Abstract
Laboratory isolates of human immunodeficiency virus type 1 (HIV-1) that utilize CXCR4 as a coreceptor infect primary human macrophages inefficiently even though these express a low but detectable level of cell surface CXCR4. In contrast, infection of primary macrophages by primary CXCR4-tropic HIV-1 isolates is readily detectable. Here, we provide evidence suggesting that this difference in cell tropism results from a higher requirement for cell surface CXCR4 for infection by laboratory HIV-1 isolates. Transfected COS7 cells that express a high level of CD4 but a low level of CXCR4 were infected significantly more efficiently by two primary CXCR4-tropic HIV-1 isolates compared to the prototypic laboratory HIV-1 isolate IIIB. More importantly, overexpression of either wild-type or signaling-defective CXCR4 on primary macrophages dramatically enhanced the efficiency of infection by the laboratory HIV-1 isolate yet only modestly enhanced infection by either primary CXCR4-tropic virus. Overexpression of CD4 had, in contrast, only a limited effect on macrophage infection by the laboratory HIV-1, although infection by the primary isolates was markedly enhanced. We therefore conclude that the laboratory CXCR4-tropic HIV-1 isolate exhibits a significantly higher CXCR4 requirement for efficient infection than do the primary CXCR4-tropic isolates and that this difference can explain the poor ability of the laboratory HIV-1 isolate to replicate in primary macrophages. More generally, we propose that the cell tropisms displayed by different strains of HIV-1 in culture can largely be explained on the basis of differential requirements for cell surface CD4 and/or coreceptor expression levels.
Research into the molecular biology of human immunodeficiency virus type 1 (HIV-1) has generally used proviral clones derived from laboratory T-cell line-adapted (TCLA) strains of HIV-1, due to the considerable practical advantage of being able to propagate these viruses in CD4+ T-cell lines. However, it has been known for some time that TCLA variants of HIV-1 differ from primary (PR) isolates in a number of key, and related, ways (reviewed in references 11 and 33). Specifically, TCLA isolates generally differ from PR isolates not only in their ability to grow in transformed CD4+ T-cell lines but also in their inability to infect primary macrophages and their increased sensitivity to neutralization by soluble CD4 (sCD4) and to certain monoclonal antibodies (MAbs) (9, 27, 29, 36, 42). These differences have been shown to map to the viral env gene and particularly to the env V3 loop region (20, 21, 28, 37).
The discovery that HIV-1 infection requires not only the CD4 receptor but also a coreceptor molecule (2, 7, 10, 14, 16) provided a partial explanation for these phenotypic differences. Specifically, it was discovered that TCLA isolates use CXCR4 as a coreceptor (X4 isolates), while the large majority of PR isolates utilize CCR5 (R5 isolates). The finding that T-cell lines generally do not express CCR5 appeared to clarify why these cells fail to support the replication of PR isolates. However, coreceptor utilization did not clearly explain why TCLA isolates fail to replicate on primary macrophages, as these CD4+ cells express low but readily detectable levels of cell surface CXCR4 (24, 44). Subsequently, several PR-X4 isolates have been obtained and these isolates, like PR-R5 isolates, generally replicate poorly on transformed T-cell lines yet can infect primary macrophages (29, 38, 39, 43).
Efforts to understand the inability of PR-X4 isolates to grow effectively in T-cell lines led to the demonstration that overexpression of CD4 in these cells could rescue PR-X4 replication (29). It has also been demonstrated that PR isolates differ from TCLA isolates in that the latter have a significantly higher affinity for CD4 and, concomitantly, that adaptation of PR isolates to growth on T-cell lines involves the acquisition of a significantly higher affinity for CD4 (22).
Based on these results, it seemed possible that TCLA isolates had lost the ability to infect macrophages due to a reduced affinity for the cell surface CXCR4 coreceptor. However, several other hypotheses to explain this phenomenon have been proposed. For example, it has been demonstrated that binding of the HIV-1 Env protein to CXCR4 can activate ionic signaling responses in primary macrophages in culture (25). This Env-induced signaling has been proposed to be potentially critical for productive infection of macrophages by HIV-1, perhaps acting at a step in the viral life cycle that occurs after entry (3, 25, 35, 40). Consistent with this model, Env proteins from TCLA isolates were found to differ from PR Env proteins in that they failed to induce mobilization of intracellular calcium in treated macrophages (3). Conversely, it has also been suggested that CXCR4 may undergo distinct posttranslational processing in macrophages that precludes its use as a coreceptor by TCLA HIV-1 isolates (23).
In this study, we analyzed the ability of the prototypic TCLA-X4 HIV-1 isolate IIIB and two novel PR-X4 isolates to infect primary macrophages and also other cells that express low levels of either CD4 or CXCR4. We show that this TCLA-X4 isolate differs from the PR-X4 isolates in that it is significantly more efficient at infecting cells with low CD4 levels yet significantly less effective at infecting cells with low cell surface CXCR4. Consistent with the hypothesis that low CXCR4 levels on primary macrophages are a key determinant of TCLA HIV-1 infection efficiency, we show that overexpression of wild-type CXCR4 or of signaling-defective CXCR4 mutants effectively rescues primary macrophage infection by this TCLA-X4 isolate.
MATERIALS AND METHODS
Primary lymphocyte and monocyte culture.
Peripheral blood mononuclear cells (PBMC) from healthy HIV-1-negative donors were isolated by Ficoll-Hypaque gradient centrifugation. The cells were then resuspended in Dulbecco modified Eagle medium (GIBCO BRL) and plated at 8 × 105 cells per well in 24-well tissue culture plates. After 3 h of culture, the adherent cells were washed extensively with phosphate-buffered saline (PBS) and cultured in Dulbecco modified Eagle medium supplemented with 10% human AB serum (Sigma) and 1,000 U of macrophage colony-stimulating factor (M-CSF; R&D Systems) for 1 week to allow differentiation into monocyte-derived macrophages (MDM). Nonadherent cells were collected by centrifugation, resuspended in RPMI 1640 (GIBCO BRL) supplemented with 10% heat-inactivated fetal calf serum, and stimulated with phytohemagglutinin (Sigma) at 3 μg/ml for 2 days. Cells were then washed and cultured for another 5 days in medium supplemented with 10 U of interleukin-2 (R&D Systems) per ml.
Plasmid construction.
Complete envelope gp120-coding sequences were PCR amplified from proviral clones encoding the PR-R5 isolate ADA (28) and the TCLA-X4 isolate HXB3 (20) or from full-length env clones derived from the PR-X4 isolates QH1549 and QH1558 (19). The primers used were targeted to a SalI site within the first coding exon of tat and to a BamHI site located within envelope gp41 sequences. These env fragments were then used to generate the infectious proviral clones pNL-ADA, pNL-HXB, pNL-1549, and pNL-1558 by replacement of the corresponding SalI-BamHI fragment of pNL4-3 (1). For viral infectivity assays, similar proviral clones carrying the luciferase gene in place of nef, termed pNL-Luc-ADA, pNL-Luc-HXB, pNL-Luc-1549, and pNL-Luc-1558, respectively, were created by cloning the env fragments described above into the same sites in pNL-Luc-E−R+ (8).
The HIV-1-based lentiviral CD4 and CXCR4 expression vectors, pNL-CD4 and pNL-CXCR4, were generated from pNL-Luc-E−R+ as follows. First, to prevent all late HIV-1 protein expression in target cells (26), a stop codon was introduced into the BamHI site located in the second exon of rev to generate pNL-Luc/Rev−. Then, the NotI-XhoI fragment encoding luciferase was replaced with a PCR-generated NotI-XhoI fragment encoding either CD4 or CXCR4 (5). pNL-CXCR4-D187A (6) was derived from pNL-CXCR4 by mutation of residue 187 in CXCR4 from aspartic acid to alanine using a QuickChange mutagenesis kit (Stratagene). Similarly, pNL-CXCR4-Δi3A (6) was constructed by deletion of four residues within the third intracellular loop of CXCR4 (227-SHSK-230) by QuickChange mutagenesis. The negative control vector, pNL-con, was generated by deleting the luciferase gene from pNL-Luc/Rev−.
