Abstract
Background
Populations of Olea europaea subsp. europaea var. sylvestris, the ancestor of cultivated olives, are scattered across the Mediterranean Basin. However, after millennia of possible hybridization with cultivated varieties, the genetic identity of many of these populations remain questionable. In the southern Levant, the plausible primary domestication center of olives, many of the naturally growing olive (NGOs) are considered feral, having developed from nearby olive groves. Here, we investigated the genetic identity of NGOs population in the Carmel region, hypothesizing that their specific location, which limit anemophily, provided an opportunity for the persistence of genuine var. sylvestris.
Results
We mapped more than 1,000 NGOs on the Kurkar ridge along the Carmel coast, within and outside the residential area of Atlit and used simple sequence repeats of 14 loci to assess the spatial genetic structure of 129 NGOs. Genetic diversity parameters and genetic distances between NGO and cultivated olives, as well as phenotypic and morphometric analyses of their oil content and pits, respectively, indicated the presence of a genuine var. sylvestris population. However, NGOs within the residential area of Atlit and old settlements showed an intermediate admix genetic structure, indicating on hybridization with local varieties, a consequence of their proximity to cultivated trees.
Conclusions
Integrating the results of genetic and phenotypic analyses we provide crucial evidence of the presence of a genuine var. sylvestris population in the southern Levant, in close geographical proximity to archaeological sites with the earliest evidence of olive exploitation in the ancient world. We supplement the results with recommendations for a conservation program that combines municipal requirements and the urgent need to preserve the largest population of var. sylvestris in the southern Levant.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12870-024-05575-7.
Keywords: Conservation, Genetic variation, Morphometry, Oil quality, Olea europaea subsp. europaea var. sylvestris, Spatial structure
Background
Cultivated olive trees (Olea europaea L. subsp. europaea) are an integral part of the rural landscape of the Mediterranean Basin. However, as early as the Middle Pleistocene (780 kya BP), records of fossil olive pollen and evidence of charred olive wood and endocarps (pits) appear in the southern Levant, long before olives were cultivated in the region [1, 2]. Thus, it seems that the wild ancestor of cultivated olives, O. europaea subsp. europaea var. sylvestris, has long been an important member of the southern Levant plant community. Moreover, several studies used the presence of fossil olive pollen in sediments as an indicator of past climatic conditions in the Mediterranean [3–5]. In addition, changes in olive pollen abundance in historical periods were used to deduce past human activity [6, 7] and the agricultural landscape of the southern Levant [8, 9]. A palynological study that compared records of fossil olive pollen in 23 sites around the Mediterranean Basin, suggested that the earliest rise in olive pollen in the southern Levant region, around 7,000 y BP, coincided with the onset of olive cultivation [10]. This evidence also coincides with other archaeobotanical and archaeological evidence from around that period [10–13], pointing to the southern Levant region as the primary area of olive domestication. However, the genetic analysis of a large number of samples of wild olive populations and cultivated varieties from around the Mediterranean Basin suggested that olive domestication started in the east Mediterranean, but not specifically in the southern Levant subregion [14, 15]. Diez et al. (2015) [16] suggested an alternative scenario in which olive domestication separately occurred in two independent centers, in the eastern and central parts of the Mediterranean Basin.
Long before the onset of cultivation, olive fruits were collected from wild trees for consumption (table olives) and oil [11, 17, 18]. The natural gene-pool of the wild relatives of olives is considered a source of useful adaptive traits [19–21]. However, the long co-existence of cultivated and wild olive populations, and generations of frequent gene-flow between them, raise questions concerning the existence of genuine var. sylvestris in the Mediterranean Basin. Indeed, by utilizing molecular fingerprint markers, many populations of naturally growing olive trees (NGOs) were identified as feral [15, 21–26]. However, NGOs showing contrasting genetic structures from cultivated and feral individuals are commonly considered genuine var. sylvestris [15, 23, 26, 27]. Thus, it is reasonable to believe that populations of genuine var. sylvestris remained in habitats which restrict gene flow via anemophily or zoochory seed dispersal. Phenotypically, NGOs of var. sylvestris possess a bushy form, and a long juvenile stage. In addition, their fruits are remarkably smaller [28], contain a lower percentage of oil [29] than cultivated trees, and possess high variation in fruit morphology [18]. Feral olives resemble those of wild var. sylvestris, thus making the distinction between feral and wild var. sylvestris through morphological characteristics difficult.
