invertebrates.
The MICAL proteins contain several functional domains, including a flavoprotein monooxygenase domain that is activated upon binding to actin filaments, resulting in the oxidation of actin methionine residues 44 and 47 16 and consequent actin‐filament disassembly. 17 MICAL monooxygenase activity is regulated through protein–protein interactions that relieve the auto‐inhibitory effects of the protein carboxyl‐terminal region, 18 as well as by phosphorylation in response to growth factor stimulation. 19 The effects of MICAL actions on actin filaments in neurons are manifested in changes in growth cone morphology, axon guidance, and synapse formation, organization, and activity. 20 In Drosophila, mical deleted flies were severely compromised in their ability to fly associated with their aberrant neuromuscular junctions. 13 However, there are no data linking deletion or inactivation of MICAL genes with behavioral effects in mice or humans. Interestingly, Mical1 deletion led to increased mortality after myocardial stress due to hyperactivation of Ca2+/calmodulin‐dependent protein kinase II, indicating important roles in cardiac function. 21
To determine if Mical1 expression in neurons was behaviourally important in vivo, we conditionally deleted the Mical1 gene in tyrosinase‐expressing cells and examined the effects on motor functions. Tyrosinase is the rate‐limiting enzyme that catalyzes the oxidation of tyrosine in the biosynthetic pathway that leads to the production of melanin in the pigmented melanocyte cells that are derived from pluripotent neural crest cells. 22 The tyrosinase gene Tyr is also expressed in multiple regions of the brain, including the cortex, olfactory system, hippocampus, amygdala, basal ganglia, thalamus, hypothalamus, midbrain, pons, medulla oblongata, substantia nigra and cerebellum. 23 , 24 , 25 However, it has been proposed that tyrosinase expressed in non‐pigmented brain tissue may be inactive or participating in other enzymatic reactions that do not contribute to pigmentation. 23 By crossing homozygous Mical1 floxed mice 15 with mice expressing an X chromosome localized transgene encoding Cre‐recombinase regulated by the Tyr‐enhancer/promoter (Tyr::Cre), 25 we sought to determine if there were motor defects that would indicate a behavioral consequence of Mical1 deletion in Tyr expressing cells. While immunohistochemistry showed prominent expression of Mical1 in the cerebellum in the brains of adult mice, expression was reduced in Mical1 fl/fl , Tyr::Cre mice. Male mice ranging in age from 12 to 19 weeks that were wild‐type (WT) or Mical1 fl/fl , Tyr::Cre were analyzed for their gait, motor coordination, and balance. Relative to WT mice, male Mical1 fl/fl , Tyr::Cre mice ran significantly more slowly on a moving treadmill, which was associated with lower stride frequency and reduced coordination between left and right feet in the front or rear halves. Female Mical1 fl/fl , Tyr::Cre mice, ranging in age from 12 to 19 weeks, were also affected in their gait relative to WT mice, with alterations in their stance during running on a treadmill. Motor coordination and balance parameters on rotarod, beam, and vertical pole tests were not significantly different between WT and Mical1 fl/fl , Tyr::Cre mice. These observations indicate that there is an essential role of Mical1 in the control of some aspects of locomotion.
2. MATERIALS AND METHODS
2.1. Mouse model
Mical1 fl/fl mice on a predominantly C57Bl/6 background (B6J.129‐Mical1tm1Past from Mark E. Anderson, Johns Hopkins University School of Medicine) were crossed with Tyr::Cre mice (B6.Cg‐Tg(Tyr‐cre)1Lru/J), which had been backcrossed for 27 generations onto the C57Bl/6 background obtained from The Jackson Laboratories, at The Centre for Phenogenomics (TCP). Animal husbandry, ethical handling of mice, and all animal work were carried out according to guidelines approved by the Canadian Council on Animal Care and under protocols approved by the TCP Animal Care Committee (AUP 25‐0391H). For routine genotyping, all animals were ear notched at weaning, and samples were sent to the Transnetyx genotyping service for analysis. Knockout (KO) male mice were Mical1 fl/fl , Tyr::Cre/Y and WT male mice were Mical1 +/+ , Tyr::Cre/Y (5), Mical1 +/+ , +/Y (1), Mical1 fl/+ , +/Y (1) and Mical1 fl/fl , +/Y (2). Female KO mice were Mical1 fl/fl , Tyr::Cre/Tyr::Cre and WT female mice were Mical1 +/+ , +/+. Nine male mice of each genotype, or ten female wild‐type mice and eleven Mical1 KO mice, which had been housed in groups, and which were not necessarily littermates and ranged in ages from 12 to 19 weeks at the time of testing, were used for motor function testing during the daylight hours performed by the TCP Clinical Phenotyping service according to established Standard Operating Procedures. At the end of each test, mice were returned to their home cages.
2.2. Mical1 expression
Following euthanasia by carbon dioxide inhalation, brains were removed, fixed in formalin, and embedded in paraffin. Sagittal sections were cut at 4 μM thicknesses. All sections were deparaffinized and rehydrated, then stained with hematoxylin and eosin (H&E). Slides were subjected to epitope retrieval with BOND epitope retrieval solution 1 (AR9961, Leica Biosystems) for 10 min at 100°C, then stained with rabbit anti‐MICAL1 primary antibody (41112, Cell Signaling Technology) at 1:300 dilution for 1 h at room temperature using BOND Polymer Refine Detection kit (DS9800, Leica Biosystems) in a Leica BOND MAX instrument (Leica Biosystems).
2.3. Gait analysis
Animals were left in their home cages inside the testing room, anteroom, or on the day‐rack for at least 30 min prior to testing. Mice were placed on a motorized transparent ExerGait treadmill belt within a clear plexiglass corral (20 × 4 × 16.5 cm). With the treadmill speed set to 19 cm/s, mice were lowered onto the belt and upon achieving a constant running speed, a Basler high‐speed digital video camera was used to capture 100 frames per second for up to 20 s from the ventral side of each mouse. The 2000 frames collected for each mouse were analyzed using TreadScan v4.00 software according to the manufacturer's instructions. The mice were returned to their home cage after video capture. Clidox‐S (1:5:1) and 70% ethanol solutions were used for disinfecting surfaces between testing mice from different cages.
2.4. Rotarod
Animals were left sitting in their home cages inside the testing room, anteroom, or on the day‐rack for at least 30 min prior to testing. Following the acclimatization period, each mouse was placed onto a rod rotating at 4 rpm constant speed. A maximum of five mice were tested at once on the five available separate lanes. Once all animals managed to keep their balance at the constant speed for at least 10 s, the accelerator button was pressed to initiate continuous acceleration from 4 to the maximum 40 rpm in 5 min. The latency time for each mouse to fall off the rod or to allow themselves to passively rotate was recorded. Three separate trials were performed for each mouse in a single day with 15‐min breaks between each trial with mice being placed back into their home cage to rest. To assess motor learning ability, the procedure was repeated on two additional days. Clidox‐S (1:5:1) and 70% ethanol solutions were used for disinfecting surfaces between testing mice from different cages.
