Abstract
The intracellular trafficking and subsequent incorporation of Gag-Pol into human immunodeficiency virus type 1 (HIV-1) remains poorly defined. Gag-Pol is encoded by the same mRNA as Gag and is generated by ribosomal frameshifting. The multimerization of Gag and Gag-Pol is an essential step in the formation of infectious viral particles. In this study, we examined whether the interaction between Gag and Gag-Pol is initiated during protein translation in order to facilitate the trafficking and subsequent packaging of Gag-Pol into the virion. A conditional cotransfection system was developed in which virion formation required the coexpression of two HIV-1-based plasmids, one that produces both Gag and Gag-Pol and one that only produces Gag-Pol. The Gag-Pol proteins were either immunotagged with a His epitope or functionally tagged with a mutation (K65R) in reverse transcriptase that is associated with drug resistance. Gag-Pol packaging was assessed to determine whether the Gag-Pol incorporated into the virion was preferentially packaged from the plasmid that expressed both Gag and Gag-Pol or whether it could be packaged from either plasmid. Our data show that translation of Gag and Gag-Pol from the same mRNA is not critical for virion packaging of the Gag-Pol polyprotein or for viral function.
The gag and pol genes of human immunodeficiency virus type 1 (HIV-1) are initially translated as polyproteins, which are later cleaved by the viral protease to produce mature virions. Gag is expressed as a 55-kDa precursor (Pr55Gag), and Pol is expressed in the form of a 160-kDa Gag-Pol fusion protein (Pr160Gag-Pol). These precursor polyproteins are encoded by the same mRNA but are not synthesized at the same rate. The pol gene lacks its own initiation codon and is in the −1 reading frame with respect to gag. The Gag-Pol fusion protein is synthesized as a result of an infrequent ribosomal frameshifting event, wherein the ribosomes translating gag slip back one nucleotide into the pol reading frame (20). Ribosomal frameshifting generates 20 times as much Gag as Gag-Pol, supporting the greater requirement for structural proteins over enzymatic proteins during viral assembly. Maintenance of the 20:1 ratio of Gag to Gag-Pol is critical for viral infectivity and RNA dimerization (29).
Several lines of indirect evidence suggest that the Gag and Gag-Pol precursor proteins and the viral genomic RNA form an assembly complex in the cytoplasm that is transported as a unit to the cell membrane for viral assembly and budding. Gag and Gag-Pol are translated from the unspliced viral genomic RNA on free polyribosomes in the cell cytoplasm, creating a high local concentration of nascent Gag and Gag-Pol molecules alongside the viral RNA. The finding that the assembly of HIV-1 and other retroviruses is facilitated by nucleic acids (4, 6, 7, 10, 26), combined with the nucleic acid binding properties of the nucleocapsid (NC) region of Gag (for a review, see reference 13), indicates an important role for interactions between Gag and viral genomic RNA in the assembly process. Although retroviral assembly can occur independently of genomic RNA packaging (as shown by HIV-1 packaging signal [Psi] deletion mutants that generate empty particles [for a review, see reference 1]), in these cases assembly may have been promoted by the host cell's nucleic acids (10).
It is generally believed that Gag-Pol is incorporated into the virion via interactions with Gag (19, 31, 33), and there is evidence to suggest that the multimerization of Gag and Gag-Pol occurs prior to their association with the cell membrane. For HIV-1 assembly to occur, Gag must undergo myristoylation (14), a step that is critical for the transport of Gag to the plasma membrane. However, both nonmyristoylated Gag and nonmyristoylated Gag-Pol can be packaged into progeny virions when coexpressed with myristoylated Gag (9, 28, 30). Velocity sedimentation analysis of Gag-only particles has shown that both myristoylated and nonmyristoylated Gag proteins can assemble into multimeric assembly intermediates, but myristoylation is required to complete virus-like particle formation (24, 27). Furthermore, Lee et al. (22, 23) have reported the formation of Gag and Gag-Pol precursor complexes in the cytoplasm of HIV-1-infected CD4+ T cells.
Experiments using Moloney murine leukemia virus have shown that the Gag portion of Gag-Pol is not required for the packaging of Pol proteins. However, a lower level of reverse transcriptase (RT) activity was observed in the resulting virions than in wild-type virions (3), suggesting a deficient packaging of Pol. In HIV-1, the provision of Gag in trans to Gag-Pol results in the formation of virus-like particles containing both Gag and Gag-Pol (28, 30). Taken together, these investigations show that the packaging of Gag-Pol does not require Gag and Gag-Pol to be synthesized from the same mRNA. However, these studies do not establish if the multimerization of Gag and Gag-Pol preferentially occurs during protein translation as a means of facilitating the transport and subsequent packaging of Gag-Pol into progeny virions. In this study, we have assessed whether the incorporation of Gag-Pol that is supplied in trans is as efficient as the incorporation of Gag-Pol that is synthesized from the same mRNA as Gag. Using a conditional cotransfection system designed to distinguish between packaged Gag-Pol proteins expressed either from the same construct that expresses Gag or from a separate construct, we observed that Gag and Gag-Pol multimerization during translation from a single mRNA molecule is not critical for virion packaging of the Gag-Pol polyprotein. Furthermore, we found that an interaction between Gag and Gag-Pol proteins generated from the same mRNA is not necessary to generate functional virions.