Cell maintenance and transfection.
Sup-T1, CEM-SS, COS7, and 293T cells were maintained as described elsewhere (20, 26). To prepare HIV-1 virus stocks, 293T cells were transfected with 2 μg of a proviral expression plasmid by using FuGENE 6 (Roche). HIV-1-based lentivirus vectors were generated by cotransfecting 293T cells with 0.5 μg of the Rev expression vector pcRev (26) and 0.5 μg of the vesicular stomatitis virus glycoprotein (VSV-G) expression vector pHIT/G (17) with 1 μg of a lentivirus vector plasmid, using FuGENE 6. The culture medium was replaced 16 h later, and the culture supernatants were harvested 40 h after transfection, and filtered through 0.45-μm-pore-size filters, and virus yield was measured by p24 Gag antigen capture enzyme-linked immunosorbent assay (ELISA) (NEN Life Science). Virus stocks were stored at −80°C until needed.
Virus replication assay.
PBMC, Sup-T1 cells, and CEM-SS cells (106 of each) were infected overnight with 50 ng of p24 antigen of NL-ADA, NL-HXB, NL-1549, or NL-1558 in the presence or absence of the CXCR4 inhibitor AMD3100 (12) at a concentration of 1 μg/ml, washed extensively with PBS, and then cultured in fresh medium. Supernatants were sampled every 2 days, and p24 Gag antigen production was quantified by ELISA.
Luciferase reporter virus assays.
COS7 cells were transfected with 100 ng of pCMV5/CD4 (5) alone or together with either pCMV5/CCR5 or pCMV5/CXCR4 (5), using FuGENE 6; 48 h later, the cells were infected with 20 ng of p24 antigen of a luciferase reporter virus. After 48 h, the cells were lysed in 200 μl of lysis buffer (Promega), and luciferase activities were determined (34) with a Lumat LB 9501 luminometer. For infection experiments in macrophages, 7-day-old cultures of MDM were infected overnight with 20 ng of p24 antigen of a luciferase reporter virus in the presence or absence of AMD3100 (1 μg/ml), washed with PBS, and cultured in fresh medium; 72 h after infection, the cells were harvested for luciferase assay as described above.
Flow cytometry.
COS7 cells transfected with pCMV5/CD4 and pCMV5/CXCR4 (5) were stained with the anti-CD4 MAb Leu-3A conjugated with fluorescein and the anti-CXCR4 MAb 12G5 conjugated with phycoerythrin (Becton Dickinson), or an isotype control antibody, for 30 min on ice. MDM were stained with the anti-CD14 MAb M5E2 conjugated with allophycocyanin (Becton Dickinson), or the MAbs described above, for 30 min on ice. The cells were then washed extensively with PBS, fixed with 4% formaldehyde in PBS, and then analyzed by fluorescence-activated cell sorting (FACS) on a FACscan cytometer. Mean fluorescence intensity (MFI) was determined using CellQuest software (Becton Dickinson).
Overexpression of CD4 or CXCR4 on macrophages.
Seven-day-old cultures of MDM were transduced overnight with 10 ng of VSV-G-pseudotyped lentiviral vector encoding CD4, wild-type or mutant CXCR4, or a control vector. The cells were then washed extensively with PBS and cultured in fresh medium for an additional 3 days. Then, the transduced MDM were infected overnight with 20 ng of p24 antigen of a luciferase reporter virus, washed with PBS, and cultured in fresh medium; 72 h after infection, the cells were harvested for luciferase assay as described above.
Calcium mobilization assay.
Calcium mobilization was measured essentially as previously described (31). Briefly, COS7 cells expressing wild-type or mutant CXCR4 were generated by retroviral transduction. Then 5 × 106 cells were loaded with the fluorescent probe indo-1/acetoxymethyl ester (1 μM, final concentration), in the presence of 1 μM pleuronic acid, for 30 min at room temperature. The cells were then washed and resuspended in 1.5 ml of HEPES-buffered saline. Intracellular calcium was measured in the presence or absence of SDF-1 (100 ng/ml; Becton Dickinson) by determination of indo-1 fluorescence in a Perkin-Elmer fluorescence spectrophotometer (model 650-19).
RESULTS
Infection of cells by PR-X4 HIV-1 isolates.
Proviral clones encoding replication-competent forms of ADA, a commonly used PR-R5 HIV-1 isolate, and HXB3, derived from the prototypic TCLA-X4 isolate IIIB, have been previously described (20, 34, 41). QH1549 and QH1558 are two recently described PR-X4 isolates, derived from two late-stage AIDS patients, for which full-length env clones exist (19). To facilitate an accurate comparison of the abilities of the env genes derived from each of these distinct isolates to support infection of different cells in culture, we initially subcloned the entire gp120 region of each of these four viruses in place of the equivalent env sequence present in the replication-competent pNL4-3 proviral clone (1) and in the indicator virus pNL-Luc-E−R+ (8). This latter virus bears the luciferase indicator gene in place of the viral nef gene and is used to quantify the level of viral infection over a single replication cycle. By varying only the env gene, leaving most of the rest of the HIV-1 provirus invariant, we hoped to avoid variability in viral gene expression due to, for example, sequence differences in the viral long terminal repeat promoter.
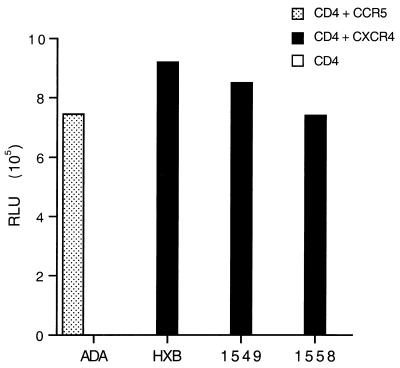
To confirm that the resultant chimeric proviral clones indeed maintained the expected coreceptor specificity, we first analyzed the abilities of the four resultant luciferase indicator viruses to infect COS7 cells that had been transfected with expression vectors encoding CD4 alone, CD4 plus CCR5, or CD4 plus CXCR4. As shown in Fig. 1, the NL-Luc-ADA virus could infect only cells expressing both CD4 and CCR5, while NL-Luc-HXB, NL-Luc-1549, and NL-Luc-1558 each proved dependent on both CD4 and CXCR4 for infection. We therefore conclude that these recombinant viruses indeed maintained their predicted R5 (ADA) or X4 (HXB3, QH1549, and QH1558) tropism.
FIG. 1.
Analysis of HIV-1 coreceptor specificity. HIV-1 proviral derivatives, bearing the luciferase indicator gene in place of nef and encoding env genes derived from viral isolate ADA, HXB3, QH1549, or QH1558 were obtained, and then used to infect COS7 cells expressing CD4 only, CD4 plus CCR5, or CD4 plus CXCR4. At ∼48 h after infection, cells were harvested and the level of expression of the virally encoded luciferase enzyme was determined. Induced luciferase enzyme activity is given in relative light units (RLU) measured in a luminometer. These data are representative of three independent experiments.
