Abstract
Background
Idiopathic Pulmonary Fibrosis (IPF) presents a severe respiratory challenge with a poor prognosis due to the lack of reliable biomarkers. Recent evidence suggests that Endoplasmic Reticulum Stress (ERS) may be associated with IPF pathogenesis. This study focuses on uncovering ERS-associated biomarkers for IPF.
Methods
Sequencing data from diverse datasets were analyzed, utilizing differential gene expression analysis and Weighted Gene Co-expression Network Analysis (WGCNA). Endoplasmic Reticulum Stress (ERS)-related genes were extracted from the GeneCards database. Hub genes were identified through Protein-Protein Interaction (PPI) analysis. Diagnostic and prognostic models were developed using machine learning algorithms and validated across both training and validation sets. Additionally, techniques such as Cell-type Identification by Estimating Relative Subsets of RNA Transcripts and single-cell RNA sequencing were employed to identify potential IPF-related cells. These findings were further investigated to elucidate their underlying mechanisms through in vitro experiments.
Results
Differentially expressed genes, WGCNA-identified blue module genes, and ERS-related genes extracted from the GeneCards database were intersected, and the resulting genes were used to construct diagnostic and prognostic models. Validation using multiple datasets indicated that both the diagnostic and prognostic models possess strong predictive capabilities. PPI analysis highlighted SPP1 as a potential hub gene in IPF. Moreover, M2 macrophages were found in higher quantities in the lung tissue of IPF patients, with a significant increase in SPP1-expressing M2 macrophages compared to the control group. In vitro experiments demonstrated that exogenous SPP1 inhibited the proliferation and migration of M2 macrophages and promoted apoptosis within a certain concentration range.
Conclusion
This study identifies ERS-related biomarkers in IPF, highlighting SPP1 and M2 macrophages. The resulting diagnostic and prognostic models offer strong predictive capabilities, unveiling new therapeutic avenues.
Keywords: Idiopathic pulmonary fibrosis, Endoplasmic Reticulum Stress, SPP1, M2 macrophage, machine-learning
Introduction
IPF is an enigmatic, chronic disease, marked by a relentless progression in interstitial lung scarring and a gradual decline in respiratory function [1]. This condition demonstrates a remarkable variability in its incidence and prevalence across the globe. In a detailed study, it was found that in Asia-Pacific countries, the adjusted incidence rate per 10,000 people lies between 0.35 and 1.30, while in Europe it ranges from 0.09 to 0.49, and in North America, it spans from 0.75 to 0.93. Correspondingly, the prevalence rates per 10,000 people in these regions are 0.57 to 4.51 in Asia-Pacific, 0.33 to 2.51 in Europe, and 2.40 to 2.98 in North America [2]. It is important to note that the IPF exhibits considerable variation across different countries and regions, a disparity influenced by a range of factors such as healthcare conditions and statistical methodologies. A systematic literature review has even pointed out that the differences between various studies can amount to several hundredfold [3].
IPF shares clinical features with other interstitial lung disease (ILD) but requires different treatment approaches, underscoring the importance of accurate diagnosis. High-resolution computed tomography (HRCT) is critical for diagnosing IPF. A typical HRCT pattern of usual interstitial pneumonia, with other ILD causes excluded, confirms IPF [4]. If HRCT is inconclusive but the diagnostic benefits outweigh biopsy risks, a lung biopsy may be necessary for clarification [4]. In some cases of IPF, patients encounter significant difficulties in obtaining an accurate diagnosis, and likewise, their prognosis often remains unfavorable. A study indicate that the median survival time from the diagnosis of IPF to death is merely 2 to 3 years [5]. The elevated mortality rate among individuals with IPF can largely be attributed to acute exacerbations of the condition. Statistical analyses reveal that approximately 46% of deaths related to IPF are precipitated by these sudden deteriorations [6]. Furthermore, a significant proportion of affected patients succumb within the initial month following an exacerbation, with the vast majority of the remaining cases resulting in mortality within the span of one year [6].
The Endoplasmic Reticulum (ER), a specialized organelle pivotal in eukaryotic cells, plays a central role in protein biosynthesis, folding, lipid biosynthesis, apoptosis, and calcium homeostasis [7]. The stability of the ER environment is essential for maintaining its physiological functions and ensuring overall cellular health. Disruptions to this stability can lead to endoplasmic reticulum stress (ERS), characterized by the accumulation of misfolded proteins within the ER. ERS is essentially a cellular mechanism to adapt to external environmental challenges, striving to reduce the buildup of unfolded proteins and restore normal cellular functions [7]. Nonetheless, persistent and excessive ERS can precipitate cellular dysfunction and apoptosis. Recent advancements in research have illuminated the role of ERS in influencing the progression of pulmonary fibrosis. It does so by affecting various cellular processes, including alveolar epithelial cell apoptosis, epithelial-mesenchymal transition, differentiation of myofibroblasts, and the polarization of M2 macrophages [8]. Consequently, ERS is increasingly recognized as a novel risk factor in the development of pulmonary fibrosis.
In this investigation, we aim to integrate transcriptome and single-cell sequencing data with sophisticated bioinformatics and machine learning algorithms to uncover potential pathogenic genes in IPF. By conducting cellular assays, we seek to elucidate the impacts of these genes on specific cell types. Identified within the framework of Predictive, Preventive, and Personalized Medicine, these novel biomarkers have the potential to significantly enhance IPF diagnostics and prognostics, presenting possible targets for targeted prevention and personalized treatment strategies. This approach is expected to enrich our understanding of IPF pathogenesis and facilitate the development of patient-specific therapeutic interventions, potentially reducing the clinical impact of this complex disease.
Materials and methods
Data processing
For this investigation, transcriptomic data from patients diagnosed with IPF and healthy controls were meticulously curated from the Gene Expression Omnibus (GEO) database. The selection criteria included: (1) a minimum sample size of 20 to ensure statistical robustness, (2) a clear clinical diagnosis of IPF, excluding other forms of interstitial lung diseases, and (3) samples originating from human lung tissues. Following the acquisition of raw datasets meeting these criteria, the ‘limma’ package [9] was used for normalization, with a log2 transformation applied to stabilize variance across expression levels. In instances where multiple probes corresponded to a single gene, average expression levels were computed to ensure data integrity. To mitigate batch effects inherent in integrating data from multiple sources, the combat function [10] in the ‘sva’ package was applied, minimizing inter-batch discrepancies. This detailed approach ensured a reliable dataset specifically tailored to exploring the molecular intricacies of IPF in human lung tissues.
Differentially expressed genes (DEGs)
Genes were designated as differentially expressed genes (DEGs) based on the criterion of an absolute log fold change (logFC) value of 1 and an adjusted P-value less than 0.05.
Weighted gene co-expression network analysis (WGCNA)
WGCNA [11] was applied to identify hub genes relevant to IPF. The process began with the identification and removal of missing values and outliers. A soft thresholding power was determined according to the scale-free topology criterion, essential for network construction. Hierarchical clustering was then used to identify gene modules, which were subsequently correlated with phenotype data. This allowed for the differentiation of modules related to IPF from those of healthy controls. The association of individual genes with IPF was quantified by calculating gene significance (GS) based on their correlation with the phenotype. Moreover, module membership (MM) was assessed by evaluating the correlation between the module eigengenes and gene expression patterns, thereby highlighting significant genes within key modules.
Protein-protein interaction (PPI) networks
Genes related to ERS were obtained from GeneCards: The Human Gene Database (https://www.genecards.org/) [12]. An intersection analysis was performed between these ERS-related genes, genes from WGCNA, and DEGs. The intersecting genes were subjected to PPI analysis using the STRING database (https://string-db.org/) [13]. Interaction networks were visualized using Cytoscape software (version 3.8.2). Degree centrality for each gene within the network was calculated, identifying those with the highest degree values as central hub genes pertinent to IPF.
Functional enrichment analysis
Disease Ontology (DO) [14], Gene Ontology (GO) [15], and Kyoto Encyclopedia of Genes and Genomes (KEGG) [16] analyses were conducted to explore the biological implications of identified genes. DO analysis revealed disease associations, offering insights into pathological significance. GO categorization elucidated gene roles across biological processes, cellular components, and molecular functions. KEGG pathway mapping highlighted participation in metabolic and signaling pathways. These integrated analyses provided a comprehensive understanding of the genes’ functional and disease-related contexts.
Immune infiltration analysis
Cell-type Identification By Estimating Relative Subsets Of RNA Transcripts (CIBERSORT) [17] employs linear support vector regression to deconvolute expression matrices, estimating the abundance of 22 distinct immune cell types. This method was applied to analyze the immune cell composition in lung tissues from patients with IPF and healthy controls, facilitating a detailed exploration of immune involvement in IPF.