MATERIALS AND METHODS
Construction of DNA plasmids.
The HIV-1 DNA constructs used in this study were derived from either the full-length wild-type HIV-1 plasmid HXB2-BH10 (32) or the luciferase reporter plasmid pNL4-3.Luc.R- E- (12). pNL4-3.Luc.R- E- was obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, National Institutes of Health, from N. Landau. For all experiments we used two fundamental HIV constructs, one that expresses both Gag and Gag-Pol but with a functional deletion in rev (G:GP), and one that expresses only Gag-Pol due to a frameshift mutation that allows continuous expression of Gag-Pol and bypasses the Gag termination codon (GP). G:GP was created by deleting the majority of the second half of exon 2 of rev in HXB2-BH10 as previously described (29). GP was constructed in HXB2-BH10 as previously described (29) to alter the heptanucleotide sequence that is responsible for −1 ribosomal frameshifting, by direct base changes and the addition of an extra nucleotide (5′-TTTTTTA-3′→5′-CTTCCTCA-3′).
The histidine (His) epitope tag was introduced into the 3′ end of integrase (IN) in the HXB2-BH10-based G:GP and GP plasmids. An MluI site was introduced into IN to replace the existing BspMI site located near the 3′ end of IN (5′-TGGCAG-3′→5′-ACGCGT-3′), using PCR stitch mutagenesis, a technique that introduces mutations via the PCR primers as previously described (16). Subsequent digestion with MluI and ligation with a double-stranded DNA adapter complementary to the MluI cleavage site and containing the His epitope (5′-CAT CAC CAT CAC CAT CAC-3′) generated the G:GPHis and GPHis plasmids. The K65R mutation is a lysine-to-arginine change at amino acid 65 in HIV-1 RT that is associated with resistance to the antiretroviral drug (−)-β-2′,3′-dideoxy-3′-thiacytidine (3TC, lamivudine) (15). This mutation (AAA→AGA) was introduced into the HXB2-BH10-based G:GP and the GP plasmids by PCR stitch mutagenesis to generate G:GPK65R and GPK65R. A functional deletion in rev was introduced into pNL4-3-Luc.R- E- by digestion with BamHI, which is located in the second exon of rev, followed by S1 nuclease treatment according to the manufacturer's instructions (Boehringer Mannheim) to remove 25 bp and generate Luc.Rev-. The frameshift mutation and/or the K65R mutation were introduced by subcloning from the G:GP, G:GPK65R, GP, and GPK65R into Luc.Rev- or pNL4-3-Luc.R- E-, generating Luc.G:GP, Luc.G:GPK65R, Luc.GP, and Luc.GPK65R. All PCRs were conducted using the high-fidelity DNA polymerase enzyme Pfx as specified by the manufacturer (Gibco BRL). All constructs were sequenced to confirm the presence of the desired mutations.
Cotransfection and virus production.
The G:GP and GP plasmids were cotransfected at a ratio of 20 μg of G:GP to 1 μg of GP in order to generate equal levels of Gag-Pol proteins derived from all constructs. The Luc.G:GP and Luc.GP plasmids were also cotransfected at a ratio of 20 to 1, and 5 μg of pVSV-G (a kind gift from J. C. Burns, University of California, San Diego) was included to provide a viral envelope that would enable infection (5). The calcium phosphate coprecipitation method was used for the transient transfection of 293T cells. Supernatants were collected at 36 h posttransfection and centrifuged for 30 min at 3,000 rpm (Beckman model GS-6) to remove cellular debris. Virions were purified and concentrated by ultracentrifugation (Beckman model L-90, SW41 rotor) at 35,000 rpm for 1 h at 4°C through a 20% sucrose cushion. For protein isolation, viral pellets were resuspended in 50 μl of 2× Tris-buffered saline lysis buffer containing 1% Nonidet P-40, 20 mM phenylmethylsulfonyl fluoride, 1 μM pepstatin, and 1 μM leupeptin. For use in the reverse transcription assay and the natural endogenous reverse transcription (NERT) assay, viral pellets were resuspended in Tris-EDTA. Virus production from transfections was quantified by measuring p24 antigen levels in the pelleted virion stocks and the cell culture supernatants (HIV-1 p24 assay; Abbott Laboratories).
Western blot analysis.
Protein samples were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (10% gel) and transferred to a nitrocellulose membrane (Hybond C; Amersham Pharmacia Biotech). Western blot analysis was carried out as previously described (29), using an α-RT antibody (NEN) to detect p66 RT, an α-His antibody (Qiagen) to detect the His-tagged Gag-Pol polyproteins, and pooled sera from HIV-infected patients to detect total HIV-1 viral proteins. An ECL (enhanced chemiluminescence) detection kit (Amersham Pharmacia Biotech) was used to visualize the proteins for assessment by laser densitometry.
