Abstract
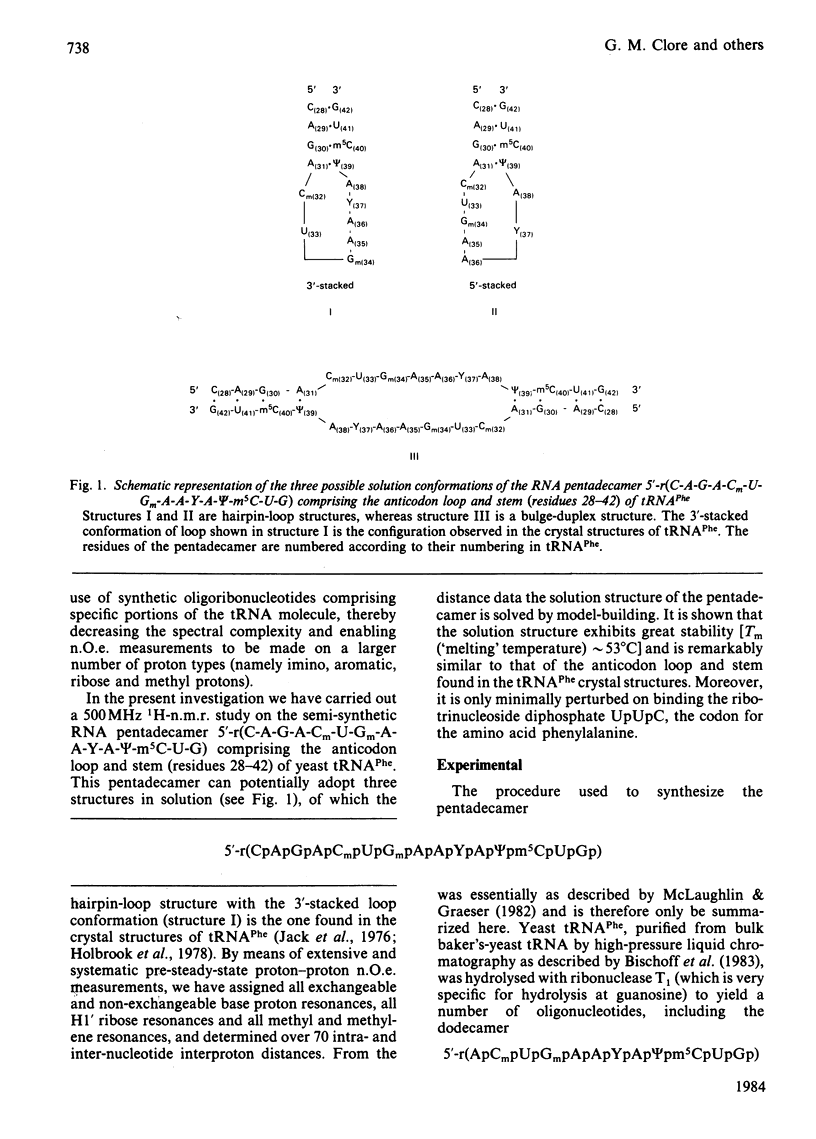
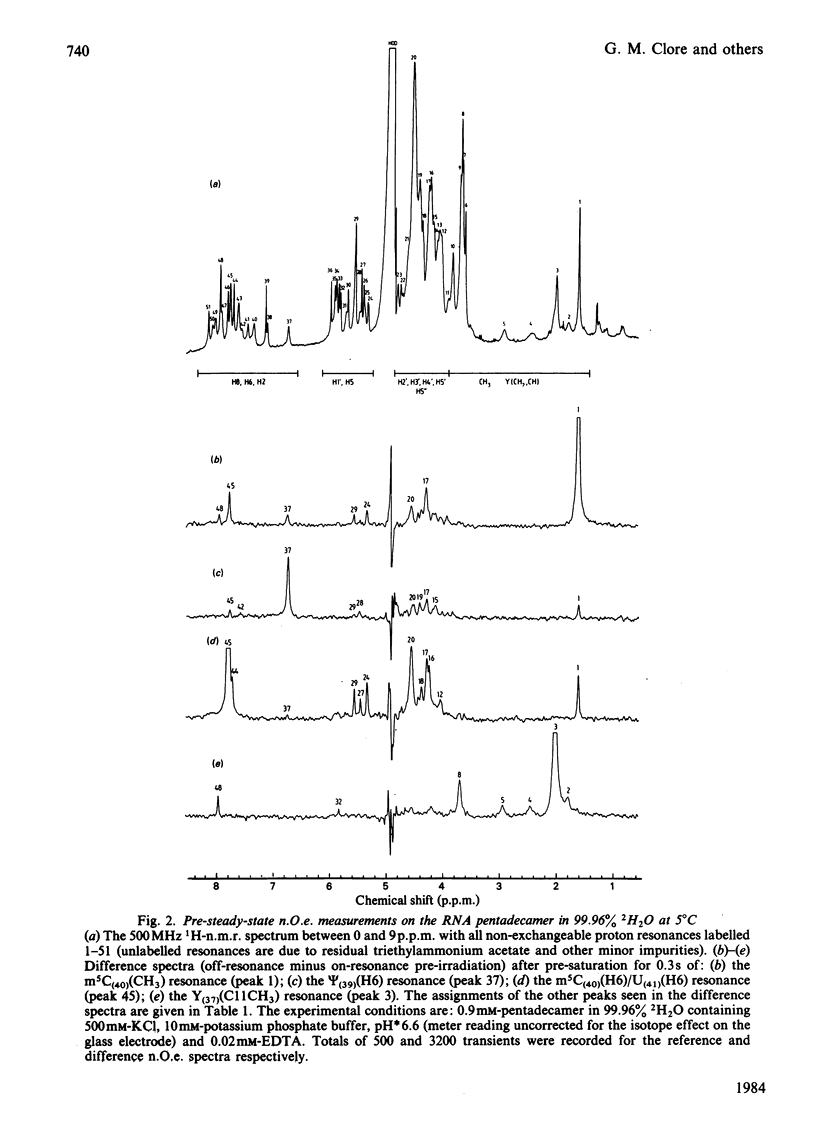
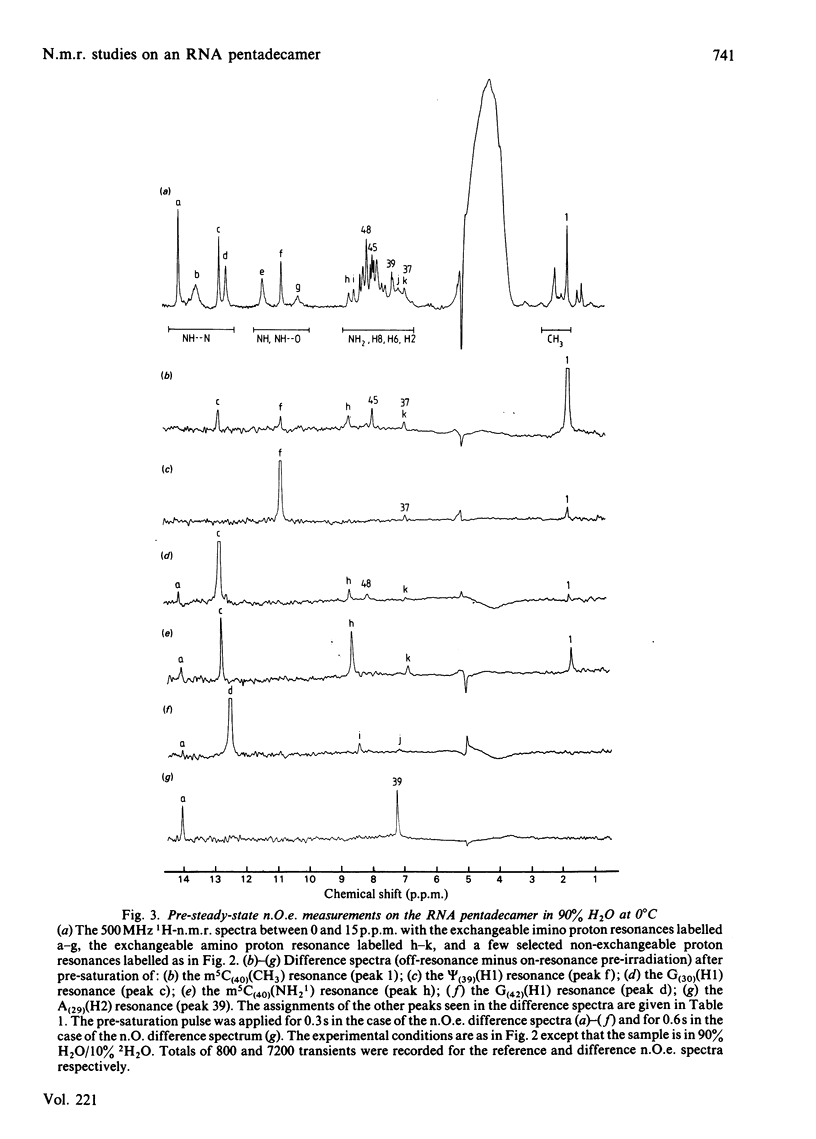
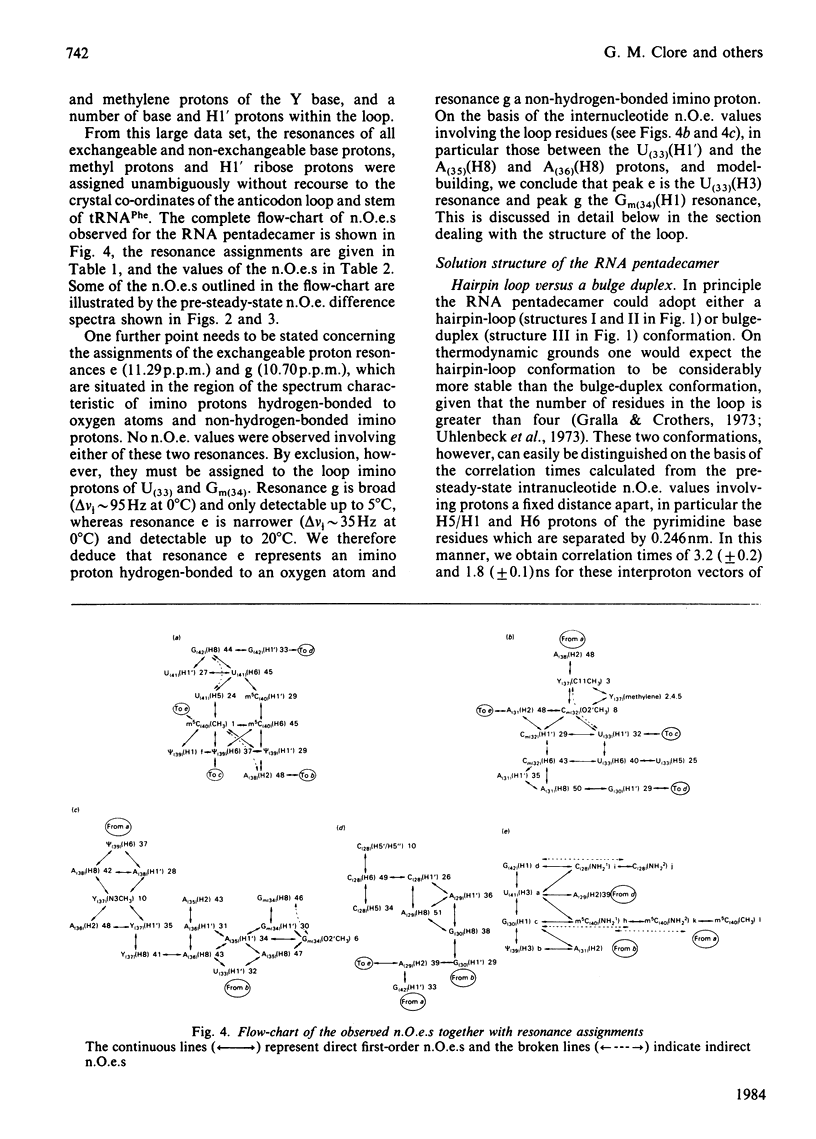
A 500 MHz 1H-n.m.r. study on the semi-synthetic RNA pentadecamer 5'-r(C-A-G-A-Cm-U-Gm-A-A-Y-A-psi-m5C-U-G) comprising the anticodon loop and stem (residues 28-42) of yeast tRNAPhe is presented. By using pre-steady-state nuclear-Overhauser-effect measurements all exchangeable and non-exchangeable base proton resonances, all H1' ribose resonances and all methyl proton resonances are assigned and over 70 intra- and inter-nucleotide interproton distances determined. From the distance data the solution structure of the pentadecamer is solved by model-building. It is shown that the pentadecamer adopts a hairpin-loop structure in solution with the loop in a 3'-stacked conformation. This structure is both qualitatively and quantitatively remarkably similar to that of the anticodon loop and stem found in the crystal structures of tRNAPhe with an overall root-mean-square difference of 0.12 nm between the interproton distances determined by n.m.r. and X-ray crystallography. The hairpin-loop solution structure of the pentadecamer is very stable with a 'melting' temperature of 53 degrees C in 500 mM-KCl, and the structural features responsible for this high stability are discussed. Interaction of the pentadecamer with the ribotrinucleoside diphosphate UpUpC, one of the codons for the amino acid phenylalanine, results only in minor perturbations in the structure of the