Abstract
It has been suggested that sequences located within the 5′ noncoding region of human foamy virus (HFV) are critical for expression of the viral Gag and Pol structural proteins. Here, we identify a discrete ∼151-nucleotide sequence, located within the R region of the HFV long terminal repeat, that activates HFV Gag and Pol expression when present in the 5′ noncoding region but that is inactive when inverted or when placed in the 3′ noncoding region. Sequences that are critical for the expression of both Gag and Pol include not only the 5′ splice site positioned at +51 in the R region, which is used to generate the spliced pol mRNA, but also intronic R sequences located well 3′ to this splice site. Analysis of total cellular gag and pol mRNA expression demonstrates that deletion of the R region has little effect on gag mRNA levels but that R deletions that would be predicted to leave the pol 5′ splice site intact nevertheless inhibit the production of the spliced pol mRNA. Gag expression can be largely rescued by the introduction of an intron into the 5′ noncoding sequence in place of the R region but not by an intron or any one of several distinct retroviral nuclear RNA export sequences inserted into the mRNA 3′ noncoding sequence. Neither the R element nor the introduced 5′ intron markedly affects the cytoplasmic level of HFV gag mRNA. The poor translational utilization of these cytoplasmic mRNAs when the R region is not present in cis also extended to a cat indicator gene linked to an internal ribosome entry site introduced into the 3′ noncoding region. Together these data imply that the HFV R region acts in the nucleus to modify the cytoplasmic fate of target HFV mRNA. The close similarity between the role of the HFV R region revealed in this study and previous data (M. Butsch, S. Hull, Y. Wang, T. M. Roberts, and K. Boris-Lawrie, J. Virol. 73:4847–4855, 1999) demonstrating a critical role for the R region in activating gene expression in the unrelated retrovirus spleen necrosis virus suggests that several distinct retrovirus families may utilize a common yet novel mechanism for the posttranscriptional activation of viral structural protein expression.
The foamy viruses (FVs) are a distinct family of complex retroviruses that are only very distantly related to the pathogenic lentiviruses and oncoviruses. While they infect a wide variety of cell types, producing a characteristic foamy cytopathology in cultures, they appear to be entirely nonpathogenic in vivo (11, 22). Although the FV replication cycle remains incompletely understood, what is known so far distinguishes them from other retroviruses in a number of ways. For example, both DNA and RNA have been isolated from virions (32); the structural proteins Gag and Pol are translated from separate mRNAs, not as a fusion protein (5, 12); and two elements, one located in the 5′ region and another located in pol, contribute to packaging of the viral RNA (6, 9). These differences have led to the consideration that in evolutionary terms, FVs may lie somewhere between retroviruses and hepadnaviruses (18).
The long terminal repeat (LTR) sequences of FVs are among the longest found in retroviruses, and key features have been assigned to the LTR of the prototype human FV (HFV). For example, several DNA response elements for the viral transactivator of transcription, Tas, are located within the LTR U3 region (7, 14), while the major splice donor (SD) for the production of spliced mRNAs is located at position +51 in the R region (23). Moreover, it has been proposed that the R region of the HFV LTR is indispensable for efficient production of the viral structural proteins Gag and Pol (10). How the R region of HFV exerts this effect is not known, although it has been reported that this region has little effect on the level of gag mRNA and is therefore likely to act at a posttranscriptional level.
One intriguing possibility is that the R region of HFV induces the nuclear export of the incompletely spliced HFV mRNAs that encode Gag and Pol. All retroviruses require the cytoplasmic translation of not only fully spliced but also unspliced and sometimes singly spliced forms of the initial, genome-length viral transcript (4). However, cells have developed mechanisms to prevent the nuclear export of immature, incompletely spliced cellular mRNAs, and these mechanisms also restrict the nuclear export of incompletely spliced retroviral mRNAs. To overcome this nuclear retention, different retroviruses have developed distinct mechanisms. For example, lentiviruses such as human immunodeficiency virus type 1 (HIV-1) encode a regulatory protein, termed Rev, that directly binds not only to a cis-acting RNA target sequence present in target viral mRNAs but also to the nuclear export factor Crm1 (8, 21, 24). Because these Rev proteins are encoded by fully spliced viral mRNA, they are produced early in the viral life cycle and only then induce the expression of the incompletely spliced, late viral mRNAs that encode the viral structural proteins. An alternative strategy is used by certain simple retroviruses, such as Mason-Pfizer monkey virus (MPMV) and avian leukemia virus (ALV). Both of these viruses encode a constitutive transport element (CTE) that directly recruits a cellular nuclear export factor distinct from Crm1 to unspliced retroviral transcripts, thereby inducing their nuclear export (1, 31). However, for the majority of retrovirus families, including the FVs, it remains unclear how the incompletely spliced or unspliced viral RNAs reach the cytoplasm despite the predicted nuclear retention by cellular factors.
In this study, we have confirmed the critical importance of the R region of HFV for the expression of Gag and have defined the minimal sequence required for this activity. Our data demonstrate that the R region does not act at the transcriptional level or at the level of retroviral nuclear RNA export but rather suggest that this sequence primarily acts to enhance the cytoplasmic utilization of mRNAs encoding Gag.
MATERIALS AND METHODS
Plasmid construction.
