Abstract
Kaposi's sarcoma-associated herpesvirus (KSHV; also known as human herpesvirus-8) establishes latent and lytic infections in both lymphoid and endothelial cells and has been associated with diseases of both cell types. The KSHV open reading frame 50 (ORF50) protein is a transcriptional activator that plays a central role in the reactivation of lytic viral replication from latency. Here we identify and characterize a DNA binding site for the ORF50 protein that is shared by the promoters of two delayed early genes (ORF57 and K-bZIP). Transfer of this element to heterologous promoters confers on them high-level responsiveness to ORF50, indicating that the element is both necessary and sufficient for activation. The element consists of a conserved 12-bp palindromic sequence and less conserved sequences immediately 3′ to it. Mutational analysis reveals that sequences within the palindrome are critical for binding and activation by ORF50, but the presence of a palindrome itself is not absolutely required. The 3′ flanking sequences also play a critical role in DNA binding and transactivation. The strong concordance of DNA binding in vitro with transcriptional activation in vivo strongly implies that sequence-specific DNA binding is necessary for ORF50-mediated activation through this element. Expression of truncated versions of the ORF50 protein reveals that DNA binding is mediated by the amino-terminal 272 amino acids of the polypeptide.
Infection by Kaposi's sarcoma-associated herpesvirus (KSHV) also known as human herpesvirus 8 (HHV-8) is associated with malignancies of both endothelial and lymphoid cells in humans. KSHV has been well established as the etiologic agent responsible for Kaposi's sarcoma (KS), an endothelial neoplasm frequent in homosexual men with AIDS and highly prevalent in sub-Saharan Africa (reviewed in reference 25). KSHV is also linked to two other AIDS-related malignancies, primary effusion lymphoma and multicentric Castleman's disease (5, 26). The presence of viral DNA in CD19+ B cells and other mononuclear cells of the peripheral blood of KS/AIDS patients (1, 2, 31), even prior to full-blown KS, suggests that infection of the lymphoid compartment is antecedent to the development of the endothelial disease.
KSHV infection, similar to infection by other herpesviruses, displays two life cycle modes, latency and lytic replication. Latency is established by the virus both in endothelial and B cells and is detectable in such cell types both in vitro and in infected hosts (1, 3, 7, 19, 22, 27, 31). KSHV latency-associated genes are expressed in most spindle cells of KS tumors and are thought to contribute to their survival and proliferation (25). However, many viral genes (e.g., vGCR and vMIPs I and II) encoding homologs of cellular signaling proteins which have been suspected of roles in the histogenesis of KS are expressed strictly as lytic cycle products (13, 20, 23, 24, 28). This suggests that the KSHV lytic cycle may also contribute to KS lesion formation. Additional support for this notion comes from studies showing that treatment of high-risk patients with the antiviral agents ganciclovir, which blocks lytic KSHV replication, reduces KS risk (18). Of course, lytic reactivation also likely contributes to KS progression by allowing spread of the virus from the lymphoid reservoir to the endothelial sites of KS tumor formation. A complete understanding of the molecular events which control and direct viral reactivation from latency is thus critical to completely understanding KSHV pathogenesis.
We and others have previously shown that ectopic overexpression of KSHV open reading frame ORF 50 (ORF50) induces lytic reactivation of the virus in B-cell models of latency (8, 15, 16, 29). Furthermore, we identified a potent carboxy-terminal activation domain in the ORF50 protein which shares homology with its counterparts in Epstein-Barr virus (EBV) and herpesvirus saimiri; deletion of this domain from the cognate KSHV protein abrogates its ability to reactivate the virus in BCBL-1 cells (15). We also identified an amino-terminal oligomerization domain which allowed us to design a dominant-negative mutant of ORF50 that strongly suppresses transcriptional activation by the wild-type (wt) protein and inhibits both spontaneous and chemically induced lytic reactivation. Thus, transcriptional activation by ORF50 is absolutely required for lytic reactivation of KSHV induced by all known reactivating signals (15).
We initiated our investigation into ORF50's transactivation mechanism by utilizing transient cotransfections of ORF50 expression vectors together with reporter plasmids in which various KSHV promoters drive expression of firefly luciferase. These studies demonstrated that ORF50 transactivates the promoters of the ORF57, K-bZIP, nut-1 (PAN), thymidine kinase (TK), kaposin, and DNA binding protein genes in CV-1, BJAB (human B-lymphoblast), and SLK (human endothelial) cells (15, 16). However, ORF50's inability to transactivate the DNA polymerase, assembly protein (AP), and glycoprotein B promoters suggested that ORF50 displays selectivity in transactivation and is not simply a promiscuous activator.
In this report, we mapped the DNA sequence required for ORF50's activation of the ORF57 promoter and show that this element is homologous to an element in the K-bZIP promoter. The element can be directly bound by recombinant ORF50 protein; the binding site consists of a 12-bp partially palindromic sequence, 5′-AACAATAATGTT-3′, together with several 3′ flanking nucleotides. Transfer of this element to heterologous cellular and viral TATA boxes which alone are not activated by ORF50 confers dramatic ORF50 responsiveness, and this activation is independent of orientation of the element. Mutations in the element which inhibit ORF50 binding to either promoter also inhibit ORF50-dependent activation in vivo, indicating the importance of DNA binding for activation.
MATERIALS AND METHODS
Plasmids.
All plasmids were propagated as described elsewhere (15, 16). pcDNA3-gORF50, described elsewhere (16), is a KSHV genomic clone which expresses the full-length ORF50 protein.
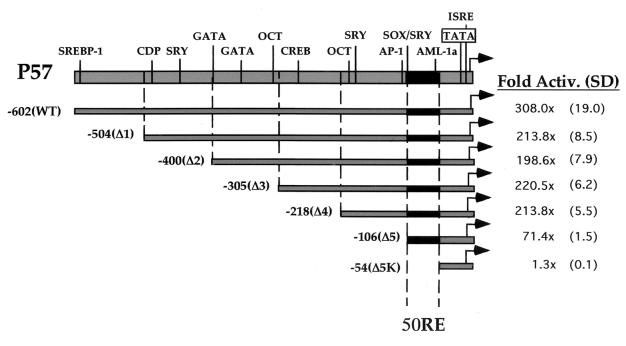
The reporter plasmid pORF57-GL3, described elsewhere (15), contains approximately 1 kb of ORF57's promoter and herein is referred to as pORF57FL-GL3. pORF57wt-GL3 was constructed by deleting the 400-bp BamHI-to-SpeI fragment from pORF57FL-GL3 and served as the template for PCR used to construct the deletion series of the ORF57 promoter (Fig. 1). Briefly, each PCR utilized the same reverse primer, 57REV (sequence 5′ GCGCGCTAGCGGTTCTTATATTGTGC), a 26-mer which lies between the ORF57 start site and TATA box and introduces an NheI site to the PCR product. 57REV was combined individually with each of five 24- to 26-bp forward primers, named 57P1 to 57P5 (sequences provided upon request), which introduced a SacI site to each PCR product. The left (TATA-distal) end of each PCR product thus corresponds to each of the promoter sequences diagrammed in Fig. 1, named according to their distance relative to the ORF57 transcription start site. Each PCR product was then digested with SacI and NheI and cloned into pGL3-basic (Promega) which had been digested with the same enzymes, to generate the promoter deletion series (p57Δ1-GL3, p57Δ2-GL3, p57Δ3-GL3, p57Δ4-GL3, and p57Δ5-GL3). p57Δ5K was constructed by digesting p57Δ5-GL3 with KpnI to remove the SacI-to-KpnI fragment of the ORF57Δ5 promoter, followed by religation. The location of this KpnI site is diagrammed in Fig. 2B.
FIG. 1.
Deletion analysis of the ORF57 promoter reveals the minimal sequences necessary for transactivation by ORF50 (50RE). This schematic depicts the features of the ORF57 promoter lying between the transcriptional start site (genomic position 82003 [12]) and 602 bp upstream of it [here named -602(WT)]. Each deletion of the promoter is depicted by the grey bars and named Δ1 to Δ5K (described in Materials and Methods); the endpoint of each deletion is also described by its distance in base pairs from the start site, listed by a negative number next to the name of the deletion. Each was cotransfected into CV-1 cells with increasing amounts of pcDNA3-FLg50 or empty pcDNA3, using Superfect, as described in Materials and Methods, and fold transactivation was calculated. The maximal point of transactivation in each titration curve is listed (as well as the standard deviation [SD], in parentheses) for each deletion. The black bar denotes promoter positions -106 to -54, referred to as 50RE. The abbreviations above the wt promoter indicate consensus binding sites for cellular transcription factors as predicted by TRANSFAC (32). Abbreviations: SREBP-1, sterol regulatory element binding protein 1; CDP, CAAT displacement protein; SRY, sex-determining region Y protein; GATA, GATA family factors; OCT, octamer proteins; CREB, cyclic AMP-responsive binding element protein; AP-1, activating protein 1; SOX, SRY-like high-mobility-group box-containing protein family; AML-1a, acute myeloid leukemia 1a protein; TATA, TATA box; ISRE, interferon-stimulated response element.
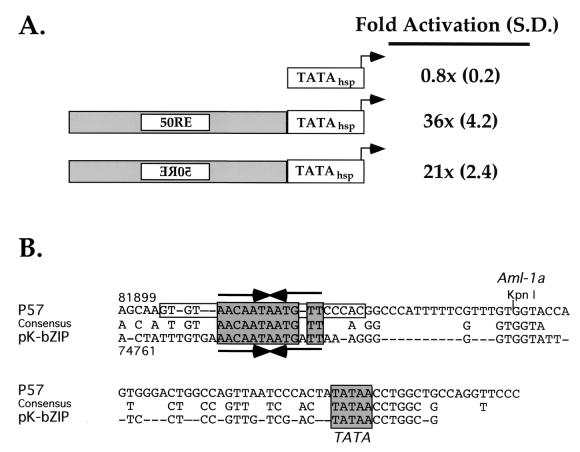
FIG. 2.
Enhancer-like function of 50RE and conservation with the K-bZIP promoter. (A) 50RE confers ORF50 responsiveness on a heterologous TATA box. 50RE was fused to the hsp70 TATA box (30) in forward and reverse orientations to generate the two reporter plasmids depicted. Each was cotransfected into CV-1 cells with various amounts of ORF50 expression vector using Fugene-6, and fold activation was calculated as for Fig. 1. Maximal activation of each reporter is given at the right, followed by standard deviations (S.D.) in parentheses. (B) The TATA-proximal promoters of the ORF57 and K-bZIP genes share three conserved sequences. The -106/-5 sequence of the ORF57 promoter is given at the top, the sequence of the TATA-proximal promoter of K-bZIP is shown at the bottom, and the consensus between them is in the center. The grey boxes denote the conserved functional blocks shared by the two promoters. The convergent arrows denote the 12-bp palindromic sequence shared by the promoters, and the open box denotes the extended, palindromic flanking sequences found only in p57. The numbers above and below the sequences refer to the genomic positions of the first nucleotide depicted in each promoter sequence. Kpn I refers to the KpnI restriction endonuclease site which demarcates the right-hand side of 50RE at nt -54 from the ORF57 start site.