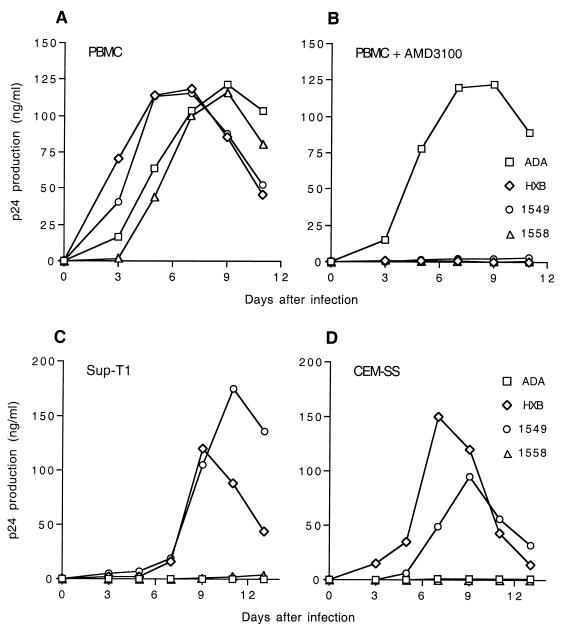
The ability of the four full-length HIV-1 proviral clones to replicate in PBMCs and in T-cell lines was analyzed. The data in Fig. 2A show that each of these four env genes is fully able to support a spreading infection in PBMCs. As predicted, the replication in PBMCs of NL-HXB, NL-1549, and NL-1558 proved highly sensitive to AMD3100, a CXCR4-specific inhibitor of HIV-1 infection (12), while replication of NL-ADA preceded normally in the presence of AMD3100 (Fig. 2B). We have also examined the abilities of these four chimeric viruses to replicate in two transformed T-cell lines, Sup-T1 (Fig. 2C) and CEM-SS (Fig. 2D). As predicted, the PR-R5-derived virus NL-ADA failed to replicate in these cells, which lack cell surface CCR5. In contrast, and as predicted, the TCLA-X4 virus NL-HXB replicated efficiently in both T-cell lines. The two chimeric PR-X4-derived viruses NL-1549 and NL-1558, gave different results, with NL-1549 yielding a readily detectable level of virus replication in both T-cell lines, while NL-1558 proved unable to replicate in either cell type (Fig. 2C and D). Therefore, in this assay, NL-1549 behaves more like a TCLA-X4 isolate, while NL-1558 appears more similar to PR-X4 isolates reported previously (29) in that it can replicate effectively in PBMC but not in T-cell lines in culture.
FIG. 2.
HIV-1 infection of T cells in culture. HIV-1 preparations were obtained by transfection of 293T cells with full-length proviral clones. Equal levels of virus, as determined by measurement of p24 Gag levels, were then used to infect PBMC (A and B), Sup-T1 cells (C), or CEM-SS cells (D). PBMC were infected both in the absence (A) and in the presence (B) of the CXCR4-specific inhibitor AMD3100. Samples of supernatant media were harvested at 2 day intervals, and virus replication was measured by determination of supernatant p24 Gag levels. Data shown are representative of several independent experiments.
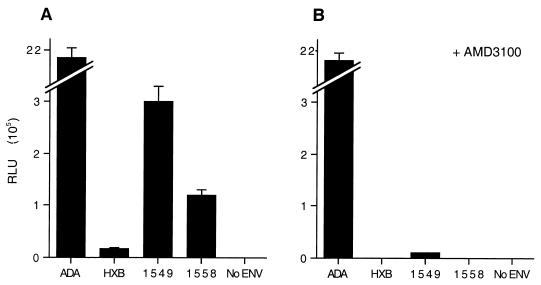
We next measured the ability of each of these four viruses to infect primary MDM, using the luciferase indicator virus single replication cycle assay. As shown in Fig. 3A, and as previously reported (28), the NL-Luc-ADA virus infected MDM very efficiently. In contrast, infection by the IIIB-derived NL-Luc-HXB virus occurred with only ∼1% of the efficiency of ADA, although this low activity was still readily detectable. The PR-X4 isolates displayed intermediate phenotypes, with NL-Luc-1549 infecting at a level ∼12% of that seen with ADA, while NL-Luc-1558 infection was ∼5% of the level seen with ADA. Infection of MDM by all three X4 viruses, including NL-Luc-HXB, proved to be highly sensitive to inhibition by the CXCR4-specific drug AMD3100 (12), while MDM infection by the PR-R5 isolate ADA was unaffected by AMD3100 treatment (Fig. 3B). Essentially identical results were obtained with MDM obtained from three different donors (data not shown). We therefore conclude that the two PR-X4-derived viruses NL-1549 and NL-1558 are indeed able to infect MDM significantly more efficiently than can the TCLA-X4 virus NL-HXB, as also reported previously for other PR-X4 viruses (38, 39, 43), and further that infection of PBMC and MDM by both NL-1549 and NL-1558 is dependent on the CXCR4 coreceptor.
FIG. 3.
Single-cycle HIV-1 replication assay in MDM. MDM were cultured for 7 days in the presence of M-CSF and then infected with equal levels of the indicated luciferase indicator viruses in the absence (A) or presence (B) of AMD3100. At ∼72 h after infection, the MDM were lysed and induced luciferase enzyme levels determined as described for Fig. 1. No ENV refers to HIV-1 virions generated in the absence of an env gene. Averages of three independent experiments with standard deviations are indicated.
PR-X4 HIV-1 isolates efficiently infect cells expressing low levels of CXCR4.
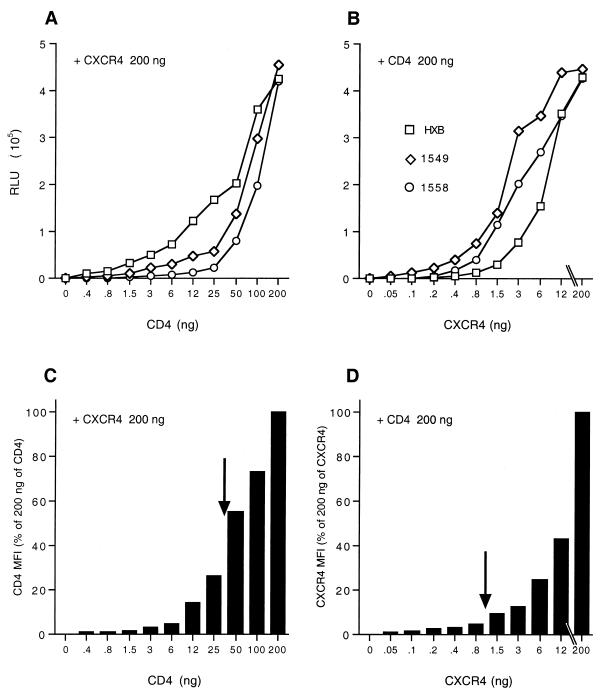
We next wished to address whether the TCLA-X4 virus NL-Luc-HXB and the PR-X4 viruses NL-Luc-1549 and NL-Luc-1558 might differ in the ability to use a given level of cell surface CD4 or CXCR4. To perform this experiment, we transfected COS7 cells, which express neither CD4 nor CXCR4, with a high and constant level of an expression vector encoding CD4 together with a range of levels of a CXCR4 expression plasmid. In parallel, we performed the converse experiment; i.e., COS7 cells were transfected with a high and constant level of the CXCR4 expression plasmid and various levels of a CD4 expression plasmid. Two days after transfection, these COS7 cells were used either for infection with the indicator virus NL-Luc-HXB, NL-Luc-1549, or NL-Luc-1558 (Fig. 4A and B) or subjected to FACS analysis using MAbs specific for CD4 and CXCR4 (Fig. 4C and D).
FIG. 4.