Cell-free RT assay.
The cell-free RT assay was adapted from a method described by Boyer et al. (2). For each sample, 0.2 μl of a 20-μg/ml stock of M13-47 sequencing primer (New England Biolabs) was annealed to 0.25 μg of single-stranded M13mp18 DNA (New England Biolabs) by heating to 95°C for 5 min and then slowly cooling to room temperature. In a final volume of 60 μl, each sample contained 10 μl of Nonidet P-40, pelleted virus (5 ng of p24), 25 mM Tris-Cl (pH 8.0), 75 mM KCl, 8.0 mM MgCl2, 2 mM dithiothreitol, 100 μg of bovine serum albumin per ml, 10 mM CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate}, 10 μM each dATP, dGTP, and dCTP, 5 μM dTTP, 0.5 μCi of [α-33P]dTTP (NEN), and 3TC-triphosphate (Moravek Biochemicals), which was routinely used at a concentration of 2.5, 5, or 15 μM. The samples were incubated for 1 h at 37°C, and then 8 μl of each reaction was spotted onto DE81 filter paper, which was washed six times in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate and then analyzed on a microbeta counter (Wallac).
NERT assay.
The NERT assay was carried out as previously described (17). Briefly, sucrose-purified virus (typically 100 pg of p24) was supplemented with MgCl2 and DNase I and incubated at 37°C for 90 min in the presence and absence of deoxynucleoside triphosphates. Reverse transcription was terminated by the addition of EDTA and proteinase K followed by boiling for 10 min. Stopped reactions were assayed for negative-strand strong-stop DNA by PCR. Standard curves were generated by amplifying serial dilutions of an HIV-1 proviral plasmid, and all PCRs were performed in the linear range of the assay.
Single-round infection analyses with the pNL4-3-Luc.R- E- constructs.
Cleared viral supernatants containing 10 ng of p24 were adjusted to 200 μl with culture medium and incubated with 106 MT-2 cells for 2 h. The virus was then removed by washing the infected cells once with phosphate-buffered saline without calcium chloride and magnesium chloride and resuspending them in 1 ml of culture medium. The luciferase activity in the cells was determined 48 h postinfection using a luciferase assay kit (Promega). In experiments conducted in the presence of 3TC (obtained from the AIDS Research and Reference Reagent Program, Division of AIDS, National Institutes of Health), the MT-2 cells were incubated with 50 nM 3TC for 4 h prior to infection, 2 h during infection, and 48 h postinfection. The mean 50% inhibitory concentration of 3TC, which was shown to inhibit several different strains of HIV-1 in primary cells, ranges between 2.5 nM and 0.67 μM (11). Trypan blue staining was used to monitor cytotoxic effects of 3TC, and no differences were observed between control and 3TC-treated cultures.
RESULTS AND DISCUSSION
G:GP and GP were cotransfected to determine whether the interaction between Gag and Gag-Pol is initiated during protein translation. If Gag and Gag-Pol interact during their synthesis from a single mRNA molecule to facilitate Gag-Pol packaging, the progeny virions would be expected to contain primarily Gag-Pol from the plasmid synthesizing both Gag and Gag-Pol. Conversely, the progeny virions will contain similar levels of Gag-Pol proteins from both plasmids if the Gag/Gag-Pol interaction can occur efficiently when Gag-Pol is supplied in trans.
Development of a conditional cotransfection system.
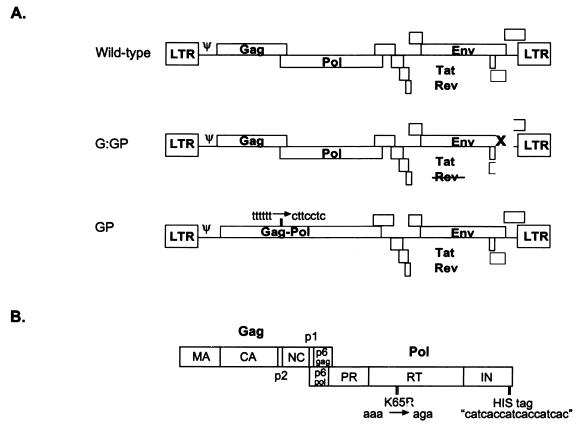
The conditional cotransfection system allows viral particle formation only when both cotransfected plasmids are simultaneously expressed in the same cell. This system takes advantage of the fact that both Gag and Rev are required for viral particle formation. As Rev is required for the transport of genomic RNA from the nucleus to the cytoplasm (for a review, see reference 18), transfection of a Rev-deficient HIV-1 plasmid (G:GP) alone results in viral mRNA being trapped in the nucleus. Similarly, viral particles will not be formed when GP is transfected alone, as Gag is required for virion production. Accordingly, when either G:GP or GP was transfected into 293T cells, no pelletable HIV-1 proteins were detected (data not shown). As the natural ratio of Gag to Gag-Pol synthesis is 20:1, the two plasmids were cotransfected at a ratio of 20 G:GP to 1 GP (based on DNA concentration) so that equivalent amounts of Gag-Pol proteins would be derived from all constructs. This enables a direct comparison of packaged Gag-Pol synthesized from either G:GP or GP. The HIV-1 constructs used in these analyses are depicted in Fig. 1A.