The reporter plasmid hsp-luc contains the TATA box and surrounding basal promoter sequences of the cellular hsp70 gene (30) cloned into the BglII/HindIII sites of pGL3-basic. The oligonucleotides hspTA-FII (5′-GATCTGCGATCTAAGTCGTGACGACTTA TAAAGA) and hspTA-RII (5′-AGCTTCTTTATAAGTCGTCACGACTTA GATCGCA) were annealed by boiling equal amounts of both oligonucleotides in annealing buffer (50 mM KCl, 10 mM Tris-HCl [pH 8.3], 1.5 mM MgCl2) for 5 min, followed by slow equilibration to 25°C overnight. Annealed oligonucleotides were then successively extracted with phenol-CHCl3 and CHCl3 and precipitated. After pelleting and resuspension in distilled H2O (dH2O), the annealed oligonucleotides were cloned into pGL3-basic which had been digested with BglII and HindIII to construct hsp-luc and confirmed by sequencing. The resultant plasmid conserves the spacing between the TATA box and the BglII site which is found in the cognate hsp70 promoter between the TATA box and CAAT box (30).
p57-Δ5F-hsp-luc and p57-Δ5_-hsp-luc (where _ is A, B, C, or D) were constructed by PCR amplification using p57Δ4 as a template and primers identical to 57REV and 57P5 but designed to introduce BglII sites at both ends of the PCR product. The resultant PCR product was ligated into hsp-luc which had been digested with BglII and treated with shrimp alkaline phosphatase (U.S. Biochemical). The orientations of the inserts were confirmed by sequencing.
p57Δ5-5A-hsp-luc, p57Δ5-5B-hsp-luc, p57Δ5-5C-hsp-luc, and p57Δ5-5D-hsp-luc were constructed by annealing oligonucleotides representing each of the sequences diagrammed in Fig. 4A with the respective reverse complement oligonucleotide for each, in a manner similar to that for the hsp70 TATA box oligonucleotides (described above). Both members of each oligonucleotide pair were designed such that the resulting double-stranded probe contained an overhanging BglII site at each end. Half of each annealed and purified probe was then set aside for electrophoretic mobility shift analysis (EMSA; see below), and the other half of the probe was multimerized using T4 DNA ligase (New England Biolabs). Following multimerization, the nucleic acid was purified by organic extraction and precipitation and then cloned into hsp-luc which had been digested with BglII and treated with shrimp alkaline phosphatase. Orientation and multimerization status were confirmed by sequencing.
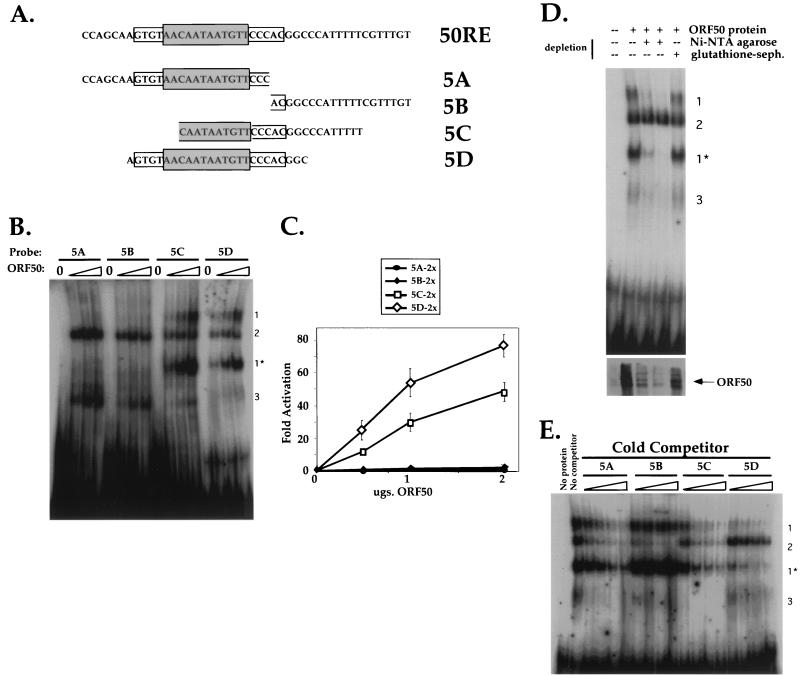
FIG. 4.
Both the palindrome and flanking sequences contribute to DNA binding and transactivation by ORF50. (A) Sequences of four 26-bp oligonucleotides used as probes in EMSA and cloned into reporter plasmids. The sequence of the top strand of 50RE is shown at top, with the grey box depicting the 12-bp palindrome and the open box depicting the extended, flanking palindromic sequences. Each sequence below that represents the top strand of each of four oligonucleotides named 5A to 5D, which were annealed to complementary oligonucleotides as described in Materials and Methods. The resultant products were radiolabeled for use in EMSA or cloned as dimers upstream of the hsp70 TATA box in plasmid hsp-luc for use in transactivation assays (generating plasmids p57-5Ahsp-luc, p57-5Bhsp-luc, p57-5Chsp-luc, and p57-5Dhsp-luc; see Materials and Methods). The palindromic and flanking sequences which are included in each oligonucleotide are indicated by the boxes. (B) Four DNA-protein complexes are formed by incubation of Bac-50-infected Sf9 cell extracts with the EMSA probes. ORF50 was partially purified from Bac-50-infected Sf9 cells as described in Materials and Methods, and 0, 2.5, 5.0, and 10.0 μg, of extract (containing ca. 0, 50, 100, and 200 ng of His6-tagged ORF50 [see Materials and Methods]) were analyzed for interaction with probes 5A to 5D by EMSA. Each set of four binding reactions is indicated above the autoradiogram according to the EMSA probe used, and resulting complexes are indicated by the numbers to the right of the autoradiogram (see text). (C) Transactivation of the 5A to 5D sequences by ORF50. Each reporter plasmid (A) was cotransfected into CV-1 cells with increasing amounts of pcDNA3-FLg50 or empty expression vector, and fold activation was assayed as for Fig. 2A. The entire titration curve is depicted for each reporter. (D) ORF50 is a component of complexes 1 and 1*. Partially purified extract from Bac-50-infected Sf9 cells (250 μg) was depleted with Ni-NTA-agarose or mock depleted with glutathione-Sepharose (glutathione-seph.) as described in Materials and Methods. The Ni-NTA-depleted extract was further depleted by a second incubation with a fresh aliquot of beads; 5 μg of untreated extract or an equivalent volume of each depleted extract was then subjected to EMSA with probe 5D as for panel B (top). The migration of complexes 1 to 3 is indicated to the right. Alternatively, 1/10 volume of each extact was analyzed by immunoblotting for the presence of ORF50 (bottom). (E) Competition EMSAs demonstrate a hierarchy of binding preferences for ORF50 binding to the 5A to 5D sequences. Partially purified extract from Bac-50-infected Sf9 cells (6 μg) was incubated with a 10-, 20-, 40-, or 80-fold molar excess of each nonradiolabelled EMSA probe before incubation with a 1-fold molar equivalent of radiolabeled EMSA probe 5D. These preparations were subject to EMSA for panel B, and each set of four binding reactions is indicated above the autoradiogram. Included for comparison are an EMSA of a binding reaction lacking protein (No protein) and a reaction containing labeled 5D probe and ORF50 without specific competitor (No competitor); the migration of complexes 1 to 3 is indicated to the right.
p57Δ5-5Dm2 and p57Δ5-5Dm3 were constructed by annealing oligonucleotides representing each of the sequences diagrammed in Fig. 5A exactly as described above. Cloning into hsp-luc and subsequent sequencing were also performed as described above. Although the 57Δ5-5Dm1 oligonucleotides were annealed successfully, we could not successfully clone them into hsp-luc.
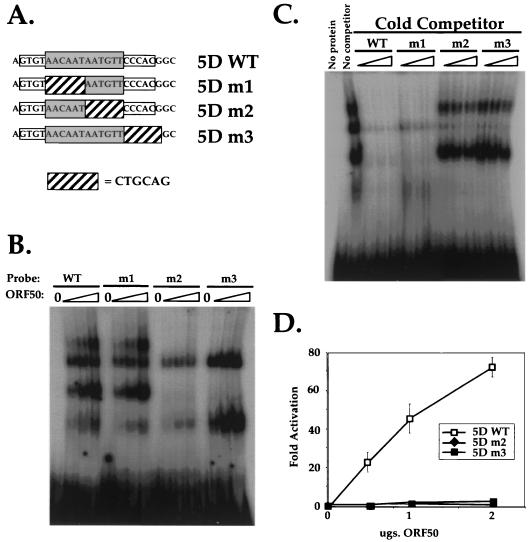
FIG. 5.
ORF50 binding to sequences within the palindrome is necessary for transcriptional activation. (A) Sequences of four 26-bp oligonucleotides used as probes in EMSA and cloned into reporter plasmids. The sequence of the top strand of p57-5Dwt is shown at the top, with the grey box depicting the 12-bp palindrome and the open box depicting the extended, flanking palindromic sequences. Each sequence below that represents the top strand of each of four mutant oligonucleotides named 5Dm1 to 5Dm3, in which the 6-bp linker substitution represented by the hatched box was substituted for the wt sequence indicated in each probe. These were annealed to a complementary oligonucleotide and then radiolabeled for use in EMSAs; 5Dwt, 5Dm2, and 5Dm3 were also cloned to produce the reporter vectors p57-5Dwthsp-luc, p57-5Dm2hsp-luc, and p57-5Dm3hsp-luc (see Materials and Methods). (B) The right half of the palindrome and flanking sequences are required for binding of ORF50 to p57-5D. Aliquots of 0, 2.5, 5.0, and 10.0 μg of partially purified extract (containing ca. 0, 50, 100, and 200 ng of His6-tagged ORF50 [see Materials and Methods]) from Bac-50-infected Sf9 cells were analyzed for interaction with probes 5Dwt and 5Dm1 to 5Dm3 by EMSA as for Fig. 4B. Each set of four binding reactions is indicated above the autoradiogram according to the EMSA probe used. (C) Competition EMSAs confirm the sequence requirements for ORF50 binding to p57-5D. Six micrograms of partially purified extract from Bac-50-infected Sf9 cells was incubated with a 50-, 100-, or 200-fold molar excess of each nonradiolabeled EMSA probe before incubation with a 1-fold molar equivalent of radiolabeled EMSA probe 5Dwt. These preparations were subject to EMSA as for Fig. 4E, and each set of four binding reactions is indicated above the autoradiogram. Included for comparison are an EMSA of a binding reaction lacking protein (No protein) and a reaction containing labeled 5Dwt probe and ORF50 without specific competitor (No competitor). (D) DNA binding of the 5D sequence by ORF50 is required for transcriptional activation. Each reporter plasmid (A) was cotransfected into CV-1 cells with increasing amounts of pcDNA3-FLg50 or empty expression vector, and fold activation was assayed and depicted as in Fig. 4C.