Analysis of the effect of cell surface CD4 or CXCR4 expression levels on HIV-1 infection efficiency. COS7 cells were transfected with 200 ng of a CXCR4 expression plasmid plus stepwise dilutions of a CD4 expression plasmid (A and C) or with 200 ng of a CD4 expression plasmid and stepwise dilutions of a CXCR4 expression plasmid (B and D). At ∼48 h after transfection, the transfected COS7 cells were infected with the luciferase indicator virus NL-Luc-HXB, NL-Luc-1549, or NL-Luc-1558 (A and B) or subjected to FACS analysis using MAbs specific for CD4 and CXCR4 (C and D). The infected COS7 cells were harvested at ∼48 h after transfection, and induced luciferase levels were determined (A and B). The MFI was determined for each transfected culture for either cell surface CD4 (C) or CXCR4 (D) and is given as a percentage of the level seen in cells transfected with 200 ng of both the CXCR4 and CD4 expression plasmids. Importantly, the CXCR4 expression levels in panel D were measured only on CD4+ cells, while conversely, the CD4 expression levels in panel C were measured only on CXCR4+ cells. In this way, COS7 cells that were not transfected, and hence could not support HIV-1 infection, were excluded from the analysis. The arrows in panels C and D show the MFI of cell surface CD4 and CXCR4 expression on MDM measured in parallel. Data shown are representative of three independent experiments.
It has previously been reported that TCLA viruses have a significantly higher affinity for CD4 than do PR isolates (22, 27), and we therefore predicted that the TCLA-X4 virus NL-Luc-HXB might give a significantly higher level of infection, at a given level of cell surface CD4 expression, than would either of the two PR-X4 viruses. As shown in Fig. 4A, this indeed proved to be true over a wide range of tested CD4 expression levels. Conversely, we also observed that infection by NL-Luc-HXB was significantly less efficient than infection by either NL-Luc-1549 or NL-Luc-1558 when CD4 expression was high but CXCR4 expression was rate limiting (Fig. 4B). Therefore, it appears that these two distinct PR-X4 viruses are indeed able to utilize low levels of cell surface CXCR4 more effectively than the IIIB-derived NL-Luc-HXB virus.
Figure 4C shows results of a FACS analysis of the average level of cell surface CD4 expression, given as MFI, of the CXCR4+ COS7 cells utilized in Fig. 4A. Conversely, in Fig. 4D, we present the average level of cell surface CXCR4 expression of the CD4+ COS7 cells used in Fig. 4B. These data demonstrate that the titrations performed in Fig. 4A and B indeed resulted in a gradual diminution in the level of, respectively, cell surface CD4 and CXCR4 expression. In parallel, we also analyzed the relative level of cell surface expression of CD4 and CXCR4 on MDM cultured as described for Fig. 3. The average cell surface levels on these MDM, indicated by arrows in Fig. 4C and D, were in the range of CD4 and CXCR4 observed on the transfected COS7 cells and were clearly significantly below a saturating level, especially in the case of CXCR4. This is particularly true when one considers that this CXCR4 dose response (Fig. 4B) measured HIV-1 infection efficiency in cells that exhibited a high level of cell surface CD4 expression. This is important, as it has previously been reported for PR-R5 isolates of HIV-1 that efficient infection requires significantly higher levels of the CCR5 coreceptor when cell surface CD4 levels are relatively low, and vice versa (30). These data imply that primary MDM express suboptimal levels of both CD4 and, particularly, CXCR4 and therefore that an increase in cell surface expression of either molecule should result in increased HIV-1 infection. Given that the TCLA-X4 variant NL-Luc-HXB is not able to effectively infect cells expressing low levels of CXCR4 (Fig. 4B), we would expect infection of primary MDM by this virus to be particularly enhanced upon overexpression of CXCR4. Conversely, the inefficient infection of cells expressing low levels of CD4 by the PR-X4 viruses NL-Luc-1549 and NL-Luc-1558 (Fig. 4A) predicts that infection of MDM by these viruses should be enhanced by overexpression of CD4.
Effect of overexpression of CXCR4 or CD4 on the level of infection of primary macrophages by HIV-1.
Macrophages are difficult to transfect and, because they are nondividing, also cannot be transduced by conventional retroviral expression vectors. We therefore chose to construct HIV-1-based lentiviral expression vectors encoding CD4 or CXCR4, as these can infect nondividing cells such as macrophages.
The vectors used were based on the luciferase indicator virus NL-Luc-E−R+ (8), which bears an inactivating frameshift mutation in the viral env gene. Initially, an inactivating frameshift mutation was also introduced into rev, thus blocking all late viral gene expression in the absence of Rev protein provided in trans (26). Then, the luc gene, which is located in place of the early HIV-1 nef gene, was replaced by either the CD4 (pNL-CD4) or the CXCR4 (pNL-CXCR4) open reading frame.
To generate infectious lentiviral virions, the pNL-CD4 or pNL-CXCR4 expression plasmid or the pNL-con control plasmid was transfected into 293T cells together with a plasmid expressing HIV-1 Rev and a second plasmid expressing VSV-G. At ∼40 h after transfection, the supernatant media were harvested and levels of released virions were quantitated by p24 ELISA. The resultant VSV-G-pseudotyped HIV-1 particles are predicted to encode only the HIV-1 early gene products after transduction of susceptible cells, due to the lack of a functional rev gene. Nef is also not expressed, as these constructs contain instead of nef either the CD4 or the CXCR4 open reading frame or, in the case of pNL-con, a deletion of nef. However, these viruses are all predicted to encode a functional tat gene product. Their ability to express tat allowed us to derive an approximate infectious titer for the released lentiviral virions by using the indicator cell line MAGI, which encodes a chromosomal β-galactosidase indicator gene, under the control of the HIV-1 long terminal repeat promoter, that is expressed only in the presence of Tat (32). Using the MAGI assay, we estimate that these released virion particles exhibit a titer of ∼8 × 105 infectious units per 10 ng of p24 protein (data not shown).
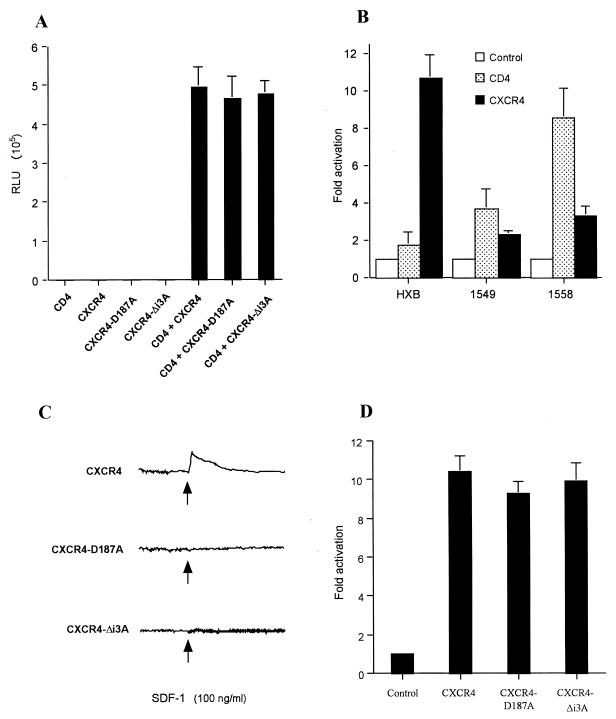
To confirm that these lentiviral vectors indeed encoded functional CD4 and CXCR4 proteins, we first transduced COS7 cells with pNL-CD4 and pNL-CXCR4 either alone or in combination. After 48 h, the cells were then infected with the NL-Luc-HXB indicator virus, and induced luciferase levels were determined after an additional 48 h. As shown in Fig. 5A, we observed efficient infection of COS7 cells transduced by both the NL-CD4 and the NL-CXCR4 lentiviral vector but no infection of cells transduced with only one of these two vectors. Therefore, we conclude that these HIV-1-based lentiviral vectors are capable of inducing the expression of biologically active CD4 and CXCR4 receptors in transduced cells.