2.5. Vertical pole test
Animals were left in their home cages inside the testing room, anteroom, or on the day‐rack at least 30 min prior to testing. Each mouse was placed head upwards at the centre of a round 1 cm diameter 60 cm long metal rough‐surfaced pole. The pole was inclined at 90° and the time taken for each mouse to turn downwards and completely descend the pole was recorded. The entire test lasted for a few seconds only and mice were returned to their home cages. Clidox 1:5:1 and 70% Ethanol solutions are used for disinfecting surfaces between testing mice from different cages.
2.6. Balance beam
Animals were left in their home cages inside the testing room, anteroom, or on the day‐rack at least 30 min prior to testing. The beam apparatus consisted of two platforms (25 cm × 35 cm) connected with 100 cm long rectangular beams of 48, 24, 12, or 6 mm diameters that were suspended 50 cm above the floor. One platform was brightly illuminated while the opposite platform was dark and contained a box to provide a safe enclosure for mice. Each mouse was placed onto the brightly lit platform and allowed to explore for 2 min. Four consecutive training trials on the 48 mm bean were performed on the first day. On the following day, two test trials were performed for each of the three test width beams (24, 12, 6 mm). If the mouse fell off the beam, it was scored as the maximum latency time (5 min). The latency to traverse the beams and the number of times the hind feet slipped off were recorded. Following the test session, the mice were returned to their home cage. Clidox‐S (1:5:1) and 70% ethanol solutions were used for disinfecting surfaces between testing mice from different cages.
2.7. Statistics
Two‐tailed Student's t‐tests were used to compare between the indicated experimental groups using GraphPad Prism 10.0 software.
3. RESULTS
3.1. Mical1fl/fl, Tyr::Cre mutant mice have altered treadmill running gaits
To conditionally delete Mical1 in tyrosinase‐expressing cells, genetically modified mice homozygous for a floxed version of the Mical1 gene (Mical1 fl/fl ) 15 were crossed with mice expressing a Tyr::Cre transgene. 25 Immunohistochemical analysis showed expression in representative sagittal sections of brains from adult WT mice in the cerebellum (Figure 1A), part of the brain associated with motor control. 26 In contrast, Mical1 expression was reduced in the cerebellum in sections of representative Mical1 fl/fl , Tyr::Cre mice (Figure 1B).
FIGURE 1.
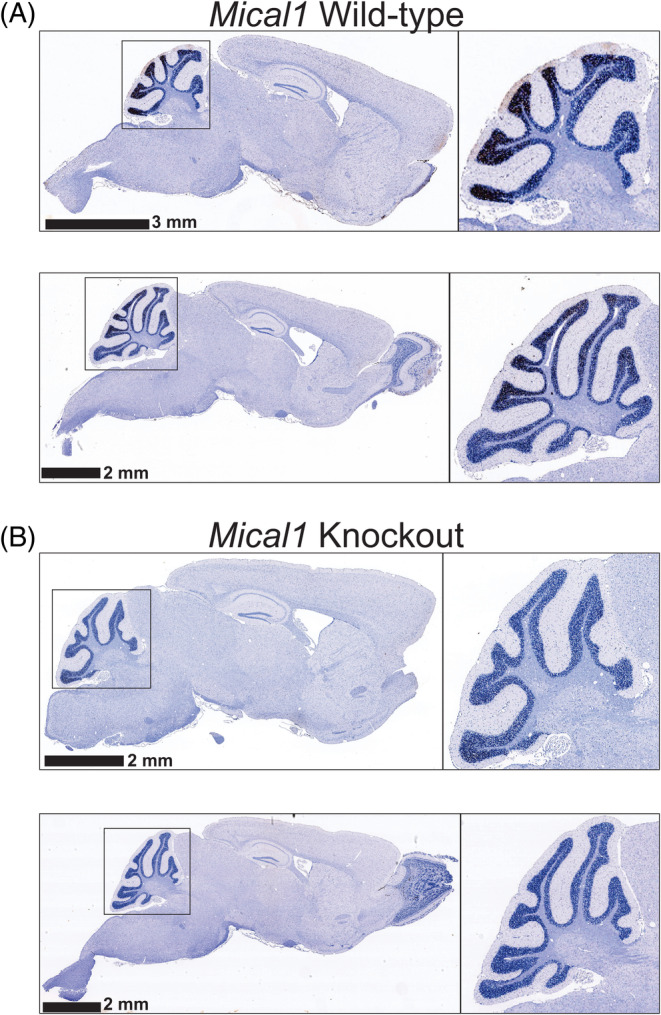
Immunohistochemistry of Mical1 expression in wild‐type and knockout mice. Sagittal sections of mouse brains from two each of (A) Wild‐type or (B) Mical1 knockout mice were analyzed by immunohistochemistry for Mical1 expression. The size of the scale bars is indicated in each image. Regions of whole brains used for the higher magnificent insets are indicated in the black squares.
Previous studies have anecdotally reported that conditional deletion mediated by Tyr::Cre of the actin cytoskeleton regulatory Rac1 27 or Cdc42 28 GTPases, as well as the Arp3 gene that promotes de novo F‐actin branching, 29 gave rise to mice with tremors, consistent with a key role for proper F‐actin regulation in motor control. In addition, conditional expression of oncogenic NRas G12D induced by Tyr::Cre‐induced recombination was associated with tremors and incoordination that impaired gait. 30 Given that previous research has shown that male and female mice have different running patterns, 31 , 32 the sexes were tested separately. Nine male wild‐type mice without possible Mical1 recombination (designated WT; average age 17.0 weeks) and nine Mical1 fl/fl Tyr::Cre/Y mice (designated KO; average age 15.9 weeks) were analyzed for their gaits on an ExerGait treadmill. With the treadmill speed set to 19 cm/s, mice were lowered onto the belt, and upon achieving a constant running speed, a Basler high‐speed digital video camera was used to capture 100 frames per second for up to 20 s. The 2000 frames collected for each mouse were analyzed using TreadScan v4.00 software. The overall average running speed, obtained by dividing the total distance traveled by the center of the animal by the time taken to travel that distance, of the Mical1 KO mice was significantly (t(16) = 2.940, p < 0.01) reduced relative to WT mice (Figure 2A). Overall running speed can be broken down into two components: the instantaneous running speed of each stride (the ratio of the stride length to the stride time); and the stride frequency (the ratio of the number of strides to the sum of the individual stride times). While there was no statistically significant (t(16) = 1.727, p > 0.05) difference in the average instant run speed (Figure 2B), the average stride frequency for all 4 ft was significantly (t(70) = 2.919, p < 0.01) reduced for Mical1 KO mice (Figure 2C). These results indicated that the Mical1 KO mice were slower than WT mice, predominantly due to their making fewer strides per unit time.
FIGURE 2.