Construction of the diagnostic model
A series of sophisticated machine learning algorithms were employed to construct a diagnostic model capable of distinguishing IPF patients from healthy controls. The model development utilized techniques such as Lasso with Stepwise Generalized Linear Models (Stepglm), Support Vector Machines (SVM), glmBoost, Ridge Regression, Elastic Net (Enet), Random Forests (RF), and their various combinations. Each algorithm underwent rigorous cross-validation to ensure robustness and accuracy.
Construction of the prognostic model
A comprehensive array of statistical and machine learning methodologies was employed to develop a prognostic model for assessing the outcomes of patients with IPF. The model integrated advanced techniques such as Survival Support Vector Machine (survival-SVM), CoxBoost, Ridge Regression, Lasso, Supervised Principal Components (SuperPC), Elastic Net (Enet), Partial Least Squares Cox regression (plsRcox), Random Survival Forests (RSF), and Stepwise Cox regression with various selection strategies. These methods were strategically combined to enhance the model’s accuracy and reliability.
Single cell sequencing analysis
The GSE122960 dataset [18], which includes single-cell sequencing data from patients with IPF and control groups, was utilized to explore potential pathogenic genes and their interactions within specific cell types associated with IPF. Initial quality control measures established thresholds for the minimum number of cells and features per cell, ensuring data integrity, while evaluating the expression of mitochondrial and ribosomal genes to identify low-quality cells. Following quality control, the LogNormalize method in Seurat [19] was applied for data normalization, and highly variable genes indicating substantial biological variation were identified. To integrate data from various samples and reduce batch effects, dimensionality reduction was conducted using the Harmony algorithm [20, 21]. Cell clustering analysis was performed utilizing the FindNeighbors and FindClusters functions with a range of resolution parameters. Clustree visualization was used to determine the optimal clustering resolution [22]. Specific marker genes for each cell cluster were then identified, and the distribution of cell clusters was visualized in t-distributed Stochastic Neighbor Embedding (t-SNE) low-dimensional space. The analysis concluded with manual annotation based on known marker genes to refine cell type identification. This comprehensive examination of the cellular and molecular landscape of IPF facilitated insights into the disease’s pathogenesis.
Cell culture
In this investigation, the RAW 264.7 macrophage cell line, purchased from the Cell Resource Center, Peking Union Medical College (Resource Number: 1101MOU-PUMC000146), was employed. Cultivation was conducted in DMEM-H supplemented with 10% fetal bovine serum (FBS). To generate M2 polarized macrophages, cells were treated with IL-4 (20 ng/mL) for 48 h, resulting in a predominantly round morphology [23]. Post-polarization, cells were maintained in serum-free RPMI-1640 for up to 72 h to preserve the M2 phenotype. This protocol facilitated the examination of M2 macrophage functions in our study, offering valuable insights into their role in immune responses.
Cell Counting Kit-8 (CKK8) assay
The Cell Counting Kit-8 (CCK-8) assay (Beyotime, Shanghai, China) was employed with rigorous adherence to protocol details to ensure a precise assessment of cell viability. The procedure commenced with essential preparatory steps, including the pre-warming of DPBS (Servicebio, Wuhan, China), high-glucose DMEM culture medium (Hyclone, Logan, UT, USA), and trypsin (Gibco, Grand Island, NY, USA), alongside preheating the cell culture incubator and sterilizing the workspace with UV light. Observations under an inverted microscope confirmed the cells were in an optimal state for trypsin-EDTA digestion, creating a single-cell suspension. Following centrifugation and resuspension in culture medium supplemented with fetal bovine serum (BI, Beit HaEmek, Israel), the cells were accurately counted and seeded into 96-well plates. The CCK-8 solution was carefully added to each well, avoiding bubble formation that could interfere with optical density (OD) measurements. After incubation to facilitate color development, the OD at 450 nm was measured, providing a quantitative evaluation of cell viability.
5-Ethynyl-2’- deoxyuridine (EDU)
The BeyoClick™ EdU-555 Cell Proliferation Assay Kit (Beyotime, Shanghai, China) was used to evaluate cell proliferation. Cells were seeded in 6-well plates, with cover slips added as needed. After overnight incubation, specific treatments were applied. A 2X EdU working solution was prepared by diluting the 10 mM EdU stock with culture medium at a 1:500 ratio to achieve a final concentration of 10 μM. The pre-warmed 2X EdU solution was then added to the wells, and cells were incubated for approximately 2 h. Following EdU incorporation, cells were fixed, washed, permeabilized, and washed again. The Click reaction mixture was applied, incubated in darkness for 30 min, and followed by additional washing. If necessary, cell nuclei were stained with Hoechst 33342. Fluorescence detection using microscopy or flow cytometry assessed cell proliferation based on the intensity of EdU incorporation, indicated by Azide 555 fluorescence.
Transwell assay
Cell migration was assessed using a transwell assay. Preparation began with warming DPBS (Servicebio, Wuhan, China), culture medium (Hyclone, Logan, UT, USA), and trypsin (Gibco, Grand Island, NY, USA) in a 37 °C incubator, followed by UV sterilization of the workspace for 30 min. Under an inverted microscope, cells were confirmed to be in optimal condition before being digested into a single-cell suspension with 0.25% trypsin-EDTA (Gibco, Grand Island, NY, USA). After centrifugation, cells were resuspended in serum-free medium and counted. Cell concentrations were adjusted for seeding in 24-well plates with transwell inserts (Chemicon, Temecula, CA, USA) at various densities. After 18-24 h of incubation, non-migrated cells were removed from the upper membrane. Migrated cells on the lower membrane were fixed with methanol, stained with crystal violet, and washed with PBS. Migration was quantitatively analyzed by photographing and counting the cells on the lower membrane using a specific microscope setup.
Apoptosis detection
In this apoptosis assay, the Annexin V-APC/PI Double Staining Kit (Invitrogen, 88-8007, Carlsbad, CA, USA) was utilized to identify apoptotic cells. The procedure began with diluting 10X Binding Buffer to 1X with ultrapure water. Cells were then trypsinized (Gibco, 25200-072, Grand Island, NY, USA) to form a single-cell suspension, washed with PBS (Servicebio, Wuhan, China), and subsequently with 1X Binding Buffer. Cells were resuspended in 1X Binding Buffer to achieve a concentration of 1–5 x 106 cells/mL. To each 100 μL of cell suspension, 5 μL of Annexin V conjugated with a fluorescent dye was added, followed by incubation in the dark at room temperature for 10 min. After a washing step with 1X Binding Buffer, cells were resuspended in 200 μL of the same buffer, and 5 μL of PI staining solution was added. The prepared cell suspensions were kept at 2-8 °C in the dark and analyzed within 4 h using a flow cytometer (Beckman CytoFLEX, Beckman Coulter, Brea, CA, USA). Data analysis was conducted with Beckman’s CytExpert software, providing a detailed assessment of cell apoptosis levels.
Statistical analysis
All statistical analyses in this study were performed using R software version 4.3.1. To compare differences between two groups, the Student’s t-test was used. For comparisons involving more than two groups, one-way ANOVA was applied, followed by post hoc tests to assess pairwise differences if the ANOVA was significant. Survival curves were generated using the Kaplan-Meier method, with prognosis-related differences assessed via the log-rank test. X-tile software [24] was used to determine cut-off values for survival groups. Diagnostic efficacy was evaluated using the Area Under the Receiver Operating Characteristic (ROC) Curve (AUC) and accuracy. Prognostic efficacy was assessed using time-dependent ROC curves and the concordance index (C-index). Graphs were created using both R software and GraphPad Prism 6 (GraphPad Software, La Jolla, CA, USA). A p-value of less than 0.05 was considered statistically significant.