FIG. 5.
Lentivirus transduction of the human CD4 and CXCR4 genes. HIV-1-based lentiviral vectors encoding CD4, wild-type CXCR4, or mutant CXCR4, or lacking an inserted heterologous gene (Control), were transfected into 293T cells together with plasmids encoding HIV-1 Rev and VSV-G. Supernatant media containing the pseudotyped HIV-1 virions were then used to infect COS7 cells (A and C) or MDM (B and D). (A) At ∼48 h after retroviral transduction, the COS7 cells were also infected with the TCLA-X4 indicator virus NL-Luc-HXB. Induced levels of luciferase activity were determined ∼48 h after infection. (B) About 72 h after transduction of the MDM with lentiviral vectors encoding the indicated receptor or with a control vector, these primary cells were also infected with HIV-1 variants encoding the luciferase indicator gene and bearing the indicated HIV-1 Env protein. Induced levels of luciferase enzyme activity were determined at ∼72 h after infection. (C) The effect of SDF-1 (100 ng/ml) on intracellular calcium was monitored using COS7 cells transduced with lentiviral vectors encoding the wild-type, D187A, or Δi3A mutant form of CXCR4 after loading the cells with the fluorescent probe indo-1/acetoxymethyl ester. Representative data are shown. (D) MDM infection experiment performed as for panel B, using lentiviral vectors expressing either wild-type CXCR4 or the D187A or Δi3A CXCR4 mutant. Panels A, B, and D show the averages of three independent experiments with standard deviations indicated.
Next, we asked whether these same lentiviral vectors would have any effect on the level of HIV-1 infection of primary MDM. Cultured MDM were transduced with ∼1 infectious unit of NL-CD4, NL-CXCR4, or the control vector NL-con per cell and then cultured for ∼72 h before being challenged with the TCLA-X4 indicator virus NL-Luc-HXB or the PR-X4 viruses NL-Luc-1549 and NL-Luc-1558. Three days later, the cultures were harvested and induced luciferase levels were determined (Fig. 5B). In parallel, MDM cultures transduced with the various lentiviral vectors were harvested ∼72 h after transduction and subjected to FACS analysis for cell surface CD4 or CXCR4. This analysis revealed that the MDM transduced with NL-CD4 expressed ∼6-fold more cell surface CD4 than did cells transduced with NL-con, while transduction of cells with NL-CXCR4 increased cell surface expression of CXCR4 ∼11-fold compared to control cells (data not shown).
As shown in Fig. 5B, overexpression of CXCR4 on MDM dramatically enhanced the level of infection observed with the TCLA-X4 virus NL-Luc-HXB, giving an ∼11-fold increase in viral gene expression. The level of infection by NL-Luc-HXB of MDM overexpressing CXCR4 was therefore almost equivalent to the level of infection of nontransduced MDM by the primary virus NL-Luc-1549 (Fig. 3). Consistent with the lower requirement for cell surface CXCR4 observed for the PR-X4 isolates in transfected COS7 cells (Fig. 4B), overexpression of CXCR4 only modestly enhanced the level of MDM infection observed with NL-Luc-1549 (∼2-fold) or NL-Luc-1558 (∼3-fold).
A very different result was observed upon overexpression of CD4. Thus, the PR-X4 virus NL-Luc-1558, which was least efficient at infecting COS7 cells with low levels of cell surface CD4 (Fig. 4A), displayed the most marked enhancement in MDM infection upon overexpression of cell surface CD4 (∼9-fold). Infection of MDM by the TCLA-X4 isolate NL-Luc-HXB, which was most efficient at infecting COS cells bearing low levels of CD4 (Fig. 4A), was in contrast only minimally (∼2-fold) enhanced by overexpression of CD4 on the MDM (Fig. 5B), while NL-Luc-1549, which was intermediate in the CD4 dose response shown in Fig. 4A, was also intermediate in terms of the enhancement in infection observed upon overexpression of CD4 on MDM (∼4-fold higher). We therefore conclude that infection of MDM by the TCLA-X4 isolate NL-Luc-HXB is inefficient primarily because the level of CXCR4 expressed on MDM is substantially lower than the optimal level. Conversely, infection of MDM by PR-X4 strains, while fairly efficient, is nevertheless constrained by the suboptimal level of CD4 expressed on these primary cells. Importantly, these data are accurately predicted by titration experiments in the transformed cell line COS7, which revealed that efficient infection by the TCLA-X4 isolate NL-Luc-HXB requires high levels of cell surface CXCR4, while efficient infection by the PR-X4 isolates requires elevated levels of cell surface CD4.
Signaling-defective CXCR4 mutants support HIV-1 infection of MDM.
It has been proposed that productive infection of MDM via the CXCR4 coreceptor requires a signaling event induced by Env binding to CXCR4 (3, 25, 40). To test this hypothesis, we constructed lentiviral vectors that express two distinct CXCR4 mutants, termed D187A and Δi3A, that have both previously been shown to be defective for SDF-1-induced signaling (6). However, both the D187A missense mutant, in which residue Asp187 has been mutated to Ala, and the Δi3A deletion mutant, which lacks four residues from the third intracellular loop of CXCR4 (227-SHSK-230), remain fully able to bind the SDF-1 chemokine (6). Both CXCR4 mutants have also been previously reported to support infection of transfected cell lines by TCLA-X4 HIV-1 (6).
To confirm these earlier data, we first asked whether the CXCR4-D187A and CXCR4-Δi3A mutants would support NL-Luc-HXB infection of COS7 cells. In fact, as shown in Fig. 5A, infection of COS7 cells expressing CD4 and either CXCR4-D187A or CXCR4-Δi3A proceeded as efficiently as did infection of COS7 cells expressing CD4 and wild-type CXCR4. Next, we asked whether the CXCR4-D187A and CXCR4-Δi3A mutants were capable of signaling by measuring the ability of SDF-1 to induce calcium mobilization in COS7 cells expressing either wild-type or mutant CXCR4. As shown in Fig. 5C, calcium mobilization was readily detected in COS7 cells expressing wild-type CXCR4, but no signaling was observed in CXCR4-D187A- or CXCR4-Δi3A-expressing cells.
To examine whether the ability of CXCR4 to signal plays a role in HIV-1 infection of primary macrophages, we next transduced MDM with lentiviral vectors expressing wild-type CXCR4, CXCR4-D187A, or CXCR4-Δi3A and then measured the level of infection by the TCLA-X4 indicator virus NL-Luc-HXB. As shown in Fig. 5D, overexpression of the signaling-defective CXCR4-D187A or CXCR4-Δi3A mutant enhanced TCLA-X4 infection to the same extent (∼10-fold) as wild-type CXCR4. These data strongly suggest that it is the level of cell surface CXCR4 expression, not signaling via CXCR4, that determines the level of productive infection.
DISCUSSION
The majority of HIV-1 research continues to utilize a small number of closely related TCLA-X4 viruses belonging to clade B. These virus isolates, i.e., LAV and the derived proviral clones LAI and NL4-3, and IIIB and the derived proviral clones HXB2 and HXB3, differ from PR isolates in a number of key ways. Probably the most significant difference is the fact that most PR isolates of HIV-1 utilize CCR5 either instead of, or sometimes in addition to, CXCR4 (2, 7, 10, 14, 16, 44, 45). However, a small number of PR-X4 isolates have also been reported; these appear generally similar to PR-R5 isolates, and dissimilar to TCLA-X4 viruses, in that they replicate relatively poorly in CD4+ T-cell lines, can infect primary macrophages, and are resistant to neutralization by sCD4 (29, 38, 39, 43) (Fig. 2 and 3). Therefore, different coreceptor specificities provide at best a partial explanation for the observed differences between the TCLA-X4 viruses on the one hand and PR isolates on the other.