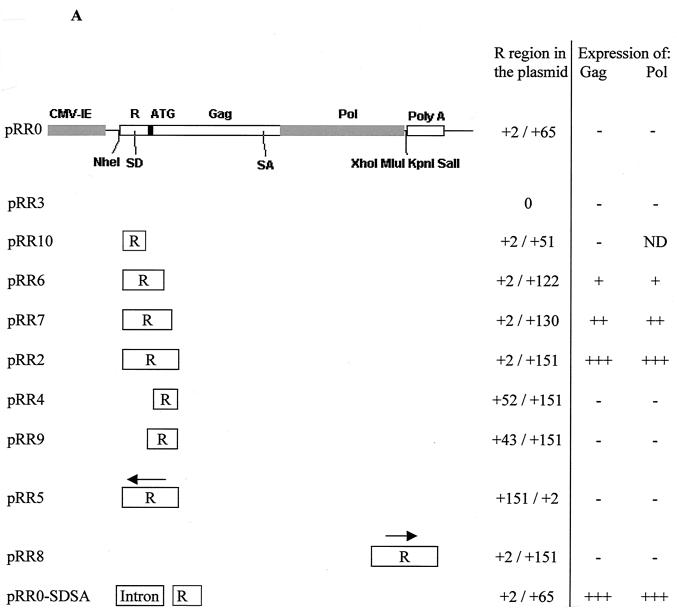
The mammalian expression plasmid pCI (Promega) contains the cytomegalovirus (CMV) immediate-early (IE) promoter linked to a chimeric intron followed by a polylinker and the simian virus 40 late polyadenylation site. In all constructs, unless specifically stated, the chimeric intron was deleted via the AflII sites at positions 820 and 1017, producing vector pCiΔ. Plasmid pRR0 contains the first 65 bp of the R region and an optimized translation initiation codon (5′GCCGCCGCCACCATGG3′) (15) linked to the HFV gag and pol genes. This plasmid was inserted into pCiΔ between the CMV IE promoter and the polylinker via the NheI and XhoI sites (Fig. 1A). Plasmid pRR2 (Fig. 1A), containing the first 151 bp of the R region and gag and pol of HFV, was generated from pRR0 in such a way that it was possible to remove the entire R region via an EagI site inserted downstream of the R region and the upstream NheI site. This plasmid was then used to produce a series of constructs containing different sections of the R region (Fig. 1A). Plasmid pRR8 was constructed by inserting the 151-bp R region into pRR3 downstream of gag and pol via the KpnI and SalI sites.
FIG. 1.
Plasmids containing different segments of the R region differ in the levels of Gag and Pol production. (A) Plasmid pRR0 consists of the CMV IE promoter followed by the first 65 bp of the R region upstream of an optimized translation initiation sequence (represented by the black box) linked to the HFV gag and pol genes upstream of a polylinker and the simian virus 40 late polyadenylation site. The SD and SA for Pol are indicated, as are relevant restriction sites. An EagI site was added after the R region in pRR2 to permit the insertion of different sections of the R region, shown below pRR0. An arrow indicates the orientation of inserted HFV sequences. The relative levels of Gag and Pol expression observed following transient transfection of 293T cells are shown. ND, not determined. (B) Western blot showing HFV Gag production following transient transfection of 293T cells with the indicated expression plasmids. The arrow indicates the 74-kDa–70-kDa Gag doublet. Unlabeled additional bands are believed to be the result of Gag protein degradation. (C) Western blot showing Pol production following transient transfection of 293T cells with the indicated plasmids. Arrows indicate the 127-kDa Pol precursor (Pol), the 80-kDa RT, and the 40-kDa integrase protein (IN). For both panels B and C, “Blank” refers to untransfected 293T cells.
Plasmids pRR3 and pRR0 were used to create constructs containing non-HFV sequences. Plasmid pRR3-IRES was generated by introducing a poliovirus internal ribosome entry site (IRES) from pPBS (6, 25) upstream of gag and pol in pRR3 via the NheI and EagI sites. Plasmid pRR3-CTE was constructed by introducing the CTE from MPMV downstream of the gag and pol genes via the KpnI and SalI sites. The CTE was cloned from plasmid pSARMX (from Eric Hunter). Plasmid pRR3-RRE was constructed by inserting the HIV-1 Rev response element (RRE) (21) into pRR3 downstream of HFV gag and pol via the restriction sites XhoI and KpnI. Plasmid pRR0-SDSA was constructed by cloning the R region and the gag and pol genes of pRR0 into mammalian expression vector pCI retaining the chimeric intron via the NheI and MluI sites. Plasmid pRR3-intron contains the more 3′ intron from the rat preproinsulin II gene derived from pBC12/CMV (3) and cloned downstream of gag and pol via the MluI site. Excising the intron via the XhoI and SalI sites and inserting it downstream of gag and pol in pRR0, pRR2, and pRR0-SDSA generated pRR0-intron, pRR2-intron, and pRR0-SDSA-intron, respectively. Plasmids pRR3-IRES-CAT, pRR0-IRES-CAT, and pRR2-IRES-CAT were generated by excising the IRES-chloramphenicol acetyltransferase (CAT) sequence from pSLIIB/CAT (28) via the HindIII and BamHI sites and cloning it into pRR3, pRR0, and pRR2, respectively, via the MluI site.
DNA transfection.
293T cells were maintained as previously described (31) and seeded at 2 × 106 per 100-mm dish on the day prior to transfection. Transfection was carried out using the Calcium Phosphate Profection Mammalian Transfection kit (Promega) and 10 μg of plasmid DNA. Sixteen hours posttransfection, the medium was changed. Forty-eight hours posttransfection, cells were counted, pelleted by centrifugation at 400 × g for 5 min, and resuspended in protein-denaturing buffer (16) at 106 cells per 50 μl of buffer. Induced CAT activity was determined from cell lysates as previously described (31).
Analysis of viral proteins.