p57Δ5-5Dm4, p57Δ5-5Dm4-2, p57Δ5-5Dm5, p57Δ5-5Dm6, p57Δ5-5Dm7, p57Δ5-5Dm8, p57Δ5-5Dm9, p57Δ5-5Dm9-2, p57Δ5-5Dm9-3, and p57Δ5-5DwtII were constructed similarly to the 5Dm1 to 5Dm3 series described above, with the following exceptions: to facilitate directional cloning of each linker scanning mutant as dimers, oligonucleotides were designed to contain each of the sequences represented in Fig. 6 as dimers flanked by different restriction sites. In this way, if each 5Dwt or mutant variant shown in Fig. 6 is represented by “5D,” each oligonucleotide would consist of the following elements: NheI-5D-XhoI-5D-BglII. Each of these oligonucleotides was annealed to its respective reverse complement, resulting in an annealed product which was flanked by overhanging NheI and BglII sites. These products were then cloned into hsp-luc which had been digested with those two enzymes, and constructs were confirmed by sequencing.
FIG. 6.
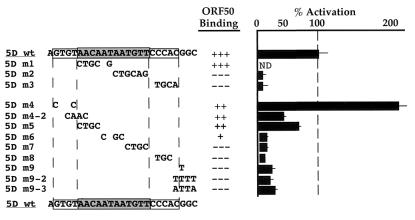
Fine mutational analysis of 50RE. A second series of nine linker substitution mutants across the 5D element was generated as EMSA probes and cloned into hsp-luc using oligonucleotides (as described in Materials and Methods). All of the mutants are named at the left; the new series of mutants, containing 1- to 4-bp changes of the wt sequence, are listed in lines 5 to 13. The specific base pairs changed in each clone of both series are listed in the center column underneath the sequence of 5Dwt. Each was tested both for DNA binding by the Bac-50-generated ORF50 and for transcriptional activation as in Fig. 4 and 5. DNA binding data were semiquantitated for each mutant by comparison to the binding of ORF50 to the 5Dwt probe, which was assigned the value of +++; similarly, activation was quantitated for each mutant by comparison to ORF50's activation of the 5Dwt reporter construct, which was assigned the value of 100% (the first four lines summarize the data from Fig. 5).
p57wt m1-GL3 was constructed by introducing the m1 linke; scanning mutation (see Fig. 5A) into the core palindrome of the ORF57 promoter by PCR. To do so, we performed two PCRs in which one primer of each primer pair overlapped the left side of the core palindrome but substituted the wt sequences with a PstI site. These primers were called p57wt m1F (5′-GCGCTGCAGAATGTT CCCACGGCCCATTT) and p57wt m1R (5′-GCGCTGCAGACACTTGTGGC AAAACACT). The two PCRs used p57wt-GL3 as a template and the following primer pairs: (i) p57wt m1F plus a reverse primer (pGL3REV) which was complementary to pGL3 vector sequences and flanked the NarI site of the vector and (ii) p57wt m1R plus a forward primer (B5A) which was complementary to pGL3 vector sequences and flanked the NotI site of the vector. Following PCR, product 2 was digested with NotI and PstI and cloned into pBluescript II KS which had been digested with the same enzymes to generate plasmid pBSIIKS-p57wt m1Left. PCR product 1 was then digested with NarI, treated with Klenow DNA polymerase (Life Technologies), and then digested with PstI. The resulting fragment was then cloned into pBSIIKS-p57wt m1Left which had been digested with EcoRV and PstI to generate the plasmid pBSIIKS-p57wt m1. The mutant promoter sequence was then transfered to pGL3-basic by digesting both plasmids with SacI and NcoI and purifying the appropriate fragments, followed by ligation.
p57wt m1+2-GL3 was constructed by substituting a BspEI site for the right side of the core palindrome in the p57wt m1 promoter. PCR was performed using p57wt-GL3 as a template and primers p57wt m1+2F (5′-GCGCTGCAGTCCGGACCCAC GGCCCATTTTTCGTT) and pGL3REV. The resultant product was digested with NarI and PstI and the cloned into p57wt m1-GL3 which had been digested with the same enzymes.
The reporter plasmid for the K-bZIP promoter, pK-bZIP-GL3, was described previously (15). pK-bZIPm1-GL3 was constructed identically to p57wt m1-GL3 except that the two PCRs used pK-bZIP-GL3 as a template and the following primer pairs: (i) pK-bZIPm1F (5′-GCGCTGCAGAATGATTAAAGGGGGTGGTA) plus pGL3REV and (ii) pK-bZIPm1R (5′-GCGCTGCAGTCACAAATAGTCACAATCAA) plus B5A. The final subcloning step was performed by transfer of the NotI/NcoI fragment from pBSIIKS-pK-bZIPm1 into pGL3-basic.
pK-bZIPm1+2-GL3 was constructed identically to p57wt m1+2-GL3, except that the PCR was performed with pK-bZIPm1 as a template and primers pK-bZIPm1+2F (5′-GCGCTGCAGTCCGGATAAAGGGGGTGGTATTTCCT) and pGL3REV. The resultant product was cloned into pK-bZIPm1-GL3.
Plasmid pcDNA3.1lacZ has been described elsewhere (15).
pBlueBacHis-50 was constructed by PCR amplification using pGem3-FLc50 (15) as a template and primers which introduced EcoRI and EcoRV sites to the full-length ORF of ORF50 during amplification. The resultant product was digested with these two enzymes and cloned into pBlueBacHis2C (Invitrogen) which had been digested with the same enzymes, to create an in-frame fusion to the six-histidine (His6) epitope tag.
Plasmid pRSET 0.8, which expresses the His6-tagged polypeptide C50 (diagrammed in Fig. 9A), has been described previously (16). Plasmid pET28b-N50 expresses the His6-tagged protein N50 and was constructed as follows. The ORF50-containing EcoRI-to-AvaI fragment of pBlueBacHis-50 was subcloned into pGex5X-3 which had been digested with EcoRI and XhoI to generate pGex-N50. pGex-N50 was digested with NotI and NcoI, treated with Klenow DNA polymerase, and religated to create pGex-N50ΔNco. pGexN50ΔNco was digested with AatII and then polished with T4 DNA polymerase (New England Biolabs) as recommended by the manufacturer. The N-terminal ORF50 sequences were then liberated by digestion with EcoRI. The resultant fragment was cloned into pET28b (Novagen) which had been digested with BlpI, treated with Klenow DNA polymerase, and then digested with EcoRI.
FIG. 9.
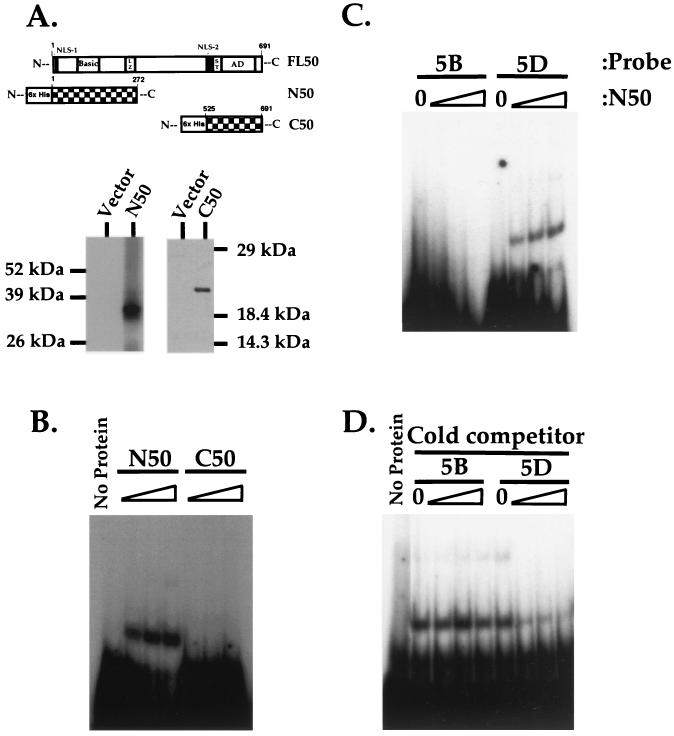
The amino terminus of ORF50 generated in E. coli binds to the response element. (A) Expression of N- and C-terminal truncations of ORF50 in E. coli. The top displays a schematic of full-length ORF50 (FL50). Below are schematics of the truncations of ORF50 expressed as fusions to the His6 epitope tag: aa 1 to 272 (N50) and aa 525 to 691 (C50 [16]). The bottom shows Western blot analysis of the proteins expressed and purified from E. coli transfected with pET28b-N50 (N50) and pRSET 0.8 (C50 [16]), detected using anti-His6 (Clontech) and anti-ORF50 (16) antibodies, respectively. Positions of migration of protein molecular weight standards (Life Technologies) are shown to the left and right. Abbreviations: NLS, putative nuclear localization signal; Basic, highly basic domain (described in the text); LZ, leucine repeats; ST, serine/threonine-rich region; AD, activation domain (15). (B) N50 but not C50 binds stably to the 5D sequence. N50 and C50 were expressed and purified from E. coli as described in Materials and Methods and then diluted to equal final concentrations; 0, 0.5, 1.0, and 2.0 μg of each protein (containing ca. 0, 0.1, 0.2, and 0.4 μg of truncated ORF50 polypeptide; see Materials and Methods) was incubated with radiolabeled 5Dwt probe and analyzed by EMSA as described in Materials and Methods. Each set of four binding reactions is indicated above the autoradiogram according to the polypeptide used. (C) N50 binds to the ORF57 5D but not 5B sequence. EMSA was performed as for panel B using the indicated radiolabeled probes and 0, 0.5, 1.0, and 2.0 μg of N50 polypeptide. (D) Unlabeled competitors confirm N50's binding specificity. One microgram of N50 polypeptide was incubated with a 50-, 100- or 200-fold molar excess of nonradiolabeled 5B or 5Dwt EMSA probe and then incubated with a 1-fold molar equivalent of radiolabeled 5Dwt probe. Included for comparison are an EMSA of a binding reaction lacking protein (B, No Protein) and a reaction containing labeled 5Dwt probe and ORF50 without specific competitor (D, No Protein).
