Abstract
Respiratory syncytial virus (RSV) produces three envelope glycoproteins, the attachment glycoprotein (G), the fusion (F) protein, and the small hydrophobic (SH) protein. It had been assumed, by analogy with other paramyxoviruses, that the G and F proteins would be required for the first two steps of viral entry, attachment and fusion. However, following repeated passage in cell culture, a viable mutant RSV that lacked both the G and SH genes was isolated (R. A. Karron, D. A. Buonagurio, A. F. Georgiu, S. S. Whitehead, J. E. Adamus, M. L. Clements-Mann, D. O. Harris, V. B. Randolph, S. A. Udem, B. R. Murphy, and M. S. Sidhu, Proc. Natl. Acad. Sci. USA 94:13961–13966, 1997). To explore the roles of the G, F, and SH proteins in virion assembly, function, and cytopathology, we have modified the full-length RSV cDNA and used it to rescue infectious RSV lacking the G and/or SH genes. The three resulting viruses and the parental virus all contain the green fluorescent protein (GFP) gene that serves to identify infected cells. We have used purified, radiolabeled virions to examine virus production and function, in conjunction with GFP to quantify infected cells. We found that the G protein enhances virion binding to target cells but plays no role in penetration after attachment. The G protein also enhances cell-to-cell fusion, presumably via cell-to-cell binding, and enhances virion assembly or release. The presence or absence of the G protein in virions has no obvious effect on the content of F protein or host cell proteins in the virion. In growth curve experiments, the viruses lacking the G protein produced viral titers that were at least 10-fold lower than titers of viruses containing the G protein. This reduction is due in large part to the less efficient release of virions and the lower infectivity of the released virions. In the absence of the G protein, virus expressing both the F and SH proteins displayed somewhat smaller plaques, lower fusion activity, and slower viral entry than the virus expressing the F protein alone, suggesting that the SH protein has a negative effect on virus fusion in cell culture.
Respiratory syncytial virus (RSV) is a major cause of serious lower respiratory tract infection in infants and young children worldwide. It is in the subfamily Pneumovirinae within the Paramyxoviridae family of enveloped, single-stranded, negative-sense RNA viruses. RSV expresses three glycoproteins on its surface. The F (fusion) protein is postsynthetically cleaved and has other structural similarities to the F proteins of other paramyxoviruses. In addition, some monoclonal antibodies directed to the F protein inhibit syncytium formation (41), confirming its role in fusion. The G (glycoprotein) protein is believed to be an attachment protein since it is the other large glycoprotein on the virion surface and antibodies specific for the G protein inhibit the adsorption of virus to cells (25). The SH protein is a small hydrophobic protein of unknown function. All other paramyxoviruses have an F protein and an attachment protein and some, but not all, have an SH protein.
Recent studies have found that RSV initially binds to target cells in culture via glycosaminoglycans (GAGs) (4) on the cell surface (12, 13, 22). A conserved region of the G protein contains several basic amino acid-rich, potential binding sites for heparin, a model GAG. A peptide from this conserved region of the G protein binds to heparin and to target cells, partially inhibiting viral infection (11). Furthermore, the G protein has been shown to bind immobilized heparin (22). These findings further support the idea that the G protein is the RSV attachment protein and point to cell surface GAGs as its ligand.
Studies with viruses in the other subfamily of the Paramyxoviridae, the Paramyxovirinae, have clearly demonstrated that the fusion function performed by the F protein requires the attachment HN (hemagglutinin-neuraminidase) protein for something other than simple attachment to the target cell. Coexpression of the HN and F proteins of one virus causes cell-to-cell fusion, but coexpression of the HN protein from one virus and the F protein of a disparate virus does not. For instance, the parainfluenza virus type 2 (PIV-2) HN protein does not enable the PIV-3 F protein to cause fusion (17), or vice versa, and the Newcastle disease virus HN protein does not enable the PIV-3 F protein to cause fusion, or vice versa (8). There appears to be a critical interaction between these two surface proteins that is required to trigger fusion and initiate infection.
It was unexpected, therefore, when a viable RSV mutant that expresses only the F protein due to a deletion of the SH and G genes was isolated (21). This mutant, cp-52, grew to relatively high titers in cultured cells but was poorly infectious in mice and humans. At least in cell culture, it seems that the F protein alone is able to carry out the important initial functions in the RSV life cycle, cell binding and fusion, as well as the final steps of assembly, budding, and release. If so, it is not clear whether or how the G and SH proteins contribute to virus production and spread.
The importance of the viral glycoproteins in RSV assembly has been demonstrated in a minigenome system (38). The F protein was required, along with the RSV replication proteins and the matrix (M) protein, to produce minivirions capable of infection. Coexpression of the G protein enhanced this passage efficiency 10- to 20-fold, while coexpression of the SH protein had no effect. Since passage requires assembly, budding, release, target cell binding, and fusion, it is difficult to determine from these experiments which steps were enhanced by the G protein.
The ability to rescue infectious RSV from full-length cDNA (7) has become a powerful tool for studying the function of individual viral proteins. To study more thoroughly the functions of the G and SH proteins, we generated a set of three RSV deletion mutants that lack the G gene, the SH gene, or both. The parent of these mutants is the recombinant green fluorescent protein (GFP)-expressing rgRSV (13). We have compared these four viruses with respect to binding, entry, assembly and release, infectivity, growth rate, fusion, and syncytium formation.
MATERIALS AND METHODS
Plasmid construction.
SN3 (Fig. 1), an RSV cDNA clone containing GFP as its first gene but lacking all three glycoprotein genes, SH, G, and F, was constructed in three steps. First, the SH gene was deleted from D13, a plasmid containing the 3′ genomic sequence of the RSV genome (leader, NS1, NS2, N [nucleoprotein], P, M, and SH). The deletion was accomplished by PCR amplification of the plasmid (6), using two primers that flank the SH gene. Unique XhoI and PvuI sites were introduced by including their sequences in the primers. This 8-kb PCR product was cut with XhoI and BamHI (a site that coincides with the start of the L gene) and ligated with a similarly digested M2 gene amplified from D46 (7), a full-length cDNA clone for the A2 strain of RSV, to yield the SN1 plasmid. This manipulation inserted the authentic M2 gene. Second, the L gene was removed from D46 and inserted into SN1 at the BamHI and MluI sites of SN1 to generate the SN2 plasmid. Third, the GFP gene from MP175, a derivative of D13 containing GFP as its first gene (13), was inserted into SN2 as an AgeI/SpeI fragment. The SpeI digestion of SN2 was partial since there are two such sites in this plasmid.
FIG. 1.
Diagram of the SN3 plasmid and its three derivative plasmids, SN10, SN11, and SN12 (not to scale). SN3 is a partial RSV antigenome plasmid, containing all of the RSV genes except for the three viral glycoprotein genes. Like its complete RSV parent cDNA, MP224, SN3 contains the GFP gene in the first position. The new intergenic sequence (IG) between the M and M2 genes contains two unique restriction sites, PvuI and XhoI, as well as native IG sequence before and after these sites, as indicated. These sites were used to subclone RSV genes back into SN3: the G and F genes to generate SN11, and the F gene to generate SN10. Le, leader; TR, trailer; nt, nucleotides. SN11 and SN10 were used to rescue rgRSV-GF and rgRSV-F, respectively. The SH gene was subcloned into the PvuI and SacII sites of SN10 to generate SN12, which was used to rescue rgRSV-SF. All inserted genes (large open rectangles) and gene start and gene end sequences (shaded boxes) are authentic. The sequence and derivation of all modified IG regions are shown, including the only nonnative sequences and the added restriction sites (bold italics). Transcription from all plasmids was driven by the T7 promoter (indicated on SN3) to initiate synthesis of the antigenome.