Results
Data processing and identification of DEGs
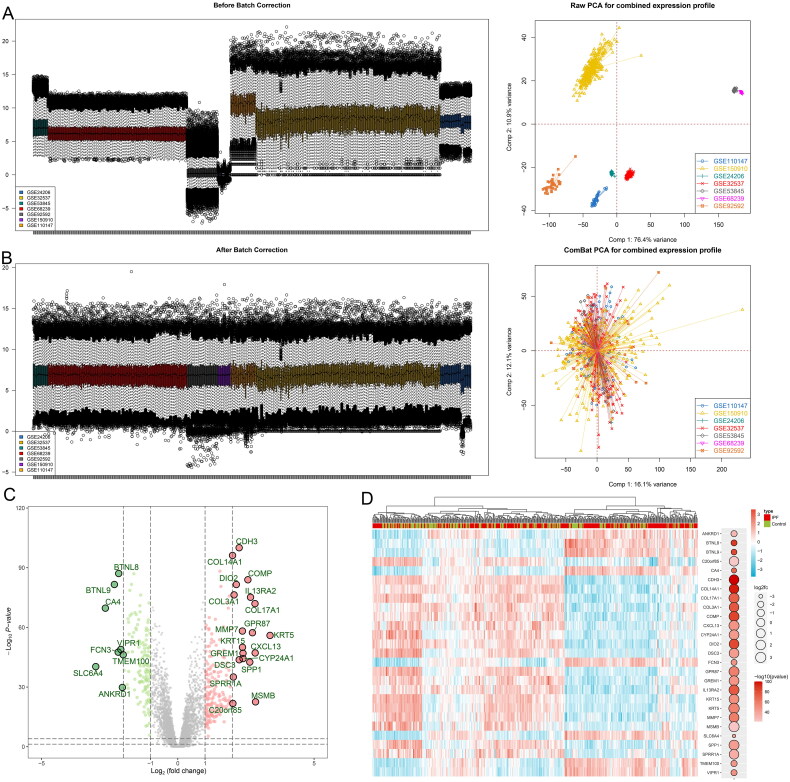
In this investigation, sequencing data were meticulously curated from a comprehensive dataset within the GEO database, encompassing seven individual series that feature lung tissue from both IPF patients and matched controls: GSE24206, GSE32537, GSE53845, GSE68239, GSE92592, GSE150910, and GSE110147. Furthermore, our study expanded to include datasets endowed with prognostic insights, namely GSE70866, GSE28221, and GSE93606, with sample origins traced to bronchoalveolar lavage fluid and blood. Survival outcomes, including overall survival (OS), were gathered from the GSE70866 datasets. Data concerning transplant-free survival (TFS) were collected from the GSE28221 datasets, and progression-free survival (PFS) metrics were retrieved from the GSE93606 datasets. The basic information of the included datasets is shown in Table S1. A rigorous normalization process was applied to the datasets prior to their amalgamation to effectively eliminate discrepancies attributable to batch effects. This meticulous approach is visually substantiated in Figure 1A and B, where pre-merger data dispersion underscored the presence of significant batch-related variance, which was conspicuously resolved post-integration. The ensuing differential expression analysis culminated in the identification of 320 distinct DEGs (Figure 1C and D).
Figure 1.
Data preprocessing and identification of differentially expressed genes. The box plot and principal component analysis elucidate the overall gene expression profiles (A) before and (B) after the normalization process. These results substantiate the effective removal of batch effects. Volcano plot (C) and heatmap (D) display the differentially expressed genes, highlighting significant variations in gene expression.
WGCNA and identification of hub genes
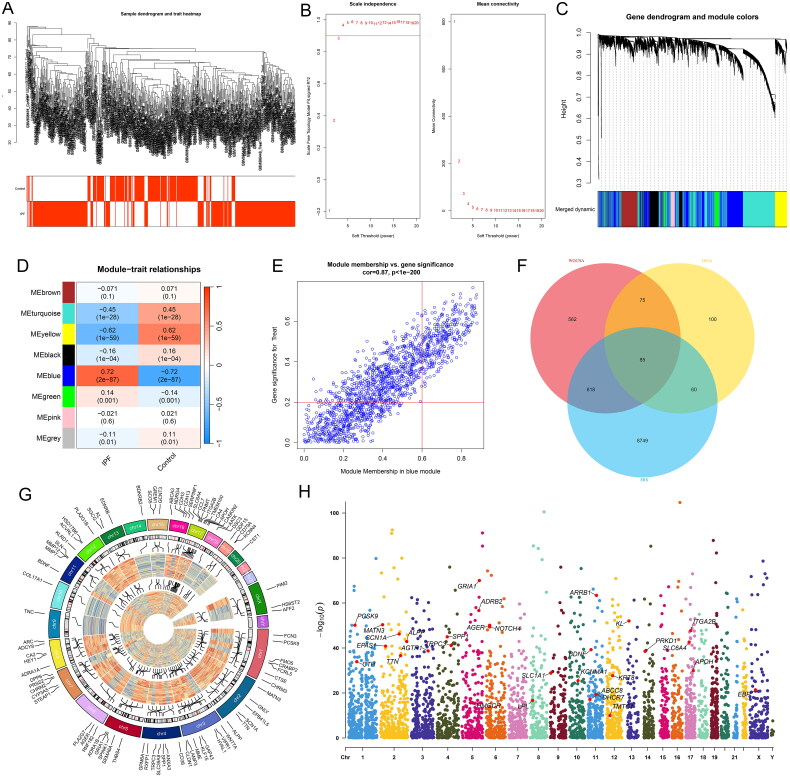
Prior to conducting WGCNA on the merged dataset, we initially confirmed the absence of outliers at a dendrogram height of 90 (Figure 2A). Subsequently, we determined a soft-thresholding power (β) of 3 for the WGCNA (Figure 2B), identifying a total of eight modules (Figure 2C). Notably, the blue module exhibited a significant positive correlation with IPF (correlation coefficient = 0.72, p-value = 2e-87) (Figure 2D). Furthermore, within this module, a significant correlation was observed between GS and MM (correlation coefficient = 0.87, p-value < 1e-200) (Figure 2E). Subsequently, we intersected genes from the blue module identified through WGCNA, DEGs, and genes associated with ERS sourced from the GeneCards database. This comprehensive approach yielded 85 hub genes (Figure 2F). The gene expression patterns and chromosomal locations of these hub genes between IPF and control groups were visualized and contrasted in Figure 2G and H. This analysis revealed that these genes are distributed across all chromosomes except for Y, 21, and 22.
Figure 2.
WGCNA results. (A) Dendrogram of sample clustering from the combined dataset alongside corresponding clinical information (indicating IPF or control group). (B) Determination of the soft-thresholding power in WGCNA, with the left side presenting scale-free index analysis for various soft-thresholding powers (β), and the right side depicting the analysis of mean connectivity for various soft-thresholding powers. (C) Dendrogram of genes based on clustering using the topological overlap matrix measure, with the color band displaying results obtained from automatic single-block analysis. (D) Heatmap illustrating the correlation between module eigengenes and clinical traits (IPF or healthy control group), with the turquoise module selected for further analysis. (E) Scatter plot of gene significance versus module membership in the blue module. (F) Venn diagram showing the intersection of genes among the three analyses. (G) Chromosomal locations of candidate hub genes. (H) Manhattan plot of candidate genes.
Functional enrichment analysis
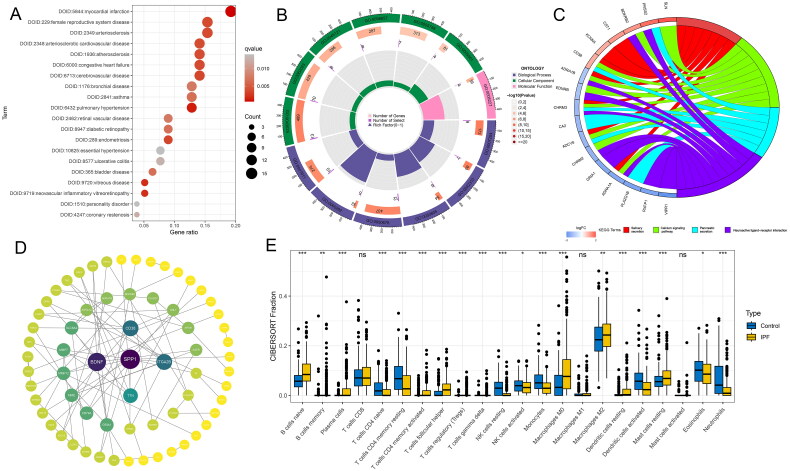
Our study’s functional enrichment analysis on 85 hub genes (Table S2) uncovered significant links to a range of diseases per DO, including cardiovascular disorders, pulmonary conditions, and systemic diseases like diabetes (Figure 3A). GO findings show these genes are involved in crucial BP including cytosolic calcium ion concentration regulation and cellular proliferation, affecting CC like neuronal cell bodies and extracellular matrix, MF analysis highlighted their role in G protein-coupled amine receptor activity (Figure 3B). KEGG pathway analysis connected these genes to essential signaling and metabolic pathways, underscoring their influence across various biological functions and disease mechanisms (Figure 3C). The PPI network, depicted in Figure 3D, identifies five genes with the highest degree of connectivity, indicating key roles within the network. These genes—titin (TTN), integrin subunit alpha 2b (ITGA2B), CD38 molecule (CD38), brain-derived neurotrophic factor (BDNF), and secreted phosphoprotein 1 (SPP1)—all exhibit degree values exceeding five, with SPP1 registering as the most interconnected gene.
Figure 3.