Proteins were analyzed on denaturing 10% polyacrylamide gels by electrophoresis and Western blotting. HFV proteins were detected using anti-HFV human serum and then goat anti-human immunoglobulin G (IgG) F(ab)2 conjugated to horseradish peroxidase (Sigma). For specific Pol detection, a monoclonal antibody (provided by Axel Rethwilm) and then rabbit anti-mouse IgG conjugated to horseradish peroxidase (Sigma) were used. Blots were developed in diaminobenzidine solution (phosphate-buffered saline containing 0.1% [vol/vol] hydrogen peroxide [Sigma] and 0.005% [wt/vol] diaminobenzidine). For weaker signals, we used a chemiluminescence system (Amersham Pharmacia Biotech) as directed by the manufacturer.
RPA.
RNase protection analysis (RPA) was performed using a Hyspeed RPA kit (Ambion). For analysis of total cellular gag and pol mRNA expression, 293T cells were transfected with pRR0, pRR2, or pRR3; total cellular RNA was isolated 2 days after transfection. The levels of gag and pol mRNA expression were then determined by RPA (31) using a single-stranded 229-nucleotide (nt) probe spanning the pol 3′ splice site located at position 1848 within the HFV genome (12).
For determination of the relative levels of cytoplasmic and nuclear mRNA expression, 293T cells were transfected with an HFV Gag-Pol expression plasmid supplemented with pBC12ΔI (3) as an internal control. At 48 h after transfection, cells were harvested and aliquots were used for Western blot analysis as described above. The remainder of the cells were then separated into nuclear and cytoplasmic fractions after treatment with a Nonidet P-40 lysis buffer as previously described (31). Total RNA was isolated from the nuclear and cytoplasmic fractions and subjected to RPA using a 181-nt RNA probe that traverses the 3′ splice site of the rat preproinsulin II gene intron.
RESULTS
The R region is necessary for both Gag and Pol expression and acts in a position- and orientation-dependent manner.
Transfection of cells with a Gag-Pol expression construct containing the full-length HFV LTR R and U5 regions results in efficient protein production (9). It has been proposed that deletion of the U5 region can be tolerated but that deletion of the R region results in a loss of both Gag and Pol expression while exerting little effect on the level of Gag-encoding mRNA present in the cell (10). To confirm this observation, a set of HFV Gag-Pol expression plasmids containing different sections of the R region in a variety of positions and orientations was generated (Fig. 1A). These constructs, which are all based on mammalian expression plasmid pCI, contain the HFV gag and pol genes under the transcriptional control of the heterologous CMV IE promoter. The translation initiation codon of HFV gag was replaced with a consensus initiation codon (15), and HFV R sequences were then introduced between the CMV IE promoter and this artificial initiation codon. The ability of these constructs to direct the synthesis of Gag or Pol protein was then assayed by transient transfection of 293T cells followed by gel electrophoresis and Western blotting of cell extracts (Fig. 1B and C).
Using this assay, the minimal, discrete R sequence that proved to be both necessary and sufficient for the induction of efficient Gag or Pol protein production was found to coincide with the first 130 to 151 bp of the R region (pRR7 and pRR2; Fig. 1B and C). This result is particularly surprising for Pol, as Pol is expressed from a spliced mRNA lacking all sequences between the major SD at position +51 and a splice acceptor (SA) site located near the 3′ end of gag (Fig. 1A) (12). Total removal of the R sequence (pRR3) or further deletion from either the 3′ or the 5′ end of this 151-bp sequence (pRR0, pRR10, pRR4, and pRR9) resulted in reduced Gag or Pol production, at times to undetectable levels (Fig. 1B and C and data not shown). Inversion of the R region (pRR5) or repositioning of the entire R region downstream of gag and pol (pRR8) failed to rescue either Gag or Pol expression (Fig. 1B and data not shown). However, removal of a short palindromic sequence from positions +122 to +130 within the R region (pRR6) resulted in only a slight decrease in protein production, suggesting that this sequence was not a critical element (Fig. 1B and C).
To confirm that these data did not reflect a species- or tissue-specific phenomenon, we also examined the level of Gag expression obtained after transfection of hamster cell line BHK-21. These data (not shown) were closely comparable to the data obtained with human 293T cells (Fig. 1). In addition, we examined whether expression in trans of the HFV regulatory protein Tas or Bet would exert any detectable effect on the level of Gag expression; however, no such effect was observed (data not shown).
The R region does not act at the level of transcription or RNA stability.
The results presented in Fig. 1 demonstrate that sequences located within the first 151 nts of the HFV LTR R region act in a location- and orientation-dependent manner to boost the production of both viral Gag and Pol proteins. We considered four possible hypotheses to explain this effect. (i) The R region was acting as a transcriptional enhancer; (ii) the R region was affecting RNA stability in the nucleus and/or in the cytoplasm; (iii) the R region was acting at a posttranscriptional level, perhaps as a CTE; or (iv) the R region was affecting translation. These hypotheses are, of course, not mutually exclusive. The observation that R sequences located between the SD at position +51 and position +130, which are not present in the pol mRNA, nevertheless could dramatically enhance Pol protein expression (Fig. 1C) seemed inconsistent with a mechanism acting entirely at the level of nuclear export or cytoplasmic translation, i.e., hypotheses iii and iv. Conversely, the earlier data of Heinkelein et al. (10) arguing that deletion of the R region did not affect total cellular levels of gag mRNA, even though Gag protein production was blocked, argued against an effect at the level of transcription or RNA stability, i.e., hypotheses i and ii.