Three SN3 derivatives, SN10, SN11, and SN12, were constructed as shown in Fig. 1 and used to rescue rgRSV carrying as their glycoproteins genes the F gene (rgRSV-F), the G and F genes (rgRSV-GF), or the SH and F genes (rgRSV-SF). The F gene was amplified by PCR from the D46 template using a positive-sense primer containing PvuI and SacII sites (5′TTAATCGATCGATTCCGCGGAGAATCAAAATAAACTGGGGCAAATAACAATGGAGTTGCTAATCCTCA; restriction sites are underlined) and a negative-sense primer containing an XhoI site (5′CATACCTCGAGTGGCATGCAATTGTGTTTTATATAACTATAAACTA). The F gene was cloned into SN3 at its PvuI and XhoI sites to generate SN10. The G and F genes were amplified as a single fragment from D46 by using a positive-sense primer containing a PvuI site (5′TTAATCGATCGAGACTAACAATAACATTGGGGCAAATGCAAACATGTCCAAAAACAAG) and the same negative-sense primer as used to amplify the F gene, cloned into SN3 at its PvuI and XhoI sites to create SN11. The SH gene was amplified from D46 by using a positive-sense primer containing a PvuI site (5′TTAATACCGATCGACATGGGGCAAATAATCATTGGAGGAAATCCAACTAATCAC) and a negative-sense primer containing a SacII site (5′ATATATCCGCGGATCCTAGTTCATTGTTATGAC), inserted into SN10 to generate SN12. All PCRs used Vent polymerase with 15 cycles and a limited amount of enzyme to decrease the chances of introducing unintended mutations (6). Construction of the full-length cDNA, MP224, and rescue of rgRSV224 was reported previously (13). In the present report, rgRSV224 is called rgRSV-SGF to be consistent with the nomenclature used for the deletion viruses.
Recovery of recombinant viruses.
To rescue virus from the cDNA clones, antigenomic plasmids SN10, SN11, and SN12, as well as the parent MP224, were transfected into HEp-2 cells along with pTM-1 plasmids expressing the RSV N, P, L, and M2-1 genes from the T7 promoter. The cells were also infected with a modified vaccinia virus, MVA-T7 (43), to provide T7 polymerase. The procedure used was as described previously (7), with one modification. After adding 1 ml of LipofectACE (Life Technologies)-DNA complex and MVA-T7, the six-well plates were spun at 900 rpm for 3 min in a tabletop centrifuge (CS-6R; Beckman) to enhance the transfection and infection efficiency (40). Cells were incubated at 32°C for two passages to reduce the cytotoxicity from vaccinia virus. The recovered viruses were amplified in HEp-2 cells at a multiplicity of infection (MOI) of less than 1, and low-passage stocks were used in all experiments.
Cells, medium, virus growth, and titration.
HEp-2 cells were maintained in OPTI-MEM (GIBCO BRL) supplemented with 2% heat-inactivated fetal bovine serum (FBS) and incubated in 5% CO2 at 37°C. For growth curve analysis, HEp-2 cells grown in 25-cm2 culture flasks were inoculated at 37°C for 3 h with each virus (MOI ∼ 1). After adsorption, the inocula were removed and cells were washed three times with medium before addition of 4 ml of fresh growth medium and incubation at 37°C. At the indicated times, 200-μl aliquots were taken and replaced with equal volumes of fresh medium. Samples of the four viruses at each time point were centrifuged briefly to remove debris, and supernatants were stored at −70°C. All samples were titrated at the same time.
Titration was performed in duplicate 96-well plates. Briefly, 90% confluent HEp-2 cells were inoculated with 30 μl of 10-fold dilutions of each sample. After 3 h of adsorption and rocking every 15 min, the inocula were replaced with 100 μl of growth medium. The plates were incubated at 37°C for 20 h before the number of green cells in each well was determined under an inverted fluorescence microscope (Nikon Diaphot). The dilution factor and inoculum volume were also used to calculate the infectious units per milliliter. Values from wells containing between 50 and 500 green cells were used in these calculations. This method compared favorably with the standard plaque assay, producing titers slightly (not more than twofold) higher. For most experiments in this report, since we tested for infection and production of GFP rather than virus spread and plaque formation, this infectivity assay was more accurate than a titer determined by plaque assay.
For plaque size measurement, inoculated cells were overlaid with OPTI-MEM–2% FBS–0.9% methylcellulose and incubated at 37°C for 6 days. The cells were fixed with methanol and stained with a mixture of monoclonal antibodies against the N, F, and G proteins of RSV (Chemicon), then by horseradish peroxidase-coupled goat anti-mouse immunoglobulin G (Kirkegaard & Perry Laboratories), followed by a mixture of equal parts of 4-chloro-1-naphthol (Kirkegaard & Perry Laboratories) and 3% hydrogen peroxide. Plaque sizes were compared by the analysis of variance test.
RT-PCR.
Total intracellular RNA from purified, radioactively labeled virions were isolated with Trizol (Life Technologies) according to the manufacturer's protocol. The RNA was subjected to reverse transcription (RT) with Superscript II (Life Technologies), using random hexamer primers. An aliquot of cDNA was then amplified in a PCR using three sets of primers to generate cDNA fragments of the F, G, and SH genes. PCR was carried out at 94°C for 3 min, followed by 30 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 1 min, followed by a 7-min extension at 72°C. PCR products were analyzed on a 1.5% agarose gel.
Immunoprecipitation.
HEp-2 cells growing in a six-well plate were inoculated with each virus (MOI ∼ 2). Each well of infected and mock-infected cells was metabolically labeled beginning at 27 h postinoculation (p.i.) for 18 h by incubation with cysteine- and methionine-free Dulbecco modified Eagle medium (DMEM; GIBCO BRL) supplemented with 3% dialyzed FBS, 10% growth medium (OPTI-MEM–2% FBS), and 50 μCi of of Tran35 S-label (ICN) per ml. Cells were lysed at 4°C for 20 min with 0.5 ml of lysis buffer (10 mM Tris [pH 7.5], 10 mM NaCl, 1.5 mM MgCl2, 1% Triton X-100, 1% sodium deoxycholate, protease inhibitors [1 mM phenylmethylsulfonyl fluoride, 2 μg of aprotinin per ml, 10 μg of leupeptin per ml]). Cell lysates were clarified by a 1-min centrifugation at the top speed of an Eppendorf microcentrifuge, and two-thirds of the supernatants were incubated at 20°C for 1 h with rabbit anti-RSV serum preabsorbed to protein G-agarose beads (Boehringer Mannheim). The beads were washed three times with lysis buffer by centrifugation and boiled immediately for 4 min in 2× sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer containing 10% β-mercaptoethanol. Polypeptides were separated on a 10 to 20% gradient minigel (ICN) by SDS-PAGE. Prestained protein ladder (BenchMark; GIBCO BRL) was used to calibrate molecular mass. Images of polypeptide profiles on the SDS-polyacrylamide gel were collected by a STORM PhosphorImager (Molecular Dynamics) and quantified using the ImageQuaNT software.
Preparation and use of radiolabeled viruses.
Two 150-cm2 plates of 80% confluent HEp-2 cells were inoculated with each virus (MOI ∼ 2). At 30 h p.i., cells were radiolabeled as described above for 24 h. At that time, the medium was removed from each plate and centrifuged at 4,500 × g for 5 min to remove cells and cell debris. Viral particles were purified from both sets of media by two sequential isolations on a linear sucrose gradient. Briefly, the supernatant from two plates was layered onto a 25 to 55% sucrose linear gradient and centrifuged in a Beckman SW28 rotor at maximum speed for 18 h at 4°C (23). One-milliliter fractions were collected manually from the top of the gradient, and the fractions containing the peak of radioactivity at sucrose density ranging from 37 to 41% were pooled and diluted in Hanks balanced salt solution. The second linear sucrose gradient centrifugation was performed as above in SW28.1 buckets, and 0.5-ml fractions were collected. Purified radiolabeled viruses were aliquoted and frozen at −70°C. All four viruses were labeled at the same time.