FIG. 1.
(A) The HIV-based constructs used for all cotransfections. (B) Schematic representation of the Gag and Gag-Pol polyproteins showing the major domains and the sites of labeling with either the His epitope or the K65R mutation. LTR, long terminal repeat; MA, matrix; CA, capsid; NC, nucleocapsid; PR, protease.
The Gag-Pol proteins were tagged with a His epitope that was inserted into the C-terminal region of IN (Fig. 1B) to allow semiquantitative assessment of Gag-Pol incorporation by Western blot analysis. Two types of viral particles, G:GPHis/GP and G:GP/GPHis, were obtained by cotransfection. If Gag and Gag-Pol interact during translation to facilitate Gag-Pol packaging, the G:GPHis/GP virion should incorporate greater levels of His-tagged Gag-Pol than G:GP/GPHis. As the His tag may alter Gag-Pol packaging by changing the conformation of the Gag-Pol protein, an alternative labeling system in which Gag-Pol proteins were functionally tagged with a K65R mutation in RT was also used (Fig. 1B). This mutation mediates resistance to 3TC, enabling wild-type Gag-Pol and K65R Gag-Pol to be differentiated in the presence of the drug. The K65R mutation was chosen because it maintains a drug-resistant phenotype when used in a cell-free RT assay (15). Four types of virus were produced through cotransfection: the drug-sensitive control G:GP/GP, the drug-resistant control G:GPK65R/GPK65R, and the two test cases G:GP/GPK65R and G:GPK65R/GP. If Gag and Gag-Pol interact during translation, Gag and Gag-Pol in progeny virions will be preferentially derived from the G:GP plasmid that expresses both proteins, so that one would expect G:GP/GPK65R to be drug sensitive and G:GPK65R/GP to be drug resistant. If Gag-Pol can be packaged with equivalent efficiency when translated with Gag or when translated alone, both types of virion should be similarly susceptible to 3TC-triphosphate.
Gag and Gag-Pol do not interact during translation from the same mRNA to facilitate Gag-Pol packaging.
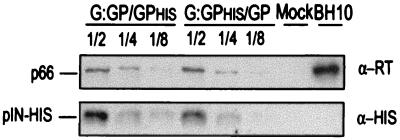
Epitope tagging of the Gag-Pol proteins was used to ascertain the amounts of packaged Gag-Pol derived from either the G:GP or the GP plasmid. G:GPHis/GP and G:GP/GPHis viral particles obtained by cotransfection of 293T cells were concentrated by ultracentrifugation and resuspended in protein lysis buffer. A protein dilution series of each virus was resolved by SDS-PAGE for Western blotting, and the resultant blots were hybridized successively with α-RT, α-His, and pooled sera from HIV-1-infected patients. RT levels provided a comparative measure of the total amount of HIV-1 Gag-Pol polyprotein in each virus stock, and the concentration of α-His reactive protein reflects the amount of Gag-Pol packaged from a particular plasmid (Fig. 2). For the representative experiment depicted in Fig. 2, the His signals (standardized to equivalent levels of RT) were 993, 421, and 309 (densitometry units) for the three dilutions of G:GP/GPHis and 906, 478, and 270 for the three dilutions of G:GPHis/GP, indicating similar efficiencies of Gag-Pol packaging from the two plasmids. Hybridization with the pooled patient sera indicated that there were no obvious differences in total HIV-1 protein expression levels or patterns between the two viruses (data not shown).
FIG. 2.
Western blot analysis of virion lysates produced from cotransfections in 293T cells to produce two virion types, G:GPHis/GP and G:GP/GPHis. Virion lysates were analyzed by SDS-PAGE with twofold dilutions of each lysate alongside the mock and the wild-type HXB2-BH10 control. RT (p66) is indicative of the overall level of Gag-Pol protein. His-tagged IN reflects the level of one particular Gag-Pol population in each virus type.