Cell lines and transfections.
CV-1 cells were propagated and maintained as previously described (15). Transfections for analysis of the initial ORF57 promoter deletion series were performed using Superfect reagent (Qiagen) as previously described (12). All other transfections were performed using Fugene-6 (Boehringer Mannheim). Briefly, 105 CV-1 cells were plated in 2 ml of medium in each well of a six-well plate and then allowed to adhere overnight under standard growth conditions. Plasmid DNA for each transfection was aliquoted into 0.1 ml of minimal Dulbecco modified Eagle medium, using 0.5 μg of pcDNA3.1lacZ, 0.5 μg of the appropriate reporter vector (as described in the figure legends and text), and 0.5 to 2 μg of pcDNA-g50 (as described in the figure legends and text). In all cases, pcDNA3 was used as a filler plasmid to bring the total DNA amount to 3 μg for each transfection; all transfections likewise were performed in triplicate. Next, 9 μl of Fugene-6 reagent was added to each DNA-medium mixture, mixed by vigorous flicking, and allowed to incubate at room temperature for 30 min. Cells were refed with 2 ml of fresh growth medium, and the DNA–medium–Fugene-6 mixture was added dropwise to each well. Transfections were harvested at 40 to 48 h posttransfection and analyzed for luciferase and β-galactosidase.
The B-cell lymphoma cell line BJAB was propagated, maintained, and transfected as previously described (15).
The Spodoptera frugiperda pupal ovarian cell line Sf9 was propagated and maintained in complete TNM-FH medium (Life Technologies) supplemented with 20 mM l-glutamine, 100 U of penicillin G sodium per ml, and 0.1 mg of streptomycin sulfate per ml at 27°C in a nonhumdified incubator. Cells were maintained primarily as adherent cultures; when suspension cultures were used, growth medium was supplemented with the surfactant Pluronic F-68 (Life Technologies) at a final dilution of 0.1%.
Luciferase and β-galactosidase assays.
Performed as described previously (15). β-Galactosidase assays were performed as an internal control for each transfection to normalize for transfection efficiency and variability in the cell extract harvest.
Construction and propagation of recombinant baculovirus.
Briefly, using a Bac-N-Blue transfection kit (Invitrogen) according to the manufacturer's recommendations, Sf9 cells (UCSF Cell Culture Facility) were cotransfected with Bac-N-Blue viral DNA and pBlueBacHis-50, using Insectin-Plus liposomes (Invitrogen). Recombinant virus was harvested by collection of the cell medium at 5 days posttransfection and purified by subsequent plaque assay using baculovirus agarose (Invitrogen) containing 5-bromo-4-chloro-3-indolyl-β-d-galactosidase (X-Gal) to identify recombinants. Plaques containing recombinant virus were transferred to fresh Sf9 monolayers in 12-well plates to generate P1 viral stocks. These viral stocks were subsequently screened for ORF50 expression by infection of fresh Sf9 monolayers in six-well plates, which were harvested in 10s buffer (4) at 48 h postinfection (hpi) and analyzed by Western blotting using our previously described anti-ORF50 rabbit serum (16). The best-expressing viral clone (named Bac-50) was then amplified into a high-titer, P2 stock by infection of a 500-ml suspension culture of Sf9 cells, which was harvested when approximately 95% of the cells had lysed. The supernatant-containing virus was subsequently separated from cell debris by centrifugation, and virus was titered by plaque assay on Sf9 cells.
Overexpression and purification of ORF50 from Bac-50-infected Sf9 cells.
A 500-ml spinner culture of exponentially dividing Sf9 cells at a density of 2 × 106 cells/ml was infected with Bac-50 at a multiplicity of infection of 10. At 48 hpi, cells were collected by centrifugation, washed twice in 1× phosphate-buffered saline, and lysed in 100 ml of 1× urea HIS binding buffer (500 mM NaCl, 20 mM Tris [pH 8.0], 5 mM imidazole, 6 M urea, 25 mM phenylalanine, 25 mM isoleucine) supplemented with the Protease Inhibitor Cocktail Set III (Calbiochem). The resultant extract was then passed through an 18-gauge needle four times to ensure complete cell lysis and to shear the genomic DNA. Cell debris was removed by centrifugation in a GSA rotor (Sorvall) at 11,000 rpm for 30 min. The supernatant was decanted and then filtered using a 0.45-μm-pore-size filter (Nalgene) to remove any remaining particular matter.
To purify the His6-tagged ORF50 protein from the clarified lysate, a 5-ml HiTrap metal chelating column (Amersham/Pharmacia) was prepared by washing the column with 15 ml of dH2O, charging the column with 20 ml of 50 mM NiSO4, and then equilibrating with 25 ml of 1× urea HIS binding buffer. The lysate was then applied to the column three times using a flow rate of 2 ml/min, after which the column was washed with 50 ml of 1× urea HIS binding buffer. The column was then eluted stepwise using 4 2.5-ml fractions, sequentially, of 1× urea HIS binding buffer containing 50, 200, 350, and 500 mM imidazole, respectively. Twenty microliters of each fraction was then analyzed for the presence of ORF50 by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) followed by Western blotting, and fractions containing ORF50 were pooled and dialyzed stepwise (to allow refolding) against 1× DNA binding buffer (50 mM Tris-HCl [pH 8.0], 100 mM KCl, 12.5 mM MgCl2, 1 mM EDTA, 20% glycerol) containing 3, 1.5, 0.75, 0.3, and 0.1 M urea, respectively. This was followed by dialysis against two changes of 1× DNA binding buffer alone. The wide elution profile (in 8 of the 16 fractions) of ORF50 allowed only partial purification from cellular proteins: by Western blotting and Coomassie blue staining, we estimated that His6-ORF50 comprised approximately 2% of the resulting total protein.
Overexpression and purification of ORF50 from Escherichia coli.
BL21(DE3) competent E. coli cells (100 μl; Life Technologies) were transformed with 3 μg of pRSET 0.8 or pET28b-N50, transferred to 1 liter of Luria broth containing 50 μg of ampicillin per ml, and grown overnight at 37°C with shaking. After about 12 h of growth, when cells reached an optical density of 0.6 to 0.8, recombinant protein expression was induced using 1 mM isopropylthio-β-d-galactosidase (IPTG), and cells were maintained at 37°C with shaking for 6 h. Cells were then pelleted and resuspended in 1× urea HIS lysis buffer as above and allowed to lyse overnight with nutation at 4°C. Next, NP-40 was added to 0.1%, and cells were sonicated using a large tip at a power setting of 8 for 30 s four times to complete cell lysis and to shear chromosomal DNA. The extract was clarified as above and loaded onto a 5-ml chelating column prepared as above. After washing as above, protein was eluted stepwise in six 2.5-ml fractions using 500 mM imidizole in 1× urea HIS binding buffer. Fractions were analyzed as above, and ORF50-containing fractions were pooled and dialyzed against DNA binding buffer as above. Using Coomassie blue staining, we estimated that His6-N50 and -C50 generated in this manner comprised approximately 20% of the resulting total protein.
SDS-PAGE/Western blotting.
Procedures were as previously described (15), using gels containing 10% polyacrylamide. Anti-ORF50 rabbit serum was used as previously described (15). Anti-His6 antibody (Clontech) was used at a dilution of 1:1,000.
EMSA.
Probes for EMSA (top strands are diagrammed in Fig. 4A, 5A, 6, and 8A) were assembled by annealing complementary as described above (see “Plasmids”) and resuspended in dH2O at a concentration of 100 ng/μl. Each EMSA probe was designed to contain an overhanging BglII site at each end to facilitate either cloning or labeling by Klenow DNA polymerase. One hundred nanograms of probe was labeled in a reaction containing 1× REact 2 buffer (Life Technologies), 0.017 mM each dATP, dGTP, and dTTP, 0.5 U of Klenow DNA polymerase (Life Technologies), and 20 μCi of [α-32P]dCTP (6,000 Ci/mmol; Amersham/Pharmacia). After a 30-min incubation at 25°C, the reaction was chased by the addition of 0.017 mM dCTP. Unincorporated nucleotides were removed by purification using QuickSpin TE G-25 columns (Boehringer Mannheim), followed by sequential extraction with phenol-CHCl3 and CHCl3. The probe was then precipitated along with 200 ng of yeast tRNA (Life Technologies) as carrier, pelleted, and resuspended at a specific activity of 105 cpm/μl (usually 100 to 200 μl, quantitated by scintillation counting).
FIG. 8.
ORF50 binds the TATA-proximal K-bZIP promoter with specificity that corresponds with ORF50's ability to activate transcription. (A) Sequence of the top strand of two 28-bp oligonucleotides used as probes in EMSA. Both were annealed to their complementary oligonucleotides as described in Materials and Methods. The box depicts the palindrome shared with the ORF57 promoter, and the grey nucleotides correspond to positions which are homologous to the ORF57 promoter (as in Fig. 2B). (B) Partially purified extract from Bac-50-infected Sf9 cells (6 μg) was incubated with a 50- or 200-fold molar excess of each nonradiolabeled 5Dwt or 5Dm2 EMSA probe (Fig. 4A) or 5- or 10-fold molar excess of each nonradiolabeled ZIPwt or ZIPm1+2 EMSA probe (A) and then incubated with a 1-fold molar equivalent of radiolabeled EMSA probe ZIPwt. These preparations were subject to EMSA as for Fig. 4B, and each set of four binding reactions is indicated above the autoradiogram. (C) Partially purified ORF50 protein was incubated with a 50- or 200-fold molar excess of nonradiolabeled ZIPwt or ZIPm1+2 EMSA probe and then incubated with 1-fold molar equivalent of radiolabeled 5Dwt probe. Included for comparison are an EMSA of a binding reaction lacking protein (No protein) and a reaction containing labeled 5Dwt probe and ORF50 without specific competitor (No competitor).
DNA binding reactions were performed by mixing Sf9 proteins (amounts indicated in the text and figure legends) with 600 ng of poly(dI-dC) (Boehringer Mannheim) in 1× DNA binding buffer [poly(dI-dC) was omitted for binding reactions using E. coli-generated proteins]. This mixture was preincubated on ice for 15 min, after which 1 μl of probe was added, bringing the total reaction volume to 20 μl; for experiments in which protein was omitted, volume was maintained using 1× DNA binding buffer. Incubation proceeded at 18°C for 15 min. and resulting DNA-protein complexes were resolved by PAGE on gels containing 7.5% polyacrylamide, 0.19% bisacrylamide, and 0.5× Tris-borate-EDTA (TBE; 44.5 mM Tris-borate, 44.5 mM boric acid, 50 mM EDTA), at 4°C. Gels were equilibrated to 0.5× TBE electrophoresis buffer by prerunning them for approximately 1 h at 25 mA, constant current; following loading of binding reactions, electrophoresis was continued at 25 mA, constant current. Following electrophoresis, gels were transferred to Whatman paper, dried, and exposed to Bio-Max MS autoradiography film (Kodak).