Functional enrichment and immune infiltration analysis. (A) Disease ontology analysis of hub genes. (B) Gene ontology analysis of hub genes, covering biological processes, cellular components, and molecular functions. (C) Kyoto Encyclopedia of Genes and Genomes pathway analysis for core genes, identifying significant pathways involved. (D) The Protein-Protein Interaction network displaying the top five genes with the highest degree of connectivity and their interactions with other genes. (E) Differential analysis of immune cell infiltration between IPF and control group lung tissues, illustrating variations in immune cell presence.
Immune infiltration analysis
Utilizing CIBERSORT to analyze the merged dataset, we obtained the proportional representation of 22 immune cell types in both IPF patients and the control group (Figure 3E). The analysis revealed a higher percentage of naïve B cells, memory B cells, plasma cells, activated CD4 memory T cells, follicular helper T cells, regulatory T cells (Tregs), gamma delta T cells, M0 macrophages, M2 macrophages, resting dendritic cells, and resting mast cells in the lung tissues of IPF patients compared to the control group. Conversely, naïve CD4 T cells, resting CD4 memory T cells, resting NK cells, activated NK cells, monocytes, activated dendritic cells, eosinophils, and neutrophils were found in higher quantities in the control group. No statistical differences were observed in CD8 T cells, M1 macrophages, and activated mast cells between the two groups.
Construction of the diagnostic model
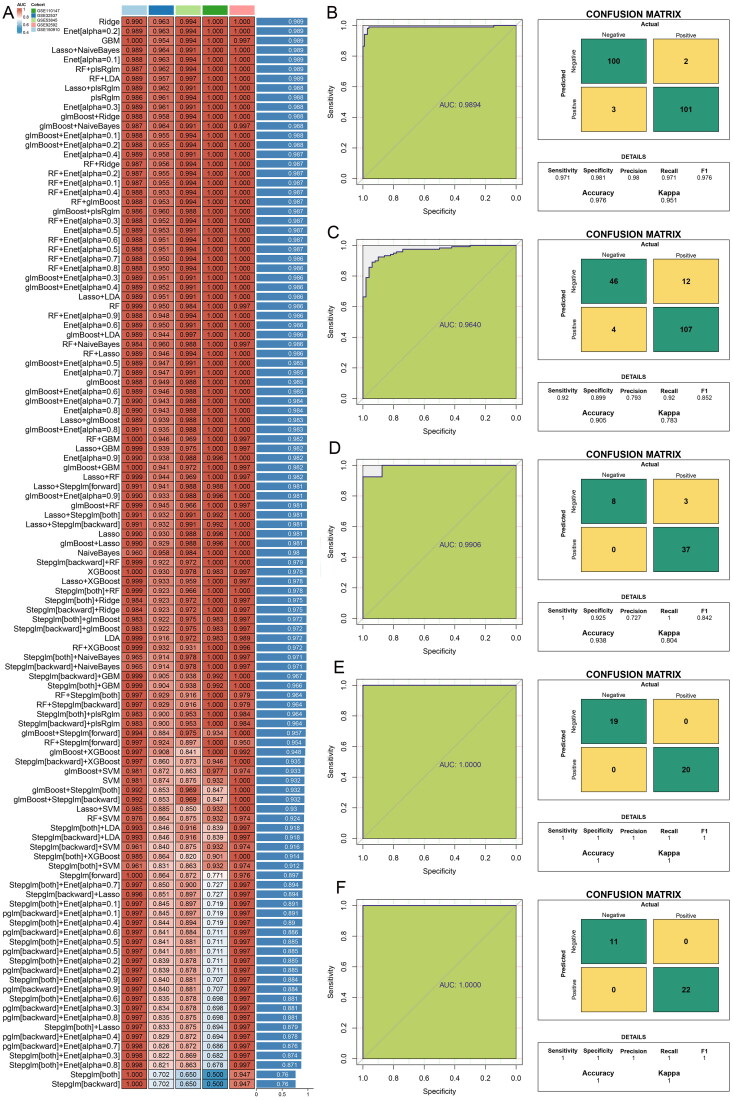
In this study, a diagnostic model was constructed utilizing five datasets, each with a sample size exceeding 30. The variable combinations for each algorithm are listed in Table S3. The GSE150910 dataset served as the training set, while GSE32537, GSE53845, GSE110147, and GSE92592 were used as the test sets. The models constructed using seven different algorithms achieved an average AUC of 0.989. However, since the Lasso + NaiveBayes algorithm required the fewest variables, with only 19, it was selected for constructing the diagnostic model. This methodology resulted in a perfect AUC of 0.989 for the training set and an impressive average accuracy of 0.989 for the five sets (Figure 4A). Moreover, the AUC for both the training and test datasets consistently surpassed the 0.95 threshold, reflecting the robustness of the model (Figure 4B–F). A comprehensive analysis was further facilitated by the confusion matrix, which delineated the distribution of true positives, false positives, true negatives, and false negatives across the training and test cohorts (Figure 4B–F), thereby providing an in-depth evaluation of the predictive capabilities of the model.
Figure 4.
Developed and validated through a machine learning-integrated approach for constructing diagnostic models. (A) a comprehensive suite of 113 diagnostic models was meticulously evaluated for accuracy across all training and test datasets. Receiver operating characteristic curves and confusion matrices for the models were generated for (B) GSE150910, (C) GSE32537, (D) GSE53845, (E) GSE92592, and (F) GSE110147 datasets, illustrating the performance and predictive validity of each diagnostic model in distinguishing IPF cases from controls.
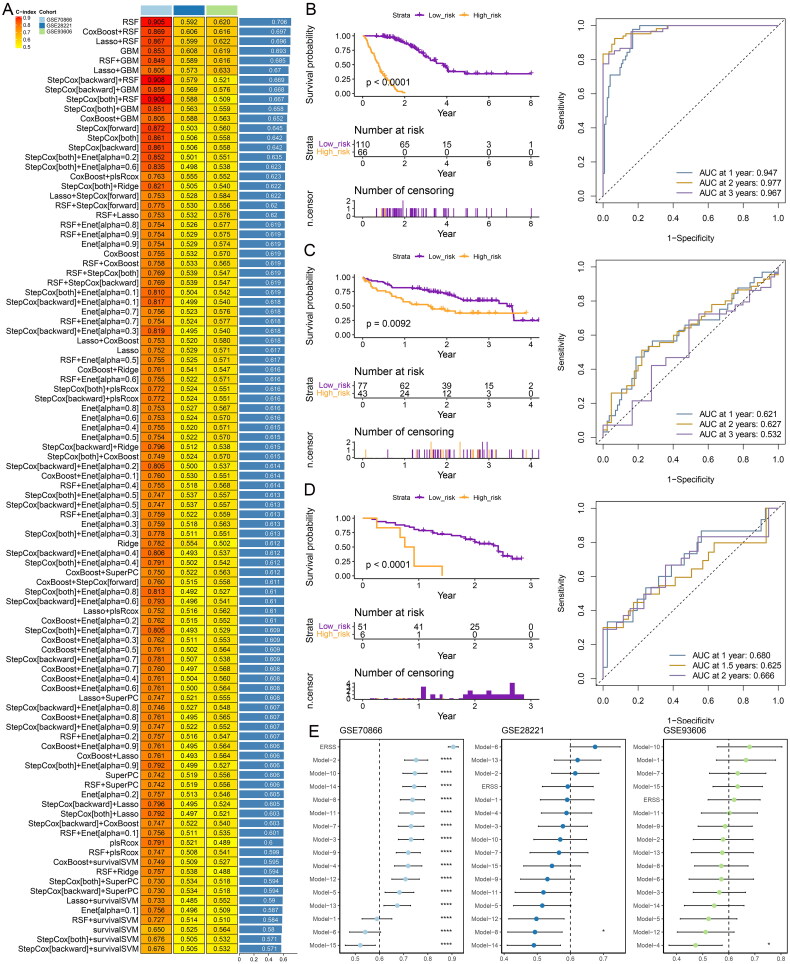
Construction of the prognostic model
The GSE70866 dataset was utilized as the training set, while GSE28221 and GSE93606 served as test sets, to construct prognostic models for IPF using various individual and combined machine learning algorithms. The variable combinations for each algorithm are listed in Table S4. It was found that RSF algorithms yielded a notably high average c-index of over 0.706 (Figure 5A). Therefore, the RSF algorithm was selected to construct the prognostic model and compute endoplasmic reticulum stress score (ERSS) for each sample. Utilizing X-tile software to stratify IPF patients from all three datasets into high and low-risk groups (Figure 5B–D), we observed that patients in the high-risk group had significantly worse prognoses than those in the low-risk group in three datasets. The risk index demonstrated a potent ability to predict prognoses, showing particular strength in the GSE70866 dataset for forecasting 1-, 2-, and 3-year outcomes. However, its predictive power was somewhat limited in the test sets GSE28221 and GSE93606 (Figure 5B–D).