In the southern Levant, a region that includes Israel, the Palestinian Authority and Jordan [30], the Carmel ridge is considered the southernmost distribution edge of O. europaea subsp. europaea var. sylvestris in the East Mediterranean [31, 32]. Therefore, NGOs south of the Carmel region are considered feral trees [23]. However, feral populations are also common within the original distribution range, and very often NGOs populations include relicts of cultivated trees and their progenies [23]. In this study, we investigated two well-established populations of NGOs: (1) in the Western Galilee (Idmit), where distinct genetic divergence between NGOs and cultivated olives was previously described [23]; and (2) at the southernmost distribution edge of var. sylvstris, on kurkar ridges (calcareous cemented quartz sandstone of aeolian origin) along the Carmel coast, near Atlit (Fig. 1). The two investigated populations are located in close geographical proximity to cultivated groves, no more than 2 km apart. However, these populations, located along the Mediterranean coast, are subjected mostly to a west-to-east wind regime [33] that minimizes the likelihood of pollination from cultivated olives growing east, south or north of these sites. We therefore hypothesized that NGOs in Atlit may represent a wild gene-pool of var. sylvestris that has been less subjected to genetic erosion by gene flow from cultivated olives. Recently, Tourvas et al. (2023) [26] stressed the immediate need for in situ protection of var. sylvestris populations. Here, an integrative eco-genetic approach was designed to understand the spatial genetic structure of two southern Levant NGOs populations, in order to provide recommendations for conservation programs.
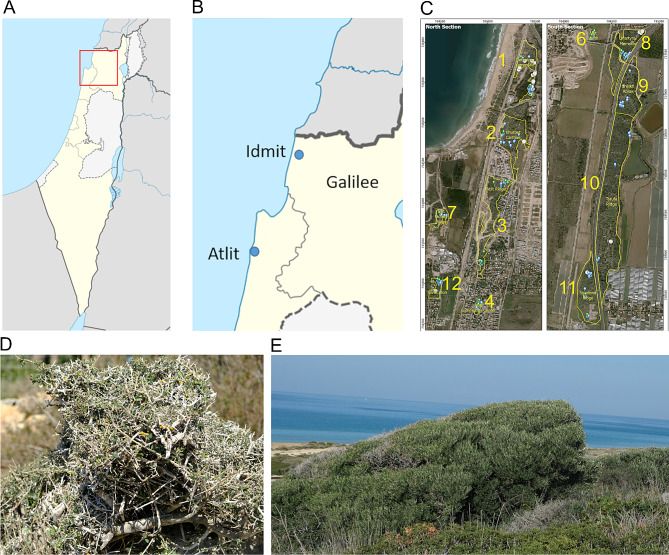
Fig. 1.
Mapping naturally growing olives in the Carmel region and Galilee, the presumed distribution range of O. europaea subsp. europaea var. sylvestris in the southern Levant. (A) Naturally growing olive trees were sampled in two populations, Idmit, and Atlit. (B) In Atlit, naturally growing olives were mapped and sampled in 12 polygons. (C) NGOs appearance in Idmit (D) and Atlit (E) indicate to the seniority of the plants, and thus the long existence of NGOs populations in the region
Methods
The study area
In Idmit (35°11’41.86"E, 33°04’35.54"N), NGOs growing on cliffs facing the Mediterranean coast were resampled and included in this study as a reference population, representing NGOs of var. sylvestris [23]. NGOs in Idmit possess a shrubby juvenile form, a result of continuous grazing by goats and rock hyrax (Procavia capensis) (Fig. 1). NGOs in the Idmit were mapped and leaves were sampled for DNA extraction from 17 individuals (Supplementary File #1). In Atlit (34°57′07″E, 32°42′41″N), the largest known NGOs population in the southern Levant, over 1,000 NGOs are distributed in unique coastal habitats in close proximity to the Mediterranean Sea, along three parallel saline Levantine kurkar ridges (Fig. 1). There, a more profound botanical survey was conducted to map and investigate the spatial distribution of NGOs in a 120-hectare area that was divided into 12 spatially separated polygons of sub-populations (Table 1 and Supplementary File #1). Supposedly, Atlit is at the southwest border of var. sylvestris distribution range (Carmel coast) in the East Mediterranean [31].
Table 1.
The number of NGOs mapped in the 12 polygons at Atlit, and those sampled for genetic and phenotypic analyses (cf. maps in supplementary file #1)
| Polygon | #NGOs | Genetic analysis | Phenotypic analysis |
|---|---|---|---|
| 1 | 311 | 10 | 3 |
| 2 | 102 | 10 | 1 |
| 3 | 100 | 10 | 6 |
| 4 | 31 | 10 | 3 |
| 5 | 42 | 10 | 3 |
| 6 | 11 | 8 | 3 |
| 7 | 67 | 7 | 1 |
| 8 | 100 | 10 | 2 |
| 9 | 76 | 10 | 1 |
| 10 | 117 | 10 | 0 |
| 11 | 39 | 8 | 2 |
| 12 | 59 | 9 | 2 |
| Total | 1055 | 112 | 27 |
Naturally growing olive trees at Atlit
NGOs in the 12 polygons in Atlit were mapped, and their phenotypic variation was assessed by characterizing their appearance (well-developed mature vs. young trees) and shape (flagged, bushy and whether it possesses an apparent trunk and suckers - as a proxy of their age). Qualitative estimations included trunk perimeter, canopy size (diameter) and plant height. Fruit load was also evaluated between none (not bearing fruits) and high load. Up to 10 individual NGOs in each polygon (a total of 112 NGOs) were arbitrarily assigned on the resulting maps for sampling canopy leaves for DNA extraction. In November 2022, fruits were harvested from a total of 27 individuals possessing high yields (more than 1 kg from each NGOs) and used for phenotypic assessment of the fruits: average weight and oil content (25 NGOs), fatty acids profiling (20 NGOs) and for morphometric analysis of their pits (23 NGOs); not all NGOs that were sampled for DNA were also sampled for the phenotypic analyses. The number of NGOs in each polygon that were sampled for the genetic and phenotypic characterization is listed in Table 1, and their location is marked on the maps presented in Supplementary File #1.