Competition EMSA was performed as above except that the unlabeled competitor probes (indicated in the text and figure legends) were added to the initial 15-min incubation on ice, together with the protein, before addition of labeled probe.
For depletion experiments (Fig. 4D), nickel-nitrilotriacetic acid (Ni-NTA)-agarose (Qiagen) and glutathione-Sepharose 4B (Amersham/Pharmacia) were equilibrated in 1× DNA binding buffer and then suspended as a 50% (vol/vol) slurry in 1× DNA binding buffer. Fifty microliters of either bead slurry was added to 250 μg of Sf9 protein in 410 μl, of 1× DNA binding buffer and incubated with nutating for 1 h at 4°C. Beads were pelleted by a 10-s pulse (12,000 rpm) in a microcentrifuge, 7.5 μl was removed for EMSA or SDS-PAGE/Western blotting, and 200 μl was transferred to a fresh 1.5-ml tube. This was diluted 1:1.375 with 1× DNA binding buffer, and another 50-μl aliquot of bead slurry was added to each. Incubation was carried out as above, beads were pelleted, and 11.5-μl aliquots were removed for EMSA or SDS-PAGE/Western blotting. EMSA was then performed as above, using the depleted extracts or 5 μl (5 μg) of the starting (undepleted) extract (all Sf9 extract volumes were normalized according the dilution of each step of the depletion). SDS-PAGE/Western blotting was performed as above.
RESULTS
Deletion analysis of the ORF57 promoter reveals the minimal sequences necessary for transactivation by ORF50 50RE.
We have previously mapped the transcripts for the KSHV ORF57 gene and showed that its promoter is responsive to ORF50 expression (12, 15). To map the elements of the ORF57 promoter that confer responsiveness to ORF50 activation, we first cloned 600 bp of the ORF57 promoter 5′ to a luciferase reporter and confirmed that this fragment contained all necessary sequences. Cotransfection of this reporter with an expression vector for ORF50 revealed a 300-fold enhancement of luciferase expression in the presence of ORF50 (Fig. 1). Next, we constructed the series of deletion mutants shown in Fig. 1 and assayed each vector in the presence or absence of ORF50. As shown in Fig. 1, deletions to nucleotide (nt) -218 (relative to the start site of ORF57 mRNA) only trivially influenced ORF50 upregulation, and further deletion to nt -106 resulted in only a threefold additional loss of inducibility. However, further deletion to nt -54 completely ablated ORF50 induction (Fig. 1, -Δ5K reporter) without affecting the basal level of luciferase expression (not shown). These data demonstrate that (i) ORF50 cannot activate transcription through the ORF57 TATA box alone and (ii) the 52 bp unique to Δ5, lying between nt -106 and -54, contain an element(s) required for ORF50 transactivation of the ORF57 promoter. We refer to this 52-bp DNA sequence hereafter as the ORF50 response element (50RE).
The 50RE confers ORF50 responsiveness on heterologous TATA boxes in an enhancer-like fashion.
To determine whether the ORF57 upstream promoter sequences can confer ORF50 responsiveness on a heterologous TATA box, we next fused the 50RE sequence (-106/-54 [Fig. 1]) to the well-characterized TATA box for the cellular gene hsp70 (30), driving firefly luciferase as a reporter. Importantly, Fig. 2A shows that, similar to the ORF57 TATA box alone (Δ5K deletion in Fig. 1), ORF50 is incapable of transactivating the hsp70 TATA box alone (Fig. 2A, line 1). However, fusion of the 50RE in either orientation relative to the TATA box allows robust ORF50 transactivation in both CV-1 cells (Fig. 2A) and BJAB cells (not shown). Furthermore, although ORF50 cannot activate transcription directed by the promoter of the late KSHV AP gene (16), the ORF57 50RE promoter sequences also confer strong ORF50-dependent activation on the AP TATA box (data not shown). This suggests that ORF50 shows little TATA box specificity. Together, Fig. 1 and 2 establish that the 50RE, between -106 and -54 of the ORF57 promoter, is both necessary and sufficient for ORF 50 transactivation.
The TATA-proximal promoters of the ORF57 and K-bZIP genes share three conserved sequences.
We have previously demonstrated that among the promoters transiently transactivated by ORF50, the ORF57 and K-bZIP promoters were most strongly activated (15). Therefore, we compared the -106/-5 sequence of the ORF57 promoter (containing the 50RE and the TATA box) with the TATA-proximal K-bZIP promoter to look for similarities.
The aligned sequences are shown in Fig. 2B. This alignment revealed that the two promoters have remarkable structural similarity in sharing three highly homologous blocks of sequence. The first conserved block contains a 12-bp partially palindromic sequence which is identical in both promoters (save for an extra A residue in a TATA-proximal position in the K-bZIP promoter). The homology between the two promoters extends weakly outward to both sides of this conserved palindrome, including sequences which extend the palindrome in the ORF57 but not the K-bZIP promoter (Fig. 2A). Next, a 6-bp block which is part of a consensus 7-bp Aml-1a element (in the ORF57 promoter) is shared by both promoters; the G in most 5′ position of the top DNA strand of the K-bZIP promoter does not agree with the Aml-1a consensus but does occur in two Aml-1a sites that have been documented in TRANSFAC (32). Finally, the last conserved block not only covers the TATA box of both genes but also extends 6 bp farther in the start site-proximal direction. This TATA-containing 13-bp block of sequence is not shared with any other loci in the remainder of the sequenced KSHV genome.
Generation of a recombinant baculovirus expressing ORF50 for use in DNA binding assays.
To determine whether or not the ORF50 protein interacted directly with the ORF57 promoter, we initially attempted to detect an interaction between the 50RE (diagrammed in Fig. 4A) and crude ORF50 protein present in (i) nuclear extracts of transiently transfected Cos9 cells, (ii) rabbit reticulocyte lysate (RRL) or wheat germ lysate in which ORF50 cDNA had been transcribed and translated in vitro, or (iii) nuclear extracts of BCBL-1 cells induced with tetradecanoyl phorbol acetate. Lysates containing ORF50 from each of these sources failed to interact with ORF57 promoter sequences in a manner that corresponded with ORF50's ability to activate transcription (not shown). This suggested that detection of this interaction might require more highly purified and concentrated material.
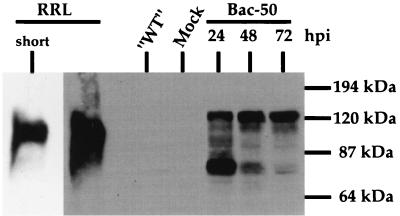
Therefore, we generated a recombinant baculovirus (Bac-50) which expresses His6-tagged ORF50 in Sf9 insect cells. Immunoblotting of Sf9 extracts with anti-ORF50 antibody (Fig. 3) shows that at 48 to 72 hpi, the majority of the protein expressed by the baculovirus has an approximate apparent molecular mass of 120 kDa, corresponding to the migration of the fully phosphorylated protein (15). Since we also determined that protein yield was highest at 48 hpi, we harvested nuclear extracts at this time point and partially purified recombinant ORF50 by nickel-Sepharose affinity chromatography. All Bac-50-infected lysates also produced a second species with an apparent molecular mass ∼80 kDa, considerably smaller than even the underphosphorylated protein produced in RRL. We do not yet know the origin of this species. While it might represent a proteolytic fragment of the ORF50 protein, the fact that it disappears as infection progresses and is progressively replaced by the 120-kDa isoform (Fig. 3) argues against this possibility and is more consistent with it representing a precursor that undergoes conversion to the 120-kDa form over time. Such a conversion must involve extensive phosphorylation (15) but could also include additional posttranslational modifications as well. The nature of this species is now under study.
FIG. 3.
Time course of ORF50 protein expression in Sf9 cells infected with recombinant baculovirus. A recombinant baculovirus in which ORF50 is expressed under control of the polyhedrin promoter (Bac-50) was constructed and grown to high titer as described in Materials and Methods. Sf9 cells were incubated with medium alone (Mock) or infected with Bac-50 at a multiplicity of infection of 10; whole-cell extracts were prepared at the indicated hours postinfection and analyzed by immunoblotting with anti-ORF50 antibody. Extract from Sf9 cells infected with a recombinant baculovirus expressing an irrelevant, non-KSHV protein (“WT”; a gift from David Morgan) and mock extract were harvested at 48 hpi and similarly analyzed. ORF50 was also expressed by transcription and translation in vitro in RRL and detected together with the Sf9 proteins or with a shorter exposure to film (short).
Both the palindrome and flanking sequences contribute to DNA binding and transactivation by ORF50.
To analyze the interactions of baculovirus-generated ORF50 with the ORF57 promoter using EMSA, we created four 26-bp probes (named 5A to 5D) which span the 50RE and are shown in Fig. 4A. Each probe was labeled with 32P and then incubated with binding buffer alone or increasing amounts of the partially purified baculovirus-derived protein. These DNA-protein mixtures were then separated on nondenaturing polyacrylamide gels; a typical example is shown in Fig. 4B. These data demonstrate the formation of four different complexes, designated 1, 1*, 2, and 3. Complexes 1 and 1* form only on probes 5C and 5D. Complex 2, however, forms on all of the probes, suggesting that it may contain nonspecific DNA binding proteins. In this experiment, complex 3 was observed only on probes 5A and 5B; however, complex 3 can occasionally be detected on probes 5C and 5D as well (Fig. 5B), suggesting that it, too, may be sequence nonspecific. The low intensity and variable appearance of complex 3 precluded further rigorous characterization.
To determine the functional significance of these protein-DNA complexes, we next cloned each EMSA probe as a dimer upstream of the hsp70 TATA box in the aforementioned reporter vector and analyzed each for transactivation by ORF50 in cotransfections of CV-1 cells. As shown in Fig. 4C, the 5C and 5D sequences, but not the 5A or 5B sequence, were activated by ORF50. In fact, 5D was activated by ORF50 nearly twice as well as 5C over the tested range of input plasmid amounts, suggesting a preference in activation of the promoter sequences by ORF50. Similar transfection of BJAB cells demonstrated that ORF50 transactivated the 5D reporter about 100-fold (data not shown), in agreement with our previous observation of little cell type specificity of transactivation by ORF50 (15). Comparison of this transactivation data to the EMSA results of Fig. 4B indicates a positive correlation between transcriptional activation by ORF50 and formation of complexes 1 and 1*, suggesting that complexes 1 and 1* may contain the ORF50 polypeptide bound to the 5C and 5D probes, respectively.