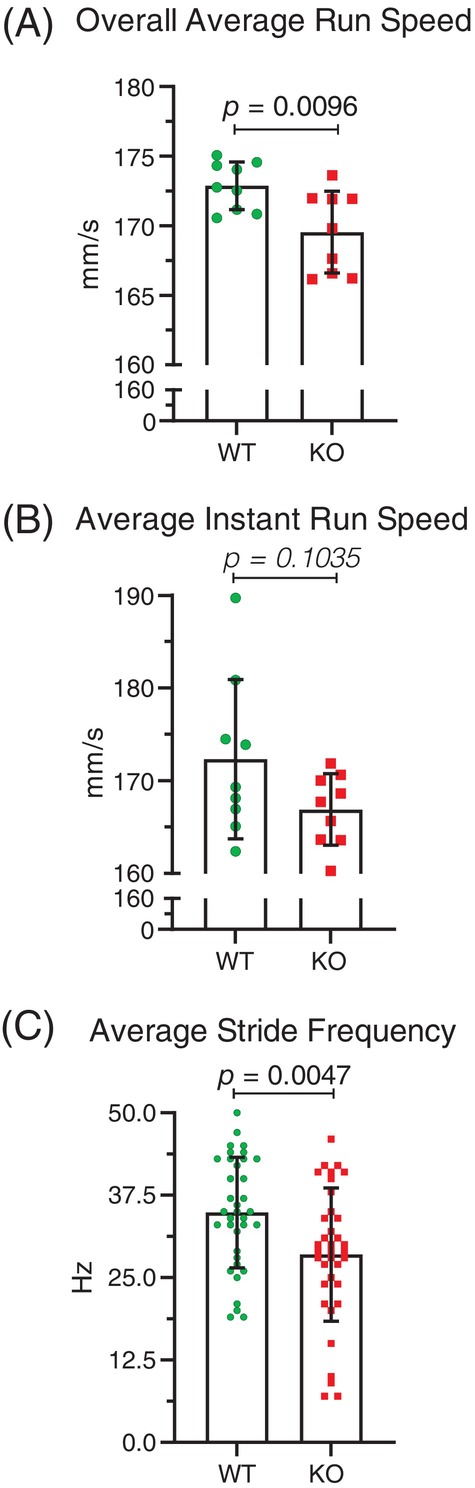
Mical1 fl/fl , Tyr::Cre mutant mice had reduced run speed and stride frequency in male mice. (A) Overall average run speeds were determined by analyzing high‐speed videos of male mice running on a treadmill for 20 s. Overall running speed (mm/s) was obtained by dividing the total distance traveled by the center of the animal by the time it took to travel that distance. Results presented are for each mouse. (B) Instantaneous running speed (mm/s) of a stride is the ratio of the stride length to the stride time. Results presented are for each mouse. (C) Stride frequency is the ratio of the number of strides to the sum of the stride times of these strides, which yields the number of strides per second (Hz) for a given foot. Results presented are for all 4 ft. Graphs indicate means ± standard deviation. Statistical tests with Student's t‐test. p values <0.05 indicated in bold typeface, p values ≥0.05 indicated in italic typeface. N = 9 mice per genotype. KO, knockout; WT, wild‐type.
By comparing the coordination of stride patterns between front and rear feet, or left and right feet on the same side, Mical1 KO mice were found to have a significantly (t(70) = 2.597, p < 0.05) reduced homologous coupling ratio between left and right feet on the front or rear half relative to WT mice (Figure 3A). In contrast, there was no significant (t(70) = 0.2362, p > 0.05) difference in the homolateral coupling ratio of both left or both right feet on each side of the mice (Figure 3B). By analyzing the number of feet from 0 to 4 supporting their running as a percentage of the total analysis time during active striding, Mical1 KO mice displayed a significantly altered gait, with proportionally longer time with no (t(16) = 3.333, p < 0.01) or 1 ft (t(16) = 3.178, p < 0.01) in contact and less time with 2 ft (t(16) = 4.233, p < 0.001) in contact with the treadmill relative to WT mice (Figure 4A–C). There were no significant differences in the proportion of time that either genotype was supported by 3(t(16) = 0.9992, p > 0.05) or 4 ft (t(16) = 0.05348, p > 0.05) during active striding (Figure 4D,E). These studies indicate that the Mical1 deletion in Tyr‐expressing cells affected stride patterns and frequency associated with overall reduced running speeds in male mice.
FIGURE 3.
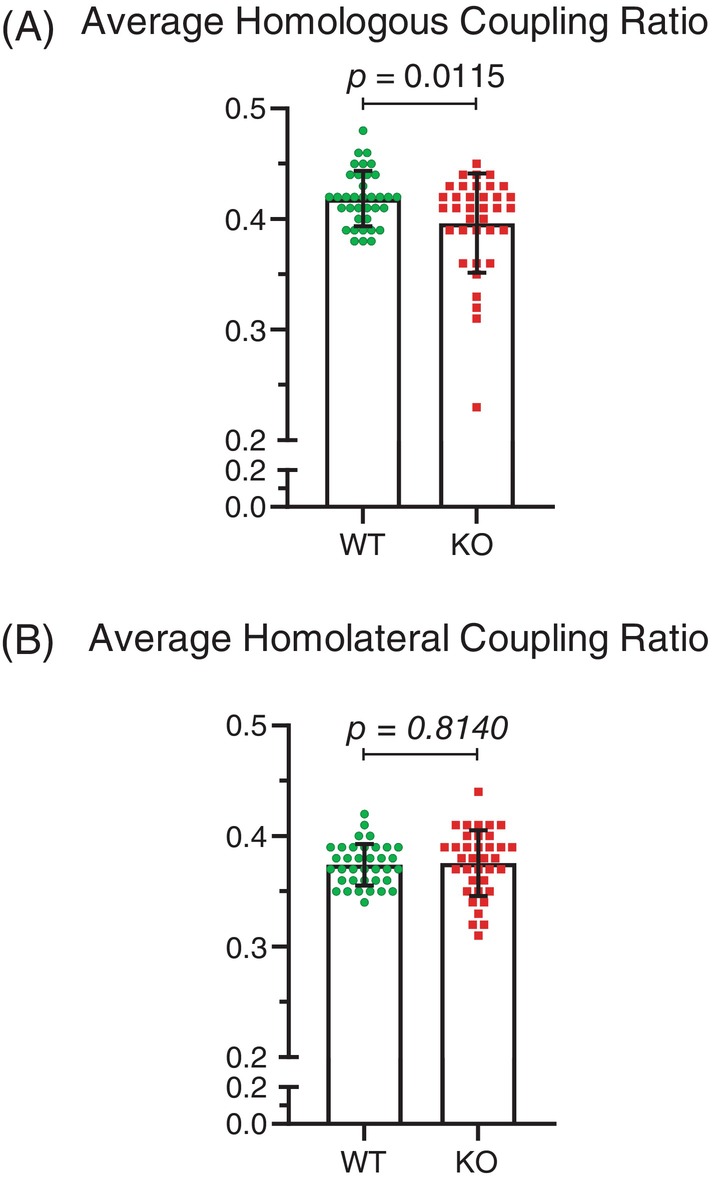
Mical1 fl/fl , Tyr::Cre mutant mice showed alterations in the coordination of left and right foot movements in front or right halves of male mice. (A) The homologous coupling ratio parameter is the fraction of the stride of a reference foot when the opposite foot on the same half (front half or rear half) starts its stride, revealing the coordination between left and right feet on the same half. (B) The homolateral coupling parameter is the fraction of the stride of a reference foot when the other foot on the same side (left or right side) starts its stride, indicating the coordination between front and rear feet on the same side. Results presented are for all 4 ft. Graphs indicate means ± standard deviation. Statistical tests with Student's t‐test. p values <0.05 indicated in bold typeface, p values ≥0.05 indicated in italic typeface. N = 9 mice per genotype. KO, knockout; WT, wild‐type.