DNA extraction and genetic analysis
DNA was extracted using the CTAB method. After a quality check on agarose gel, DNA was quantified using a NanoDrop spectrophotometer (DeNovix DS-11, NC, USA), diluted and PCR reactions were performed using 14 SSR loci (simple sequence repeats) (Supplementary Table S1) that were previously tested and showed polymorphic, clear, and scorable profiles [23, 34]. The analysis included DNA samples of two multi locus analysis (MLL) genetic groups, representing two traditional local cultivars (TCs) that exist among local old living olive trees, MLL1 and MLL7 [34]. DNA was extracted from leaves that were collected from trees of each of MLL1 and MLL7 groups (13 and 11 samples, respectively), growing in a live germplasm collection at the Gilat Research Center (34°39’57.3"E, 31°20’08.3"N) (Supplementary Fig. S1), which includes olives developed from propagated material of local TCs [34].
PCR products were separated on an ABI automated sequencer (Applied Biosystems, 3730xl DNA Analyzer 96-Capillary Array (Thermo Fisher, USA); Center of Genomic Technologies, the Hebrew University of Jerusalem) as a multiplex of several loci labeled with three different fluorescent dyes (FAM-6, TAMARA and HEX, Applied Biosystems; see Supplementary Table S1 for loci used in each Multiplex pool), using the size standard GeneScan 500 LIZ. Electropherograms were then scored manually using Genmarker 1.75 (SoftGenetics, State College, Pennsylvania, USA).
Genetic analysis included the analysis of multi-locus genotypes (MLGs) and grouping those into MLLs using Genotype 1.2 [34]. One sample of each MLL was then used to calculate genetic diversity values (allele number, expected, observed and unbiased heterozygosity) using GenAlEx v6.5 [35]. FST genetic distances were calculated using GenAlEx v6.5 to infer the genetic distances between populations and local cultivars. In addition, gene flow was assessed by calculating GST values by GenAlEx v6.5 [26]. Principal coordinate analysis (PCoA) was performed on the standardized covariance matrix of genetic distances calculated according to Smouse & Peakall (1999) [36] using GenAlEx. Finally, Structure V.2.3.4 [37] was used for Bayesian clustering with the admixture model, to assign each MLG from each of the populations to K clusters. Ten independent runs for given Ks (2 to 9) were performed with a burning length of 10,000, followed by 20,000 repetitions [38]. The log likelihoods for a given K were used to choose the best given K based on an ad hoc quantity of ΔK [39]. Visualization of the results was made with distruct ver. 1.1 (https://rosenberglab.stanford.edu/distruct.html).
Fruit phenotype assessment
One hundred fruits, a subsample randomly selected from the above-mentioned harvested fruits from each of the 25 individual NGOs harvested in Atlit, were used to determine the average weight of a single fruit. Following Naor et al. (2013) [40], the oil content was determined on the fruit paste using the OliveScann near infrared (Foss, Denmark). A laboratory oil mill (Abencor, MC2 Ingenieria y Sistemas, Spain) was used to extract virgin olive oil from 1 kg of fruit from 20 NGOs individuals (Atlit) and 11 trees of MLL1 TC, used as a reference of a local cultivar. Oil was not extracted from fruits of MLL7 as they ripe early in the season and exhibit a different maturity index than those of NGOs and MLL1. Fatty acid (FA) composition in the oil was determined after cold methylation using an Agilent 7890 gas chromatograph (Agilent Technologies, Santa Clara, CA, USA), coupled to a 5799 mass spectrometer (Agilent Technologies, Santa Clara, CA, USA) [41]. FAs were identified according to standard compounds and their relative composition in the oil was determined as the percentage of total FA. The resulting data set was used for PCA (principal component analysis) and statistical analysis using JMP 16 (SAS Institute, Inc.).