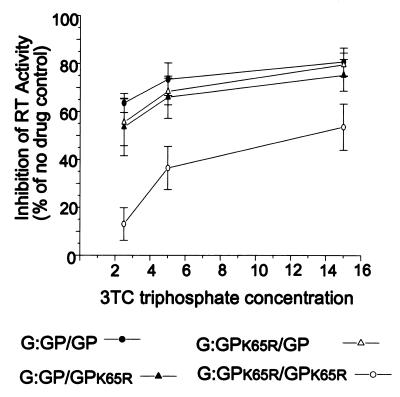
G:GP/GP, G:GPK65R/GPK65R, G:GP/GPK65R, and G:GPK65R/GP virions were also generated by cotransfection of 293T cells. The four virion samples were normalized on the basis of p24 levels, and a cell-free RT assay was used to determine RT activity in the presence and absence of 3TC-triphosphate, a competitive inhibitor of dCTP incorporation into the nascent DNA chain. Initially three concentrations (2.5, 5, and 15 μM) of 3TC-triphosphate were selected that gave clearly distinguishable differences between the drug-sensitive control virion G:GP/GP and the drug-resistant control virion G:GPK65R/GPK65R. Averaged data from three assays of virions produced from three separate transfections are shown in Fig. 3. The levels of 3TC-triphosphate inhibition of RT activity in the two test cases (G:GP/GPK65R and G:GPK65R/GP) were very similar to each other. While not readily distinguishable from the drug-susceptible control, the level of inhibition of RT activity for G:GP/GPK65R and G:GPK65R/GP was clearly distinct from that of the drug-resistant control. 3TC-triphosphate concentrations between 0.5 and 16 μM were used to address the possibility that lower drug concentrations may better determine a preference for Gag-Pol packaging in a population of both wild-type and drug-resistant Gag-Pol polyproteins. RT enzymes from G:GP/GPK65R and G:GPK65R/GP displayed very similar levels of 3TC-triphosphate susceptibility. A virus dilution series demonstrated that our results were not influenced by saturating concentrations of RT enzyme (data not shown). Overall, the RT assay results showed that both drug-sensitive and drug-resistant Gag-Pol proteins were packaged into both G:GP/GPK65R and G:GPK65R/GP.
FIG. 3.
Cell-free RT assay performed in the presence and absence of 3TC-triphosphate on four virion types: the drug-sensitive G:GP/GP and drug-resistant G:GPK65R/GPK65R controls and the two test cases, G:GP/GPK65R and G:GPK65R/GP. Virions were produced by co-transfection in 293T cells and were normalized on the basis of p24 levels. Inhibition of RT activity is presented relative to a no-drug control for three 3TC-triphosphate concentrations. The data shown are the averages of assays performed with virions produced from three separate transfections.
Consequently, no preference involving Gag-Pol encapsidation could be distinguished by either Western blot analysis with His-tagged Gag-Pol or RT assays with functionally labeled Gag-Pol. These results are in agreement with those of Park and Morrow (28) and Smith et al. (30), who demonstrated that Gag-Pol could be incorporated into the virion when supplied in trans to Gag. Moreover, our data show that the Gag-Pol polyprotein is efficiently packaged into the virion irrespective of being translated with Gag or translated alone.
Gag and Gag-Pol interaction during translation from the same mRNA is not required for endogenous reverse transcription or viral infectivity.
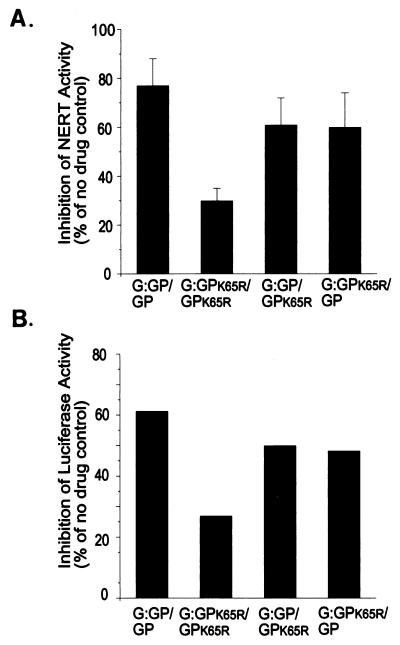
To determine whether or not an interaction between Gag and Gag-Pol at the time of translation is important for correct protein folding and the proper molecular arrangement of proteins within the virion, functional competence of the K65R labeled viruses was assessed by NERT and viral infectivity assays. The NERT assay measures the endogenous RT activity within intact virions, which is thought to reflect the viruses' ability to successfully initiate infection (35, 36). The NERT assays (Fig. 4A) show that the drug-sensitive (G:GP/GP) and the drug-resistant (G:GPK65R/GPK65R) controls are affected by 3TC-triphosphate as anticipated, with G:GPK65R/GPK65R being two- to threefold more resistant to 3TC-triphosphate than G:GP/GP (Fig. 4A). However, the two test cases, G:GP/GPK65R and G:GPK65R/GP, displayed similar levels of NERT activity; both were approximately 1.3-fold more resistant than the drug-sensitive control and 0.5-fold less resistant than the drug-resistant control (Fig. 4A). Viral infectivity was assessed from the infection of MT-2 cells in the presence and absence of 3TC via the luciferase reporter gene. The controls were clearly separable on the basis of drug response, with the Luc.G:GP/Luc.GP virions being 2- to 2.5-fold more susceptible to 3TC than the Luc.G:GPK65R/Luc.GPK65R virions. However, the two test samples, Luc.G:GP/Luc.GPK65R Luc.G:GPK65R/Luc.GP, consistently generated similar levels of luciferase activity, with both approximately 1.3-fold more resistant than the drug-sensitive control and 0.5-fold less resistant than the drug-resistant control (Fig. 4B).