FIGURE 4.
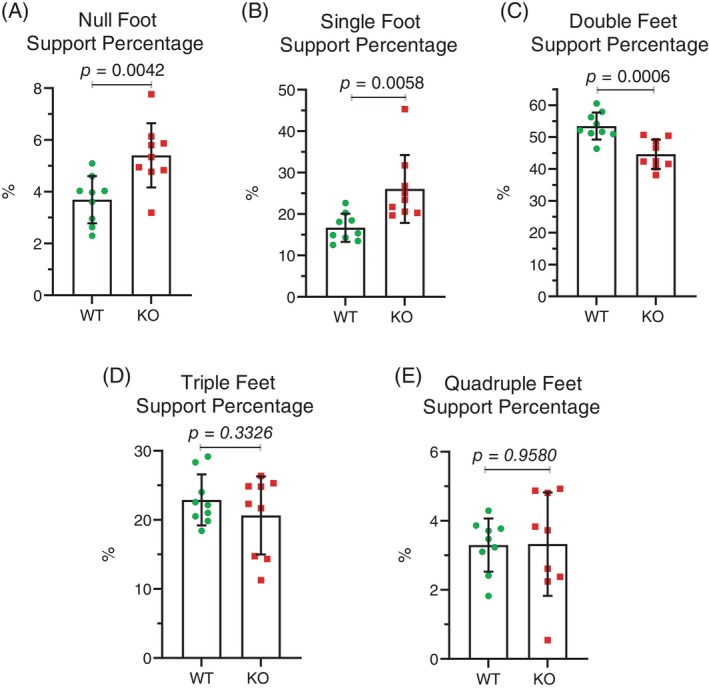
Mical1 fl/fl , Tyr::Cre mutant male mice had altered stride patterns during running. The percentage of the analysis time during which the number of feet touching the treadmill surface during running were: (A) none; (B) one; (C) two; (D) three; (E) four. Results presented are for each mouse. Graphs indicate means ± standard deviation. Statistical tests with Student's t‐test. p values <0.05 indicated in bold typeface, p values ≥0.05 indicated in italic typeface. N = 9 mice per genotype. KO, knockout; WT, wild‐type.
Ten female wild type mice without possible Mical1 recombination (average age 15.0 weeks) and eleven Mical1 fl/fl, Tyr::Cre/Tyr::Cre KO mice (average age 15.3 weeks) were also analyzed for their gaits on an ExerGait treadmill. In contrast to the male mice, there was no significant difference (t(19) = 3.405 p > 0.05) in overall average run speed between WT and Mical1 KO female mice (Figure 5). However, there were significant differences in the stride patterns with which the female mice ran. During each successive stride, leg position can be classified as either being in contact with the treadmill surface (stance) or in the process of being moved to the next contact point (swing). By comparing the percentage of each stride spent in the stance position, Mical1 KO were significantly (t(82) = 0.0166, p < 0.01) more often in the stance position overall relative to WT mice (Figure 6A). When each leg was examined individually, there were no significant differences for the front left (t(19) = 0.6755 p > 0.05) (Figure 6B) or front right legs (t(19) = 0.2179 p > 0.05) (Figure 6C) relative to WT mice. In contrast, both the left rear legs (t(19) = 0.1975 p < 0.05) (Figure 6D) and right rear legs (t(19) = 0.1825 p < 0.001) (Figure 6E) were significantly more often in the stance position in the Mical1 KO mice relative to WT. Accompanying the differences in stance, the legs of Mical1 KO mice were significantly (t(82) = 0.0166, p < 0.01) less often in the swing phase (Figure 7A) than in WT mice. While both front left (t(19) = 0.6755 p > 0.05) (Figure 7B) or front right legs (t(19) = 0.2179 p > 0.05) (Figure 7C) were equivalently in the swing phase in both genotype mice, the rear left (t(19) = 0.1975 p < 0.05) (Figure 7D) and rear right legs (t(19) = 0.1825 p < 0.001) (Figure 7E) were less often in the swing phase of each stride in Mical1 KO mice relative to WT.
FIGURE 5.
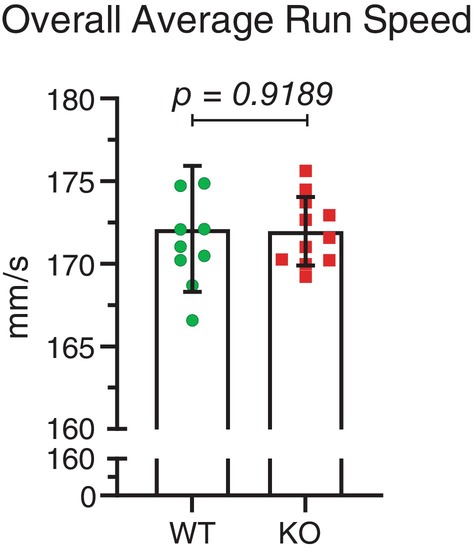
Mical1 fl/fl , Tyr::Cre mutant female mice did not have changes in overall running speed. Overall average runs speeds were determined by analyzing high‐speed videos of female mice running on a treadmill for 20 s. Overall running speed (mm/s) was obtained by dividing the total distance traveled by the center of the animal by the time it took to travel that distance. Results presented are for each mouse. Graphs indicate means ± standard deviation. Statistical tests with Student's t‐test. p value ≥0.05 indicated in italic typeface. N = 10 wild‐type mice, N = 11 Mical1 knockout (KO) mice. WT, wild‐type.
FIGURE 6.

Mical1 fl/fl , Tyr::Cre mutant female mice had increased stance times. (A) The percentage of time spent during a single stride in which feet were in contact with the treadmill surface (stance position) for all four legs for all mice (N = 40 wild‐type mice, N = 44 Mical1 knockout [KO] mice). For each foot, results are presented for the (B) front left, (C) front right, (D) rear left, (E) rear right stance positions. Graphs indicate means ± standard deviation. Statistical tests with Student's t‐test. p values <0.05 indicated in bold typeface, p values ≥0.05 indicated in italic typeface. N = 10 wild‐type mice, N = 11 Mical1 KO mice. WT, wild‐type.
FIGURE 7.