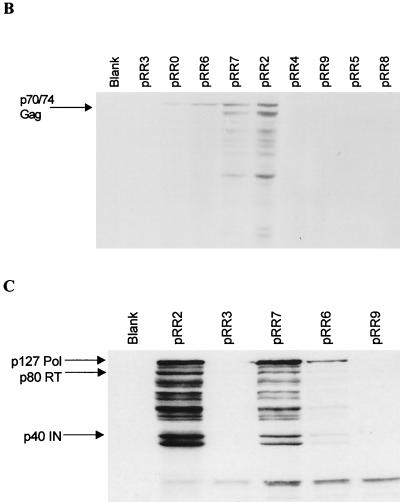
As a first test of the above hypotheses, we performed RPA to measure the total steady-state expression of the unspliced gag and spliced pol mRNAs in cells transfected with the active construct pRR2 or the inactive construct pRR0 or pRR3 (Fig. 1A). The probe used in this assay traverses the pol SA and can therefore simultaneously quantitate the levels of expression of both mRNA species. As shown in Fig. 2 and as previously reported by Heinkelein et al. (10), the presence or absence of the R region did not have a significant effect on the level of expression of gag mRNA. In contrast, pol mRNA was detected only in cells transfected with the active plasmid pRR2. The lack of expression of pol mRNA was predicted for pRR3, as this plasmid lacks the SD used to generate this mRNA, but was unexpected for pRR0, as this plasmid retains the pol SD at position +51 as well as 12 additional 3′-flanking nts that fully suffice to encode a consensus splice site (Fig. 1A). These data therefore imply that sequences located in the HFV R region, 3′ to position +65, are required for effective utilization of the SD located at position +51 in the R region.
FIG. 2.
RNase protection analysis of total cellular gag and pol mRNAs. 293T cells were transfected with the indicated plasmids, and total RNA was harvested 2 days after transfection. The probe used in this RPA is 229 nts long and spans the pol SA at position 1848 in gag (12). Protection by unspliced gag mRNA should yield a 210-nt probe fragment, while the less prevalent pol mRNA should rescue a fragment of 116 nts. Lane 1, 1/30 the input probe; lane 2, mock-transfected cells. Ten micrograms of total cellular RNA was used for each lane. The relative mobilities of RNA size markers are indicated at the left.
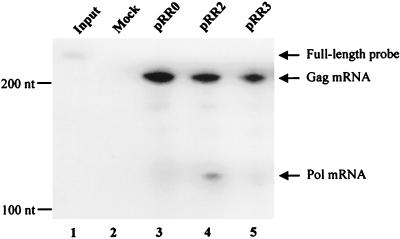
Recent data have suggested that splicing can significantly enhance the expression of certain mRNA species (20); therefore, we decided next to examine whether the presence of a functional intron in cis might enhance Gag protein expression. For this purpose, we constructed derivatives of pRR0, pRR2, and pRR3 containing an intron introduced into the 5′ noncoding region and/or into the 3′ noncoding region of the predicted gag mRNA molecule (Fig. 3A). The intron introduced into the 5′ noncoding region of pRR0, to give pRR0-SDSA, is a chimeric intron that is present in parental expression plasmid pCI and that contains a 5′ splice site derived from a human β-globin intron linked to a 3′ splice site derived from an IgG intron. In contrast, the intron introduced into the 3′ noncoding region of pRR0, pRR2, pRR3, and pRR0-SDSA is the complete second intron derived from the rat preproinsulin II gene (19). The resultant clones are shown in Fig. 3A. As shown in Fig. 3B, insertion of an intron into the 5′ noncoding region of pRR0, to give pRR0-SDSA, markedly activated Gag protein expression, while insertion of an intron into the 3′ noncoding region neither activated (pRR0-intron and pRR3-intron) nor enhanced (pRR2-intron) Gag protein expression. When an intron was present in both the 5′ and the 3′ noncoding regions (pRR0-SDSA-intron), the level of Gag protein expression observed was closely similar to that seen in pRR0-SDSA (Fig. 3A). Therefore, while the intron introduced into the 5′ noncoding region was able to activate Gag protein expression, an intron introduced into the 3′ noncoding region had no detectable effect, either positive or negative, on the level of Gag synthesis.
FIG. 3.
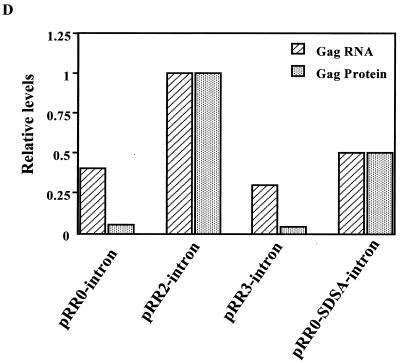
Effect of the R region on nuclear and cytoplasmic mRNA expression. (A) Schematic representation of the structures of clones bearing inserted introns. (B) Western analysis of Gag protein production in 293T cells transfected with the indicated HFV Gag expression plasmids. Blank, untransfected cells. (C) RPA of nuclear and cytoplasmic RNAs. 293T cells were transfected with 1.8 μg of pRR0-intron, pRR2-intron, pRR3-intron, or pRR0-SDSA-intron. As an internal RNA control, each culture also received 50 ng of pBC12ΔI (3). Two days after transfection, cells were harvested and separated into cytoplasmic and nuclear fractions and RNA was isolated (31). The RPA was performed using a full-length probe (F) of 181 nts, designed such that the unspliced gag-containing RNA (U) would give a protected probe fragment of 158 nts, the spliced gag-containing transcript (S) would give a fragment of 96 nts, while the internal control (IC) would protect a fragment of 76 nts. Lane 1, 1/30 the input probe; lanes 2 to 12, 45 μg of yeast cell RNA and 5 μg of 293T cell RNA as starting materials; lane 2, total 293T RNA from mock-transfected cells; lanes 3, 5, 7, 9, and 11, RNA from the nuclear (N) fractions; lanes 4, 6, 8, 10, and 12, RNA from the cytoplasmic (C) fractions. In lanes 3 and 4, cells were transfected with the pBC12ΔI internal control DNA only, while in lanes 5 through 12, cells were transfected with pBC12ΔI plus the indicated plasmid. (D) Comparison of the relative levels of cytoplasmic spliced gag RNA and Gag protein expression from the various constructs. RNA levels were determined by quantifying the band intensity in panel C using a Storm 860 Phosphorimager (Molecular Dynamics). The cytoplasmic gag RNA was normalized against the internal control RNA. Gag protein production was analyzed by quantitation of the intensities of bands detected by Western analysis using chemiluminescence, as shown in panel B, with LabImage 2.51 software (Labsoft). The gag RNA and protein levels from the pRR2-intron transfection were both arbitrarily set at one.