Morphometric analysis of olive pits
Pits were recovered from at least 20 randomly selected fruits from each of the 23 NGOs (Atlit), and fruits of the MLL1 and MLL7 TCs, as references of local cultivars. Olive pits were recovered by manually scrubbing the pulp (mesocarp) with a plastic abrasive sponge, and the pits were then washed in a 2% hypochlorite solution to digest the remains of the pulp. Each olive pit was photographed on a blue background with a mm scale with a Canon EOS 200D camera equipped with EFS 18–55 mm lens. Separate photos were taken vertically for a dorsal and lateral view of each pit. Several cleaning steps were conducted using the GIMP image editing software as required (version 2.4.1.3, https://www.gimp.org/release-notes/gimp-2.4.html), and a dedicated MATLAB code was used for cropping, filtering and rescaling the images. The images were then filtered and sharpened using a two-dimensional median filter and standard unsharp masking method, by applying a Gaussian lowpass filter. The filtered and sharpened images were then filtered using a 2-dimensional convolution filter with an adjustable kernel window size; binarization, connectivity and erosion factors, that were manually adjusted to best trace the silhouette of each pit. The boundaries of the pits were traced using the Canny approach for edge detection. The analyzed features included area, circularity, convex area, eccentricity, equivalent diameter, major axis length, maximum ferret diameter, minor axis length, minimum ferret diameter, orthogonal diameter, perimeter, solidity, roundness, aspect ratio, form factor and compactness for the dorsal and lateral view of each pit. PCA was performed on the resulting morphometric dataset using R, after scaling each feature using Z-score normalization by column.
Results
Naturally growing olive trees in Atlit
A total of 1,055 NGOs were mapped in Atlit (Table 1 and see maps in Supplementary file #1). These represent only well-established NGOs, whereas young saplings were excluded. The majority of the mapped individuals (94%) were well-developed senior trees that included trees with apparent trunks and substantial canopies but maintaining a bush form (Supplementary Fig. S2 and File #2). Fruit load varied among the well-developed olive trees, ranging from trees bearing no fruit at all to high fruit load (Supplementary Fig. S2 and Files #1 and #2). Fruits were collected from NGOs from the latter group for phenotypic assessment (below). A considerable number of NGOs in Atlit had only juvenile leaves or both juvenile and mature leaves (104 and 475 trees, respectively). Young NGOs, n = 59, showed an early juvenile stage, thus additional data were not collected (Supplementary File #2). Field observations indicate that of the 444 NGOs mapped in a west aspect facing the Mediterranean Sea, 85 possess a Krummholz (flag) formation (Supplementary Fig. S2 and File #2, cf. Figure 1E). Those were found in polygons 1, 2, 7, 8, 10 and 11, on the kurkar ridge facing the Mediterranean Sea.
Genetic analysis
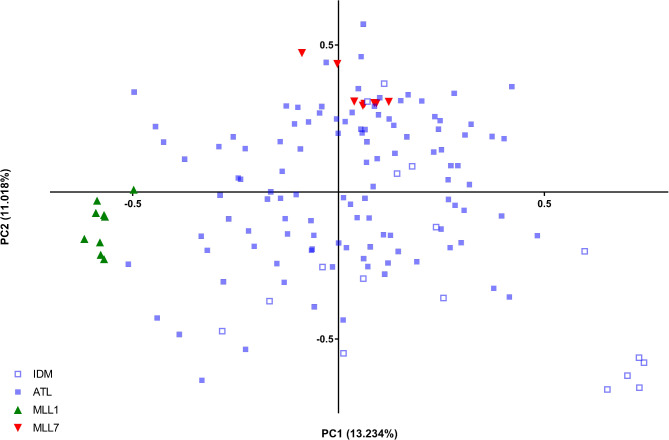
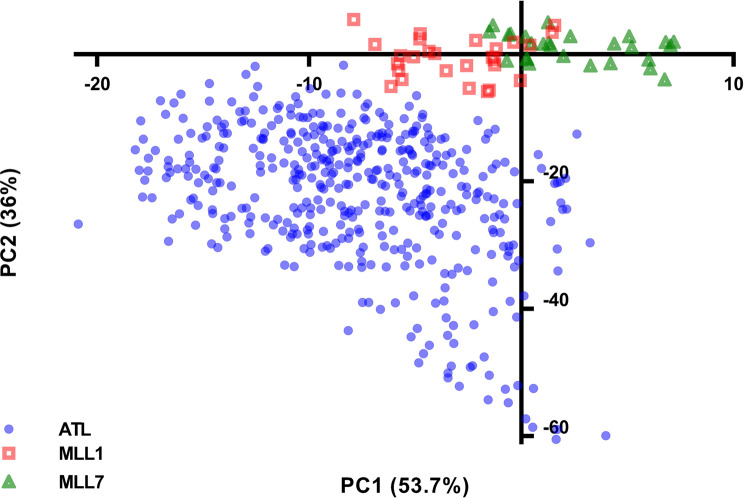
The analysis of 14 SSR loci resulted in allele numbers ranging from 3 to 29 (Supplementary Table S1). The number of alleles in the two studied populations was twice as high as their number in the local MLL1 and MLL7 TC (Table 2). Noticeable differences between the NGOs and TCs also appeared in their heterozygosity values, with higher values of unbiased heterozygosity (uHe) than observed heterozygosity (Ho) in NGOs and the opposite in the two MLL groups. However, NGOs sampled in polygon #5 in Atlit, within the residential area, showed substantially higher Ho and uHe values (0.730 and 0.748, respectively) than other NGOs (Table 2). PCoA analysis provided further evidence of higher genetic diversity among the NGOs in comparison with the TCs, which were grouped closely together in two separate clusters (Fig. 2).
Table 2.