To determine whether or not ORF50 was a component of complexes 1 and 1*, we attempted to supershift the EMSA complexes by incubation with a polyclonal ORF50 antiserum directed toward the C-terminal activation domain (16). Unfortunately, however, this was unsuccessful. Therefore, we attempted to deplete the His-tagged ORF50 polypeptide from the partially purified Sf9 extracts by incubation with Ni-NTA-agarose beads, followed by EMSA using the depleted extracts. Two consecutive rounds of depletion were performed (as in Materials and Methods) using Ni-NTA-agarose beads, or using glutathione-Sepharose beads for mock depletion as a negative control. Undepleted extract and an equivalent volume of depleted extract from each round of depletion were incubated individually with labeled probe 5D and analyzed by EMSA as before. Figure 4D demonstrates that each round of depletion progressively reduced the formation of complexes 1 and 1* but not complex 2. Importantly, mock depletion using the control glutathione beads does not eliminate formation of any of the protein complexes forming on probe 5D. Furthermore, analysis of the depleted supernatants for the presence of ORF50 demonstrated that each round of depletion with the Ni-NTA beads progressively depleted His-tagged ORF50 from the extracts, while depletion with the glutathione beads did not (Fig. 4D, bottom). Together, these data strongly suggest that protein-DNA complexes 1 and 1*, but not complex 2, contain ORF50, and formation of these complexes correlates with the ability of ORF50 to activate transcription directed by these sequences. Given that the baculovirus-generated ORF50 protein exists as two major species of 120 and 80 kDa (Fig. 3), it is tempting to speculate that complexes 1 and 1* reflect the binding of the 120- and 80-kDa isoforms, respectively, to the probe. However, additional work is required to validate this inference.
We have subsequently attempted to generate more highly purified ORF50 protein from Bac-50-infected Sf9 cells by using alternative purification schemes (data not shown); we have found that as the purity of the recombinant ORF50 protein increases, its solubility decreases in physiologic buffers. These attempts have thus yielded little usable protein for EMSAs. However, when a Ni2+ affinity column is loaded with limiting amounts of crude Sf9 extract from Bac-50-infected cells, complex 3 constituents do not bind the column but remain in the flowthrough fraction, and formation of complexes 1 and 1* corresponds exclusively with the presence of ORF50 in column fractions.
Although Fig. 4B demonstrated that ORF50 binds to both the 5C and 5D probes, Fig. 4C suggested a preference of ORF50 for transactivation of 5D over 5C. To determine whether or not this functional preference of transfected ORF50 for 5D would be reflected in quantitative differences in DNA binding in vitro, we preincubated the ORF50 polypeptide with increasing concentrations of each unlabeled EMSA probe (5A to 5D) before introducing labeled 5D probe. In this way, ORF50 was exposed to successive 10-, 20-, 40-, and 80-fold molar excesses of each unlabeled probe over labeled 5D probe. The resulting mixtures were subjected to EMSA, and the results are shown in Fig. 4E. As expected, both unlabeled probes 5C and 5D could compete for binding of labeled probe 5D to ORF50; however, these experiments demonstrate that probe 5D is a more avid competitor for ORF50 binding than is probe 5C (especially evident by comparison of the 10-fold excess lanes). The quantitative interaction of ORF50 in vitro with probes 5D and 5C thus reflects the preference of ORF50 for in vivo activation.
Similarly, probe 5B, which was not activated by ORF50 in vivo, was likewise unable to compete for ORF50 binding to labeled probe 5D, extending the correlation between DNA binding and transcriptional activation. However, we found that unlabeled probe 5A was able to weakly compete for binding of ORF50, with significant competition only at 40-fold molar excess and above. Apparently, this weak interaction, which was not reflected under the conditions used for EMSA in Fig. 4B, is not sufficient to allow activation by ORF50 in vivo (Fig. 4C).
Taken together, these competition EMSAs allowed us to determine a hierarchy for binding preferences of ORF50 to the 50RE sequences: 5D > 5C > > 5A > > > 5B. This suggests that the palindromic sequences shared by probes 5A, 5B, and 5D are required for binding by ORF50, but that addition of the sequences flanking the core palindrome on its 3′ (TATA-proximal) side (only in probes 5C and 5D) is required for an avid interaction of ORF50 with this element. Furthermore, the 3′ sequences seem to be absolutely required for transcriptional activation by ORF50.
ORF50 binding to sequences within the palindrome is necessary for transcriptional activation.
To determine the role of the palindrome and flanking sequences in DNA binding and transcriptional activation by ORF50 in the context of the optimized 5D EMSA probe sequence, we generated the three linker scanning mutations shown in Fig. 5A. The 6-bp sequence CTGCAG (a PstI restriction endonuclease site) was substituted either for half of the core palindrome (5Dm1 or 5Dm2) or for the 6 bp on the TATA-proximal side of the core palindrome (5Dm3). The wt probe and each mutant probe were then labeled and analyzed by EMSA as before. Figure 5B shows that ORF50 binds to 5Dm1 in a quantitatively similar manner as to the 5Dwt probe. However, ORF50 binds to neither 5Dm2 nor 5Dm3. These data therefore suggest that the right half of the palindrome and the 3′ flanking sequences are the primary determinants for ORF50 binding to this element. In fact, these data agree well with the ORF50 binding preferences determined for probes 5A through 5D (compare probe 5C [Fig. 4A and B] with 5Dm1 [Fig. 5A and 5B]).
To confirm these binding data, we preincubated the partially purified ORF50 protein with unlabeled 5Dwt or each mutant prior to addition of labeled 5Dwt probe. These mixtures were then analyzed by EMSA and showed the expected results: only the unlabeled 5Dwt and 5Dm1 probes could compete for binding of ORF50 to labeled 5Dwt probe. Neither unlabeled 5Dm2 or 5Dm3 could compete for ORF50 binding to 5Dwt.
We next analyzed the functional significance of the DNA binding activity of ORF50 by testing the ability of transiently expressed ORF50 to activate transcription from 5Dm2 or 5Dm3 in the context of the hsp70 TATA reporter construct. Figure 4D demonstrates that although ORF50 activates transcription from the 5Dwt construct, it is unable to activate transcription from either of the mutant constructs. This provides conclusive evidence that DNA binding by ORF50 is required for transcriptional activation through this response element.
For unclear technical reasons, we have been unable to construct the corresponding reporter construct for 5Dm1 in the context of the hsp70 TATA reporter. However, introduction of 5Dm1 in the context of the full-length, wt ORF57 promoter reduces activation by ORF50 only two- to threefold, consistent with its minimal impact on DNA binding in vitro (not shown). These results, affirmed by testing of additional mutants (see below and Fig. 6), indicate that while sequences within the palindrome are essential, the inverted repeat itself may not be required for binding and activation by ORF50.
Fine mutational analysis of the 50RE.
To confirm that the negative effects of the 5Dm1 to 5Dm3 mutations were not specific to the chosen linker sequence or to the large size of the substitutions, we generated a second series of nine linker substitution mutations across the 5D element in which 1 to 4 bp were changed in each mutation; these are shown in Fig. 6 and named 5Dm4 to 5Dm9 (for 5Dm4 and 5Dm9, we constructed one and two additional variants, respectively).
Each was tested for both DNA binding by the baculovirus-generated ORF50 and for transcriptional activation by transiently expressed ORF50. DNA binding data was semiquantitated for each mutant by comparison to the binding of ORF50 to the 5Dwt probe, which was assigned the value of +++; similarly, activation was quantitated for each mutant by comparison to ORF50's activation of the 5Dwt reporter construct, which was assigned the value of 100% (the first four lines of Fig. 6 summarize the data from Fig. 5).
This new series of mutants (Fig. 6, lines 5 to 13) extend the correlation between DNA binding by ORF50 in vitro and transcriptional activation in vivo. All mutations in the center/right portion of the palindrome and in the 3′ flanking sequences strongly impair both DNA binding and activation (5Dm6 to 5Dm9-3). Like 5Dm1, 5Dm5 in the left side of the palindrome has only modest effects on DNA binding and reduces activation only twofold. This affirms that the presence of a palindromic sequence, per se, is not the key determinant for binding to ORF50. Flanking mutations 5Dm4 and 5Dm4-2 again show only modest effects on DNA binding and inconsistent twofold effects on activation. These findings suggest that while DNA binding is necessary for activation, mutations that alter but do not ablate DNA binding allow formation of complexes that can retain significant activation potential.
The homologous sequences within the K-bZIP promoter are required for full activation by ORF50.
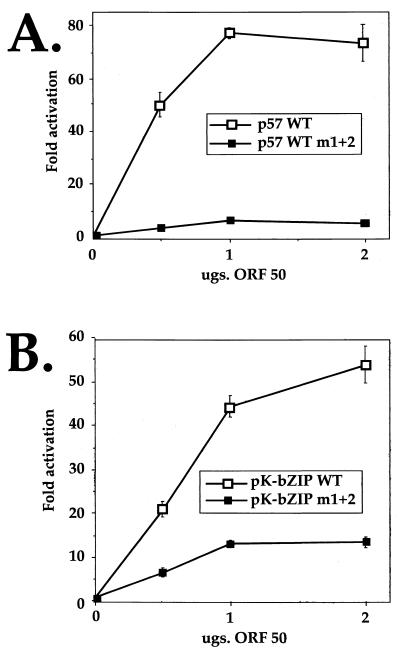
Thus far, we have analyzed the functional consequences of DNA binding by ORF50 in the context of the isolated, 25-bp 5D sequences from 50RE assembled as a dimer upstream of the heterologous TATA box from the cellular hsp70 gene. Such an analysis places this response element in an artificial position relative to the TATA box and eliminates the potential influences of natural flanking sequences in DNA binding by ORF50, heterologous cellular proteins, or both. Therefore, we introduced these linker substitution mutations into the context of the wt, full-length ORF57 or K-bZIP promoter. In both promoters, we mutated the entire shared palindrome by introducing m1 (to substitute the left side of the palindrome) together with the sequence TCCGGA (a BspEI restriction site) to substitute for the right side of the palindrome. The resultant mutant reporters were named m1+2.
Each mutant was then compared to its respective wt promoter in cotransfections of CV-1 cells. Figure 7 demonstrates that mutation of the entire palindrome in both the ORF57 and K-bZIP promoters severely debilitates ORF50's ability to activate transcription from either cognate promoter. In the case of the ORF57 promoter, activation by ORF50 is completely abrogated by the mutation, while in the K-bZIP promoter, activation is reduced by about 70%. Together, these data demonstrate the critical requirement for the shared sequence in activation of both promoters by ORF50. The residual activity of the mutant K-bZIP promoter suggests that additional ORF50 response elements may exist in that promoter (see Discussion).
FIG. 7.