pentadecamer, and the 3'-stacked conformation of the loop is preserved. The stability of the pentadecamer-UpUpC complex (K approximately 2.5 X 10(4) M-1 at 0 degrees C) is approximately an order of magnitude greater than that of the tRNAPhe-UpUpC complex.
Full text
PDF







Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arnott S., Hukins D. W., Dover S. D., Fuller W., Hodgson A. R. Structures of synthetic polynucleotides in the A-RNA and A'-RNA conformations: x-ray diffraction analyses of the molecular conformations of polyadenylic acid--polyuridylic acid and polyinosinic acid--polycytidylic acid. J Mol Biol. 1973 Dec 5;81(2):107–122. doi: 10.1016/0022-2836(73)90183-6. [DOI] [PubMed] [Google Scholar]
- Arnott S., Hukins D. W. Optimised parameters for A-DNA and B-DNA. Biochem Biophys Res Commun. 1972 Jun 28;47(6):1504–1509. doi: 10.1016/0006-291X(72)90243-4. [DOI] [PubMed] [Google Scholar]
- Balasubramaian R., Seetharamulu P. A conformational rationale for the wobble behaviour of the first base of the anticodon triplet in tRNA. J Theor Biol. 1983 Mar 7;101(1):77–86. doi: 10.1016/0022-5193(83)90273-4. [DOI] [PubMed] [Google Scholar]
- Bischoff R., Graeser E., McLaughlin L. W. tRNA separation by high-performance liquid chromatography using an aggregate of ODS-Hypersil and trioctylmethylammonium chloride. J Chromatogr. 1983 Mar 4;257(2):305–315. doi: 10.1016/s0021-9673(01)88186-3. [DOI] [PubMed] [Google Scholar]