A possible explanation for the inability of most PR-X4 viruses to replicate efficiently in CD4+ T-cell lines was suggested by the observation that PR viruses have a relatively low affinity for CD4 compared to TCLA-X4 viruses and that adaptation of PR-X4 isolates for growth on CD4+ T cells in culture selects for a significantly higher affinity for CD4 (22). In fact, it has been demonstrated that the ability of some, but not all, PR-X4 isolates to replicate in T-cell lines can be rescued by overexpression of CD4 (29).
Based on these data, it seemed possible that the inability of TCLA-X4 viruses to infect primary macrophages that are susceptible to infection by PR-X4 isolates might simply reflect an inability of the viruses to effectively utilize low levels of cell surface CXCR4. This hypothesis makes three clear predictions. First, the requirement of TCLA-X4 viruses, compared to PR-X4 viruses, for a higher level of cell surface CXCR4 should not be unique to macrophages. TCLA-X4 viruses should therefore also be less effective than PR-X4 viruses at infecting transformed CD4+ cells that express low levels of CXCR4. Second, overexpression of CXCR4 on primary macrophages should boost infection by TCLA-X4 viruses but have at most a moderate effect on infection by PR-X4 viruses. Third, mutations that block signaling via CXCR4 should not affect infection via CXCR4 on either cell lines or primary macrophages.
In this study, we used a prototypic TCLA-X4 virus, expressing an env gene derived from the HXB3 proviral clone (20), and two novel PR-X4 viruses, expressing env genes derived from the recently described patient isolates QH1549 and QH1558 (19), to test each of these three predictions. Specifically, we have shown that the TCLA-X4 virus was able to infect COS7 cells expressing high levels of CXCR4 and low levels of CD4 more effectively than either PR-X4 virus (Fig. 4A), yet the TCLA-X4 virus was significantly less effective than either PR-X4 virus at infecting COS7 cells expressing low levels of CXCR4 and high levels of CD4 (Fig. 4B). Consistent with the hypothesis that cell surface CXCR4 levels are a major determinant of infection efficiency by TCLA-X4, but not PR-X4, viruses, we showed that overexpression of CXCR4 on MDM, using a lentiviral vector, dramatically enhanced the efficiency of infection by the TCLA-X4 virus while exerting only a modest positive effect on PR-X4 virus infection efficiency (Fig. 5B). The hypothesis that PR-X4 virus infection is, in contrast, more subject to variation in the level in CD4 expression (22) was supported by the finding that overexpression of CD4 on macrophages significantly enhanced infection by both PR-X4 isolates yet had little effect on infection by the TCLA-X4 virus (Fig. 5B). Finally, we present data showing that two distinct, previously described (6) mutants of CXCR4, termed D187A and Δi3A, that fail to signal upon binding of SDF-1 (Fig. 5C) are nevertheless fully able to support MDM infection by TCLA-X4 HIV-1 (Fig. 5D).
Two alternative hypotheses have previously been proposed to explain the inability of TCLA-X4 viruses to infect primary macrophages even though these express low but readily detectable levels of CXCR4. One hypothesis suggests that productive infection of macrophages by X4 HIV-1 isolates is unusual in requiring virion-induced signaling via the CXCR4 chemokine receptor (3, 25, 40). This hypothesis therefore suggests that the interaction of a TCLA-X4 Env protein with CXCR4 is unable to generate this signaling event, while PR-X4 Env binding to CXCR4 does activate signaling. However, this hypothesis cannot explain why simply overexpressing CXCR4 on macrophages would greatly enhance TCLA-X4 virus infection (Fig. 5B), as the predicted inability of TCLA-X4 Env proteins to activate CXCR4 signaling would remain unchanged. Moreover, the fact that two distinct CXCR4 mutants that are not able to signal can also effectively support infection of MDM by TCLA-X4 HIV-1 (Fig. 5C and D) is clearly inconsistent with the proposal that CXCR4-mediated signaling is key for productive macrophage infection, although it does remain formally possible that both mutations inhibit only SDF-1-dependent, not HIV-1 Env-dependent, signaling via CXCR4. Finally, the observation that low levels of CXCR4 expressed on the transformed COS7 cell line can effectively support infection by PR-X4, but not TCLA-X4, viruses (Fig. 4) suggests that the selective tropism of PR-X4 viruses for macrophages can be accurately recreated in unrelated cells that express comparable levels of CXCR4 but that clearly do not depend on signaling for productive infection (6, 13, 15, 18).
A second proposal to explain the inability of TCLA-X4 viruses to infect primary macrophages suggests that CXCR4 in macrophages is expressed in a distinct, high-molecular-weight form that is selectively nonpermissive for TCLA-X4, but not PR-X4, virus infection (23). However, if CXCR4 is subject to distinct posttranslational processing in macrophages, then it is again hard to explain why simply overexpressing CXCR4 would rescue infection of macrophages by TCLA-X4 viruses (Fig. 5B) and also why it is possible to reproduce the inefficient infection of MDM by TCLA-X4 viruses, but not PR-X4 viruses, using transfected COS7 cells expressing a comparable level of CXCR4 (Fig. 4). We therefore conclude that the primary determinant of the inefficient infection of macrophages by TCLA-X4 viruses such as IIIB and LAI is the relatively low level of CXCR4 expressed on the surface of these primary cells in culture.
As noted above, one could suggest that the inability of TCLA-X4 viruses to efficiently infect low CXCR4-expressing cells reflects the low affinity of TCLA-X4 Env proteins for CXCR4, just as the inability of PR-X4 viruses to efficiently infect low-CD4-expressing cells (Fig. 4A) appears to result from a low affinity for CD4 (22, 29). However, it is also possible that it is the lability of the TCLA Env-CD4 heterodimer, compared to the highly stable PR Env-CD4 heterodimer, that leads to a requirement for a high level of cell surface CXCR4. This difference in the stability of these heterodimeric complexes can be readily revealed by treatment of virus preparations with sCD4, which rapidly neutralizes TCLA-X4 virions but has little or no inhibitory effect on virions bearing a PR-X4 Env protein (references 9, 21, 27, and 29 and data not shown). It is therefore possible that only a very short window of opportunity exists for the labile TCLA Env-CD4 complex to recruit CXCR4 and form a more stable Env-CD4-CXCR4 ternary complex. Clearly, the likelihood that this recruitment would occur successfully would depend on the level of CXCR4 on the surface of the CD4+ target cell. Conversely, PR-X4 isolates may form a highly stable Env-CD4 complex, and even a low level of CXCR4 would then be predicted to suffice to support the eventual formation of the final ternary complex.
It is of interest to compare the data presented in this report, arguing that low CXCR4 expression can explain the inability of TCLA-X4 HIV-1 to infect primary human macrophages, with recently published data examining why certain primary T-cell-tropic (T-tropic) simian immunodeficiency virus (SIV) variants are unable to infect primary simian macrophages (4). Remarkably, these workers were able to show that the ability of T-tropic SIV to infect primary simian macrophages could be effectively rescued by overexpression of CD4 after transduction of these primary cells with a lentiviral CD4 expression vector. Overexpression of the CCR5 coreceptor had, in contrast, relatively little effect on the level of infection (4). Therefore, it would appear that inefficient infection of simian macrophages by T-tropic SIV simply results from a suboptimal level of cell surface CD4. Similarly, Platt et al. (29) have recently presented evidence arguing that inefficient infection of CD4+ T-cell lines by PR-X4 isolates of HIV-1 also largely reflects a suboptimal level of cell surface CD4. Finally, in this report, we present evidence that the inefficient infection of human macrophages by the TCLA HIV-1 isolate IIIB results from a low level of cell surface CXCR4. Together, these data support the general hypothesis that the various tissue tropisms displayed by different primate lentivirus isolates are likely to largely reflect different minimal requirements for both CD4 and coreceptor expression on the surface of target cells.