For infectivity and binding of radiolabeled viruses, confluent HEp-2 cells in 12-well plates were inoculated with 3 × 104 cpm of each purified virus. Adsorption was performed at 4°C for 1 h in 100 μl of minimal essential medium (MEM) containing 1% bovine serum albumin with rocking every 10 min. After binding, the inocula were removed and the monolayers were washed four times with ice-cold MEM. For infectivity assays, growth medium was added and the plates were moved to 37°C. After incubation for 18 h, cells were trypsinized, washed once with phosphate-buffered saline (PBS), and fixed with 3% paraformaldehyde, and the infected (green fluorescent) cells were counted by a flow cytometer (Ortho Cytoronabsolute). For cell binding assays, cells were lysed with radioimmunoprecipitation assay buffer (10 mM Tris [pH 7.5], 150 mM NaCl, 1% sodium deoxycholate, 1% Triton X-100, 0.1% SDS), and the amount of labeled viruses bound was determined by scintillation counting (Bio-Safe II; Research Products International Corp.) in a 1215 Rack Beta II liquid scintillation counter (Pharmacia LKB).
Rate of entry assay.
Confluent monolayers of HEp-2 cells growing in 24-well plates were inoculated with each virus for 1 h at 4°C, the inocula were removed, and the monolayers were rinsed once with ice-cold MEM. Prewarmed OPTI-MEM–2% FBS was added, and the cells were incubated at 37°C to allow virus penetration. At the indicated times after this temperature shift, virus-infected wells were treated for 1 min with 0.5 ml of citrate-buffered saline (77 mM citric acid, 40 mM Na2HPO4, 146 mM NaCl [pH 3.0]) (18) or 0.5 ml of PBS and washed three times with MEM, prewarmed OPTI-MEM–2% FBS was added, and the dishes were incubated at 37°C. At 18 h after the temperature shift, green cells were counted by flow cytometry.
Fusion assay.
A modification of the assay used by Nussbaum et al. (30) was used to assess fusion activity. HEp-2 cells in OPTI-MEM–2% FBS seeded in 48-well plates the previous day were inoculated (MOI ∼ 2) and incubated at 37°C. Twenty hours before the fusion assay, the RSV-infected cells were infected with a recombinant vaccinia virus which encodes T7 polymerase under the control of a Lac-inducible promoter (vT7lacOI) (1) (MOI ∼ 10). The inoculum was replaced with OPTI-MEM–2% FBS containing 0.1 mM isopropyl-1-thio-β-d-galactoside (IPTG). At the same time, a 100-mm2 plate of HEp-2 cells was inoculated with another recombinant vaccinia virus carrying the β-galactosidase gene under the control of a T7 promoter (vCB21RlacZ) (2) (MOI ∼ 10). Both sets of vaccinia virus-infected cells were incubated overnight at 32°C. At 36 or 42 h p.i. with RSV, the vCB21RlacZ-infected reporter cells were removed from the plate with EDTA (Versene; Life Technologies) and rinsed once with OPTI-MEM–2% FBS–2.5 mM CaCl2 and twice with OPTI-MEM–2% FBS. They were resuspended in OPTI-MEM–2% FBS containing 40 μg of arabinoside C (Sigma) per ml at a density of 5 × 105 cells/200 μl. The RSV-infected cells were rinsed once with PBS before addition of the 200 μl of reporter cell suspension to each well. The plates were incubated for 3 h at 37°C. Three wells for each condition were assayed by the colorimetric chlorophenol red–β-d-galactopyranoside (CRGP; Boehringer Mannheim) assay. Briefly, cells were lysed by the addition of 10 μl of 10% NP-40 to the medium. Fifty microliters of this lysate was mixed with 50 μl of CRGP reagent (16 mM CRGP, 0.12 M Na2HPO4 · 7H2O, 0.08 M NaH2PO4 · H2O, 0.02 M KCl, 0.002 M MgSO4 · 7H2O, 0.01 M β-mercaptoethanol) in a well of a 96-well plate. After incubation at room temperature for 30 min, the plate was read at A570 in a plate reader (Ceres UV 900 Hdi; Bio-Tek Instruments Inc.).
RESULTS
Generation of GFP-expressing RSV that lack one or two viral glycoproteins.
Previously, we inserted a gene unit containing the GFP open reading frame as the first gene in a full-length RSV cDNA (13) and used it to rescue rgRSV. In the present study, this virus is designated rgRSV-SGF because it contains the genes for all three of the viral glycoproteins, SH, G, and F. To generate rgRSV that carries only the F protein gene, the F and G protein genes, or F and SH protein genes (rgRSV-F, rgRSV-GF, or rgRSV-SF, respectively), we first constructed a plasmid, SN3, that lacks all three of the glycoprotein genes (Fig. 1). Instead, two restriction sites, PvuI and XhoI, were placed in the hybrid intergenic region between the M and M2 genes.
Since the F protein is required for infectivity, the F gene was included in all constructs. The three possible combinations of RSV glycoprotein genes were amplified by PCR from the full-length RSV cDNA and subcloned into SN3 between the PvuI and XhoI sites (Fig. 1). Infectious virus was recovered from these plasmids by transfection (7) and amplified in HEp-2 cells. Viruses were passaged (MOI < 1) the minimum number of times necessary to generate high-titered stocks. rgRSV-SGF and rgRSV-GF reached titers of 5 × 107 to 8 × 107 by four passages, and rgRSV-F and rgRSV-SF reached titers of 2 × 107 to 9 × 106 by six and seven passages, respectively. These stocks were used in all experiments.
The glycoprotein genes and their expression.
The recombinant virus stocks used for all experiments were examined by RT-PCR to confirm the glycoprotein genes they contain. RNA isolated from virions was subjected to RT using random hexamer primers. Three sets of specific primers, one for each glycoprotein gene, were then used to amplify these genes in a multiplex PCR. The PCR products generated from each virus (Fig. 2A) matched the predicted sizes (Fig. 2B) and the expected genes in each virus, confirming their identities.
FIG. 2.
RT-PCR verification of the glycoprotein genes present in each rescued virus. (A) RT-PCR products from RNA extracted from purified rgRSV-SGF, rgRSV-GF, rgRSV-SF, and rgRSV-F virions. The PCR products were separated by electrophoresis on a 1.5% agarose gel and stained with ethidium bromide. (B) Three sets of PCR primers (arrows) were used to amplify segments of the SH, G, and F genes in one reaction tube for each virus.
To confirm that the expected glycoproteins were expressed by each of these viruses, HEp-2 cells were infected and metabolically labeled with [35S]Met-[35S]Cys for 18 h, beginning at 27 h p.i. Cells were lysed and viral proteins were immunoprecipitated with rabbit anti-RSV serum, reduced, separated by SDS-PAGE, and visualized by phosphorimaging (Fig. 3A). The G protein is difficult to detect with [35S]Met-[35S]Cys labeling since it is relatively poor in these amino acids. Nevertheless, the G protein was detected in cells infected with rgRSV-SGF and rgRSV-GF, but not in cells infected with the other two viruses, as expected. Likewise, the SH protein is detected in cells infected with rgRSV-SGF and rgRSV-SF, as expected. Phosphorimage quantification indicated that the amount of SH produced in rgRSV-SF-infected cells was 40% of that produced in rgRSV-SGF cells when normalized to the amount of N protein produced in each case. The reason for this lower expression is not clear and was not explored.
FIG. 3.