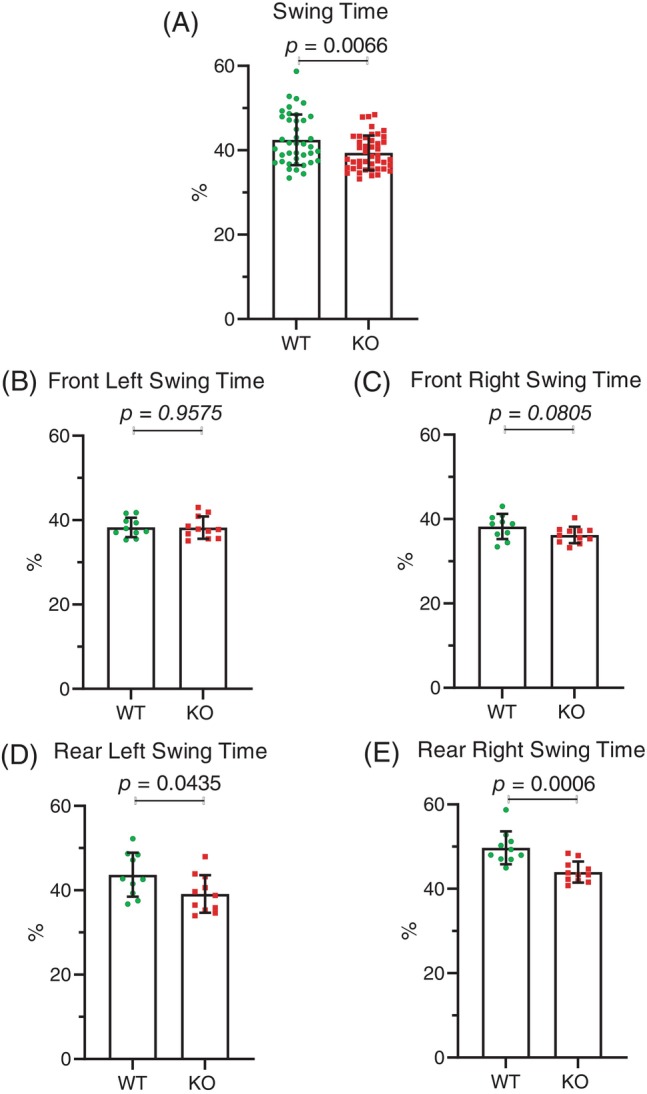
Mical1 fl/fl , Tyr::Cre mutant female mice had increased swing times. (A) The percentage of time spent during a single stride in which feet were not in contact with the treadmill surface (swing position) for all four legs for all mice (N = 40 wild‐type (WT) mice, N = 44 Mical1 knockout [KO] mice). For each foot, results are presented for the (B) front left, (C) front right, (D) rear left, (E) rear right swing positions. Graphs indicate means ± standard deviation. Statistical tests with Student's t‐test. p values <0.05 indicated in bold typeface, p values ≥0.05 indicated in italic typeface. N = 10 WT mice, N = 11 Mical1 KO mice.
In addition to the changes in the stance versus swing phases of each stride, foot positions during striding were altered in female Mical1 KO mice. In the stance phase, Mical1 KO mice positioned their front and rear left feet significantly more closely (t(19) = 0.1669 p < 0.05) (Figure 8A) than WT mice, although there was no significant difference (t(19) = 0.7377 p > 0.05) in the distance between their front and rear right feet (Figure 8B). During running on the treadmill, Mical1 KO mice tended to spend less time on two or fewer feet (null feet; t(19) = 0.5355 p > 0.05) (Figure 8C): single foot; (t(19) = 0.4697 p < 0.05) (Figure 8D): double foot; (t(19) = 0.4387 p > 0.05) (Figure 8E), with single foot support being statistically significant, and more time on 3 or 4 ft (triple foot; t(19) = 0.8079 p < 0.01) (Figure 8F): quadruple feet; (t(19) = 0.5809 p > 0.05) (Figure 8G), with triple foot support being statistically significant relative to WT mice. This contrasts with male Mical1 KO mice, which spent a proportionally more time with no or single foot support and less time with support from two or more feet relative to WT mice (Figure 4). Taken together, these results show that the Mical1 deletion induced by Tyr::Cre produced significant effects on the gaits of both male and female mice, although the specific alterations were different in each sex.
FIGURE 8.
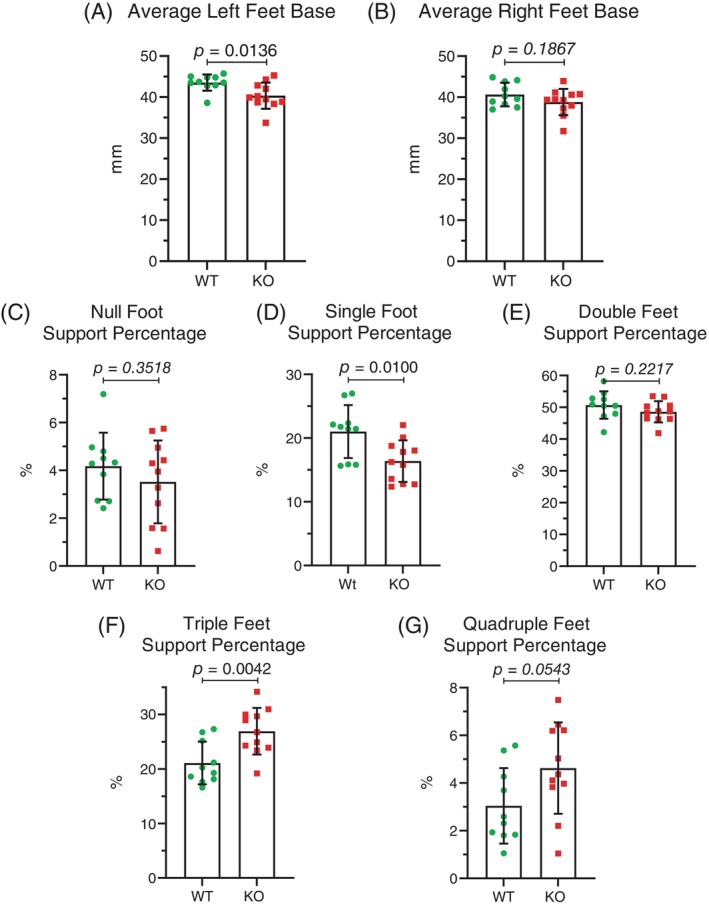
Mical1 fl/fl , Tyr::Cre mutant female mice had altered foot positioning during running. The distance between the midpoint of the trajectory of the front foot stance and the midpoint of the trajectory of the rear foot stance for the (A) Left or (B) Right foot pairs. The percentage of the analysis time during which the number of feet touching the treadmill surface during running were: (C) none; (D) one; (E) two; (F) three; (G) four. Results presented are for each mouse. Graphs indicate means ± standard deviation. Statistical tests with Student's t‐test. p values <0.05 indicated in bold typeface, p values ≥0.05 indicated in italic typeface. N = 10 wild‐type (WT) mice, N = 11 Mical1 knockout (KO) mice.