Figure 5.
Developed and validated through a machine learning-based integrative approach for constructing prognostic models. (A) a comprehensive analysis involving 97 prognostic models assessed the C-index of each model across all training and test datasets. Kaplan-Meier curves and time-dependent ROC curves were generated for datasets (B) GSE70866, (C) GSE28221, and (D) GSE93606. (E) Comparison of prognostic models and 15 published signatures.
To further compare the predictive ability of our model with other models, we reviewed 15 studies [25–40] related to IPF that provided variables and coefficients for prognostic models (Table S5). Based on the content of these 15 studies, we calculated the respective risk indices. As shown in Figure 5E, for the GSE70866 dataset, the ERSS computed by our model demonstrated the best predictive ability, with a significantly higher C-index than those of other studies. For the GSE28221 dataset, our model’s C-index ranked fourth, and for the GSE93606 dataset, our model’s C-index ranked fifth. Overall, the model developed in this study exhibits strong prognostic capability.
Single cell sequencing analysis
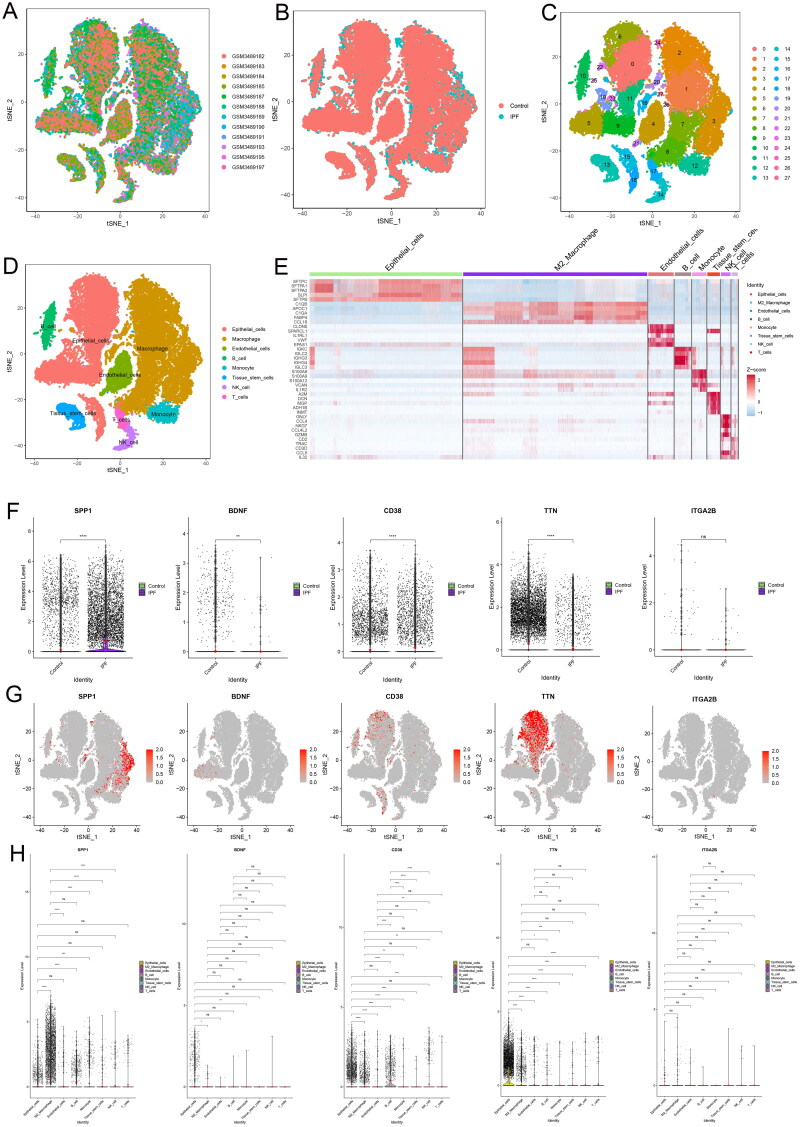
In the single-cell sequencing analysis, rigorous quality control was initially conducted to ensure that each cell had a minimum of 200 detectable features (Figure S1A). This threshold was established to confirm cell viability and exclude any dead or dying cells, as well as empty droplets, while also ensuring that the proportion of mitochondrial and ribosomal genes remained at acceptably low levels. The original t-SNE visualization is displayed in Figure S1B. Subsequently, we applied the Harmony integration method (Figure S1C) to mitigate batch effects and variability stemming from different experimental conditions and datasets. The post-harmonization results for the combined samples are presented in Figure 6A, and those specific to the IPF and control groups are depicted in Figure 6B. The single-cell data’s cluster dendrogram (Figure S1D) and the principal component (PC) stability analysis (Figure S1E) conducted via the Harmony algorithm indicated that the clustering results were particularly stable at PC 17, revealing 28 distinct clusters (Figure 6C). The 28 clusters were categorized into eight cell types: epithelial cells, macrophages, endothelial cells, B cells, monocytes, tissue stem cells, NK cells, and T cells (Figure 6D). Heat map showing the top 5 marker gene across the six cell types (Figure 6E).
Figure 6.
Single-cell sequencing results. (A) t-distributed Stochastic Neighbor Embedding (t-SNE) visualization of the 12 samples. (B) t-SNE plots comparing normal and IPF samples. (C) t-SNE representation of the 28 cell clusters. (D) Cell types were delineated based on marker gene profiles. (E) Heat map displaying the top 5 marker genes across 8 cell types. (F) Comparative analysis of 5 hub genes between IPF and control groups. (G) Feature plots illustrating the expression of the 5 hub genes across 8 cell types. (H) Differential comparison of the 5 hub genes among the 8 cell types, highlighting the variance in gene expression patterns.
Subsequent to utilizing the PPI network for initial gene prioritization, we analyzed the top five genes with the highest degree values. We discovered that SPP1 and CD38 were upregulated in the lung tissues of patients with IPF, while TTN and BDNF were downregulated (Figure 6F). However, ITGA2B did not exhibit significant differential expression between the IPF and control lung tissues. Feature plots elucidated the expression patterns of these five genes across eight identified cell types (Figure 6G). Notably, SPP1 was significantly overexpressed in M2 macrophages and exhibited substantial statistical differences in expression compared to all other cell types except B cells (Figure 6H). The expression differences among the remaining four cell types for these genes were not particularly pronounced. Additionally, by employing the median expression level of SPP1 as a cutoff, we categorized cells into high and low expression groups (Figure S1F). It was evident that M2 macrophages with high SPP1 expression were more prevalent in patients with IPF than in the control group.
Results of cell experiments
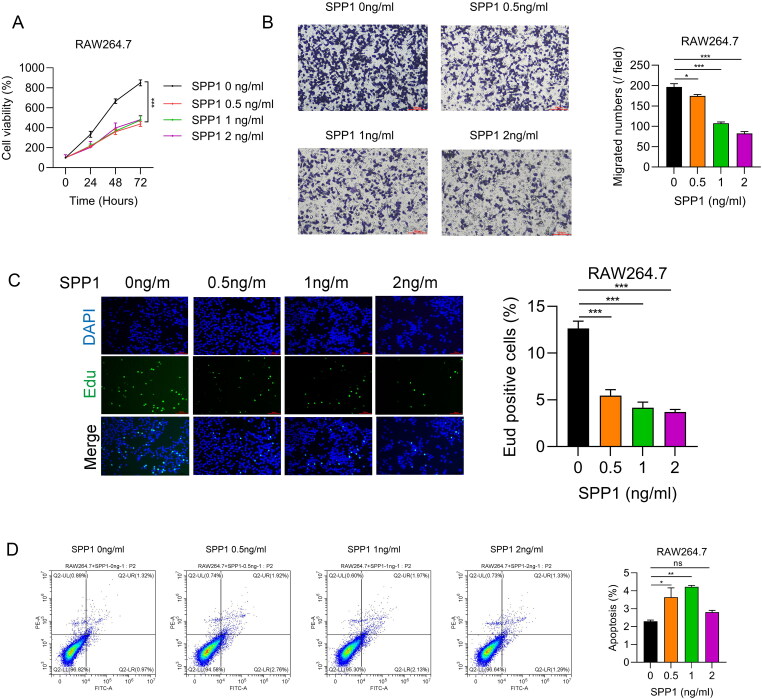
Previous research identified SPP1 as a potential biomarker for IPF patients [41]. This study aims to clarify the relationship between SPP1 and M2 macrophages through various biological experiments. Using data from the GSE32537 dataset, the relationship between SPP1 and clinical characteristics in IPF patients was analyzed. Higher SPP1 levels were found in patients under 60 (Figure S2A) and in males (Figure S2B), but smoking status (Figure S2C) was not associated with SPP1 expression. Additionally, higher SPP1 expression correlated with lower FVC % pred (Figure S2D), but not with DLCO % pred (Figure S2E). The impact of SPP1 on M2 macrophages was examined using a concentration gradient of 0, 0.5 ng/ml, 1 ng/ml, and 2 ng/ml. Results from the CCK8 assay indicated that the addition of SPP1 notably slowed the cell viability of M2 macrophages, although there was no significant difference in cell viability across the SPP1 concentration gradient (Figure 7A). In contrast, the Transwell assay revealed a decrease in M2 macrophage migration rate with increasing concentrations of SPP1 (Figure 7B). The EDU assay suggested a significant reduction in the proliferation rate of M2 macrophages upon SPP1 addition (Figure 7C). The Apoptosis Detection assay showed that the apoptosis rate of M2 macrophages significantly increased with SPP1 concentrations of 0.5 ng/ml and 1 ng/ml; however, a decline was observed at a concentration of 2 ng/ml (Figure 7D).