Protein expression in virus-infected cells and in purified virions. (A) HEp-2 cells mock infected or infected with rgRSV-SGF, rgRSV-GF, rgRSV-SF, or rgRSV-F were labeled with [35S]Met-[35S]Cys. Lysates prepared from these cells were immunoprecipitated with rabbit anti-RSV serum, reduced, and separated by SDS-PAGE. (B) [35S]Met-[35S]Cys virions were purified twice by equilibrium centrifugation; 30,000 cpm of each virus was reduced and electrophoresed in the same gel as in panel A. The RSV proteins are indicated on the right, and molecular mass markers (kilodaltons) are shown on the left.
To assess the incorporation of viral proteins into virions, HEp-2 cells were inoculated (MOI ∼ 2), and a high level of infection (∼85%) was confirmed by fluorescence microscopy at 24 h p.i. At 30 h p.i., metabolic labeling with [35S]Met-[35S]Cys was initiated. After 24 h of labeling, the culture medium was harvested and virions were purified by two sequential equilibrium centrifugations through linear sucrose gradients.
An equivalent amount of radioactivity from each purified virion preparation was reduced, electrophoresed, and visualized by phosphorimaging (Fig. 3B). As expected, the G protein was only found in rgRSV-SGF and rgRSV-GF virions, and the SH protein was only found in rgRSV-SGF and rgRSV-SF virions. As in the infected cells, the amount of SH protein in the rgRSV-SF virions was low, 25% that in the rgRSV-SGF virions. All four virion preparations contained similar amounts of F protein.
Viral growth and spread.
As described above, the titers of rgRSV-SF and rgRSV-F virus stocks were approximately 10-fold lower than those of rgRSV-SGF and rgRSV-GF. To compare the multiplication of these viruses more carefully and under parallel conditions, we performed a growth curve experiment. Monolayers of HEp-2 cells were inoculated with similar amounts of each virus (MOI ∼ 1). To directly assess the infection levels of cultures inoculated with each virus, we examined them at 24 h. Among the four cultures, the proportions of green (infected) cells were nearly equivalent. Virus was released from rgRSV-SGF- and rgRSV-GF-infected cells as early as 18 h p.i., reaching near peak titers by 30 h (Fig. 4). rgRSV-SF and rgRSV-F released infectious virus with similar kinetics, but with titers at least 10-fold lower than rgRSV-SGF and rgRSV-GF titers, confirming our earlier observations and indicating that the G protein enhances the assembly, release, or infectivity of virions.
FIG. 4.
Growth curves for rgRSV-SGF, rgRSV-GF, rgRSV-SF, and rgRSV-F. HEp-2 cells were inoculated (MOI ∼ 1) and incubated at 37°C. At the indicated times, samples of culture medium were removed and frozen at −70°C. Virus was titrated at the same time in duplicate; the results averaged and reported as infectious units (IU) per milliliter. The experiment was repeated a total of three times with similar results, a 1- to 2-log-lower titer for rgRSV-SF and rgRSV-F compared to rgRSV-SGF and rgRSV-GF.
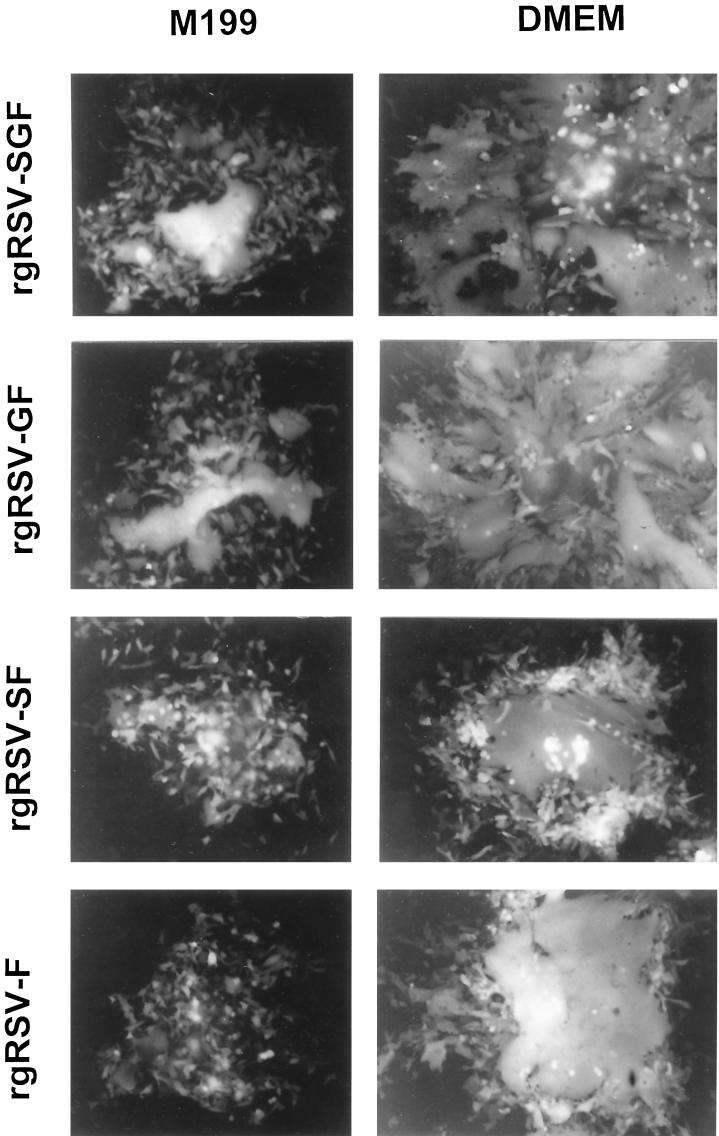
As an assay for viral spread, we examined the diameter of plaques induced by each virus after a 6-day incubation (Table 1). The rgRSV-SGF and rgRSV-GF plaques were not significantly different (P > 0.05). The rgRSV-F plaques were smaller than rgRSV-SGF plaques (P < 0.05) and rgRSV-GF plaques (P < 0.05), and the rgRSV-SF plaques were the smallest (P < 0.05). To determine if there was any gross difference in the mechanism of plaque formation, we examined plaques microscopically (Fig. 5). All four viruses clearly generated syncytia. These results indicate that the F protein alone can induce syncytia and that the G protein enhances virus spread, and they suggest that the SH protein inhibits virus spread in the absence of the G protein.
TABLE 1.
Plaque size comparisona
| Virus | Diam (mean ± SD) |
|---|---|
| RSV-A2 | 1.6 ± 0.2 |
| rgRSV-SGF | 1.4 ± 0.3 |
| rgRSV-GF | 1.4 ± 0.2 |
| rgRSV-SF | 0.9 ± 0.2 |
| rgRSV-F | 1.2 ± 0.2 |
Inoculated HEp-2 cells were overlaid with methylcellulose and incubated for 6 days. Cells were fixed and stained with a mixture of monoclonal antibodies against RSV. Plaques were measured with the aid of a dissecting microscope. The longest and narrowest axes were measured and averaged.
FIG. 5.
Typical plaques produced by rgRSV-SGF, rgRSV-GF, rgRSV-SF, and rgRSV-F in different overlay media. HEp-2 cells were inoculated, overlaid with 0.8% agarose in medium 199 or DMEM, and incubated at 37°C for 6 days. Plaques were photographed with a 10× objective.
We also found that plaque size and quality vary with the nutrient medium used. As shown in Fig. 5, medium 199 resulted in smaller plaques than DMEM. The two viruses lacking G protein, which show small syncytia in medium 199, exhibit massive fusion in DMEM, though the plaques remained smaller than the viruses containing G.
Virion assembly and release.
RSV is a highly cell-associated virus, spontaneously releasing only 10% of its infectivity into the medium (24). According to their growth curves (Fig. 4), the two viruses lacking the G protein released fewer infectious virions than the two viruses containing the G protein. Fewer infectious virions may be a reflection of either fewer virions assembled or released into the culture medium or lower infectivity of those virions. To compare the quantities of virions released by the viruses, total counts per minute in the purified virion preparations (Fig. 3B) were plotted (Fig. 6). rgRSV-SF and rgRSV-F released approximately three times fewer virions than rgRSV-SGF and rgRSV-GF. The threefold lower number of virions released in the absence of the G protein indicates that the G protein enhances virion assembly and/or release.