3.2. No effect on motor coordination, balance and motor learning ability tests in Mical1fl/fl, Tyr::Cre mutant mice
To assess motor coordination, balance and motor learning ability, male WT and Mical1 KO mice were evaluated on a rotating rod (rotarod). Mice were placed on a rod rotating at 4 rpm, and once they were able to keep their balance for at least 10 s, the rotation speed was accelerated up to 40 rpm over 5 min and the time for them to fall off or to allow themselves to passively rotate was recorded. Three trials were performed over three consecutive days, and the means at each day for each genotype were used for comparisons. There were no statistically significant differences between WT and Mical1 KO mice at day 1 (t(16) = 0.6362, p > 0.05), day 2 (t(16) = 0.02664, p > 0.05) or day 3 (t(16) = 0.5724, p > 0.05) (Figure 9A). Although mice from both genotypes stayed on the rotating rod for longer times on subsequent days, there were no differences between the genotypes in the increases on days 2 or 3 (Figure 9A). To further evaluate motor coordination and balance, mice were placed head upwards at the centre of a round 1 cm diameter metal rough‐surfaced pole. The 60 cm long pole was tilted 90° and the time taken for the mice to turn downwards and descend the pole was recorded. There were no significant differences in the time taken to turn (t(16) = 0.06017, p > 0.05) (Figure 9B) or to descend to the bottom of the pole (t(16) = 0.1863, p > 0.05) (Figure 9C) between the WT and Mical1 KO mice.
FIGURE 9.
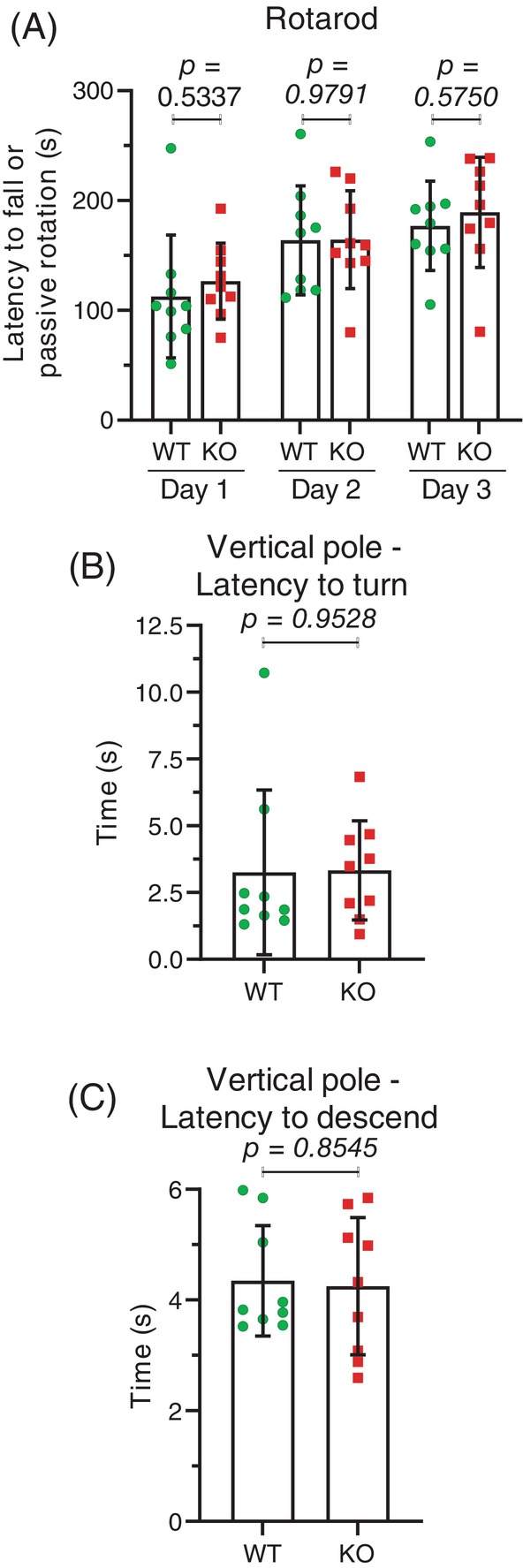
Mical1 fl/fl , Tyr::Cre mutant mice had no differences in rotarod or vertical pole tests. (A) Mice were on a rod rotating at 40 rpm and the time for them to fall off or to allow themselves to passively rotate was recorded. Averages of three independent trials performed each day are reported for each mouse. Graphs indicate means ± standard deviation. Statistical test with one‐way ANOVA and post‐hoc Tukey's multiple comparisons test. p values <0.05 indicated in bold typeface, p values ≥0.05 indicated in italic typeface. N = 9 mice per genotype. (B) The time taken for mice to turn downwards after having been placed in the centre of a 60 cm pole that was tilted 90°. (C) The time taken for mice to descend from the centre of a 60 cm pole that was tilted 90°. Results presented are the averages from three independent trials for each mouse. Graphs indicate means ± standard deviation. Statistical tests with Student's t‐test. p values ≥0.05 indicated in italic typeface. N = 9 mice per genotype. KO, knockout; WT, wild‐type.
A final test of motor coordination was a beam test, in which mice were trained four times to cross between two platforms on a 100 cm long and 48 mm wide rectangular beam. On the following test day, the average time of two trials taken to cross beams of varying widths (24, 12 and 6 mm) and the number of hind foot slips were recorded for two trials. Comparing the mean values showed no significant differences between WT and Mical1 KO mice for the time taken to traverse the 24 mm (t(16) = 0.7242, p > 0.05), 12 mm (t(16) = 0.8192, p > 0.05) or 6 mm (t(16) = 0.001137, p > 0.05) beams. Similarly, there were no significant differences in the number of hind foot slips for the 22 mm (t(16) = 0.5898, p > 0.05), 12 mm (t(16) = 0.000, p > 0.05) or 6 mm (identical values) beam widths (Figure 10A,B).
FIGURE 10.