Genetic diversity among naturally growing olives in Idmit and Atlit, and the main local MLL1 and MLL7 traditional cultivars: number of different alleles (na), and observed (Ho) and unbiased expected (uHe) heterozygosity
| Population | Polygon | N | Na | Ho | uHe |
|---|---|---|---|---|---|
| Idmit | 17 | 5.714 | 0.591 | 0.717 | |
| Atlit | 1 | 10 | 5.429 | 0.507 | 0.695 |
| Atlit | 2 | 10 | 5.643 | 0.541 | 0.709 |
| Atlit | 3 | 10 | 5.071 | 0.435 | 0.649 |
| Atlit | 4 | 10 | 5.571 | 0.562 | 0.706 |
| Atlit | 5 | 10 | 6.214 | 0.730 | 0.748 |
| Atlit | 6 | 8 | 5.143 | 0.541 | 0.702 |
| Atlit | 7 | 7 | 4.429 | 0.565 | 0.701 |
| Atlit | 8 | 10 | 6.000 | 0.609 | 0.705 |
| Atlit | 9 | 10 | 6.286 | 0.654 | 0.775 |
| Atlit | 10 | 10 | 6.429 | 0.652 | 0.749 |
| Atlit | 11 | 8 | 4.714 | 0.579 | 0.726 |
| Atlit | 12 | 9 | 5.214 | 0.495 | 0.723 |
| MLL1 | 13 | 2.357 | 0.819 | 0.469 | |
| MLL7 | 11 | 2.786 | 0.968 | 0.565 |
Fig. 2.
Principal coordinate analysis (PCoA) of the two studied NGOs populations (Idmit and Atlit), MLL1 and MLL7 represent the most common southern Levant traditional cultivars
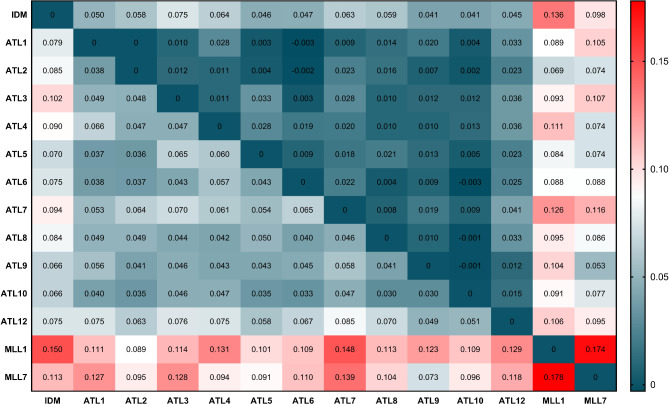
The largest GST values were detected between MLL1 and MLL7 (0.174), as expected from vegetatively propagated cultivars (Fig. 3). A large GST value was found between the NGOs in Idmit to both MLL1 and MLL7 (0.136 and 0.098, respectively). Calculating GST values separately for each polygon at Atlit showed the largest GST values between NGOs in polygon #7 and MLL1 and MLL7 (0.126 and 0.116, respectively). Relatively small GST values between NGOs and MLL7 were found in polygon #9 (0.053) and between NGOs and MLL1 in polygon #2 (0.069) (Fig. 3).
Fig. 3.
Pairwise FST and GST values between NGOs of two investigate populations, Idmit (IDM) and Atlit (ATL), and MLL1 and MLL7 traditional cultivars. GST and FST values are presented above and below the diagonal, respectively
In general, pairwise genetic distances values between populations of NGOs and MLL7 were lower than between NGOs and MLL1, as reflected in the PCoA (Figs. 2 and 3). Relatively low genetic pairwise distances were found between NGOs of the two populations and within the subpopulations at Atlit, reaching high pairwise distances between the NGOs in Idmit to those in polygon #3 in Atlit (FST =0.102). Relatively higher FST values were found between the NGOs of Atlit and Idmit and the two groups of cultivated trees. NGOs in polygon #9 in Atlit showed the lowest pairwise distances to MLL7 (0.073). Similarly, the lowest FST value was found between NGOs in polygon #2 and MLL1 (0.089) (Fig. 3). Relatively low genetic distances were also found between NGOs in polygon #4 and #5 to MLL7 (Fig. 3).
Assigning the NGOs (and samples of the two TCs as a reference group) to MLLs indicated that the majority of NGOs in Idmit and Atlit were assigned to a single occurring MLL, genetically different from any other sample included in the analysis (Table 3). The spatial distribution of MLLs in the mapped populations is presented in Supplementary File #1. However, in Idmit, 10 out of the 17 sampled NGOs were assigned to three site-specific MLLs (Table 3 and maps in Supplementary File #1), providing further support to a high frequency of clonal reproduction at this site [23], as was similarly shown in NGOs at Atlit (polygon #9, Supplementary File #1). Additionally, no evidence for local olive TCs was found in either Atlit or Idmit (Table 3), unlike in previous investigations of several other NGOs populations in the Carmel Ridge and Galilee [23].
Table 3.