- Clore G. M., Gronenborn A. M. A nuclear-Overhauser-enhancement study of the solution structure of a double-stranded DNA undecamer comprising a portion of the specific target site for the cyclic-AMP-receptor protein in the gal operon. Sequential resonance assignment. Eur J Biochem. 1984 May 15;141(1):119–129. doi: 10.1111/j.1432-1033.1984.tb08166.x. [DOI] [PubMed] [Google Scholar]
- Clore G. M., Gronenborn A. M., McLaughlin L. W. Structure of the ribotrinucleoside diphosphate codon UpUpC bound to tRNAPhe from yeast. A time-dependent transferred nuclear Overhauser enhancement study. J Mol Biol. 1984 Mar 25;174(1):163–173. doi: 10.1016/0022-2836(84)90370-x. [DOI] [PubMed] [Google Scholar]
- Clore G. M., Gronenborn A. M. Sequence-dependent structural variations in two right-handed alternating pyrimidine-purine DNA oligomers in solution determined by nuclear Overhauser enhancement measurements. EMBO J. 1983;2(12):2109–2115. doi: 10.1002/j.1460-2075.1983.tb01710.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crick F. H. Codon--anticodon pairing: the wobble hypothesis. J Mol Biol. 1966 Aug;19(2):548–555. doi: 10.1016/s0022-2836(66)80022-0. [DOI] [PubMed] [Google Scholar]
- Dickerson R. E., Drew H. R. Kinematic model for B-DNA. Proc Natl Acad Sci U S A. 1981 Dec;78(12):7318–7322. doi: 10.1073/pnas.78.12.7318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Early T. A., Kearns D. R., Hillen W., Wells R. D. A 300- and 600-MHz proton nuclear magnetic resonance investigation of a 12 base pair deoxyribonucleic acid restriction fragment: relaxation behavior of the low-field resonances in water. Biochemistry. 1981 Jun 23;20(13):3756–3764. doi: 10.1021/bi00516a014. [DOI] [PubMed] [Google Scholar]