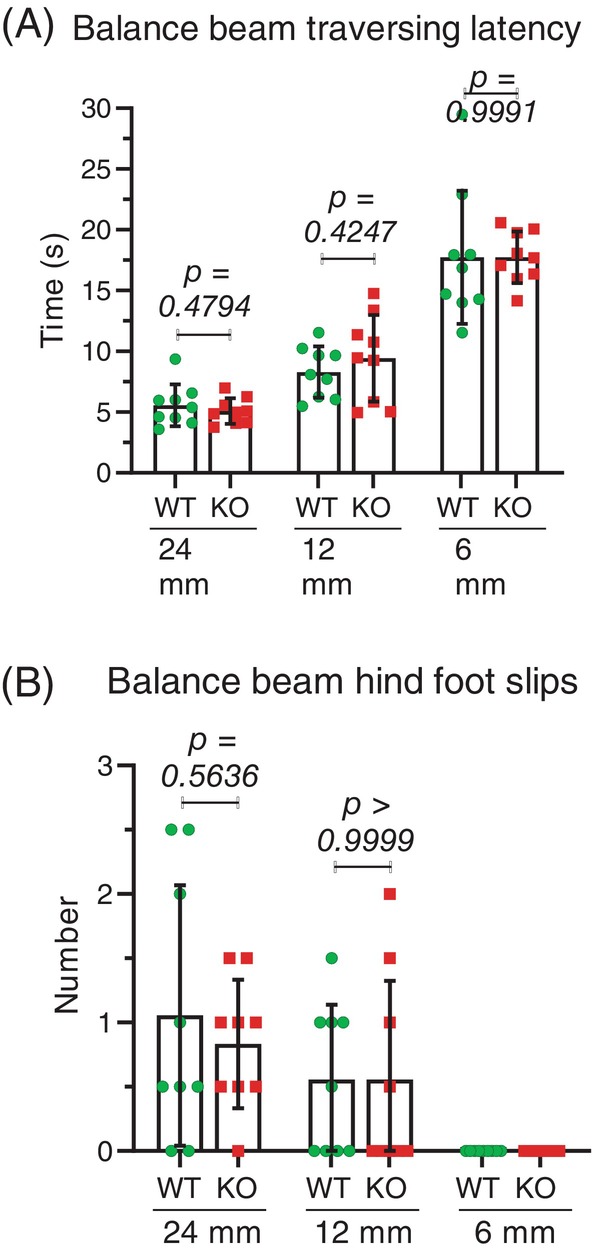
No effect in Mical1 fl/fl , Tyr::Cre mutant mice on their movement across balance beams. (A) Time taken for mice to cross 90 cm round beams of varying widths (24, 12 and 6 mm). (B) The number of hind foot slips during the crossing of the balance beams. Results presented are the averages from two independent trials for each mouse. Graphs indicate means ± standard deviation. Statistical test with one‐way ANOVA and post‐hoc Tukey's multiple comparisons test. p values ≥0.05 indicated in italic typeface. N = 9 mice per genotype. KO, knockout; WT, wild‐type.
4. DISCUSSION
The results of these studies show that the genetic modification of Mical1 mediated by the Tyr::Cre transgene leads to changes in gait during treadmill running in male and female mice. Mical1 expression as determined by immunohistochemistry in adult mice was highest in the cerebellum (Figure 1A), a part of the brain associated with motor coordination. 26 Expression of Tyr::Cre in Mical1 fl/fl mice resulted in reduced Mical1 protein expression in the cerebellum (Figure 1B). Both male and female Mical1 fl/fl , Tyr::Cre mice had significant alterations to their gaits during treadmill running, but the specific aspects of their gaits that were changed differed. In male mice, the overall average run speed was reduced (Figure 2A), which was associated with lower stride frequency (Figure 2C), and changes in how their feet were coordinated during running (Figures 3A and 4A–C). In female mice, the Mical1 deletion did not affect average run speed (Figure 5) but did change their running motion (Figures 6 and 7) and the positioning of their feet (Figure 8). Other tests of motor coordination, balance, and motor learning ability did not show significant differences between WT and Mical1 fl/fl , Tyr::Cre mice (Figures 9 and 10), which suggested that the relatively subtle movement defects were only manifested when mice were forced to perform the coordinated motions necessary for running for a sustained period of time at an elevated speed.
The mild phenotypes observed in Mical1 fl/fl , Tyr::Cre mice could be due to functional redundancy with the closely related Mical2 and Mical3 homologs, which have similar roles in the regulation of F‐actin structures. 20 The conditional deletion of the RhoA GTPase in Tyr::Cre expressing mice did not produce a coat color phenotype, possibly due to redundancy with the highly homologous RhoC gene. 28 It is also possible that not all actin regulators have a phenotype when deleted by Tyr::Cre mediated recombination. Similar to the conditional RhoA KO, there was no coat color phenotype when the Strumpellin subunit of the five‐membered WASH (WASP and SCAR homologue) complex, which acts as an actin nucleation promoting factor by directly interacting with the Arp2/3 complex to trigger the assembly of branched actin networks, was deleted in Tyr::Cre expressing mice. 33 Whether RhoA or Strumpellin KO mice had any motor control abnormalities was not examined. 28 , 33 An alternative possibility is that incomplete Mical1 deletion and consequent residual Mical1 protein expression was sufficient to provide adequate levels of enzyme activity for most requirements.
In humans, two independent activating MICAL1 mutations (Gly150Ser and Ala1065fs) were identified in autosomal‐dominant lateral temporal epilepsy (ADLTE) patients, indicating that aberrant regulation can have a profound effect on normal brain function. 34 , 35 Additional uncharacterized MICAL1 mutations have been identified in epileptic patients, including two predicted RNA transcript splice acceptor sites (Clinical Variants 1517355 and 1525979), as well as Val255Leu (Clinical Variant 1780680), Val308Leu (Clinical Variant 2534410), Ala341Thr (Clinical Variant 1016623), Gly506Arg (Clinical Variant 1671509), Pro681_Leu682 insertion/termination (Clinical Variant 721639), Leu821Pro (Clinical Variant 1016622) and Ala917Val (Clinical Variant 1671506) missense mutations. Given the evidence that the ADLTE‐associated Gly150Ser and Ala1065fs mutations both activated MICAL1, it is possible that the spice accepter variants and missense mutations also lead to elevated MICAL1 activity. However, it is also possible that some or all of these mutations are inactivating. In either case, the observation of multiple independent MICAL1 mutations suggests that properly functioning MICAL1 protein is important to limit the risk of epilepsy, and by extension, for normal brain functioning.
5. CONCLUSIONS
Genetic modification of Mical1 in Tyr expressing cells mediated by the Tyr::Cre transgene resulted in changes in striding coordination in running mice, consistent with a role for Mical1 in the development and/or function of brain regions associated with motor control.
FUNDING INFORMATION
This research was supported by funding to M.F.O from the Canadian Institutes of Health Research (PJT‐169125, PJT‐169106), Canada Research Chairs Program (950‐231,665), Natural Sciences and Engineering Research Council Discovery Grant (RGPIN‐2020‐05388) and the Toronto Metropolitan University Faculty of Science Dean's Research Fund.
CONFLICT OF INTEREST STATEMENT
The authors declare no conflict of interest.
ACKNOWLEDGMENTS
The Mical1 floxed mice were a kind gift from Mark E. Anderson (Johns Hopkins University School of Medicine). Thanks to the staff at The Centre for Phenogenomics (TCP; Toronto Canada) for their technical assistance.