Number of naturally growing and cultivated olives assigned to different multi-locus lineages (MLL). The number of trees assigned to each MLL1, MLL7, single occurring (SO) MLL, and the total number of MLLs found in each population are given. MLL1 and MLL7 represent the most common MLLs found in old cultivated trees [34]
| Idmit | Atlit | MLL1 | MLL7 | |
|---|---|---|---|---|
| N | 17 | 112 | ||
| MLL1 | 0 | 0 | 13 | |
| MLL7 | 0 | 0 | 11 | |
| SO MLL | 7 | 111 | ||
| Total MLL | 11 | 112 | 1 | 1 |
Based on genetic distances, applying the Bayesian admixture model for K = 2 to 9, showed a clear peak of ∆K at K = 8, the optimal number of subgroups (Supplementary Fig. S3). The dominance of the two TC clusters was evident in all K (Fig. 4, cf. Table 2). At Idmit, the genetic structure provided further evidence of clonal vegetative propagation of several NGOs. The maps in Supplementary File #1 show the close proximity of NGOs sharing the same MLL in Idmit, and similarly in Atlit (polygon #9). Nevertheless, at K8, NGOs of both Idmit and Atlit showed admixture genetic structure (Fig. 4).
Fig. 4.
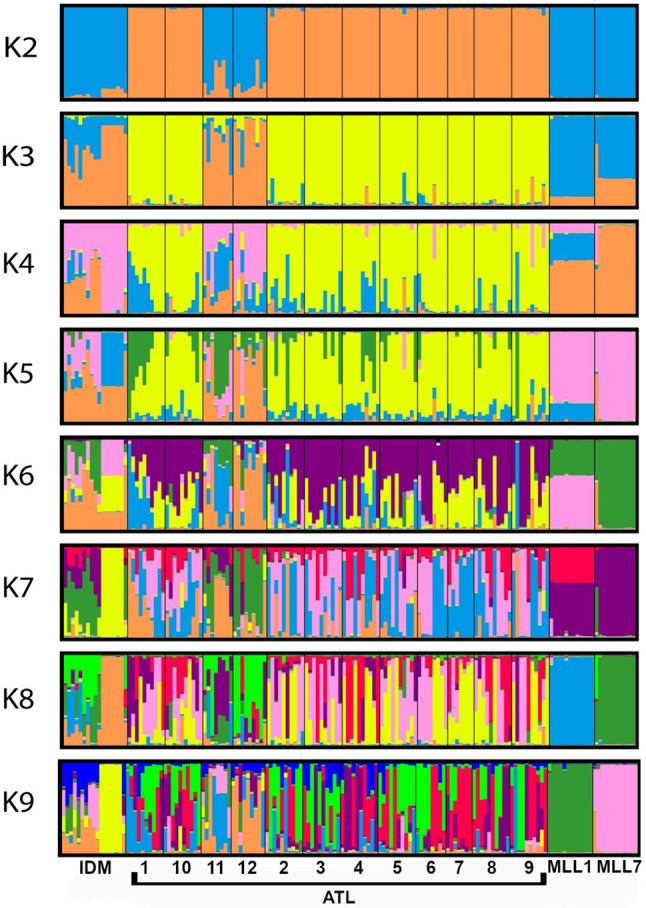
Inferred genetic structure of naturally growing olives and of the two main local traditional cultivars MLL1 and MLL7. Bayesian clustering with the admixture model implemented in Structure was used to assign individual MLGs to genetic clusters (K = 8, Supplementary Fig. S3). Individual MLGs within each group are represented by vertical bars and genetic groups are shown in different colors
Fruits characteristics
The average fruit weight of NGOs at Atlit was substantially (2.8 times) lower than those of MLL1 (0.95 ± 0.36 and 2.64 ± 0.15 g, respectively; t-test, P < 0.001), as was the case for the relative oil content in the fruit (7.4 ± 0.52 and 16.32 ± 1.05%, respectively; t-test, P < 0.01) (Fig. 5). However, no difference in relative water content was found between NGOs and MLL1 olive pastes (∼58.0%).
Fig. 5.