Figure 7.
Cellular experiment results. (A) Growth curves were monitored using the cell Counting Kit-8 (CCK-8) assay under various SPP1 concentration conditions. (B) Transwell experiments indicated the migratory capacity of cells at different SPP1 concentrations. (C) Results of 5-Ethynyl-2’-deoxyuridine (EDU) incorporation under various SPP1 concentrations. (D) Apoptotic cells were identified using the Annexin V-APC/PI Double Staining Kit across different SPP1 concentration conditions.
Discuss
Multiple bioinformatics techniques, including differential analysis and WGCNA, were utilized to analyze data from various GEO datasets. By integrating ERS-related target genes from GeneCards, candidate genes associated with IPF were identified. Over 100 combined machine learning algorithms were employed to construct gene signatures with strong diagnostic capabilities and prognostic models, which were validated using both training and test sets. Single-cell sequencing analysis revealed the gene architecture and expression patterns at the cellular level, highlighting cellular heterogeneity. A higher proportion of M2 macrophages expressing SPP1 was observed in the lung tissue of IPF patients compared to controls. To further elucidate the relationship between SPP1 and M2 macrophages, a series of cellular experiments were conducted. The findings indicate that SPP1 significantly impacts the proliferation, migration, invasion, and overall viability of M2 macrophages.
While IPF can be diagnosed through distinctive imaging findings and clinical presentations, a subset of patients still requires histopathological biopsy for definitive confirmation. Consequently, there is an ongoing quest for diagnostic biomarkers for IPF. For instance, a previous investigation revealed that SPP1 alone could distinguish between IPF patients and healthy individuals, as well as differentiate IPF patients from those with lung cancer [41]. However, the diagnostic performance of SPP1 alone was relatively low. In a recent piece of research, scholars utilized RF to create a three-gene signature associated with circadian rhythms, employing GSE150910 as the training dataset. However, the AUC was only 0.905, with another validation set yielding an AUC of merely 0.767 [42]. Another study engaged GSE32537 as the training dataset, formulating a gene signature through LASSO regression and SVM, achieving an AUC of 1, but with a relatively limited overall sample size [43]. While leveraging sequencing data alongside machine-learning algorithms presents substantial benefits in diagnosing IPF, notable challenges persist. These include issues related to data quality, model interpretation, and their translation into clinical practice. Thus, forthcoming studies should concentrate on enhancing both the interpretability and clinical applicability of these models.
The prognostic model demonstrated significant advantages compared to 15 previously established models [25–40]. The training set used bronchoalveolar lavage fluid (BALF) samples, while the validation sets used blood samples, presenting both limitations and benefits. A key limitation is the variability in biomarker expression between BALF and blood, which may affect the model’s generalizability. BALF samples, obtained directly from the lungs, may more accurately reflect local pathological processes, explaining the model’s superior performance. However, blood samples are easier to obtain and less invasive, making them more practical for routine clinical use. Notably, our model, trained on BALF, performed well in blood-based validation sets, indicating that certain biomarkers are consistently expressed in both sample types. While BALF provides a more precise medium for detecting lung-specific changes, blood-based biomarkers offer valuable prognostic information. Additionally, the previous models based on Cox regression are simpler to apply in clinical settings. In contrast, the RSF algorithm, despite its power, has poorer interpretability and requires more computational resources, which can hinder its clinical adoption. Therefore, improving the interpretability and usability of the RSF algorithm is essential for its integration into clinical practice.
Elevated SPP1 concentrations across various fibrotic organs indicate its potential as a biomarker for fibrosis. Studies consistently show that SPP1 levels rise in serum and bodily fluids during the early stages of fibrogenesis, correlating with the progression of organ fibrosis [44–48]. This highlights SPP1’s utility as an early, non-invasive indicator for diagnosing and assessing fibrosis. However, relying on a single biomarker to differentiate and predict fibrosis prognosis is impractical due to patient-specific and disease-specific factors. Therefore, a panel of biomarkers is more effective in improving diagnostic and prognostic accuracy. Integrating SPP1 into such panels could provide crucial insights. For example, a study showed that a combination of plasma MMP-7, Surfactant Protein D, and SPP1 effectively distinguished IPF from other non-IPF interstitial lung diseases [49]. Additionally, a progression index using four biomarkers (OPN, MMP-7, Intercellular Adhesion Molecule-1, and Periostin) outperformed the conventional GAP score in predicting disease progression over 12 months [33]. Another study demonstrated that plasma biomarkers, including SPP1, could predict TFS in IPF patients undergoing antifibrotic therapy, showing higher thresholds compared to those not receiving such treatments [50]. Moreover, previous research found that SPP1 promotes lung fibrosis by regulating the PI3K/AKT signaling pathway, leading fibroblasts to secrete more fibrotic proteins, and knocking down SPP1 can reduce the severity of lung fibrosis in mice [31]. Thus, SPP1 is a promising biomarker for fibrotic diseases, aiding in early identification, severity assessment, progression monitoring, and treatment evaluation.
Recent studies have identified macrophages as immune cells with diverse origins, deriving from both blood monocytes and embryonic sources, and possessing self-renewal capabilities [51, 52]. In the lungs, macrophages are categorized into Alveolar Macrophages (AMs) and Interstitial Macrophages (IMs) [53]. AMs are key in immune responses, eliminating pathogens and recruiting immune cells [54]. while IMs maintain immune homeostasis and promote fibroblast proliferation, affecting tissue fibrosis and inflammation [55]. Both AMs and IMs can differentiate into M1 or M2 macrophages, with M1 involved in inflammatory damage and M2 facilitating fibrotic remodeling [56]. M2 macrophages, known for their role in tissue repair and fibrosis, secrete anti-inflammatory cytokines like IL-10 and TGF-β, and express markers such as arginase-1 (Arg-1) [57]. Modulating the arginase-ornithine pathway can reduce fibrosis [58]. TGF-β is crucial in fibrosis, promoting fibroblast activity and collagen deposition, and creating a cycle that exacerbates fibrosis [59]. However, some studies show different results; for instance, bosutinib increased M2 macrophages but resisted fibrosis [60]. and human mesenchymal stem cell treatment increased M2 macrophages yet suppressed fibrosis progression in neonatal mice [61].
Previous research has shown that SPP1 promotes fibrosis in fibroblasts. Current single-cell sequencing data reveals a significant presence of M2 macrophages with high SPP1 expression in the lung tissue of IPF patients. Studies have also found high SPP1 expression in macrophages in the lower lobes of IPF lungs [62] and highlighted the role of CXCL4-driven SPP1+ macrophages in fibroblast activation and fibrosis progression [63]. These findings led us to hypothesize that SPP1 might promote M2 macrophage proliferation and migration while inhibiting apoptosis. However, our experiments indicated that SPP1 suppressed M2 macrophage proliferation and migration and promoted apoptosis within a certain concentration range. This unexpected result could be due to a negative feedback mechanism in the body or the differences between in vitro and in vivo environments, including receptor expression, cell activation states, and other signaling molecules. Further research is necessary to elucidate these mechanisms and better understand SPP1’s role in M2 macrophage function and pulmonary fibrosis.