The palindromic sequences within the K-bZIP promoter are required for full activation by ORF50. A 12-bp linker substitution mutation was introduced to replace the 12-bp palindromic sequence of the wt, full-length ORF57 (A) or K-bZIP (B) promoter (see text). Each mutant promoter was compared to its corresponding wt promoter in cotransfections of CV-1 cells as described for Fig. 5D.
DNA binding of the K-bZIP promoter by ORF50 protein correlates with transcriptional activation.
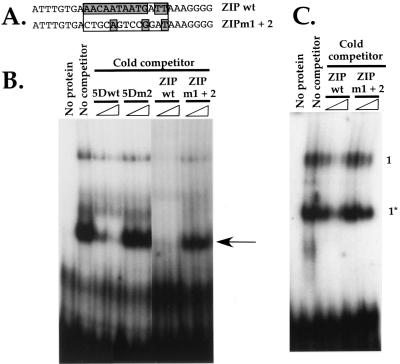
To determine whether or not the ORF50 protein could bind directly to the K-bZIP promoter in a manner that correlated with transcriptional activation, we performed EMSA using oligonucleotides representing the wt K-bZIP promoter sequence and the K-bZIP m1+2 sequence (ZIPwt and ZIPm1+2, respectively [Fig. 8A]). The second lane of Fig. 8B demonstrates that a single, major complex (indicated by an arrow) forms when the labeled ZIPwt probe is incubated with partially purified ORF50 protein. (An additional weak complex of unclear origin is also observed.)
To determine the specificity of formation of this complex relative to transcriptional activation by ORF50, we preincubated partially purified ORF50 with increasing concentrations (50- and 200-fold molar excesses) of un-labeled 5Dwt or 5Dm2 probe (sequences from Fig. 5A), respectively, before incubation with the labeled ZIPwt probe. The resulting mixtures were subjected to EMSA, and the results are shown in Fig. 8B, lanes 3 to 6. 5Dwt, but not 5Dm2, can avidly compete for formation of the complex with the ZIPwt probe. Furthermore, when the extracts were preincubated with the unlabeled ZIPwt or ZIPm1+2 probe, respectively, at 5- and 10-fold molar excesses, the complex was competed by the wt but not mutant K-bZIP element (Fig. 8B, lanes 7 to 10). This result suggests that the complex forming on the K-bZIP promoter is dependent on cognate sequences required for ORF50 transactivation of both the ORF57 and K-bZIP promoters in vivo; conversely, probes corresponding to the ORF57 5Dm2 and ZIPm1+2 mutant promoters, which are not activated by ORF50, cannot compete for binding to the complex that forms on the cognate K-bZIP element.
To determine whether or not the EMSA complex on the K-bZIP probe contained ORF50, we reversed the above approach and tested the ability of the wt and mutant K-bZIP elements to compete for ORF50 binding to the ORF57 5Dwt probe. Figure 8C shows that 50- and 200-fold molar excesses of unlabeled ZIPwt probe, but not ZIPm1+2 probe, can compete for binding to the proteins of complexes 1 and 1* which contain the ORF50 protein. This confirms that ORF50 binds to both the ORF57 and K-bZIP TATA-proximal promoters in a fashion that corresponds to its ability to transcriptionally transactivate them.
The amino terminus of ORF50 generated in E. coli binds to the ORF57 response element.
To independently confirm ORF50's ability to bind the DNA palindrome, we attempted to express full-length ORF50 polypeptide as a His6 fusion in E. coli. Unfortunately, this protein was highly insoluble and also unstable, and it yielded little full-length material after purification. Alternatively, we generated His6 fusions of ORF50 truncated at both amino and carboxy termini (amino acids [aa] 1 to 272 [N50] and aa 525 to 691 [C50]). These are displayed schematically in Fig. 9A. Each was expressed to very high levels and purified to near homogeneity as described in Materials and Methods (Fig. 9A, immunoblots).
Next, the polypeptides were diluted to identical concentrations, and increasing amounts were incubated with labeled p57-5Dwt probe. These were then analyzed by EMSA, and the results are shown in Fig. 9B. Clearly, only N50, not C50, binds stably to the 5D sequence. This demonstrates that (i) highly purified ORF50 polypeptide from an independent, noneukaryotic source binds DNA and (ii) ORF50's DNA binding activity maps to its N-terminal 272 aa.
To confirm the specificity of DNA binding by the N50 protein, we performed EMSA by incubating increasing amounts of N50 with either labeled 5B or 5D probe derived from the ORF57 promoter. The results shown in Fig. 9C confirm that N50 only binds to 5D, not to 5B, reflecting the specificity of the protein generated by baculovirus infection. These results were extended by preincubating N50 with 50-, 100-, and 200-fold molar excesses of unlabeled 5B or 5D before incubation with labeled 5D probe. As expected, Fig. 9D demonstrates that unlabeled 5D but not 5B can compete for binding to N50.
DISCUSSION
In this investigation, we have used deletion and linker scanning mutations to identify and partially characterize a binding site for the KSHV lytic switch protein ORF50. Our deletion analyses of the ORF57 promoter enabled us to reduce the ORF50-responsive region to a 25-bp sequence (5Dwt) which conferred strong activation by ORF50 on a heterologous TATA box (Fig. 4). ORF50 polypeptide produced recombinantly in both baculovirus-infected Sf9 cells (Fig. 3) and E. coli (Fig. 9) bound avidly to this element in vitro. The response element contains a 12-bp palindrome (5′-AACAATAATG-3′) which is shared between the viral ORF57 and K-bZIP promoters (Fig. 2B), two DE promoters that are potently transactivated by ORF50 (15). Substitution of the 12-bp palindrome with a 12-bp linker scanning mutation severely inhibited ORF50's ability to transactivate either cognate promoter (Fig. 7) and abrogated its ability to bind to the K-bZIP proximal promoter (Fig. 8). However, smaller mutations within the palindrome from ORF57's promoter indicate that its two halves do not make equal contributions to its activities—the left-hand side of the sequence is substantially more tolerant of mutations than the right-hand side. This suggests that it is the primary sequence within this region rather than its palindromic nature that is critical for DNA binding and activation. Together, these data conclusively demonstrate that DNA binding of this region by ORF50 is required for activation of both the ORF57 and K-bZIP promoters. Based on this evidence, we propose that the 25-bp element defined by fragment 5D be named 50RE57 (for ORF50 response element from the ORF57 promoter).
Extensive linker scanning mutations of 50RE57 revealed that additional sequences flanking the 12-bp palindrome were also required for both DNA binding and activation by ORF50 (Fig. 5 and 6). Although these flanking sequences extend the palindrome in the ORF57 but not the K-bZIP promoter, both promoters contain shared GT/CA and GG/CC doublets in the left (TATA-distal) and right (TATA-proximal) flanking sequences (Fig. 2B). In fact, a single G-to-T mutation in the GG/CC doublet completely abrogates both DNA binding and activation by ORF50 (5Dm9 in Fig. 6). In general, the linker scanning mutations of the sequences flanking the palindrome revealed that, like the palindrome itself, only the right-side (TATA-proximal) flanking sequences were strictly required for DNA binding and activation by ORF50; every mutation in this part of the element which eliminated ORF50 binding also abrogated activation. Mutations in flanking sequences to the left of the palindrome had only modest effects on DNA binding and small (and variable) impacts on transactivation in vivo. Further biochemical analyses (i.e., DNA footprinting and interference of chemical alterations of specific nucleotides) should reveal the specific contacts between ORF50 and 50RE57 and will likely be required to explain the specific sequence requirements for ORF50 binding of the element in both promoters.
Further biochemical experiments should also likely aid the derivation of a consensus binding site for ORF50, especially since sequence analysis of the entire KSHV genome (23) did not reveal a similar 12-bp palindrome in any other inter or intragenic regions of the viral nucleic acid. Direct comparison of 50RE57 to the promoters for three other KSHV genes which ORF50 transactivates (encoding nut-1, kaposin, and DNA binding protein [16]) also did not reveal significant homology, even within response elements which we have recently mapped in those promoters (50REnut-1, and 50REkap [J. Chang, D. M. Lukac, and D. Ganem, unpublished data]). This suggests significant heterogeneity between 50REs within DE promoters, indicating that other, unrelated sequences may also bind ORF50 protein. We have recently observed binding of ORF50 to an element required for transactivation of the KSHV TK promoter; this element lacks the 12-bp palindrome of 50RE57 but shares homology to its 3′ flanking sequences (Chang et al., unpublished). Interestingly, 50RETK is inverted relative to its TATA box and to 50RE57, in agreement with Fig. 2B, which demonstrates orientation independence of ORF50-mediated activation.
Alternatively, as has been well documented for the ORF50 homolog in EBV (9, 10, 14, 17, 21) and other viral transcriptional activators (6), KSHV ORF50 may activate promoters not only directly by binding DNA but also indirectly by (i) piggybacking on cellular DNA binding proteins or (ii) altering their activity and/or abundance. This indirect mode of activation by ORF50 may in fact also be operative on the ORF57 promoter, since we saw small effects of TATA-distal deletions on ORF50 activation (Fig. 1, deletions of nt -602 to -504 and -218 to -106). The fact that the K-bZIP promoter bearing a lesion in the 12-bp palindrome retained residual ORF50 inducibility is also compatible with the presence of unrelated response elements that directly or indirectly contribute to ORF50 responses. We are currently analyzing the interactions of ORF50 with cellular factors to address these effects.
The remarkable structural similarity between the TATA-proximal promoters of both ORF57 and K-bZIP included conservation of the relative placement of not only the 12-bp palindrome and TATA-containing elements but also the potential binding site for the cellular protein Aml-1a. Although our studies have not demonstrated a role for the latter site in activation by ORF50, the conservation of this element in the ORF57 and K-bZIP promoters may reflect an as yet unidentified regulatory significance for this lymphoid-specific transcription factor in viral replication.
Generation of amino and carboxy-terminal truncations of ORF50 in E. coli allowed us to confirm that ORF50 bound directly to 50RE57 and, more importantly, demonstrated that ORF50's N terminus (aa 1 to 272), but not its C terminus (aa 525 to 691), contains its primary DNA binding activity (Fig. 9). This is not surprising, since the N terminus contains an 80-aa sequence which is 18% basic. Furthermore, this extends the structure-function similarity of KSHV ORF50 with EBV Rta, which share N-terminal DNA binding and oligomerization domains and C-terminal activation domains (11, 15–17). It will be interesting to determine whether or not KSHV ORF50 binds DNA as an oligomer and if its DNA binding and oligomerization domains can be separated genetically.