FIG. 6.
Release of virus particles from HEp-2 cells infected with rgRSV-SGF, rgRSV-GF, rgRSV-SF, and rgRSV-F. Monolayers (two 150-mm-diameter plates each) were inoculated (MOI ∼ 2) and incubated at 37°C. At 24 h, approximately the same number of cells were infected (85% green) in all plates. Cells were labeled with [35S]Met-[35S]Cys from 30 to 54 h. Culture media were harvested and clarified by low-speed centrifugation. Virions were purified by equilibrium centrifugation through two sequential linear sucrose gradients. The counts per minute (CPM) plotted is the sum from the peak fractions of the second gradient.
Cell binding activity and infectivity.
To determine whether viruses lacking the G protein differ in cell binding activity and/or infectivity, the purified 35S-labeled virions were used. Equivalent radioactivity, representing an equal number of virions as indicated by SDS-PAGE (Fig. 3B) and by Western blotting using a monoclonal antibody against the N protein (data not shown), was added to each well of HEp-2 cells. After 1 h at 4°C to allow attachment, cells were washed extensively to remove unbound virions. One set of cells was lysed, and the virions bound to the cells were quantified by beta scintillation counting (Fig. 7A). The binding activity of the two viruses lacking the G protein was approximately three times lower than that of the two viruses expressing the G protein. This result indicates that the G protein is responsible for attaching two-thirds of the virions to target cells under these conditions.
FIG. 7.
Binding and infectivity of purified rgRSV-SGF, rgRSV-GF, rgRSV-SF, and rgRSV-F virions. Twice-sucrose gradient-purified [35S]Met-[35S]Cys virions (30,000 cpm) were incubated with confluent monolayers of HEp-2 cells at 4°C for 1 h and then washed four times with ice-cold medium to remove unbound virus. (A) To assess binding, cells were lysed and radioactivity was quantified by a beta counter. The average background from wells containing only medium was subtracted (∼100 cpm). (B) To assess infectivity, growth medium was added and cells were incubated at 37°C for 18 h. The percent infected (green) cells was determined by flow cytometry. The results are the average ± standard deviation of three wells. The two assays were performed at the same time. These data are representative of three separate experiments.
Following attachment at 4°C, fresh medium was added to the other set of cells, which were then incubated at 37°C for 18 h and analyzed for infected (green) cells by flow cytometry (Fig. 7B). Approximately one-third as many cells were infected by virions lacking the G protein compared to virions containing the G protein. Since equal amounts of virions were added to these cells, virions that lack the G protein were one-third as infectious as virions with the G protein. These infectivity results correspond closely to the results of the parallel binding experiment, indicating that (i) virions lacking the G protein are threefold less infectious than virions containing the G protein, this is due to a lower binding activity; (ii) once a virion binds to a cell it is just as infectious, whether or not it contains the G protein; and (iii) the SH protein does not appear to be involved in binding or infectivity, since virions with SH have binding and infection activities similar to those of virions lacking SH.
Rate of viral entry.
To directly examine and compare the rates of virus penetration, pH 3 buffer was added at various times during this process to inactivate virions remaining outside the cells. We incubated virus with HEp-2 cells at 4°C for 1 h to allow attachment, removed the virus, washed the cells, and added warmed medium. Monolayers were then shifted to 37°C, allowing penetration to proceed. At selected times, cells were treated briefly with pH 3 citrate buffer and washed, and fresh medium was added. Virions that had penetrated the cells prior to the pH 3 treatment were protected and able to replicate, resulting in green cells by 15 h p.i. (Fig. 8). The percent entry at each time was calculated relative to wells treated with PBS, to normalize for differences in MOI.
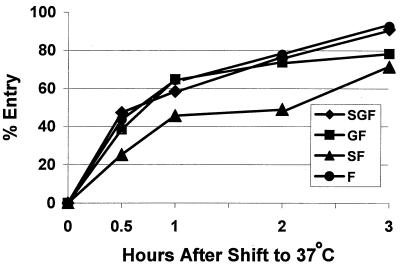
FIG. 8.
Postattachment entry rate of rgRSV-SGF, rgRSV-GF, rgRSV-SF, and rgRSV-F virions. Each virus (MOI < 1) was adsorbed to a confluent monolayer of HEp-2 cells at 4°C for 1 h. Cells were rinsed to remove unbound virus and moved to 37°C to allow penetration. At the indicated times, cells were treated with pH 3 citrate-buffered saline for 1 min to inactivate viruses remaining on the cell surface and rinsed, OPTI-MEM–2% FBS was added, and cells were incubated at 37°C. PBS instead of citrate-buffered saline was applied to control, nontreated cells. After incubation at 37°C for 15 h, infected (green) cells were counted by flow cytometry. Wells that had been treated with the pH 3 buffer before shifting to 37°C resulted in no infected (green) cells. Percent entry was calculated by dividing the percent infected cells in the pH 3-treated wells at each time by the percent infected cells in the parallel PBS-treated well and multiplying by 100. This experiment represents three separate experiments with similar results.
Three of the four viruses displayed similar entry kinetics. The finding that rgRSV-F enters as rapidly as rgRSV-SGF and rgRSV-GF indicates that once a virion has bound, neither the G nor the SH protein facilitates RSV penetration of cells, consistent with the conclusion from the previous experiment. The delay observed for rgRSV-SF penetration suggests that the SH protein may inhibit the rate of fusion, at least in the absence of the G protein.
Fusion activity.
All of these viruses are able to cause HEp-2 cells to fuse over the course of a 6-day plaque assay (Fig. 5). The viruses containing the G protein produced larger syncytia than the viruses lacking the G protein. However, the size of those syncytia might have been affected by the higher yield of viruses expressing the G protein (Fig. 4), rather than greater fusion efficiency. To directly assay fusion induced by each of these viruses over a shorter period of time that would be less affected by viral spread in the culture, we used a vaccinia virus-based β-galactosidase reporter system (30). Cells infected with RSV and recombinant vaccinia virus encoding T7 polymerase were mixed with another set of cells infected with vaccinia virus encoding the lacZ gene under the control of the T7 promoter. When RSV F protein causes fusion, the T7 polymerase from the first set of cells drives expression of β-galactosidase, which is assayed with the enzyme substrate CRPG and quantified by spectrophotometry at 36 h and 42 h p.i.
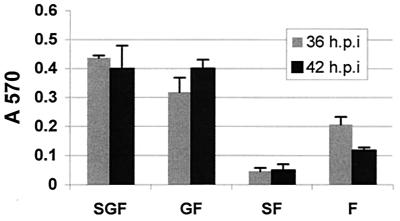
Fusion activity was detected in cells infected with each virus, though the levels differed (Fig. 9). The fusion activity of rgRSV-GF was similar to that of rgRSV-SGF, while the fusion activity of rgRSV-F was approximately half of that of rgRSV-SGF at 36 h. By 42 h, the rgRSV-F fusion activity appears to have decreased, though in other experiments it remained close to the same level. Fusion activity of the rgRSV-SF-infected cells was approximately twofold less than that of rgRSV-F-infected cells. These results indicate that the G protein does, indeed, enhance cell-to-cell fusion, presumably by enhancing cell-to-cell attachment. They also suggest that in the absence of the G protein, the SH protein inhibits fusion.
FIG. 9.