- Ezra F. S., Lee C. H., Kondo N. S., Danyluk S. S., Sarma R. H. Conformational properties of purine-pyrimidine and pyrimidine-purine dinucleoside monophosphates. Biochemistry. 1977 May 3;16(9):1977–1987. doi: 10.1021/bi00628a035. [DOI] [PubMed] [Google Scholar]
- Gralla J., Crothers D. M. Free energy of imperfect nucleic acid helices. II. Small hairpin loops. J Mol Biol. 1973 Feb 5;73(4):497–511. doi: 10.1016/0022-2836(73)90096-x. [DOI] [PubMed] [Google Scholar]
- Gronenborn A. M., Clore G. M., Kimber B. J. An investigation into the solution structures of two self-complementary DNA oligomers, 5'-d(C-G-T-A-C-G) and 5'-d(A-C-G-C-G-C-G-T), by means of nuclear-Overhauser-enhancement measurements. Biochem J. 1984 Aug 1;221(3):723–736. doi: 10.1042/bj2210723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hare D. R., Reid B. R. Direct assignment of the dihydrouridine-helix imino proton resonances in transfer ribonucleic acid nuclear magnetic resonance spectra by means of the nuclear Overhauser effect. Biochemistry. 1982 Apr 13;21(8):1835–1842. doi: 10.1021/bi00537a020. [DOI] [PubMed] [Google Scholar]
- Heerschap A., Haasnoot C. A., Hilbers C. W. Nuclear magnetic resonance studies on yeast tRNAPhe. III. Assignments of the iminoproton resonances of the tertiary structure by means of nuclear Overhauser effect experiments at 500 MHz. Nucleic Acids Res. 1983 Jul 11;11(13):4501–4520. doi: 10.1093/nar/11.13.4501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hingerty B., Brown R. S., Jack A. Further refinement of the structure of yeast tRNAPhe. J Mol Biol. 1978 Sep 25;124(3):523–534. doi: 10.1016/0022-2836(78)90185-7. [DOI] [PubMed] [Google Scholar]