Our study has its limitations. Firstly, the data used for constructing both diagnostic and prognostic models were sourced from public databases, without further validation using our own sequencing data. Additionally, the prognostic model’s predictive performance in the test set was somewhat lacking. Secondly, the data utilized in our analysis originated primarily from Western countries, lacking representation from regions such as Asia and Africa, which might limit the global applicability of our findings. Lastly, our study did not include animal experiments, and the cellular experiments conducted were relatively simplistic, not providing further evidence on the specific pathways through which SPP1 influences pulmonary fibrosis progression via M2 macrophages. Therefore, future studies will require more prospective clinical trials and detailed in vivo and in vitro experiments to substantiate our conclusions.
Conclusion
In conclusion, by leveraging sequencing data from comprehensive datasets, this study meticulously crafted and thoroughly validated custom diagnostic and prognostic models for IPF using machine learning algorithms. The research identified SPP1 as a potential key gene in IPF, with significant expression in M2 macrophages within the lung tissue of IPF patients. In vitro experiments revealed that SPP1 inhibits the proliferation and migration of M2 macrophages and promotes apoptosis. Further large-scale, well-designed experiments are needed to substantiate these findings.
Supplementary Material
Funding Statement
The author(s) reported there is no funding associated with the work featured in this article.
Ethics approval
Since the research involved the use of commercially available cell lines and did not involve any human participants or live animals, it was exempted from review by the Institutional Review Board (IRB) of Sichuan Provincial People’s Hospital. All experimental procedures were conducted in accordance with the relevant ethical guidelines and institutional policies.
Authors’ contributions
Study conception: YL, YY and QZ; manuscript draft preparation: XP and JZ; data collection, critical review, commentary, revision, and figure design: XP, GZ, LC, YY, HL, XC, and SG; and supervision of the prepared manuscript: QZ and JZ. All the authors verified the conclusion and approved the final version of the manuscript.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Code availability
All other R codes and analyses are available from the corresponding author upon request.
Data availability statement
The data that support the findings of this study are available from the corresponding author, Jun Zou, upon reasonable request.
References
- 1.Raghu G, Remy-Jardin M, Richeldi L, et al. Idiopathic pulmonary fibrosis (an update) and progressive pulmonary fibrosis in adults: an official ATS/ERS/JRS/ALAT clinical practice guideline. Am J Respir Crit Care Med. 2022;205(9):e18–e47. doi: 10.1164/rccm.202202-0399ST. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maher TM, Bendstrup E, Dron L, et al. Global incidence and prevalence of idiopathic pulmonary fibrosis. Respir Res. 2021;22(1):197. doi: 10.1186/s12931-021-01791-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shah Gupta R, Koteci A, Morgan A, et al. Incidence and prevalence of interstitial lung diseases worldwide: a systematic literature review. BMJ Open Respir Res. 2023;10(1):e001291. doi: 10.1136/bmjresp-2022-001291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lynch DA, Sverzellati N, Travis WD, et al. Diagnostic criteria for idiopathic pulmonary fibrosis: a Fleischner Society White Paper. Lancet Respir Med. 2018;6(2):138–153. doi: 10.1016/S2213-2600(17)30433-2. [DOI] [PubMed] [Google Scholar]
- 5.Raghu G, Collard HR, Egan JJ, et al. An official ATS/ERS/JRS/ALAT statement: idiopathic pulmonary fibrosis: evidence-based guidelines for diagnosis and management. Am J Respir Crit Care Med. 2011;183(6):788–824. doi: 10.1164/rccm.2009-040GL. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kondoh Y, Cottin V, Brown KK.. Recent lessons learned in the management of acute exacerbation of idiopathic pulmonary fibrosis. Eur Respir Rev. 2017;26(145):170050. doi: 10.1183/16000617.0050-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Almanza A, Carlesso A, Chintha C, et al. Endoplasmic reticulum stress signalling - from basic mechanisms to clinical applications. Febs J. 2019;286(2):241–278. doi: 10.1111/febs.14608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burman A, Tanjore H, Blackwell TS.. Endoplasmic reticulum stress in pulmonary fibrosis. Matrix Biol. 2018;68–69:355–365. doi: 10.1016/j.matbio.2018.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ritchie ME, Phipson B, Wu D, et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johnson WE, Li C, Rabinovic A.. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8(1):118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 11.Langfelder P, Horvath S.. WGCNA: an R package for weighted correlation network analysis. BMC Bioinf. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stelzer G, Rosen N, Plaschkes I, et al. The GeneCards suite: from gene data mining to disease genome sequence analyses. Curr Protoc Bioinformatics. 2016;54:1.30.1–1.30.33. doi: 10.1002/cpbi.5. [DOI] [PubMed] [Google Scholar]
- 13.Szklarczyk D, Kirsch R, Koutrouli M, et al. The STRING database in 2023: protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023;51(D1):D638–D646. doi: 10.1093/nar/gkac1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schriml LM, Munro JB, Schor M, et al. The Human Disease Ontology 2022 update. Nucleic Acids Res. 2022;50(D1):D1255–D1261. doi: 10.1093/nar/gkab1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gene Ontology Consortium . Gene Ontology Consortium: going forward. Nucleic Acids Res. 2015;43(Database issue):D1049–1056. doi: 10.1093/nar/gku1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kanehisa M, Furumichi M, Tanabe M, et al. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017;45(D1):D353–D361. doi: 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12(5):453–457. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reyfman PA, Walter JM, Joshi N, et al. Single-cell transcriptomic analysis of human lung provides insights into the pathobiology of pulmonary fibrosis. Am J Respir Crit Care Med. 2019;199(12):1517–1536. doi: 10.1164/rccm.201712-2410OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Butler A, Hoffman P, Smibert P, et al. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol. 2018;36(5):411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kang DW, Mo LP, Wang FL, et al. Adaptive harmony search algorithm utilizing differential evolution and opposition-based learning. Math Biosci Eng. 2021;18(4):4226–4246. doi: 10.3934/mbe.2021212. [DOI] [PubMed] [Google Scholar]
- 21.Korsunsky I, Millard N, Fan J, et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat Methods. 2019;16(12):1289–1296. doi: 10.1038/s41592-019-0619-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zappia L, Oshlack A.. Clustering trees: a visualization for evaluating clusterings at multiple resolutions. GigaScience. 2018;7(7):giy083. doi: 10.1093/gigascience/giy083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu J, Ding D, Jiang Z, et al. Long non-coding RNA CCAT1/miR-148a/PKCζ prevents cell migration of prostate cancer by altering macrophage polarization. Prostate. 2019;79(1):105–112. doi: 10.1002/pros.23716. [DOI] [PubMed] [Google Scholar]
- 24.Camp RL, Dolled-Filhart M, Rimm DL.. X-tile: a new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clin Cancer Res. 2004;10(21):7252–7259. doi: 10.1158/1078-0432.CCR-04-0713. [DOI] [PubMed] [Google Scholar]
- 25.Fan G, Liu J, Wu Z, et al. Development and validation of the prognostic model based on autophagy-associated genes in idiopathic pulmonary fibrosis. Front Immunol. 2022;13:1049361. doi: 10.3389/fimmu.2022.1049361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Feng A, Caro YM, Gardner C, et al. PTK2-associated gene signature could predict the prognosis of IPF. Respir Res. 2023;24(1):304. doi: 10.1186/s12931-023-02582-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guo R, Zhou Y, Lin F, et al. A novel gene signature based on the hub genes of COVID-19 predicts the prognosis of idiopathic pulmonary fibrosis. Front Pharmacol. 2022;13:981604. doi: 10.3389/fphar.2022.981604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He Y, Shang Y, Li Y, et al. An 8-ferroptosis-related genes signature from Bronchoalveolar Lavage Fluid for prognosis in patients with idiopathic pulmonary fibrosis. BMC Pulm Med. 2022;22(1):15. doi: 10.1186/s12890-021-01799-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang T, He WY.. Construction and validation of a novel prognostic signature of idiopathic pulmonary fibrosis by identifying subtypes based on genes related to 7-methylguanosine modification. Front Genet. 2022;13:890530. doi: 10.3389/fgene.2022.890530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li X, Cai H, Cai Y, et al. Investigation of a hypoxia-immune-related microenvironment gene signature and prediction model for idiopathic pulmonary fibrosis. Front Immunol. 2021;12:629854. doi: 10.3389/fimmu.2021.629854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liao Y, Yang Y, Zhou G, et al. Anoikis and SPP1 in idiopathic pulmonary fibrosis: integrating bioinformatics, cell, and animal studies to explore prognostic biomarkers and PI3K/AKT signaling regulation. Expert Rev Clin Immunol. 2024;20(6):679–693. doi: 10.1080/1744666X.2024.2315218. [DOI] [PubMed] [Google Scholar]