Fusogenic activity of rgRSV-SGF, rgRSV-GF, rgRSV-SF, and rgRSV-F. HEp-2 cell monolayers were inoculated with each virus (MOI ∼2) and then with vT7lacOI encoding T7 polymerase. At 36 or 42 h p.i., these cells were overlaid with a second population of HEp-2 cells infected with vCB21RlacZ, carrying a β-galactosidase gene preceded by a T7 promoter. Fusion of these two cell types allows the T7 polymerase to drive expression of the β-galactosidase gene. After 3 h at 37°C, fusion was quantified by measuring β-galactosidase activity with the colorimetric CRPG assay. The infection level of each monolayer was assessed (65 to 80% green cells), and fusion values were normalized to account for these small differences. The mean values of three wells ± standard deviations are shown. These data are representative of two separate experiments with similar results.
DISCUSSION
Isolation of cp-52, the RSV mutant that is infectious despite the lack of two of its three glycoprotein genes (21), was not anticipated. One formal possibility is that the single mutation in the cp-52 F protein (Glu218Gly) may have been selected by growth in cultured cells for its ability to attach to cells or to assemble virus. Since that time, viable rRSV mutants lacking the SH gene (5, 19) or the G gene (39) have been rescued. Infection with these mutants reinforce the conclusion from cp-52 infection that, at least separately, neither the SH nor the G protein is required for infectivity. In this study we have constructed and rescued a virus that, like cp-52, lacks both its SH and G genes. rgRSV-F is infectious, confirming the results of Karron et al. (21) and indicating that a mutation in the F protein is not required for it to function as the sole glycoprotein in an infectious RSV.
The simple fact that cp-52 is infectious indicates that the F protein can perform all of the necessary functions in virus assembly and release and in virus entry. It brings into question the need for the G and SH proteins for replication in vitro. Our growth curve experiments indicated that the G protein has a profound effect on virus production: viruses lacking the G protein released at least 10-fold less infectivity. By comparing the activities of purified, radiolabeled virions, we determined that viruses lacking the G protein attached to cells with one-third the efficiency of those containing the G protein. Likewise, virions lacking the G protein were one-third as infectious as those containing the G protein. Since the lower infectivity in the absence of G protein can be completely explained by less efficient attachment to cells, the G protein is not necessary for virus fusion and entry. That is, the F protein alone is responsible for efficient fusion. The idea that the G protein plays no role in fusion and entry, beyond enhancing attachment, was reinforced in the entry experiment: after binding to cells, rgRSV-F entered as rapidly as rgRSV-SGF and rgRSV-GF. Both of these results indicate that, unlike other paramyxoviruses, the two major glycoproteins of RSV do not interact to initiate the fusion event.
The entry rate of rgRSV is much slower (half-life = 30 min) than that of herpesvirus type 1 (half-life = 7 min) (29), which also initiates infection by binding to cell surface GAGs and fusing with the plasma membrane. The entry rates of other paramyxoviruses have not been reported. Since the G protein does not appear to play a role beyond enhancing binding, the F protein's fusion activity may be the rate-limiting step in entry. Perhaps the F protein is slow to change conformation and initiate fusion because it lacks an interacting partner to trigger the change that has been proposed for other paramyxoviruses. Alternatively, after its initial binding to the cell, the delay in entry may reflect the need for RSV to search for a specific F receptor before initiating fusion.
Our results are consistent with those of Teng et al. (38) in the infectious minivirion system. They found that the F protein was essential for production of infectious minivirions, but the addition of the G protein enhanced production 10- to 20-fold. They could not determine whether the lower level of minivirions was due to less efficient assembly or less efficient infection in the absence of the G protein. We were able to determine that the 10-fold reduction in yield of viruses lacking the G protein is due to a combination of both problems, the product of threefold less efficient assembly or release and threefold less efficient binding and therefore infectivity.
The only virus from the Paramyxovirinae subfamily whose F protein has been shown to cause fusion in the absence of its partner attachment protein is simian virus 5 (SV5) (16), though its fusion activity is enhanced by coexpression of the SV5 HN attachment protein (9). Interestingly, attachment proteins of other paramyxoviruses or influenza virus also enhance SV5 F protein fusion, though not to levels as high as found with the homologous SV5 HN protein (3). The fusion enhancement induced by heterologous attachment proteins would appear to be simply a result of enhanced binding. We have found that the RSV G protein behaves similarly in the context of infectious rgRSV virions. Likewise, a recombinant vesicular stomatitis virus (VSV) expressing the RSV F protein in addition to its usual proteins induces cell-to-cell fusion, while the parental VSV does not (20). In this situation, the VSV G protein may also have enhanced the RSV F protein fusion activity by providing an attachment function.
Previous transient expression studies of the roles of the three RSV glycoproteins in fusion found that the F protein alone caused little (31) or no (15) cell-to-cell fusion. Here, in the context of virions and RSV-infected cells, we have found that the F protein can cause fusion in the absence of the G and SH proteins. In both transient expression reports, addition of the SH protein or the G protein enhanced fusion somewhat. We found in the context of virions that the G protein enhanced both fusion and infectivity, and that this was due solely to enhanced binding. However, in the context of virions, the SH protein actually inhibited fusion and entry, resulting in smaller plaques. The reason for this discrepancy may lie in the cells used, unexpected contributions made by the vaccinia virus in the transient studies, the relative expression levels of the SH protein, or the contribution of another RSV protein, perhaps the M protein, in our virion system. In any case, it would seem that results obtained with infectious virus would be more likely to reflect the true physiological function of these proteins.
We have found that plaque size varies with the nutrient medium used. Plaques produced in medium 199 were smaller than those in DMEM. In addition, the two viruses lacking G protein, which usually show small syncytia in both medium 199 and OPTI-MEM, exhibit not only larger plaques but also much greater fusion in DMEM. The amounts of glutamine and calcium in the culture medium have been reported to affect syncytium formation (28, 35). DMEM and medium 199 contain the same amount of calcium, but DMEM has twice the amount of glutamine. DMEM also contains 4.5 times the amount of glucose found in medium 199, which might increase metabolic activity of the cells.
Virion assembly in the absence of the viral attachment glycoprotein has been reported for one other virus in the Paramyxovirinae subfamily, Sendai virus (SV). The temperature-sensitive SV mutant, ts271, produces an HN protein that is unstable at nonpermissive temperature such that virions released under these conditions do not contain either detectable HN protein (33) or detectable HN fragments (36). The ability of SV to produce virions without HN was confirmed by the plasmid-driven production of virus-like particles containing an SV minigenome and all of the SV proteins except for HN (26). In that study, virions were produced as efficiently when the HN plasmid was included in the transfection as in its absence. Interestingly, neither ts271 nor the HN-less virus-like particles were infectious, unless the target cells expressed the asialoglycoprotein receptor. Apparently, the asialoglycoprotein receptor provides an alternative method of attachment by binding to terminal galactose on the virion (26, 27). As demonstrated in the present report with RSV, SV containing the F protein as its only glycoprotein is able to produce virions. Furthermore, these virions can be infectious.
Truncations in the cytoplasmic tail of the SV5 HN protein leads to an enhancement in the amount of nonviral proteins incorporated into virions (34). Deletion of an entire RSV glycoprotein might have been expected to have a similar or even more pronounced effect. However, there was no appreciable enhancement of host cell protein incorporation in any of the RSV mutant virions (Fig. 3B).
The absence of the G and/or SH proteins from the virion might also allow space in the viral envelope for incorporation of more F protein. Alternatively, if the G and/or SH proteins enhance incorporation of the F protein into virions, there would be less F protein in the virion in their absence. Neither seemed to be the case, since a similar amount of F protein was found in purified viral particles that lack the G protein, the SH protein, or both, compared to complete virus. Therefore, an altered amount of F protein in the viral particles cannot account for the viability of an RSV mutant containing the F protein as its only glycoprotein, nor can it account for the lower binding activity and infectivity of these viruses. This situation is different in SV: in the absence of HN, there is an enhanced incorporation of F protein into virions (26).