- Holbrook S. R., Sussman J. L., Warrant R. W., Kim S. H. Crystal structure of yeast phenylalanine transfer RNA. II. Structural features and functional implications. J Mol Biol. 1978 Aug 25;123(4):631–660. doi: 10.1016/0022-2836(78)90210-3. [DOI] [PubMed] [Google Scholar]
- Jack A., Ladner J. E., Klug A. Crystallographic refinement of yeast phenylalanine transfer RNA at 2-5A resolution. J Mol Biol. 1976 Dec 25;108(4):619–649. doi: 10.1016/s0022-2836(76)80109-x. [DOI] [PubMed] [Google Scholar]
- Kim S. H., Sussman J. L. pi turn is a conformational pattern in RNA loops and bends. Nature. 1976 Apr 15;260(5552):645–646. doi: 10.1038/260645a0. [DOI] [PubMed] [Google Scholar]
- Labuda D., Pörschke D. Multistep mechanism of codon recognition by transfer ribonucleic acid. Biochemistry. 1980 Aug 5;19(16):3799–3805. doi: 10.1021/bi00557a023. [DOI] [PubMed] [Google Scholar]
- Pardi A., Morden K. M., Patel D. J., Tinoco I., Jr Kinetics for exchange of imino protons in the d(C-G-C-G-A-A-T-T-C-G-C-G) double helix and in two similar helices that contain a G . T base pair, d(C-G-T-G-A-A-T-T-C-G-C-G), and an extra adenine, d(C-G-C-A-G-A-A-T-T-C-G-C-G). Biochemistry. 1982 Dec 7;21(25):6567–6574. doi: 10.1021/bi00268a038. [DOI] [PubMed] [Google Scholar]
- Pörschke D., Labuda D. Codon-induced transfer ribonucleic acid association: quantitative analysis by sedimentation equilibrium. Biochemistry. 1982 Jan 5;21(1):53–56. doi: 10.1021/bi00530a010. [DOI] [PubMed] [Google Scholar]