FIG. 4.
(A) Inhibition of NERT activity in the presence of 10 μM 3TC-triphosphate, relative to no-drug controls, in the four virus types, G:GP/GP, G:GPK65R/GPK65R, G:GP/GPK65R, and G:GPK65R/GP, that were produced by cotransfection in 293T cells and normalized on the basis of p24 levels. The results obtained in duplicate assays performed with two independent virus stocks. Standard deviation from the mean is indicated. (B) Inhibition of luciferase activity by 3TC when the four virion types, Luc.G:GP/Luc.GP, Luc.G:GPK65R/Luc.GPK65R, Luc.G:GP/Luc.GPK65R, and Luc.G:GPK65R/Luc.GP, were produced by cotransfection in 293T cells, normalized on the basis of p24 levels, and used to infect MT-2 cells in the presence and absence of 50 nM 3TC. The results are presented relative to a no-drug control.
The results of both the NERT and the viral infectivity assays were consistent with the Gag-Pol packaging data described above, which established that Gag-Pol was efficiently packaged when supplied in trans. Moreover, the NERT and viral infectivity assays show that an interaction between Gag and Gag-Pol during translation is not essential for the function of Gag-Pol proteins in virions. The packaging of the retroviral enzymes in the form of a precursor protein is in itself suggestive of a role for Gag-Pol in coordinating the placement of the viral enzymes within the mature virion. Functional RT and IN proteins can be packaged when supplied in trans as Vpr fusion proteins to generate infectious virions (25, 34). However, Wu et al. (34) found that the levels of infection achieved when the proteins were supplied individually as Vpr-RT and Vpr-IN did not reach those of virions complemented by RT-IN supplied in fusion, Vpr-RT-IN, which in turn were not as infectious as wild-type virions. Thus, it remains likely that the expression of the Pol proteins in the form of a precursor is important both for the control of expression in required ratios and the coordinated arrangement of viral enzymes in the virion.
The precise steps and timing of virion assembly and maturation are not clearly understood. Together with the 1,500 or so Gag molecules present in an HIV-1 virion, 70 Gag-Pol proteins must also be incorporated. While our results indicate that the multimerization of Gag and Gag-Pol is not reliant on Gag-Pol being synthesized from the same mRNA as Gag, they do not, however, rule out the possibility that the Gag and Gag-Pol interaction occurs during translation between protein molecules on adjacent separate polysomes. Consequently, Gag-Pol may still be incorporated into a viral assembly complex formed in the context of the polysome. While Gag drives viral assembly and Gag alone will form virus-like particles, expression of Gag-Pol in the absence of Gag generates processed Gag and Pol proteins; however, no progeny virions are formed for study in systems equivalent to those of Gag-only particles (30). Kaye and Lever (21) have reported that Gag-Pol expressed alone in a T-cell line will produce pelletable Gag-Pol and negligible amounts of viral genomic RNA, but there is no further evidence to suggest that virus-like particles are formed in the absence of Gag. The difficulty involved in separating the functions of the shared regions of Gag and Gag-Pol provides a further obstacle in addressing the process of Gag-Pol assembly. The cotransfection and Gag-Pol labeling system described here enable independent examination of Gag-Pol packaging, as alterations to the Gag-Pol-only expression vector (GP) that affect Gag-Pol packaging can be monitored by changes in the Gag-Pol packaging profiles. This system could also be used to investigate Gag-Pol interactions not only with Gag but also with RNA and host cellular factors involved in viral assembly.
ACKNOWLEDGMENTS
We thank John Mills for critical review of the manuscript. We thank Paul Boyer and Stephen Hughes for helpful advice on developing the cell-free RT assay. We also thank Shahan Campbell, Miranda Shehu-Xhilaga, and Katherine Kedzierska for constructive advice.
This study was funded by grants from the National Health and Medical Research Council (NHMRC) and the Macfarlane Burnet Centre (MBC) Research Fund. Melissa Hill is a recipient of a Burnet Centenary postdoctoral fellowship. Johnson Mak is the recipient of a NHMRC Peter Doherty postdoctoral fellowship. Suzanne Crowe, Bill Hooker, and David Harrich are supported by the Australian National Centre in HIV Virology Research (NCHVR).