Studies of SH deletion mutants from SV5 (14) and mumps virus (37) have failed to identify an obvious phenotype in cell culture. An SH deletion mutant of RSV has been found to grow to higher titers and produce larger plaques than its parent (5). Our rgRSV-GF also appeared to produce more infectious virus than rgRSV-SGF in the growth curve experiment. However, we were unable to identify the cause of this increased viral yield. The amount of viral particles released and the binding and infection activities of these particles were all similar to, or slightly less than, those for rgRSV-SGF. We did not observe the slight increase in plaque size for rgRSV-GF that has been noted for previous SH deletion mutants of RSV (5, 19).
It may simply be more difficult to detect the minor inhibitory effect of the SH protein in the presence of the strongly enhancing G protein. In the absence of the G protein, we found that further deletion of the SH gene enhanced the rate of virion entry, cell-to-cell fusion, and plaque size. The rgRSV-SF virus was also more difficult to recover from its cDNA than rgRSV-F or either of the other two constructs, and it was more difficult to amplify (data not shown). These results would be consistent with a dampening role of the SH protein on F protein function.
The SH protein does appear to be important in vivo, since SH-deficient RSV is attenuated in the upper respiratory tracts of mice (5) and in chimpanzees (42). It is not clear what role SH plays during infection in vivo, but it seems more dramatic than that played in vitro. It will be interesting to examine the viruses we have generated here in an animal model. Such studies may help to clarify the roles played by the G and SH proteins in vivo.
Finally, the RSV F protein appears to function as an attachment protein in rgRSV-F. Feldman et al. (10) have recently reported that cp-52, the RSV mutant lacking its SH and G genes, binds to GAGs on the target cell surface in a manner similar to wild-type RSV. Furthermore, they found that the RSV F protein, like its G protein, binds to heparin-agarose, though the relative affinities were not explored. These results suggest that in the absence of the G protein, the F protein may be able to attach the virion to cellular GAGs to initiate infection. We have also found that a portion of the cell binding activity of rgRSV-F is due to GAG binding (S. Techaarpornkul and M. E. Peeples, unpublished data). However, the finding that murine leukemia virus lacking all viral glycoproteins is able to attach to target cells (32) suggests that, in some cases, virions may not need a viral glycoprotein for simple attachment. Although it is not yet feasible to produce RSV virions lacking all three viral glycoproteins due to the very low yield of virus particles from the initial transfection, perhaps increases in the efficiency of virus recovery will eventually allow us to test this possibility for RSV.
ACKNOWLEDGMENTS
We thank Peter Collins for the recombinant RSV system, reagents, discussions, and support; we thank Ada Cole, Greg Spear, and Alan Landay for use of the inverted fluorescence microscope and the flow cytometer. We also thank Bernard Moss for providing vaccinia virus MVA-T7, Christopher Broder, Paul E. Kennedy, and Edward A. Berger for providing vaccinia viruses vCB21RlacZ and vT7lacOI, and Barbara Newton and Mohammad Saifuddin for technical help.
This work was funded by a grant from the Rush University Committee on Research and the Public Health Service (AI47213). S.T. was partially supported by a scholarship from the Thai government.
REFERENCES
- 1.Alexander W A, Moss B, Fuerst T R. Regulated expression of foreign genes in vaccinia virus under the control of bacteriophage T7 RNA polymerase and the Escherichia coli lac repressor. J Virol. 1992;66:2934–2942. doi: 10.1128/jvi.66.5.2934-2942.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alkhatib G, Broder C C, Berger E A. Cell type-specific fusion cofactors determine human immunodeficiency virus type 1 tropism for T-cell lines versus primary macrophages. J Virol. 1996;70:5487–5494. doi: 10.1128/jvi.70.8.5487-5494.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bagai S, Lamb R A. Quantitative measurement of paramyxovirus fusion: differences in requirements of glycoproteins between simian virus 5 and human parainfluenza virus 3 or Newcastle disease virus. J Virol. 1995;69:6712–6719. doi: 10.1128/jvi.69.11.6712-6719.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bourgeois C, Bour J B, Lidholt K, Gauthray C, Pothier P. Heparin-like structures on respiratory syncytial virus are involved in its infectivity in vitro. J Virol. 1998;72:7221–7227. doi: 10.1128/jvi.72.9.7221-7227.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bukreyev A, Whitehead S S, Murphy B R, Collins P L. Recombinant respiratory syncytial virus from which the entire SH gene has been deleted grows efficiently in cell culture and exhibits site- specific attenuation in the respiratory tract of the mouse. J Virol. 1997;71:8973–8982. doi: 10.1128/jvi.71.12.8973-8982.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Byrappa S, Gavin D K, Gupta K C. A highly efficient procedure for site-specific mutagenesis of full-length plasmids using Vent DNA polymerase. Genome Res. 1995;5:404–407. doi: 10.1101/gr.5.4.404. [DOI] [PubMed] [Google Scholar]
- 7.Collins P L, Hill M G, Camargo E, Grosfeld H, Chanock R M, Murphy B R. Production of infectious human respiratory syncytial virus from cloned cDNA confirms an essential role for the transcription elongation factor from the 5′ proximal open reading frame of the M2 mRNA in gene expression and provides a capability for vaccine development. Proc Natl Acad Sci USA. 1995;92:11563–11567. doi: 10.1073/pnas.92.25.11563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deng R, Mirza A M, Mahon P J, Iorio R M. Functional chimeric HN glycoproteins derived from Newcastle disease virus and human parainfluenza virus-3. Arch Virol Suppl. 1997;13:115–130. doi: 10.1007/978-3-7091-6534-8_12. [DOI] [PubMed] [Google Scholar]
- 9.Dutch R E, Joshi S B, Lamb R A. Membrane fusion promoted by increasing surface densities of the paramyxovirus F and HN proteins: comparison of fusion reactions mediated by simian virus 5 F, human parainfluenza virus type 3 F, and influenza virus HA. J Virol. 1998;72:7745–7753. doi: 10.1128/jvi.72.10.7745-7753.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feldman S A, Audet S, Beeler J A. The fusion glycoprotein of human respiratory syncytial virus facilitates virus attachment and infectivity via an interaction with cellular heparan sulfate. J Virol. 2000;74:6442–6447. doi: 10.1128/jvi.74.14.6442-6447.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feldman S A, Hendry R M, Beeler J A. Identification of a linear heparin binding domain for human respiratory syncytial virus attachment glycoprotein G. J Virol. 1999;73:6610–6617. doi: 10.1128/jvi.73.8.6610-6617.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hallak L K, Collins P L, Knudson W, Peeples M E. Iduronic acid-containing glycosaminoglycans on target cells are required for efficient respiratory syncytial virus infection. Virology. 2000;271:264–275. doi: 10.1006/viro.2000.0293. [DOI] [PubMed] [Google Scholar]
- 13.Hallak L K, Spillmann D, Collins P L, Peeples M E. Glycosaminoglycan sulfation requirements for respiratory syncytial virus infection. J Virol. 2000;74:10508–10513. doi: 10.1128/jvi.74.22.10508-10513.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He B, Leser G P, Paterson R G, Lamb R A. The paramyxovirus SV5 small hydrophobic (SH) protein is not essential for virus growth in tissue culture cells. Virology. 1998;250:30–40. doi: 10.1006/viro.1998.9354. [DOI] [PubMed] [Google Scholar]