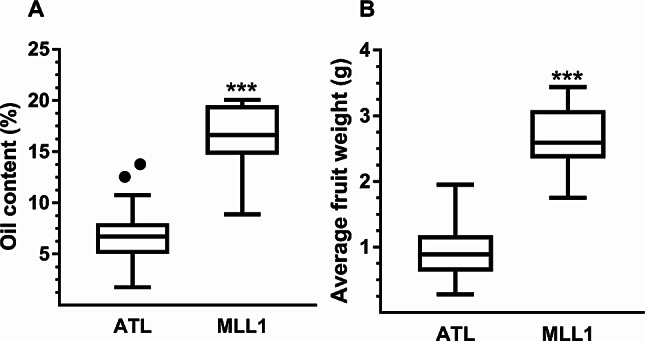
Box-and-Whisker diagrams showing the means with upper and lower variability of the oil content in the fruit (A) and average fruit weight (B) of naturally growing olives in Atlit and MLL1. Asterisks represent significant differences at P < 0.001 level
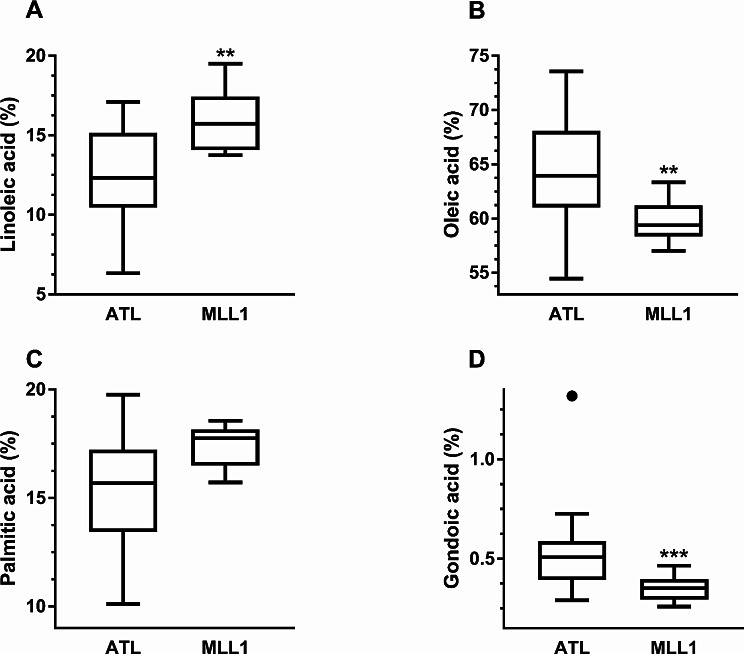
PCA separated the NGOs from cultivated olives on the basis of 12 detected FAs in the oil (Fig. 6 and Supplementary Table S2). In addition, NGOs fruit oil extracts from Atlit showed substantially larger variation in the relative content of FAs in comparison to the cultivated MLL1 oil extracts (Fig. 6). The first component of the PCA was mostly dominated by oleic, linoleic, gondoic and palmitic FAs and the second by behenic FA. Significant differences (t-test, P < 0.001) were found between the two groups of olive in the relative content of the abovementioned FAs, showing significantly higher relative oleic and gondoic FA content in the NGOs oil than in MLL1 (Fig. 7).
Fig. 6.
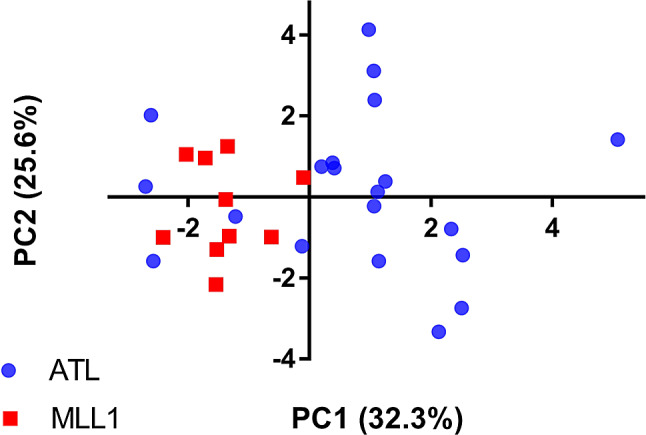
Results of the principal component analysis (PCA) of chemical compounds in the oil of the two groups of olive, cultivated MLL1 and naturally growing olives in Atlit (red and blue symbols, respectively) (see Table S2). Percent of variance explained by each axis is indicated in the axis titles
Fig. 7.
Box-and-Whisker diagrams showing the median with upper and lower variability in the relative content of linoleic (A), oleic (B), palmitic (C) and gondoic (D). FAs of naturally growing olives in Atlit and the MLL1 cultivar. Asterisks represent significance at P < 0.01(**) and P < 0.001 (***) levels
Morphometric analysis of olive pits
The PCA (Fig. 8), after normalization of the dataset, showed that most of the variation was dominated by three main pit features: (a) size (e.g., length, diameter, area); (b) shape (e.g., circularity, compactness, roundness and form factor); and (c) specific shape features, i.e., eccentricity and aspect-ratio (Supplementary Fig. S4). The PCA demonstrated clear differences between pits of the Atlit NGOs and the two TCs (Fig. 8). In addition, pits of NGOs showed considerably higher variation in size and shape (Figs. 8 and 9).
Fig. 8.
Results of the principal component analysis (PCA) of morphometric characteristics of NGOs olive pits NGOs from Atlit and those of the cultivated traditional cultivar reference groups: MLL1 and MLL7. Percent of variance explained by each axis in the PCA is indicated in the axis titles
Fig. 9.