REFERENCES
- 1.Berkowitz R D, Fisher J, Goff S P. RNA packaging. In: Kräusslich H-G, editor. Morphogenesis and maturation of retroviruses. Berlin, Germany: Springer-Verlag; 1996. pp. 177–218. [Google Scholar]
- 2.Boyer P L, Tantillo C, Jacobo-Molina A, Nanni R G, Ding J, Arnold E, Hughes S H. Sensitivity of wild-type human immunodeficiency virus type 1 reverse transcriptase to dideoxynucleotides depends on template length; the sensitivity of drug-resistant mutants does not. Proc Natl Acad Sci USA. 1994;91:4882–4886. doi: 10.1073/pnas.91.11.4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buchschacher G L, Jr, Yu L, Murai F, Friedmann T, Miyanohara A. Association of murine leukemia virus Pol with virions, independent of Gag-Pol expression. J Virol. 1999;73:9632–9637. doi: 10.1128/jvi.73.11.9632-9637.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burniston M T, Cimarelli A, Colgan J, Curtis S P, Luban J. Human immunodeficiency virus type 1 Gag polyprotein multimerization requires the nucleocapsid domain and RNA and is promoted by the capsid-dimer interface and the basic region of matrix protein. J Virol. 1999;73:8527–8540. doi: 10.1128/jvi.73.10.8527-8540.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burns J C, Friedmann T, Driever W, Burrascano M, Yee J K. Vesicular stomatitis virus G glycoprotein pseudotyped retroviral vectors: concentration to very high titer and efficient gene transfer into mammalian and nonmammalian cells. Proc Natl Acad Sci USA. 1993;90:8033–8037. doi: 10.1073/pnas.90.17.8033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Campbell S, Rein A. In vitro assembly properties of human immunodeficiency virus type 1 Gag protein lacking the p6 domain. J Virol. 1999;73:2270–2279. doi: 10.1128/jvi.73.3.2270-2279.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Campbell S, Vogt V M. Self-assembly in vitro of purified CA-NC proteins from Rous sarcoma virus and human immunodeficiency virus type 1. J Virol. 1995;69:6487–6497. doi: 10.1128/jvi.69.10.6487-6497.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen Y L, Ts'ai P W, Yang C C, Wang C T. Generation of infectious virus particles by transient co-expression of human immunodeficiency virus type 1 Gag mutants. J Gen Virol. 1997;78:2497–2501. doi: 10.1099/0022-1317-78-10-2497. [DOI] [PubMed] [Google Scholar]
- 10.Cimarelli A, Sandin S, Hoglund S, Luban J. Basic residues in human immunodeficiency virus type 1 nucleocapsid promote virion assembly via interaction with RNA. J Virol. 2000;74:3046–3057. doi: 10.1128/jvi.74.7.3046-3057.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coates J A, Cammack N, Jenkinson H J, Jowett A J, Jowett M I, Pearson B A, Penn C R, Rouse P L, Viner K C, Cameron J M. (−)-2′-Deoxy-3′-thiacytidine is a potent, highly selective inhibitor of human immunodeficiency virus type 1 and type 2 replication in vitro. Antimicrob Agents Chemother. 1993;36:733–739. doi: 10.1128/aac.36.4.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Connor R I, Chen B K, Choe S, Landau N R. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology. 1995;206:935–944. doi: 10.1006/viro.1995.1016. [DOI] [PubMed] [Google Scholar]
- 13.Darlix J L, Lapadat-Tapolsky M, de Rocquigny H, Roques B P. First glimpses at structure-function relationships of the nucleocapsid protein of retroviruses. J Mol Biol. 1995;254:523–537. doi: 10.1006/jmbi.1995.0635. [DOI] [PubMed] [Google Scholar]
- 14.Gheysen D, Jacobs E, de Foresta F, Thiriart C, Francotte M, Thines D, de Wilde M. Assembly and release of HIV-1 precursor Pr55gag virus like particles from recombinant baculovirus-infected cells. Cell. 1989;59:103–112. doi: 10.1016/0092-8674(89)90873-8. [DOI] [PubMed] [Google Scholar]
- 15.Gu Z, Fletcher R S, Arts E J, Wainberg M A, Parniak M A. The K65R mutant reverse transcriptase of HIV-1 cross-resistant to 2′,3′-dideoxycytidine, 2′,3′-dideoxy-3′-thiacytidine, and 2′,3′-dideoxyinosine shows reduced sensitivity to specific dideoxynucleoside triphosphate inhibitors in vitro. J Biol Chem. 1994;269:28118–22812. [PubMed] [Google Scholar]
- 16.Higuchi R. Recombinant PCR. In: Innis M A, Gelfand D H, Sninsky J J, White T J, editors. PCR protocols, a guide to methods and applications. New York, N.Y: Academic Press, Inc.; 1990. pp. 177–183. [Google Scholar]
- 17.Hooker C W, Lott W B, Harrich D. Inhibitors of human immunodeficiency virus type 1 reverse transcriptase target distinct phases of early reverse transcription. J Virol. 2001;75:3095–3104. doi: 10.1128/JVI.75.7.3095-3104.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hope T J. The ins and outs of HIV Rev. Arch Biochem Biophys. 1999;365:186–191. doi: 10.1006/abbi.1999.1207. [DOI] [PubMed] [Google Scholar]