- 15.Heminway B R, Yu Y, Tanaka Y, Perrine K G, Gustafson E, Bernstein J M, Galinski M S. Analysis of respiratory syncytial virus F, G, and SH proteins in cell fusion. Virology. 1994;200:801–805. doi: 10.1006/viro.1994.1245. [DOI] [PubMed] [Google Scholar]
- 16.Horvath C M, Paterson R G, Shaughnessy M A, Wood R, Lamb R A. Biological activity of paramyxovirus fusion proteins: factors influencing formation of syncytia. J Virol. 1992;66:4564–4569. doi: 10.1128/jvi.66.7.4564-4569.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu X L, Ray R, Compans R W. Functional interactions between the fusion protein and hemagglutinin-neuraminidase of human parainfluenza viruses. J Virol. 1992;66:1528–1534. doi: 10.1128/jvi.66.3.1528-1534.1992. . (Erratum, 66:5176.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang A S, Wagner R R. Penetration of herpes simplex virus into human epidermoid cells. Proc Soc Exp Biol Med. 1964;116:863–869. doi: 10.3181/00379727-116-29392. [DOI] [PubMed] [Google Scholar]
- 19.Jin H, Zhou H, Cheng X, Tang R, Munoz M, Nguyen N. Recombinant respiratory syncytial viruses with deletions in the NS1, NS2, SH, and M2–2 genes are attenuated in vitro and in vivo. Virology. 2000;273:210–218. doi: 10.1006/viro.2000.0393. [DOI] [PubMed] [Google Scholar]
- 20.Kahn J S, Schnell M J, Buonocore L, Rose J K. Recombinant vesicular stomatitis virus expressing respiratory syncytial virus (RSV) glycoproteins: RSV fusion protein can mediate infection and cell fusion. Virology. 1999;254:81–91. doi: 10.1006/viro.1998.9535. [DOI] [PubMed] [Google Scholar]
- 21.Karron R A, Buonagurio D A, Georgiu A F, Whitehead S S, Adamus J E, Clements-Mann M L, Harris D O, Randolph V B, Udem S A, Murphy B R, Sidhu M S. Respiratory syncytial virus (RSV) SH and G proteins are not essential for viral replication in vitro: clinical evaluation and molecular characterization of a cold-passaged, attenuated RSV subgroup B mutant. Proc Natl Acad Sci USA. 1997;94:13961–13966. doi: 10.1073/pnas.94.25.13961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krusat T, Streckert H J. Heparin-dependent attachment of respiratory syncytial virus (RSV) to host cells. Arch Virol. 1997;142:1247–1254. doi: 10.1007/s007050050156. [DOI] [PubMed] [Google Scholar]
- 23.Levine S. Polypeptides of respiratory syncytial virus. J Virol. 1977;21:427–31. doi: 10.1128/jvi.21.1.427-431.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Levine S, Hamilton R. Kinetics of the respiratory syncytial virus growth cycle in HeLa cells. Arch Gesamte Virusforsch. 1969;28:122–132. doi: 10.1007/BF01249378. [DOI] [PubMed] [Google Scholar]
- 25.Levine S, Klaiber-Franco R, Paradiso P R. Demonstration that glycoprotein G is the attachment protein of respiratory syncytial virus. J Gen Virol. 1987;68:2521–2524. doi: 10.1099/0022-1317-68-9-2521. [DOI] [PubMed] [Google Scholar]
- 26.Leyrer S, Bitzer M, Lauer U, Kramer J, Neubert W J, Sedlmeier R. Sendai virus-like particles devoid of haemagglutinin-neuraminidase protein infect cells via the human asialoglycoprotein receptor. J Gen Virol. 1998;79:683–687. doi: 10.1099/0022-1317-79-4-683. [DOI] [PubMed] [Google Scholar]
- 27.Markwell M A, Portner A, Schwartz A L. An alternative route of infection for viruses: entry by means of the asialoglycoprotein receptor of a Sendai virus mutant lacking its attachment protein. Proc Natl Acad Sci USA. 1985;82:978–982. doi: 10.1073/pnas.82.4.978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marquez A, Hsiung G D. Influence of glutamine on multiplication and cytopathic effect of respiratory syncytial virus. Proc Soc Exp Biol Med. 1967;124:95–99. doi: 10.3181/00379727-124-31674. [DOI] [PubMed] [Google Scholar]
- 29.McClain D S, Fuller A O. Cell-specific kinetics and efficiency of herpes simplex virus type 1 entry are determined by two distinct phases of attachment. Virology. 1994;198:690–702. doi: 10.1006/viro.1994.1081. [DOI] [PubMed] [Google Scholar]
- 30.Nussbaum O, Broder C C, Berger E A. Fusogenic mechanisms of enveloped-virus glycoproteins analyzed by a novel recombinant vaccinia virus-based assay quantitating cell fusion-dependent reporter gene activation. J Virol. 1994;68:5411–5422. doi: 10.1128/jvi.68.9.5411-5422.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pastey M K, Samal S K. Analysis of bovine respiratory syncytial virus envelope glycoproteins in cell fusion. J Gen Virol. 1997;78:1885–1889. doi: 10.1099/0022-1317-78-8-1885. [DOI] [PubMed] [Google Scholar]
- 32.Pizzato M, Marlow S A, Blair E D, Takeuchi Y. Initial binding of murine leukemia virus particles to cells does not require specific Env-receptor interaction. J Virol. 1999;73:8599–8611. doi: 10.1128/jvi.73.10.8599-8611.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Portner A, Scroggs R A, Marx P S, Kingsbury D W. A temperature-sensitive mutant of Sendai virus with an altered hemagglutinin-neuraminidase polypeptide: consequences for virus assembly and cytopathology. Virology. 1975;67:179–187. doi: 10.1016/0042-6822(75)90415-8. [DOI] [PubMed] [Google Scholar]
- 34.Schmitt A P, He B, Lamb R A. Involvement of the cytoplasmic domain of the hemagglutinin-neuraminidase protein in assembly of the paramyxovirus simian virus 5. J Virol. 1999;73:8703–8712. doi: 10.1128/jvi.73.10.8703-8712.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shahrabadi M S, Lee P W. Calcium requirement for syncytium formation in HEp-2 cells by respiratory syncytial virus. J Clin Microbiol. 1988;26:139–141. doi: 10.1128/jcm.26.1.139-141.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stricker R, Roux L. The major glycoprotein of Sendai virus is dispensable for efficient virus particle budding. J Gen Virol. 1991;72:1703–1707. doi: 10.1099/0022-1317-72-7-1703. [DOI] [PubMed] [Google Scholar]
- 37.Takeuchi K, Tanabayashi K, Hishiyama M, Yamada A. The mumps virus SH protein is a membrane protein and not essential for virus growth. Virology. 1996;225:156–162. doi: 10.1006/viro.1996.0583. [DOI] [PubMed] [Google Scholar]
- 38.Teng M N, Collins P L. Identification of the respiratory syncytial virus proteins required for formation and passage of helper-dependent infectious particles. J Virol. 1998;72:5707–5716. doi: 10.1128/jvi.72.7.5707-5716.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Teng M N, Collins P L. Studies on RSV biology using mutant recombinant viruses. Vancouver, B.C.: American Society for Virology; 1998. p. 134. [Google Scholar]
- 40.Verma R S, Giannola D, Shlomchik W, Emerson S G. Increased efficiency of liposome-mediated transfection by volume reduction and centrifugation. BioTechniques. 1998;25:46–49. doi: 10.2144/98251bm09. [DOI] [PubMed] [Google Scholar]
- 41.Walsh E E, Hruska J. Monoclonal antibodies to respiratory syncytial virus proteins: identification of the fusion protein. J Virol. 1983;47:171–177. doi: 10.1128/jvi.47.1.171-177.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Whitehead S S, Bukreyev A, Teng M N, Firestone C Y, St. Claire M, Elkins W R, Collins P L, Murphy B R. Recombinant respiratory syncytial virus bearing a deletion of either the NS2 or SH gene is attenuated in chimpanzees. J Virol. 1999;73:3438–3442. doi: 10.1128/jvi.73.4.3438-3442.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wyatt L S, Moss B, Rozenblatt S. Replication-deficient vaccinia virus encoding bacteriophage T7 RNA polymerase for transient gene expression in mammalian cells. Virology. 1995;210:202–205. doi: 10.1006/viro.1995.1332. [DOI] [PubMed] [Google Scholar]