The HFV LTR R region is not a CTE.
We next used RPA to examine whether the presence of the R region affected the level of RNA expressed in the nucleus or cytoplasm of transfected cells. The RNA probe used in this assay traverses the 3′ splice site present in the rat preproinsulin II gene-derived intron inserted into the 3′ noncoding region of selected Gag expression constructs. This RNA probe therefore allows us to distinguish between RNAs that retain this intron and those that have lost it due to splicing. In addition, construct pBC12ΔI (3) expresses an mRNA containing a short segment of the rat preproinsulin gene that can hybridize to this RNA probe, and pBC12ΔI therefore serves as an important internal control to normalize for transfection efficiency and RNA recovery.
As shown in Fig. 3C, this 181-nt probe could rescue a protected fragment of 158 nts specific for unspliced (U) mRNA, a protected fragment of 96 nts specific for spliced (S) mRNA, and a 76-nt fragment specific for the pBC12ΔI internal control (IC) mRNA. Unspliced mRNAs were readily detected in the nucleus of transfected cells, yet little or no unspliced RNA reached the cytoplasm for all constructs tested. Quantitation of the level of spliced mRNA in the cytoplasm of transfected cells, which presumably could encode either Gag or Pol, revealed a relatively small difference in RNA levels between the two constructs that express readily detectable levels of Gag (i.e., pRR2-intron and pRR0-SDSA-intron) and those that do not (i.e., pRR0-intron and pRR3-intron) (Fig. 3C and D). Specifically, steady-state levels of cytoplasmic mRNA differed by a maximum of ∼3-fold (pRR2-intron versus pRR3-intron) to a minimum of ∼1.4-fold (pRR0-intron versus pRR0-SDSA-intron). In contrast, quantitation of the level of Gag protein expression from these constructs by analysis of the intensity of the chemiluminescence signal detected by Western blotting of transfected cell extracts demonstrated that the level of Gag expression induced by pRR0-intron and pRR3-intron was ∼15-fold lower than that seen with pRR2-intron and ∼8-fold lower than that seen with pRR0-SDSA-intron (Fig. 3B and D). Therefore, whatever the mechanism by which the R region (in pRR2-intron) or the 5′ intron (in pRR0-SDSA-intron) enhances Gag protein expression, it does not exert a marked effect on the level of cytoplasmic gag mRNA.
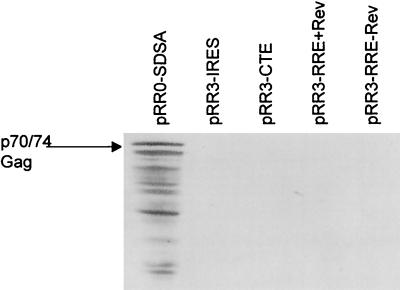
The data shown in Fig. 3C demonstrate that the removal of the introduced rat insulin gene intron was a prerequisite for nuclear RNA export (for example, compare lanes 7 and 8); therefore, it seemed unlikely that the R region was acting as an RNA export signal analogous to a CTE. As an alternative test of this hypothesis, we inserted full-length copies of the MPMV CTE (1) or the HIV-1 RRE (21) into pRR3 to give pRR3-CTE or pRR3-RRE, respectively. As shown in Fig. 4, neither of these elements was able to induce HFV Gag protein expression, in the latter case whether HIV-1 Rev was expressed or not. Similar data were also obtained upon insertion of the ALV CTE (31) into the 3′ noncoding region of pRR3 (data not shown). Therefore, the R element does not induce the export of incompletely spliced mRNAs when present in cis, nor can it be functionally substituted for by RNA elements known to act as nuclear RNA export inducers.
FIG. 4.
The R region cannot be functionally substituted for by retroviral nuclear export elements. Western blot showing Gag production from 293T cells transfected with the indicated expression plasmids containing inserted non-HFV sequences. The location of the 74-kDa–70-kDa doublet of HFV Gag protein is indicated.
Effect of the HFV LTR R region on mRNA utilization.
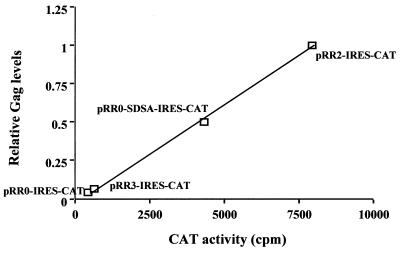
The data presented in Fig. 3 demonstrate that the R region can dramatically enhance the level of Gag protein expression without markedly affecting the level of cytoplasmic gag mRNA expression. If the R region indeed enhances the utilization of gag mRNA in the cytoplasm, then one can envisage two possible mechanisms. On the one hand, the R region might simply enhance the efficiency of ribosome recruitment to the 5′ noncoding region and/or the efficiency with which recruited ribosomes initiate translation of the Gag protein. Conversely, the R region might act in the nucleus to target the linked mRNA to a region within the cytoplasm where translation initiation is more efficient. To distinguish between these two possibilities, we constructed derivatives of pRR0, pRR3, and pRR2 as well as of pRR0-SDSA in which a poliovirus IRES (25) linked to a cat indicator gene was inserted into the 3′ noncoding region. We reasoned that if the R region or the inserted SDSA intron exerted its effect by promoting translation initiation of scanning ribosomes recruited to the 5′ end of these mRNAs, then the R region should have little or no effect on the expression of cat, which should be controlled exclusively by the inserted IRES (27). Conversely, if the R region targeted the RNA to a cytoplasmic subdomain where translation was highly efficient, then Gag and CAT expression might be closely correlated. As shown in Fig. 5, there was indeed a strong and apparently linear correlation between the level of expression of Gag and the level of expression of CAT in transfected 293T cells. We therefore conclude that the R element, if it exerts its effect at the translational level, does so by a mechanism other than by optimizing translation initiation at the Gag initiation codon.