- 32.Liu B, Zhang X, Liu Z, et al. A novel model for predicting prognosis in patients with idiopathic pulmonary fibrosis based on endoplasmic reticulum stress-related genes. Cell Biol Int. 2024;48(4):483–495. doi: 10.1002/cbin.12121. [DOI] [PubMed] [Google Scholar]
- 33.Liu J, Gu L, Li W.. The prognostic value of integrated analysis of inflammation and hypoxia-related genes in idiopathic pulmonary fibrosis. Front Immunol. 2022;13:730186. doi: 10.3389/fimmu.2022.730186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lyu Y, Guo C, Zhang H.. Fatty acid metabolism-related genes in bronchoalveolar lavage fluid unveil prognostic and immune infiltration in idiopathic pulmonary fibrosis. Front Endocrinol (Lausanne). 2022;13:1001563. doi: 10.3389/fendo.2022.1001563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qiu L, Gong G, Wu W, et al. A novel prognostic signature for idiopathic pulmonary fibrosis based on five-immune-related genes. Ann Transl Med. 2021;9(20):1570. doi: 10.21037/atm-21-4545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xia Y, Lei C, Yang D, et al. Construction and validation of a bronchoalveolar lavage cell-associated gene signature for prognosis prediction in idiopathic pulmonary fibrosis. Int Immunopharmacol. 2021;92:107369. doi: 10.1016/j.intimp.2021.107369. [DOI] [PubMed] [Google Scholar]
- 37.Zheng J, Dong H, Zhang T, et al. Development and validation of a novel gene signature for predicting the prognosis of idiopathic pulmonary fibrosis based on three epithelial-mesenchymal transition and immune-related genes. Front Genet. 2022;13:865052. doi: 10.3389/fgene.2022.865052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou M, Ouyang J, Zhang G, et al. Prognostic value of tripartite motif (TRIM) family gene signature from bronchoalveolar lavage cells in idiopathic pulmonary fibrosis. BMC Pulm Med. 2022;22(1):467. doi: 10.1186/s12890-022-02269-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhu H, Zhou A, Zhang M, et al. Comprehensive analysis of an endoplasmic reticulum stress-related gene prediction model and immune infiltration in idiopathic pulmonary fibrosis. Front Immunol. 2023;14:1305025. doi: 10.3389/fimmu.2023.1305025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li M, Wang K, Zhang Y, et al. Ferroptosis-related genes in bronchoalveolar lavage fluid serves as prognostic biomarkers for idiopathic pulmonary fibrosis. Front Med (Lausanne). 2021;8:693959. doi: 10.3389/fmed.2021.693959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liao Y, Wang R, Wen F.. Diagnostic and prognostic value of secreted phosphoprotein 1 for idiopathic pulmonary fibrosis: a systematic review and meta-analysis. Biomarkers. 2023;28(1):87–96. doi: 10.1080/1354750X.2022.2148744. [DOI] [PubMed] [Google Scholar]
- 42.He J, Hu J, Liu H.. A three-gene random forest model for diagnosing idiopathic pulmonary fibrosis based on circadian rhythm-related genes in lung tissue. Expert Rev Respir Med. 2023:17(12):1307–1320. [DOI] [PubMed] [Google Scholar]
- 43.Lin Y, Lai X, Huang S, et al. Identification of diagnostic hub genes related to neutrophils and infiltrating immune cell alterations in idiopathic pulmonary fibrosis. Front Immunol. 2023;14:1078055. doi: 10.3389/fimmu.2023.1078055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Matsue Y, Tsutsumi M, Hayashi N, et al. Serum osteopontin predicts degree of hepatic fibrosis and serves as a biomarker in patients with hepatitis C virus infection. PLoS One. 2015;10(3):e0118744. doi: 10.1371/journal.pone.0118744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pereira TA, Syn WK, Pereira FE, et al. Serum osteopontin is a biomarker of severe fibrosis and portal hypertension in human and murine schistosomiasis mansoni. Int J Parasitol. 2016;46(13–14):829–832. doi: 10.1016/j.ijpara.2016.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ruberti S, Bianchi E, Guglielmelli P, et al. Involvement of MAF/SPP1 axis in the development of bone marrow fibrosis in PMF patients. Leukemia. 2018;32(2):438–449. doi: 10.1038/leu.2017.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rubiś P, Wiśniowska-Śmiałek S, Dziewięcka E, et al. Prognostic value of fibrosis-related markers in dilated cardiomyopathy: a link between osteopontin and cardiovascular events. Adv Med Sci. 2018;63(1):160–166. doi: 10.1016/j.advms.2017.10.004. [DOI] [PubMed] [Google Scholar]
- 48.Steinbrenner I, Sekula P, Kotsis F, et al. Association of osteopontin with kidney function and kidney failure in chronic kidney disease patients: the GCKD study. Nephrol Dial Transplant. 2023;38(6):1430–1438. doi: 10.1093/ndt/gfac173. [DOI] [PubMed] [Google Scholar]
- 49.White ES, Xia M, Murray S, et al. Plasma surfactant protein-D, matrix metalloproteinase-7, and osteopontin index distinguishes idiopathic pulmonary fibrosis from other idiopathic interstitial pneumonias. Am J Respir Crit Care Med. 2016;194(10):1242–1251. doi: 10.1164/rccm.201505-0862OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Adegunsoye A, Alqalyoobi S, Linderholm A, et al. Circulating plasma biomarkers of survival in antifibrotic-treated patients with idiopathic pulmonary fibrosis. Chest. 2020;158(4):1526–1534. doi: 10.1016/j.chest.2020.04.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Epelman S, Lavine KJ, Randolph GJ.. Origin and functions of tissue macrophages. Immunity. 2014;41(1):21–35. doi: 10.1016/j.immuni.2014.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhao Y, Zou W, Du J, et al. The origins and homeostasis of monocytes and tissue-resident macrophages in physiological situation. J Cell Physiol. 2018;233(10):6425–6439. doi: 10.1002/jcp.26461. [DOI] [PubMed] [Google Scholar]
- 53.Shi T, Denney L, An H, et al. Alveolar and lung interstitial macrophages: definitions, functions, and roles in lung fibrosis. J Leukoc Biol. 2021;110(1):107–114. doi: 10.1002/JLB.3RU0720-418R. [DOI] [PubMed] [Google Scholar]
- 54.Suber T, Camiolo MJ, Ray A.. A no-Wnt situation for alveolar macrophage self-renewal. Immunity. 2021;54(6):1099–1101. doi: 10.1016/j.immuni.2021.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gu Y, Lawrence T, Mohamed R, et al. The emerging roles of interstitial macrophages in pulmonary fibrosis: a perspective from scRNA-seq analyses. Front Immunol. 2022;13:923235. doi: 10.3389/fimmu.2022.923235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Arora S, Dev K, Agarwal B, et al. Macrophages: their role, activation and polarization in pulmonary diseases. Immunobiology. 2018;223(4–5):383–396. doi: 10.1016/j.imbio.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.He Q, Zhang W, Zhang J, et al. Cannabinoid analogue WIN 55212-2 protects paraquat-induced lung injury and enhances macrophage M2 polarization. Inflammation. 2022;45(6):2256–2267. doi: 10.1007/s10753-022-01688-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang L, Qu S, Wang L, et al. Tianlongkechuanling inhibits pulmonary fibrosis through down-regulation of arginase-ornithine pathway. Front Pharmacol. 2021;12:661129. doi: 10.3389/fphar.2021.661129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Inui N, Sakai S, Kitagawa M.. Molecular pathogenesis of pulmonary fibrosis, with focus on pathways related to TGF-β and the ubiquitin-proteasome pathway. Int J Mol Sci. 2021;22(11):6107. doi: 10.3390/ijms22116107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Carneiro PJ, Clevelario AL, Padilha GA, et al. Bosutinib therapy ameliorates lung inflammation and fibrosis in experimental silicosis. Front Physiol. 2017;8:159. doi: 10.3389/fphys.2017.00159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Al-Rubaie A, Wise AF, Sozo F, et al. The therapeutic effect of mesenchymal stem cells on pulmonary myeloid cells following neonatal hyperoxic lung injury in mice. Respir Res. 2018;19(1):114. doi: 10.1186/s12931-018-0816-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Morse C, Tabib T, Sembrat J, et al. Proliferating SPP1/MERTK-expressing macrophages in idiopathic pulmonary fibrosis. Eur Respir J. 2019;54(2):1802441. doi: 10.1183/13993003.02441-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hoeft K, Schaefer G, Kim H, et al. Platelet-instructed SPP1(+) macrophages drive myofibroblast activation in fibrosis in a CXCL4-dependent manner. Cell Rep. 2023;42(2):112131. doi: 10.1016/j.celrep.2023.112131. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All other R codes and analyses are available from the corresponding author upon request.
The data that support the findings of this study are available from the corresponding author, Jun Zou, upon reasonable request.