- 19.Huang M, Martin M A. Incorporation of Pr160gag-pol into virus particles requires the presence of both the major homology region and adjacent C-terminal capsid sequences within the Gag-Pol polyprotein. J Virol. 1997;71:4472–4478. doi: 10.1128/jvi.71.6.4472-4478.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jacks T, Power M D, Masiarz F R, Luciw P A, Barr P J, Varmus H E. Characterization of ribosomal frameshifting in HIV-1 Gag-Pol expression. Nature. 1988;331:280–283. doi: 10.1038/331280a0. [DOI] [PubMed] [Google Scholar]
- 21.Kaye J F, Lever A M. trans-acting proteins involved in RNA encapsidation and viral assembly in human immunodeficiency virus type 1. J Virol. 1996;70:880–886. doi: 10.1128/jvi.70.2.880-886.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee Y-M, Yu X-F. Identification and characterization of virus assembly intermediate complexes in HIV-1 infected CD4+ T-cells. Virology. 1998;243:78–93. doi: 10.1006/viro.1998.9064. [DOI] [PubMed] [Google Scholar]
- 23.Lee Y-M, Liu B, Yu X-F. Formation of virus assembly intermediate complexes in the cytoplasm by wild-type and assembly-defective mutant human immunodeficiency virus type 1 and their association with membranes. J Virol. 1999;73:5654–5662. doi: 10.1128/jvi.73.7.5654-5662.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lingappa J R, Hill R L, Wong M L, Hegde R S. A multistep, ATP-dependent pathway for assembly of human immunodeficiency virus capsids in a cell-free system. J Cell Biol. 1997;136:567–581. doi: 10.1083/jcb.136.3.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu H, Wu X, Xiao H, Conway J A, Kappes J C. Incorporation of functional human immunodeficiency virus type 1 integrase into virions independent of the Gag-Pol precursor protein. J Virol. 1997;71:7704–7710. doi: 10.1128/jvi.71.10.7704-7710.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morikawa Y, Goto T, Sano K. In vitro assembly of human immunodeficiency virus type 1 Gag protein. J Biol Chem. 1999;274:27997–28002. doi: 10.1074/jbc.274.39.27997. [DOI] [PubMed] [Google Scholar]
- 27.Morikawa Y, Hockley D J, Nermut M V, Jones I M. Roles of matrix, p2, and N-terminal myristylation in human immunodeficiency virus type 1 Gag assembly. J Virol. 2000;74:16–23. doi: 10.1128/jvi.74.1.16-23.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park J, Morrow C D. The nonmyristylated Pr160gag-pol polyprotein of human immunodeficiency virus type 1 interacts with Pr55gag and is incorporated into virus-like particles. J Virol. 1992;66:6304–6313. doi: 10.1128/jvi.66.11.6304-6313.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shehu-Xhilaga M, Crowe S M, Mak J. The maintenance of the Gag/Gag-Pol ratio is important for HIV-1 RNA dimerization and viral infectivity. J Virol. 2001;75:1834–1841. doi: 10.1128/JVI.75.4.1834-1841.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith A J, Srinivasakumar N, Hammarskjöld M L, Rekosh D. Requirements for incorporation of Pr160gag-pol from human immunodeficiency virus type 1 into virus-like particles. J Virol. 1993;67:2266–2275. doi: 10.1128/jvi.67.4.2266-2275.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Srinivasakumar N, Hammarskjöld M L, Rekosh D. Characterization of deletion mutations in the capsid region of human immunodeficiency virus type 1 that affect particle formation and Gag-Pol precursor incorporation. J Virol. 1995;69:6106–6114. doi: 10.1128/jvi.69.10.6106-6114.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Terwilliger E, Sodroski J G, Rosen C A, Haseltine W A. Effects of mutations within the 3′ orf open reading frame region of human T-cell lymphotropic virus type III (HTLV-III/LAV) on replication and cytopathogenicity. J Virol. 1986;60:754–760. doi: 10.1128/jvi.60.2.754-760.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wills J W, Craven R C. Form, function, and use of retroviral gag proteins. AIDS. 1991;5:639–654. doi: 10.1097/00002030-199106000-00002. [DOI] [PubMed] [Google Scholar]
- 34.Wu X, Liu H, Xiao H, Conway J A, Hunter E, Kappes J C. Functional RT and IN incorporated into HIV-1 particles independently of the Gag/Pol precursor protein. EMBO J. 1997;16:5113–5122. doi: 10.1093/emboj/16.16.5113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang H, Zhang Y, Spicer T P, Abbott L Z, Abbott M, Poiesz B J. Reverse transcription takes place within extracellular HIV-1 virions: potential biological significance. AIDS Res Hum Retroviruses. 1993;9:1287–1296. doi: 10.1089/aid.1993.9.1287. [DOI] [PubMed] [Google Scholar]
- 36.Zhang H, Dornadula G, Pomerantz R J. Endogenous reverse transcription of human immunodeficiency virus type 1 in physiological microenviroments: an important stage for viral infection of nondividing cells. J Virol. 1996;70:2809–2824. doi: 10.1128/jvi.70.5.2809-2824.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]