- Quigley G. J., Rich A. Structural domains of transfer RNA molecules. Science. 1976 Nov 19;194(4267):796–806. doi: 10.1126/science.790568. [DOI] [PubMed] [Google Scholar]
- Reid D. G., Salisbury S. A., Bellard S., Shakked Z., Williams D. H. Proton nuclear Overhauser effect study of the structure of a deoxyoligonucleotide duplex in aqueous solution. Biochemistry. 1983 Apr 12;22(8):2019–2025. doi: 10.1021/bi00277a044. [DOI] [PubMed] [Google Scholar]
- Roy S., Redfield A. G. Assignment of imino proton spectra of yeast phenylalanine transfer ribonucleic acid. Biochemistry. 1983 Mar 15;22(6):1386–1390. doi: 10.1021/bi00275a010. [DOI] [PubMed] [Google Scholar]
- Shakked Z., Rabinovich D., Kennard O., Cruse W. B., Salisbury S. A., Viswamitra M. A. Sequence-dependent conformation of an A-DNA double helix. The crystal structure of the octamer d(G-G-T-A-T-A-C-C). J Mol Biol. 1983 May 15;166(2):183–201. doi: 10.1016/s0022-2836(83)80005-9. [DOI] [PubMed] [Google Scholar]
- Sussman J. L., Holbrook S. R., Warrant R. W., Church G. M., Kim S. H. Crystal structure of yeast phenylalanine transfer RNA. I. Crystallographic refinement. J Mol Biol. 1978 Aug 25;123(4):607–630. doi: 10.1016/0022-2836(78)90209-7. [DOI] [PubMed] [Google Scholar]
- Uhlenbeck O. C., Borer P. N., Dengler B., Tinoco I., Jr Stability of RNA hairpin loops: A 6 -C m -U 6 . J Mol Biol. 1973 Feb 5;73(4):483–496. doi: 10.1016/0022-2836(73)90095-8. [DOI] [PubMed] [Google Scholar]