ACKNOWLEDGMENTS
We thank R. M. Richardson for help with the calcium mobilization assay, N. R. Landau for the gift of the luciferase indicator virus pNL-Luc-E−R+, and M. H. Malim for the VSV-G expression plasmid pHIT/G.
This work was supported in part by grant AI42538 from the National Institute of Allergy and Infectious Diseases to K.T. and B.R.C., as well as by NIH grants CA78673 to H.K.L., RR00030 to M.A.M., and AI40237 to M.L.G., and also by the Duke Center for AIDS Research (AI28662). M.A.M. is a recipient of an American Society of Clinical Oncology Career Award and B.R.C. is an Investigator in the Howard Hughes Medical Institute.
REFERENCES
- 1.Adachi A, Gendelman H E, Koenig S, Folks T, Willey R, Rabson A, Martin M A. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J Virol. 1986;59:284–291. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alkhatib G, Combadiere C, Broder C C, Feng Y, Kennedy P E, Murphy P M, Berger E A. CC CKR5: a RANTES, MIP-1α, MIP-1β receptor as a fusion cofactor for macrophage-tropic HIV-1. Science. 1996;272:1955–1958. doi: 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- 3.Arthos J, Rubbert A, Rabin R L, Cicala C, Machado E, Wildt K, Hanbach M, Steenbeke T D, Swofford R, Farber J M, Fauci A S. CCR5 signal transduction in macrophages by human immunodeficiency virus and simian immunodeficiency virus envelopes. J Virol. 2000;74:6418–6424. doi: 10.1128/jvi.74.14.6418-6424.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bannert N, Schenten D, Craig S, Sodroski J. The level of CD4 expression limits infection of primary rhesus monkey macrophages by a T-tropic simian immunodeficiency virus and macrophagetropic human immunodeficiency viruses. J Virol. 2000;74:10984–10993. doi: 10.1128/jvi.74.23.10984-10993.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bieniasz P D, Fridell R A, Aramori I, Ferguson S S G, Caron M G, Cullen B R. HIV-1 induced cell fusion is mediated by multiple regions within both the viral envelope and the CCR-5 co-receptor. EMBO J. 1997;16:2599–2609. doi: 10.1093/emboj/16.10.2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brelot A, Heveker N, Montes M, Alizon M. Identification of residues of CXCR4 critical for human immunodeficiency virus coreceptor and chemokine receptor activities. J Biol Chem. 2000;275:23736–23744. doi: 10.1074/jbc.M000776200. [DOI] [PubMed] [Google Scholar]
- 7.Choe H, Farzan M, Sun Y, Sullivan N, Rollins B, Ponath P D, Wu L, Mackay C R, LaRosa G, Newman W, Gerard N, Gerard C, Sodroski J. The β-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell. 1996;85:1135–1148. doi: 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- 8.Connor R I, Chen B K, Choe S, Landau N R. Vpr is required for efficient replication of human immunodeficiency virus type-I in mononuclear phagocytes. Virology. 1995;206:935–944. doi: 10.1006/viro.1995.1016. [DOI] [PubMed] [Google Scholar]
- 9.Daar E S, Li X L, Moudgil T, Ho D D. High concentrations of recombinant soluble CD4 are required to neutralize primary human immunodeficiency virus type 1 isolates. Proc Natl Acad Sci USA. 1990;87:6574–6578. doi: 10.1073/pnas.87.17.6574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deng H, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, Di Marzio P, Marmon S, Sutton R E, Hill C M, Davis C B, Peiper S C, Schall T J, Littman D R, Landau N R. Identification of a major co-receptor for primary isolates of HIV-1. Nature. 1996;381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 11.Doms R W. Beyond receptor expression: the influence of receptor conformation, density, and affinity in HIV-1 infection. Virology. 2000;276:229–237. doi: 10.1006/viro.2000.0612. [DOI] [PubMed] [Google Scholar]
- 12.Donzella G A, Schols D, Lin S W, Este J A, Nagashima K A, Maddon P J, Allaway G P, Sakmar T P, Henson G, De Clercq E, Moore J P. AMD3100, a small molecule inhibitor of HIV-1 entry via the CXCR4 co-receptor. Nat Med. 1998;4:72–77. doi: 10.1038/nm0198-072. [DOI] [PubMed] [Google Scholar]
- 13.Doranz B J, Orsini M J, Turner J D, Hoffman T L, Berson J F, Hoxie J A, Peiper S C, Brass L F, Doms R W. Identification of CXCR4 domains that support coreceptor and chemokine receptor functions. J Virol. 1999;73:2752–2761. doi: 10.1128/jvi.73.4.2752-2761.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dragic T, Litwin V, Allaway G P, Martin S R, Huang Y, Nagashima K A, Cayanan C, Maddon P J, Koup R A, Moore J P, Paxton W A. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature. 1996;381:667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 15.Farzan M, Choe H, Martin K A, Sun Y, Sidelko M, Mackay C R, Gerard N P, Sodroski J, Gerard C. HIV-1 entry and macrophage inflammatory protein-1β-mediated signaling are independent functions of the chemokine receptor CCR5. J Biol Chem. 1997;272:6854–6857. doi: 10.1074/jbc.272.11.6854. [DOI] [PubMed] [Google Scholar]
- 16.Feng Y, Broder C C, Kennedy P E, Berger E A. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science. 1996;272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 17.Fouchier R A M, Meyer B E, Simon J H M, Fischer U, Malim M H. HIV-1 infection of non-dividing cells: evidence that the amino-terminal basic region of the viral matrix protein is important for Gag processing but not for post-entry nuclear import. EMBO J. 1997;16:4531–4539. doi: 10.1093/emboj/16.15.4531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gosling J, Monteclaro F S, Atchison R E, Arai H, Tsou C-L, Goldsmith M A, Charo I F. Molecular uncoupling of C-C chemokine receptor 5-induced chemotaxis and signal transduction from HIV-1 coreceptor activity. Proc Natl Acad Sci USA. 1997;94:5061–5066. doi: 10.1073/pnas.94.10.5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu Q-H, Barry A P, Wang Z-X, Connolly S M, Peiper S C, Greenberg M L. Evolution of the human immunodeficiency virus type I envelope during infection reveals molecular corollaries of specificity for coreceptor utilization and AIDS pathogenesis. J Virol. 2000;74:11858–11872. doi: 10.1128/jvi.74.24.11858-11872.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hwang S S, Boyle T J, Lyerly H K, Cullen B R. Identification of the envelope V3 loop as the primary determinant of cell tropism in HIV-1. Science. 1991;253:71–74. doi: 10.1126/science.1905842. [DOI] [PubMed] [Google Scholar]
- 21.Hwang S S, Boyle T J, Lyerly H K, Cullen B R. Identification of envelope V3 loop as the major determinant of CD4 neutralization sensitivity of HIV-1. Science. 1992;257:535–537. doi: 10.1126/science.1636088. [DOI] [PubMed] [Google Scholar]
- 22.Kozak S L, Platt E J, Madani N, Ferro F E, Jr, Peden K, Kabat D. CD4, CXCR-4, and CCR-5 dependencies for infections by primary patient and laboratory-adapted isolates of human immunodeficiency virus type 1. J Virol. 1997;71:873–882. doi: 10.1128/jvi.71.2.873-882.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lapham C K, Zaitseva M B, Lee S, Romanstseva T, Golding H. Fusion of monocytes and macrophages with HIV-1 correlates with biochemical properties of CXCR4 and CCR5. Nat Med. 1999;5:303–308. doi: 10.1038/6523. [DOI] [PubMed] [Google Scholar]