FIG. 5.
Effect of the R region on the translation of linked genes. 293T cells were transfected with plasmid pRR0-IRES-CAT, pRR3-IRES-CAT, pRR0-SDSA-IRES-CAT, or pRR2-IRES-CAT. At 48 h after transfection, cells were harvested. Aliquots were used for measurement of induced CAT enzyme activity or for measurement of Gag protein production by Western blotting, followed by quantitation with LabImage 2.51 software. The data were then normalized against pRR2-IRES-CAT, the value for which was arbitrarily set at one, and plotted as shown.
In a final experiment, we examined whether the R element could itself be functionally replaced by an IRES. However, as shown in Fig. 4, insertion of an IRES in place of the R region to give pRR3-IRES did not result in the activation of Gag protein expression.
DISCUSSION
Previously, Heinkelein et al. (10) reported that the R region of the HFV LTR was critical for the expression of both viral Gag and Pol proteins, even though the presence or absence of the R region had little effect on the level of expression of HFV gag mRNA. This result was of interest as it suggested that the HFV R region was activating the expression of the HFV gag and pol gene products by a posttranscriptional mechanism. This suggestion raised the possibility that the R region might be functionally comparable to the CTE RNA targets present in certain simple retroviruses, such as MPMV and ALV, that act to induce viral structural protein expression by inducing the nuclear export of the cognate viral mRNA species (1, 31).
In this study, we have confirmed the observation (10) that the R region is required for HFV Gag and Pol expression and have mapped the essential sequences to the first ∼130 nts of the R region (Fig. 1). Remarkably, further 3′ deletion to +122 or, particularly, to +65 resulted in a dramatic inhibition not only of Gag expression but also of Pol expression (Fig. 1). This was a surprising observation, in that Pol is translated from a spliced mRNA that utilizes an SD located at +51 in the R region (12). Therefore, sequences that are located within the HFV pol gene intron and that extend significantly beyond the ∼6-nt splice site consensus located immediately 3′ to the SD are critical for Pol expression. In agreement with the previous data of Heinkelein et al. (10), we observed that the R region did not have a significant effect on the total steady-state level of HFV gag mRNA expressed in transfected cells (Fig. 2). However, expression of the spliced (HFV) pol mRNA was not detectable in cells transfected with constructs, such as pRR0, that do not contain the entire HFV R element (Fig. 2), even though, as noted above, pRR0 retains the pol SD and 14 3′-flanking nts (Fig. 1A). These data therefore imply that sequences that are located in the R region and that form part of the pol mRNA intron are required in cis for the effective production of the spliced pol mRNA. In contrast, while these same R sequences also exert a strong activating effect on the expression of the unspliced gag mRNA, they do not affect the total level of gag mRNA (Fig. 1B and 2).
Given this conclusion, we therefore next examined whether the R region had any effect on the subcellular localization of unspliced HFV gag mRNAs. As shown in Fig. 3B, we did not see any marked effect of the R region on either the nuclear or the cytoplasmic level of expression of gag mRNAs. Although there was a modest 2- to 3-fold increase in the level of cytoplasmic expression of mRNAs bearing the functional R element, this effect was clearly much lower than the ∼15-fold effect of the R element on the level of Gag protein expression (Fig. 1 and 3). We next examined whether the R region might be functionally replaceable by RNA elements that have been shown in other contexts to exert a positive effect on gene expression. Consistent with the observations that the R region had little effect on the cytoplasmic level of expression of spliced mRNAs and was incapable of inducing the cytoplasmic expression of an unspliced mRNA containing an introduced heterologous intron (Fig. 3), we did not find that R function could be substituted for by any of several different retroviral RNA export elements, including the MPMV CTE and the HIV-1 RRE (Fig. 4 and data not shown). We therefore conclude that the R element does not act as a nuclear RNA export element analogous to a CTE. Surprisingly, however, insertion into the gag 5′ noncoding region of an intron in place of the R region was able to largely rescue Gag protein expression, even though insertion of an intron into the 3′ noncoding region had no detectable effect (Fig. 3A). Even though this 5′ intron, present in pRR0-SDSA, increased Gag protein expression by ∼8-fold, it also exerted no significant effect on the cytoplasmic level of expression of the cognate spliced viral mRNA (Fig. 3).
To examine whether the R region had any direct effect on the translation of linked RNA sequences, we next inserted a poliovirus IRES, linked to the cat indicator gene, into the 3′ noncoding region of selected Gag expression plasmids. IRES elements are able to directly recruit ribosomes to a linked open reading frame (25, 27, 29), and they should therefore be functionally independent of any sequence elements that are introduced into the mRNA 5′ noncoding region and that might act to facilitate cap-dependent ribosome recruitment. Nevertheless, as shown in Fig. 5, the R region exerted exactly the same positive effect on cat expression as it did on the level of expression of Gag.