Although both the baculovirus- and E. coli-generated ORF50 polypeptides could bind to 50RE57, they showed subtle differences. As demonstrated in Fig. 4E, the 5A probe (Fig. 4A) was able to weakly compete with 5D for binding to the baculovirus-generated ORF50. However, although this weak interaction of ORF50 with 5A was not evident under conditions in which 5A was labeled (Fig. 4B), we have found that the N-terminal ORF50 polypeptide from E. coli is able to interact detectably with labeled 5A under similar conditions (data not shown). Among other possibilities, this discrepancy between ORF50 from the two sources might be explained by structural differences between the full-length and truncated proteins or by posttranslational modifications. For example, the serine/threonine-rich region of ORF50, a likely target of the known extensive phosphorylations of the full-length polypeptide (15), is absent in the N50 fragment expressed in E. coli (Fig. 9). If indeed phosphorylation and/or other modifications of ORF50 affect its DNA binding or transactivation activities, this would raise the intriguing possibility that ORF50's functions in vivo may be subject to multiple levels of regulation.
ACKNOWLEDGMENTS
We thank the members of the Ganem lab for helpful discussions and advice, Rob Brazas for advice with protein purification, Andrew Polson for hints regarding sequence analysis, and Mike Rothenberg for assistance with designing subclones.
D.M.L. is a postdoctoral fellow of the Irvington Institute for Immunological Research.
REFERENCES
- 1.Ambroziak J, Blackbourn D, Herndier B, Glogan R, Gullet J, McDonald A, Lennette E, Levy J. Herpesvirus-like sequences in HIV-infected and uninfected Kaposi's sarcoma patients. Science. 1995;268:582–583. doi: 10.1126/science.7725108. [DOI] [PubMed] [Google Scholar]
- 2.Blasig C, Zietz C, Haar B, Neipel F, Esser S, Brockmeyer N H, Tschachler E, Colombini S, Ensoli B, Sturzl M. Monocytes in Kaposi's sarcoma lesions are productively infected by human herpesvirus 8. J Virol. 1997;71:7963–7968. doi: 10.1128/jvi.71.10.7963-7968.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boshoff C, Schulz T F, Kennedy M M, Graham A K, Fisher C, Thomas A, McGee J O, Weiss R A, O'Leary J J. Kaposi's sarcoma-associated herpesvirus infects endothelial and spindle cells. Nat Med. 1995;1:1274–1278. doi: 10.1038/nm1295-1274. [DOI] [PubMed] [Google Scholar]
- 4.Boyer T G, Berk A J. Functional interaction of adenovirus E1A with holo-TFIID. Genes Dev. 1993;7:1810–1823. doi: 10.1101/gad.7.9.1810. [DOI] [PubMed] [Google Scholar]
- 5.Cesarman E, Chang Y, Moore P S, Said J W, Knowles D M. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N Engl J Med. 1995;332:1186–1191. doi: 10.1056/NEJM199505043321802. [DOI] [PubMed] [Google Scholar]
- 6.Flint J, Shenk T. Viral transactivating proteins. Annu Rev Genet. 1997;31:177–212. doi: 10.1146/annurev.genet.31.1.177. [DOI] [PubMed] [Google Scholar]
- 7.Flore O, Rafii S, Ely S, O'Leary J J, Hyjek E M, Cesarman E. Transformation of primary human endothelial cells by Kaposi's sarcoma-associated herpesvirus. Nature. 1998;394:588–592. doi: 10.1038/29093. [DOI] [PubMed] [Google Scholar]
- 8.Gradoville L, Gerlach J, Grogan E, Shedd D, Nikiforow S, Metroka C, Miller G. Kaposi's sarcoma-associated herpesvirus open reading frame 50/Rta protein activates the entire lytic cycle in the HH-B2 primary effusion lymphoma cell line. J Virol. 2000;74:6207–6212. doi: 10.1128/jvi.74.13.6207-6212.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gruffat H, Duran N, Buisson M, Wild F, Buckland R, Sergeant A. Characterization of an R-binding site mediating the R-induced activation of the Epstein-Barr virus BMLF1 promoter. J Virol. 1992;66:46–52. doi: 10.1128/jvi.66.1.46-52.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gruffat H, Sergeant A. Characterization of the DNA-binding site repertoire for the Epstein-Barr virus transcription factor R. Nucleic Acids Res. 1994;22:1172–1178. doi: 10.1093/nar/22.7.1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hardwick J M, Tse L, Applegren N, Nicholas J, Veliuona M A. The Epstein-Barr Virus R transactivator (Rta) contains a complex, potent activation domain with properties different from those of VP16. J Virol. 1992;66:5500–5508. doi: 10.1128/jvi.66.9.5500-5508.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kirshner J R, Lukac D M, Chang J, Ganem D. Kaposi's sarcoma-associated herpesvirus open reading frame 57 encodes a posttranscriptional regulator with multiple distinct activities. J Virol. 2000;74:3586–3597. doi: 10.1128/jvi.74.8.3586-3597.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kirshner J R, Staskus K, Haase A, Lagunoff M, Ganem D. Expression of the open reading frame 74 (G-protein-coupled receptor) gene of Kaposi's sarcoma (KS)-associated herpesvirus: implications for KS pathogenesis. J Virol. 1999;73:6006–6014. doi: 10.1128/jvi.73.7.6006-6014.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu C, Sista N D, Pagano J S. Activation of the Epstein-Barr virus DNA polymerase promoter by the BRLF1 immediate-early protein is mediated through USF and E2F. J Virol. 1996;70:2545–2555. doi: 10.1128/jvi.70.4.2545-2555.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lukac D M, Kirshner J R, Ganem D. Transcriptional activation by the product of the open reading frame 50 of Kaposi's-associated herpesvirus is required for lytic viral reactivation in B cells. J Virol. 1999;73:9348–9361. doi: 10.1128/jvi.73.11.9348-9361.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lukac D M, Renne R, Kirshner J R, Ganem D. Reactivation of Kaposi's sarcoma-associated herpesvirus infection from latency by expression of the ORF 50 transactivator, a homolog of the EBV R protein. Virology. 1998;252:304–312. doi: 10.1006/viro.1998.9486. [DOI] [PubMed] [Google Scholar]
- 17.Manet E, Rigolet A, Gruffat H, Giot J-F, Sergeant A. Domains of the Epstein-Barr virus (EBV) transcription factor R required for dimerization, DNA binding and activation. Nucleic Acids Res. 1991;19:2661–2667. doi: 10.1093/nar/19.10.2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martin D F, Kuppermann B D, Wolitz R A, Palestine A G, Li H, Robinson C A. Oral ganciclovir for patients with cytomegalovirus retinitis treated with a ganciclovir implant. Roche Ganciclovir Study Group. N Engl J Med. 1999;340:1063–1070. doi: 10.1056/NEJM199904083401402. [DOI] [PubMed] [Google Scholar]
- 19.Moses A V, Fish K N, Ruhl R, Smith P P, Strussenberg J G, Zhu L, Chandran B, Nelson J A. Long-term infection and transformation of dermal microvascular endothelial cells by human herpesvirus 8. J Virol. 1999;73:6892–6902. doi: 10.1128/jvi.73.8.6892-6902.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Neipel F, Albrecht J C, Fleckenstein B. Cell-homologous genes in the Kaposi's sarcoma-associated rhadinovirus human herpesvirus 8: determinants of its pathogenicity? J Virol. 1997;71:4187–4192. doi: 10.1128/jvi.71.6.4187-4192.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Quinlivan E B, Holley-Guthrie E A, Norris M, Gutsch D, Bachenheimer S L, Kenney S C. Direct BRLF1 binding is required for cooperative BZLF1/BRLF1 activation of the Epstein-Barr virus early promoter, BMRF1. Nucleic Acids Res. 1993;21:1999–2007. doi: 10.1093/nar/21.8.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Renne R, Zhong W, Herndier B, McGrath M, Abbey N, Kedes D, Ganem D. Lytic growth of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) in culture. Nat Med. 1996;2:342–346. doi: 10.1038/nm0396-342. [DOI] [PubMed] [Google Scholar]
- 23.Russo J J, Bohenzky R A, Chien M C, Chen J, Yan M, Maddalena D, Parry J P, Peruzzi D, Edelman I S, Chang Y, Moore P S. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8) Proc Natl Acad Sci USA. 1996;93:14862–14867. doi: 10.1073/pnas.93.25.14862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sarid R, Flore O, Bohenzky R A, Chang Y, Moore P S. Transcription mapping of the Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) genome in a body cavity-based lymphoma cell line (BC-1) J Virol. 1998;72:1005–1012. doi: 10.1128/jvi.72.2.1005-1012.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schulz T F. Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8): epidemiology and pathogenesis. J Antimicrob Chemother. 2000;45(Suppl. T3):15–27. doi: 10.1093/jac/45.suppl_4.15. [DOI] [PubMed] [Google Scholar]
- 26.Soulier J, Grollet L, Oksenhendler E, Cacoub P, Cazals-Hatem D, Babinet P, d'Agay M F, Clauvel J P, Raphael M, Degos L, et al. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman's disease. Blood. 1995;86:1276–1280. [PubMed] [Google Scholar]
- 27.Staskus K A, Zhong W, Gebhard K, Herndier B, Wang H, Renne R, Beneke J, Pudney J, Anderson D J, Ganem D, Haase A T. Kaposi's sarcoma-associated herpesvirus gene expression in endothelial (spindle) tumor cells. J Virol. 1997;71:715–719. doi: 10.1128/jvi.71.1.715-719.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun R, Lin S-F, Staskus K, Gradoville L, Grogan E, Haase A, Miller G. Kinetics of Kaposi's sarcoma-associated herpesvirus gene expression. J Virol. 1999;73:2232–2242. doi: 10.1128/jvi.73.3.2232-2242.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun R, Lin S F, Gradoville L, Yuan Y, Zhu F, Miller G. A viral gene that activates lytic cycle expression of Kaposi's sarcoma-associated herpesvirus. Proc Natl Acad Sci USA. 1998;95:10866–10871. doi: 10.1073/pnas.95.18.10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Taylor I C, Kingston R E. Factor substitution in a human HSP70 gene promoter: TATA-dependent and TATA-independent interactions. Mol Cell Biol. 1990;10:165–175. doi: 10.1128/mcb.10.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Whitby D, Howard M R, Tenant-Flowers M, Brink N S, Copas A, Boshoff C, Hatzioannou T, Suggett F E, Aldam D M, Denton A S, et al. Detection of Kaposi sarcoma associated herpesvirus in peripheral blood of HIV-infected individuals and progression to Kaposi's sarcoma. Lancet. 1995;346:799–802. doi: 10.1016/s0140-6736(95)91619-9. [DOI] [PubMed] [Google Scholar]
- 32.Wingender E, Chen X, Hehl R, Karas H, Liebich I, Matys V, Meinhardt T, Prub M, Reuter I, Schacherer F. TRANSFAC: an integrated system for gene expression regulation. Nucleic Acids Res. 2000;28:316–319. doi: 10.1093/nar/28.1.316. [DOI] [PMC free article] [PubMed] [Google Scholar]