Micovic K, Canuel A, Remtulla A, et al. Mical1 deletion in tyrosinase expressing cells affects mouse running gaits. Genes, Brain and Behavior. 2024;23(5):e70004. doi: 10.1111/gbb.70004
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- 1. Cheever TR, Ervasti JM. Actin isoforms in neuronal development and function. Int Rev Cell Mol Biol. 2013;301:157‐213. [DOI] [PubMed] [Google Scholar]
- 2. Nelson JC, Stavoe AKH, Colón‐Ramos DA. The actin cytoskeleton in presynaptic assembly. Cell Adh Migr. 2013;7(4):379‐387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Compagnucci C, Piemonte F, Sferra A, Piermarini E, Bertini E. The cytoskeletal arrangements necessary to neurogenesis. Oncotarget. 2016;7(15):19414‐19429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Goellner B, Aberle H. The synaptic cytoskeleton in development and disease. Dev Neurobiol. 2012;72(1):111‐125. [DOI] [PubMed] [Google Scholar]
- 5. Schneider F, Metz I, Rust MB. Regulation of actin filament assembly and disassembly in growth cone motility and axon guidance. Brain Res Bull. 2023;192:21‐35. [DOI] [PubMed] [Google Scholar]
- 6. Kalil K, Dent EW. Touch and go: guidance cues signal to the growth cone cytoskeleton. Curr Opin Neurobiol. 2005;15(5):521‐526. [DOI] [PubMed] [Google Scholar]
- 7. Hlushchenko I, Koskinen M, Hotulainen P. Dendritic spine actin dynamics in neuronal maturation and synaptic plasticity. Cytoskeleton. 2016;73(9):435‐441. [DOI] [PubMed] [Google Scholar]
- 8. Yazdani U, Terman JR. The semaphorins. Genome Biol. 2006;7(3):211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tamagnone L, Comoglio PM. Signalling by semaphorin receptors: cell guidance and beyond. Trends Cell Biol. 2000;10(9):377‐383. [DOI] [PubMed] [Google Scholar]
- 10. Fujisawa H. Discovery of semaphorin receptors, neuropilin and plexin, and their functions in neural development. J Neurobiol. 2004;59(1):24‐33. [DOI] [PubMed] [Google Scholar]
- 11. de Wit J, Verhaagen J. Role of semaphorins in the adult nervous system. Prog Neurobiol. 2003;71(2–3):249‐267. [DOI] [PubMed] [Google Scholar]
- 12. Terman JR, Mao T, Pasterkamp RJ, Yu H‐H, Kolodkin AL. MICALs, a family of conserved flavoprotein oxidoreductases function in plexin‐mediated axonal repulsion. Cell. 2002;109(7):887‐900. [DOI] [PubMed] [Google Scholar]
- 13. Beuchle D, Schwarz H, Langegger M, Koch I, Aberle H. Drosophila MICAL regulates myofilament organization and synaptic structure. Mech Dev. 2007;124(5):390‐406. [DOI] [PubMed] [Google Scholar]
- 14. Pasterkamp RJ, Dai H, Terman JR, et al. MICAL flavoprotein monooxygenases: expression during neural development and following spinal cord injuries in the rat. Mol Cell Neurosci. 2006;31(1):52‐69. [DOI] [PubMed] [Google Scholar]
- 15. Van Battum EY, Gunput R‐AF, Lemstra S, et al. The intracellular redox protein MICAL‐1 regulates the development of hippocampal mossy fibre connections. Nat Commun. 2014;5(1):4317. [DOI] [PubMed] [Google Scholar]
- 16. Hung R‐J, Pak CW, Terman JR. Direct redox regulation of F‐actin assembly and disassembly by Mical. Science. 2011;334(6063):1710‐1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hung RJ, Yazdani U, Yoon J, et al. Mical links semaphorins to F‐actin disassembly. Nature. 2010;463(7282):823‐827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Schmidt EF, Shim S‐O, Strittmatter SM. Release of MICAL autoinhibition by semaphorin‐plexin signaling promotes interaction with collapsin response mediator protein. J Neurosci. 2008;28(9):2287‐2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. McGarry DJ, Castino G, Lilla S, et al. MICAL1 activation by PAK1 mediates actin filament disassembly. Cell Rep. 2022;41(1):111442. [DOI] [PubMed] [Google Scholar]
- 20. Rajan S, Terman JR, Reisler E. MICAL‐mediated oxidation of actin and its effects on cytoskeletal and cellular dynamics. Front Cell Dev Biol. 2023;11:1124202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Konstantinidis K, Bezzerides VJ, Lai L, et al. MICAL1 constrains cardiac stress responses and protects against disease by oxidizing CaMKII. J Clin Invest. 2020;130(9):4663‐4678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. D'Mello SAN, Finlay GJ, Baguley BC, Askarian‐Amiri ME. Signaling pathways in melanogenesis. Int J Mol Sci. 2016;17(7):1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tief K, Hahne M, Schmidt A, Beermann F. Tyrosinase, the key enzyme in melanin synthesis, is expressed in murine brain. Eur J Biochem. 1996;241(1):12‐16. [DOI] [PubMed] [Google Scholar]
- 24. Tief K, Schmidt A, Beermann F. New evidence for presence of tyrosinase in substantia nigra, forebrain and midbrain. Mol Brain Res. 1998;53(1):307‐310. [DOI] [PubMed] [Google Scholar]
- 25. Delmas V, Martinozzi S, Bourgeois Y, Holzenberger M, Larue L. Cre‐mediated recombination in the skin melanocyte lineage. Genesis. 2003;36(2):73‐80. [DOI] [PubMed] [Google Scholar]
- 26. Roostaei T, Nazeri A, Sahraian MA, Minagar A. The human cerebellum: a review of physiologic neuroanatomy. Neurol Clin. 2014;32(4):859‐869. [DOI] [PubMed] [Google Scholar]
- 27. Li A, Ma Y, Yu X, et al. Rac1 drives melanoblast organization during mouse development by orchestrating pseudopod‐driven motility and cell‐cycle progression. Dev Cell. 2011;21(4):722‐734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Woodham EF, Paul NR, Tyrrell B, et al. Coordination by Cdc42 of actin, contractility, and adhesion for melanoblast movement in mouse skin. Curr Biol. 2017;27(5):624‐637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Papalazarou V, Swaminathan K, Jaber‐Hijazi F, et al. The Arp2/3 complex is crucial for colonisation of the mouse skin by melanoblasts. Development. 2020;147(22):dev194555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pedersen M, Küsters‐Vandevelde HVN, Viros A, et al. Primary melanoma of the CNS in children is driven by congenital expression of oncogenic NRAS in melanocytes. Cancer Discov. 2013;3(4):458‐469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Warner EJ, Padmanabhan K. Sex differences in head‐fixed voluntary running behavior in C57BL/6J mice. Eur J Neurosci. 2020;51(3):721‐730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lightfoot JT, Turner MJ, Daves M, Vordermark A, Kleeberger SR. Genetic influence on daily wheel running activity level. Physiol Genomics. 2004;19(3):270‐276. [DOI] [PubMed] [Google Scholar]
- 33. Tyrrell BJ, Woodham EF, Spence HJ, Strathdee D, Insall RH, Machesky LM. Loss of strumpellin in the melanocytic lineage impairs the WASH complex but does not affect coat colour. Pigment Cell Melanoma Res. 2016;29(5):559‐571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Dazzo E, Rehberg K, Michelucci R, et al. Mutations in MICAL‐1cause autosomal‐dominant lateral temporal epilepsy. Ann Neurol. 2018;83(3):483‐493. [DOI] [PubMed] [Google Scholar]
- 35. Haikazian S, Olson MF. MICAL1 monooxygenase in autosomal dominant lateral temporal epilepsy: role in cytoskeletal regulation and relation to cancer. Genes (Basel). 2022;13(5):715. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