- 24.Lee B, Sharron M, Montaner L J, Weissman D, Doms R W. Quantification of CD4, CCR5, and CXCR4 levels on lymphocyte subsets, dendritic cells, and differentially conditioned monocyte-derived macrophages. Proc Natl Acad Sci USA. 1999;96:5215–5220. doi: 10.1073/pnas.96.9.5215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu Q H, Williams D A, McManus C, Baribaud F, Doms R W, Schols D, De Clercq E, Kotlikoff M I, Collman R G, Freedman B D. HIV-1 gp120 and chemokines activate ion channels in primary macrophages through CCR5 and CXCR4 stimulation. Proc Natl Acad Sci USA. 2000;97:4832–4837. doi: 10.1073/pnas.090521697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Malim M H, Hauber J, Fenrick R, Cullen B R. Immunodeficiency virus rev trans-activator modulates the expression of the viral regulatory genes. Nature. 1988;335:181–183. doi: 10.1038/335181a0. [DOI] [PubMed] [Google Scholar]
- 27.Moore J P, McKeating J A, Huang Y X, Ashkenazi A, Ho D D. Virions of primary human immunodeficiency virus type 1 isolates resistant to soluble CD4 (sCD4) neutralization differ in sCD4 binding and glycoprotein gp120 retention from sCD4-sensitive isolates. J Virol. 1992;66:235–243. doi: 10.1128/jvi.66.1.235-243.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O'Brien W A, Koyanagi Y, Namazie A, Zhao J-Q, Diagne A, Idler K, Zack J A, Chen I S Y. HIV-1 tropism for mononuclear phagocytes can be determined by regions of gp120 outside the CD4-binding domain. Nature. 1990;348:69–73. doi: 10.1038/348069a0. [DOI] [PubMed] [Google Scholar]
- 29.Platt E J, Kozak S L, Kabat D. Critical role of enhanced CD4 affinity in laboratory adaptation of human immunodeficiency virus type 1. AIDS Res Hum Retroviruses. 2000;16:871–882. doi: 10.1089/08892220050042819. [DOI] [PubMed] [Google Scholar]
- 30.Platt E J, Wehrly K, Kuhmann S E, Chesebro B, Kabat D. Effects of CCR5 and CD4 cell surface concentrations on infections by macrophagetropic isolates of human immunodeficiency virus type 1. J Virol. 1998;72:2855–2864. doi: 10.1128/jvi.72.4.2855-2864.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Richardson R M, Pridgen B C, Haribabu B, Ali H, Snyderman R. Differential cross-regulation of the human chemokine receptors CXCR1 and CXCR2. Evidence for time-dependent signal generation. 1998. J Biol Chem. 1998;273:23830–23836. doi: 10.1074/jbc.273.37.23830. [DOI] [PubMed] [Google Scholar]
- 32.Rocancourt D, Bonnerot C, Jouin H, Emerman M, Nicolas J F. Activation of a beta-galactosidase recombinant provirus: application to titration of human immunodeficiency virus (HIV) and HIV-infected cells. J Virol. 1990;64:2660–2668. doi: 10.1128/jvi.64.6.2660-2668.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ross T M, Bieniasz P D, Cullen B R. Role of chemokine receptors in HIV-1 infection and pathogenesis. Adv Virus Res. 1999;52:233–267. doi: 10.1016/s0065-3527(08)60300-0. [DOI] [PubMed] [Google Scholar]
- 34.Ross T M, Cullen B R. The ability of HIV type 1 to use CCR-3 as a coreceptor is controlled by envelope V1/V2 sequences acting in conjunction with a CCR-5 tropic V3 loop. Proc Natl Acad Sci USA. 1998;95:7682–7686. doi: 10.1073/pnas.95.13.7682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schmidtmayerova H, Alfano M, Nuovo G, Bukrinsky M. Human immunodeficiency virus type 1 T-lymphotropic strains enter macrophages via a CD4- and CXCR4-mediated pathway: replication is restricted at a postentry level. J Virol. 1998;72:4633–4642. doi: 10.1128/jvi.72.6.4633-4642.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schuitemaker H, Koot M, Kootstra N A, Dercksen M W, de Goede R E Y, van Steenwijk R P, Lange J M A, Eeftink Schattenkerk J K M, Miedema F, Tersmette M. Biological phenotype of human immunodeficiency virus type 1 clones at different stages of infection: progression of disease is associated with a shift from monocytotropic to T-cell-tropic virus populations. J Virol. 1992;66:1354–1360. doi: 10.1128/jvi.66.3.1354-1360.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shioda T, Levy J A, Cheng-Mayer C. Macrophage and T cell-line tropisms of HIV-1 are determined by specific regions of the envelope gp120 gene. Nature. 1991;349:167–169. doi: 10.1038/349167a0. [DOI] [PubMed] [Google Scholar]
- 38.Simmons G, Reeves J D, McKnight A, Dejucq N, Hibbitts S, Power C A, Aarons E, Schols D, De Clercq E, Proudfoot A E I, Clapham P R. CXCR4 as a functional coreceptor for human immunodeficiency virus type 1 infection of primary macrophages. J Virol. 1998;72:8453–8457. doi: 10.1128/jvi.72.10.8453-8457.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Verani A, Pesenti E, Polo S, Tresoldi E, Scarlatti G, Lusso P, Siccardi A G, Vercelli D. CXCR4 is a functional coreceptor for infection of human macrophages by CXCR4-dependent primary HIV-1 isolates. J Immunol. 1998;161:2084–2088. [PubMed] [Google Scholar]
- 40.Weissman D, Rabin R L, Arthos J, Rubbert A, Dybul M, Swofford R, Venkatesan S, Farber J M, Fauci A S. Macrophage-tropic HIV and SIV envelope proteins induce a signal through the CCR5 chemokine receptor. Nature. 1997;389:981–985. doi: 10.1038/40173. [DOI] [PubMed] [Google Scholar]
- 41.Westervelt P, Gendelman H E, Ratner L. Identification of a determinant within the human immunodeficiency virus 1 surface envelope glycoprotein critical for productive infection of primary monocytes. Proc Natl Acad Sci USA. 1991;88:3097–3101. doi: 10.1073/pnas.88.8.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wrin T, Loh T P, Charron Vennari J, Schuitemaker H, Nunberg J H. Adaptation to persistent growth in the H9 cell line renders a primary isolate of human immunodeficiency virus type 1 sensitive to neutralization by vaccine sera. J Virol. 1995;69:39–48. doi: 10.1128/jvi.69.1.39-48.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yi Y, Isaacs S N, Williams D A, Frank I, Schols D, De Clercq E, Kolson D L, Collman R G. Role of CXCR4 in cell-cell fusion and infection of monocyte-derived macrophages by primary human immunodeficiency virus type 1 (HIV-1) strains: two distinct mechanisms of HIV-1 dual tropism. J Virol. 1999;73:7117–7125. doi: 10.1128/jvi.73.9.7117-7125.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yi Y, Rana S, Turner J D, Gaddis N, Collman R G. CXCR-4 is expressed by primary macrophages and supports CCR5-independent infection by dual-tropic but not T-tropic isolates of human immunodeficiency virus type 1. J Virol. 1998;72:772–777. doi: 10.1128/jvi.72.1.772-777.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang L, Huang Y, He T, Cao Y, Ho D D. HIV-1 subtype and second-receptor use. Nature. 1996;383:768. doi: 10.1038/383768a0. [DOI] [PubMed] [Google Scholar]