Based on these data, it appears that sequences located within the gag and/or pol genes of HFV exert a significant inhibitory effect not only on the expression of Gag but also on the expression of other genes introduced in cis, such as the cat indicator gene (Fig. 4). Introducing the R region at the 5′ end of the mRNA overcomes this inhibition by an apparently nuclear mechanism that nevertheless does not exert a significant effect on the cytoplasmic accumulation of gag mRNA. The activating effect of the R region can be partly substituted for by insertion into the 5′ noncoding sequence of an intron in place of the R region, but neither the R region nor an intron inserted into the 3′ noncoding sequence can rescue Gag protein expression. How an R region-induced posttranscriptional event occurring in the nucleus would affect the cytoplasmic fate of HFV gag mRNA remains unclear, although it has been shown that certain nuclear posttranscriptional modifications can “imprint” mRNAs with specific mRNA binding proteins that then accompany the mRNAs into the cytoplasm of the cell (13, 17). If the R region indeed acts in the nucleus to modify the subsequent cytoplasmic utilization of gag mRNA, then mRNAs that retain the R region but have not been imprinted in the nucleus should not demonstrate any R region-dependent activation of translation. It will be of interest to test this hypothesis using in vitro translation assays or perhaps direct transfection of mRNA molecules.
It is of interest to compare the observations reported above with published data examining the importance of the R and U5 regions for gene expression in other retroviruses. For example, Trubetskoy et al. (30) reported that the R region of the murine leukemia virus LTR could dramatically boost the expression of a linked unspliced cat mRNA but that the presence or absence of the R region had little effect on cat expression if an intron was introduced into the cat 5′ noncoding region. These findings appear similar to our data showing that the HFV R region can be functionally replaced by an intron introduced into the 5′ noncoding region but not by an intron present in the 3′ noncoding region. However, Trubetskoy et al. (30) noted a major reduction in the steady-state level of the unspliced RNA lacking the R element, a result which clearly differs from our findings (Fig. 2).
Analysis of the importance of the R region in spleen necrosis virus (SNV), a retrovirus that is fairly closely related to murine leukemia virus, produced a somewhat different result. While the R region of the SNV LTR also proved critical for the expression of the linked viral gag gene, Butsch et al. (2) observed only a modest 2- to 3-fold increase in the level of cytoplasmic gag RNA even though the production of Gag protein was increased by >100-fold when the R region was present. Like the HFV R region (Fig. 1), the SNV R region was found to function in a position- and orientation-dependent manner. Based on these data, as well as subsequent work examining the effect of the R region on the expression of linked nonviral sequences (26), it was proposed that the SNV R element functioned by enhancing the efficiency of translation of linked genes. Why the SNV gag gene was incapable of being translated even though the cognate mRNA was readily detectable in the cytoplasm was not addressed in these earlier reports. The close similarity of the phenotypes exerted by the SNV and HFV R regions strongly suggests that these essentially unrelated retroviruses have developed comparable, and apparently entirely novel, mechanisms to activate the cytoplasmic expression of their viral structural proteins. However, how this activation is achieved and whether the R elements have indeed evolved as some form of mechanistically distinct substitute for the retroviral RNA export elements observed in other retrovirus families are currently unclear.
ACKNOWLEDGMENTS
Rebecca A. Russell and Yan Zeng contributed equally to this work.
This work was supported by The Wellcome Trust, The Jefferiss Research Trust, and the Howard Hughes Medical Institute.
We thank Axel Rethwilm at the University of Dresden, Dresden, Germany, for the Pol monoclonal antibody and Eric Hunter at the University of Alabama, Birmingham, for plasmid pSARMX.
REFERENCES
- 1.Bray M, Prasad S, Dubay J W, Hunter E, Jeang K T, Rekosh D, Hammarskjold M L. A small element from the Mason-Pfizer monkey virus genome makes human immunodeficiency virus type 1 expression and replication Rev-independent. Proc Natl Acad Sci USA. 1994;91:1256–1260. doi: 10.1073/pnas.91.4.1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Butsch M, Hull S, Wang Y, Roberts T M, Boris-Lawrie K. The 5′ RNA terminus of spleen necrosis virus contains a novel posttranscriptional control element that facilitates human immunodeficiency virus Rev/RRE-independent Gag production. J Virol. 1999;73:4847–4855. doi: 10.1128/jvi.73.6.4847-4855.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cullen B R. trans-Activation of human immunodeficiency virus occurs via a bimodal mechanism. Cell. 1986;46:973–982. doi: 10.1016/0092-8674(86)90696-3. [DOI] [PubMed] [Google Scholar]
- 4.Cullen B R. Retroviruses as model systems for the study of nuclear RNA export pathways. Virology. 1998;249:203–210. doi: 10.1006/viro.1998.9331. [DOI] [PubMed] [Google Scholar]
- 5.Enssle J, Jordan I, Mauer B, Rethwilm A. Foamy virus reverse transcriptase is expressed independently from the Gag protein. Proc Natl Acad Sci USA. 1996;93:4137–4141. doi: 10.1073/pnas.93.9.4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Erlwein O, Bieniasz P D, McClure M O. Sequences in pol are required for transfer of human foamy virus-based vectors. J Virol. 1998;72:5510–5516. doi: 10.1128/jvi.72.7.5510-5516.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Erlwein O, Rethwilm A. BEL-1 transactivator responsive sequences in the long terminal repeat of human foamy virus. Virology. 1993;196:256–268. doi: 10.1006/viro.1993.1474. [DOI] [PubMed] [Google Scholar]
- 8.Fornerod M, Ohno M, Yoshida M, Mattaj I W. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- 9.Heinkelein M, Schmidt M, Fischer N, Moebes A, Lindemann D, Enssle J, Rethwilm A. Characterization of a cis-acting sequence in the Pol region required to transfer human foamy virus vectors. J Virol. 1998;72:6307–6314. doi: 10.1128/jvi.72.8.6307-6314.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heinkelein M, Thurow J, Dressler M, Imrich H, Neumann-Haefelin D, McClure M O, Rethwilm A. Complex effects of deletions in the 5′ untranslated region of primate foamy virus on viral gene expression and RNA packaging. J Virol. 2000;74:3141–3148. doi: 10.1128/jvi.74.7.3141-3148.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hooks J J, Detrick-Hooks B. Spumavirinae foamy virus group infections: comparative aspects and diagnosis. In: Kurstak E, Kurstak C, editors. Comparative diagnosis of viral diseases. IV. New York, N.Y: Academic Press, Inc.; 1981. pp. 599–618. [Google Scholar]
- 12.Jordan I, Enssle J, Göttler E, Mauer B, Rethwilm A. Expression of human foamy virus reverse transcriptase involves a spliced pol mRNA. Virology. 1996;224:314–319. doi: 10.1006/viro.1996.0534. [DOI] [PubMed] [Google Scholar]
- 13.Kataoka N, Yong J, Kim V N, Velazquez F, Perkinson R A, Wang F, Dreyfuss G. Pre-mRNA splicing imprints mRNA in the nucleus with a novel RNA-binding protein that persists in the cytoplasm. MolCell. 2000;6:673–682. doi: 10.1016/s1097-2765(00)00065-4. [DOI] [PubMed] [Google Scholar]
- 14.Keller A, Partin K M, Lochelt M, Bannert H, Flugel R M, Cullen B R. Characterization of the transcriptional trans activator of human foamy retrovirus. J Virol. 1991;65:2589–2594. doi: 10.1128/jvi.65.5.2589-2594.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell. 1986;44:283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- 16.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;277:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 17.Le Hir H, Moore M J, Maquat L E. Pre-mRNA splicing alters mRNP composition: evidence for stable association of proteins at exon-exon junctions. Genes Dev. 2000;14:1098–1108. [PMC free article] [PubMed] [Google Scholar]
- 18.Linial M L. Foamy viruses are unconventional retroviruses. J Virol. 1999;73:1747–1755. doi: 10.1128/jvi.73.3.1747-1755.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lomedico P, Rosenthal N, Efstratidadis A, Gilbert W, Kolodner R, Tizard R. The structure and evolution of the two nonallelic rat preproinsulin genes. Cell. 1979;18:545–558. doi: 10.1016/0092-8674(79)90071-0. [DOI] [PubMed] [Google Scholar]
- 20.Luo M J, Reed R. Splicing is required for rapid and efficient mRNA export in metazoans. Proc Natl Acad Sci USA. 1999;96:14937–14942. doi: 10.1073/pnas.96.26.14937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Malim M H, Hauber J, Le S Y, Maizel J V, Cullen B R. The HIV-1 rev trans-activator acts through a structured target sequence to activate nuclear export of unspliced viral mRNA. Nature. 1989;338:254–257. doi: 10.1038/338254a0. [DOI] [PubMed] [Google Scholar]
- 22.Mergia A, Leung N J, Blackwell J. Cell tropism of the simian foamy virus type 1 (SFV-1) J Med Primatol. 1996;25:2–7. doi: 10.1111/j.1600-0684.1996.tb00185.x. [DOI] [PubMed] [Google Scholar]
- 23.Muranyi W, Flugel R M. Analysis of splicing patterns of human spumaretrovirus by polymerase chain reaction reveals complex RNA structures. J Virol. 1991;65:727–735. doi: 10.1128/jvi.65.2.727-735.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Neville M, Stutz F, Lee L, Davis L I, Rosbash M. The importin-beta family member Crm1p bridges the interaction between Rev and the nuclear pore complex during nuclear export. Curr Biol. 1997;7:767–775. doi: 10.1016/s0960-9822(06)00335-6. [DOI] [PubMed] [Google Scholar]
- 25.Pelletier J, Sonenberg N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature. 1988;334:320–325. doi: 10.1038/334320a0. [DOI] [PubMed] [Google Scholar]
- 26.Roberts T M, Boris-Lawrie K. The 5′ RNA terminus of spleen necrosis virus stimulates translation of nonviral mRNA. J Virol. 2000;74:8111–8118. doi: 10.1128/jvi.74.17.8111-8118.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sachs A B, Sarnow P, Hentze M W. Starting at the beginning, middle, and end: translation initiation in eukaryotes. Cell. 1997;89:831–838. doi: 10.1016/s0092-8674(00)80268-8. [DOI] [PubMed] [Google Scholar]
- 28.Tiley L S, Madore S J, Malim M H, Cullen B R. The VP16 transcription activation domain is functional when targeted to a promoter-proximal RNA sequence. Genes Dev. 1992;6:2077–2087. doi: 10.1101/gad.6.11.2077. [DOI] [PubMed] [Google Scholar]
- 29.Trono D, Pelletier J, Sonenberg N, Baltimore D. Translation in mammalian cells of a gene linked to the poliovirus 5′ noncoding region. Science. 1988;241:445–448. doi: 10.1126/science.2839901. [DOI] [PubMed] [Google Scholar]
- 30.Trubetskoy A M, Okenquist S A, Lenz J. R region sequences in the long terminal repeat of a murine retrovirus specifically increase expression of unspliced RNAs. J Virol. 1999;73:3477–3483. doi: 10.1128/jvi.73.4.3477-3483.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang J, Cullen B R. Structural and functional analysis of the avian leukemia virus constitutive transport element. RNA. 1999;5:1645–1655. doi: 10.1017/s1355838299991616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu S F, Baldwin D N, Gwynn S R, Yendapalli S, Linial M L. Human foamy virus replication: a pathway distinct from that of retroviruses and hepadnaviruses. Science. 1996;271:1579–1582. doi: 10.1126/science.271.5255.1579. [DOI] [PubMed] [Google Scholar]