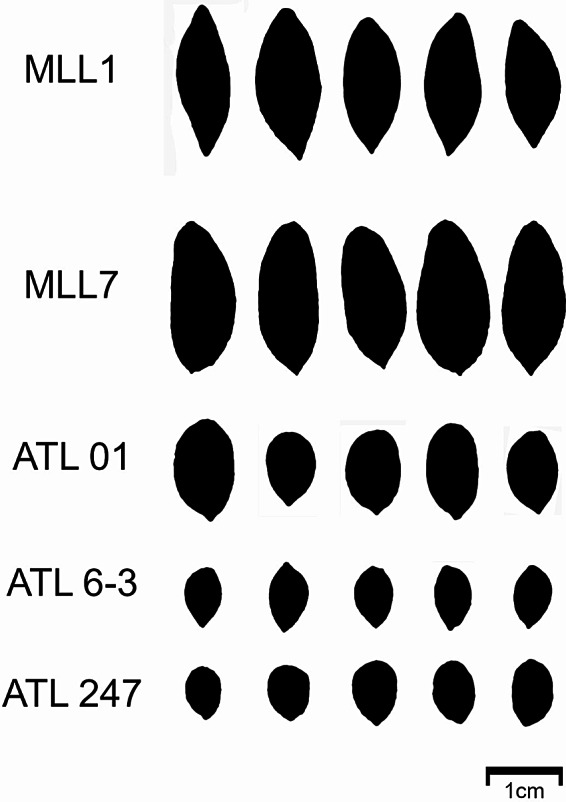
Representative pits of naturally growing trees from Atlit in comparison to those of the cultivated reference varieties
Discussion
Genetic evidence supports the existence of O. europaea subsp europaea var sylvestris in the southern Levant
The results of the genetic analysis (Figs. 2, 3 and 4; Tables 2 and 3) indicate that the NGOs in Atlit represent the largest known population of O. europaea subsp. europaea var. sylvestris in the southern Levant. In addition, the results further support the existence of the wild olive form in Idmit [23]. A similar SSR analysis that showed strong genetic differentiation between NGO populations and olive cultivars recently suggested the existence of populations of var. sylvestris in Greece [26]. In addition, by using a target gene approach in analyzing 27 NGO populations, Zunino et al. (2024) [42] proved the existence of a few populations of genuine var. sylvestris in Morocco, France and Spain. However, the majority of the investigated populations, both in Greece and in the western parts of the Mediterranean Basin, showed admixed genetic structures and were identified as feral, regardless of their distance from cultivated groves [26, 42]. Palynological studies show that olive pollen can reach distances far away from olive groves, as in the case of olive pollen records documented in sediment layers near the Dead Sea, even before the onset of olive cultivation [43]. Moreover, modeling olive pollen intensity shows that in elevations below and above 300 m a.s.l., olive trees can be pollinated by pollen from as far as 59 and 27 km, respectively [44]. Thus, considering that olive is an allogamous species that relies on anemophily, and millennia of possible gene-flow between cultivated olives and NGOs, it was surprising to find the presence of any wild olive in the southern Levant. However, considering that in this region, the Idmit and Atlit sites are at the edge of the Mediterranean coast (Fig. 1), where NGOs are subjected to a specific wind regime that minimizes pollination probability by cultivated trees, our results support the hypothesis that specific insolated niches provide enough protection to maintain genuine populations of var. sylvestris [26].
Signs of inbreeding (uHo < He, Table 2) suggested a high degree of isolation of the two populations of Atlit and Idmit [23]. However, the GST and FST values (Fig. 3), and the admixed population genetic structure (Fig. 4), suggests some influence of cultivated trees at Atlit on the genetic structure of the NGOs in polygons #2 and #9 and to a lesser extent in polygons #4 and #5. A recent land-use survey conducted in the Carmel region indicated that cultivated olives and olive groves are among the main components of the Carmel landscape, encompassing up to 22% of the land along the Carmel coastal plain and the Carmel mountain ridge [45]. Moreover, polygon #4 and #5 are situated within the Atlit residential area, where cultivated trees are present in private and public gardening. In addition, the NGOs in polygons #2 and #9 grow around two old settlements: the ancient medieval fortified ‘Casel Destreiz’ (or ‘le Destroit’) in polygon #2 [46], and an Armenian village established in 1920 and finally deserted in 1981 in polygon #9 [47]. It can thus be assumed that NGOs in these four polygons show an intermediate genetic structure in the transition between the wild var. sylvestris and cultivated olives, and exemplify the fate of the population if immediate action is not taken.
Phenotypic differences between Olea europaea subsp europaea var sylvestris and cultivated olivest
The NGOs in Atlit enabled further investigation of the phenotypic traits of var. sylvestris. The NGOs in Atlit varied in size, shape, appearance and formation (Supplementary File #2 and Fig. S2), all indicating a well-established population. Fruits of NGOs were considerably smaller and contained lower oil content compared to cultivated trees (Fig. 5), traits that are typical to var. sylvestris [28, 48]. In addition, PCA showed a clear divergence between MLL1 and NGOs in the relative FAs content (Fig. 6), although the oil of both groups was dominated by oleic, palmitic and linoleic FAs (Supplementary Table S2). Considering that the fruits of NGOs and MLL1 were collected in two distinctly different environments, the results may reflect environmental factors on olive quantity and quality, particularly the effect of irrigation [49]. However, the relatively low oil content measured in fruits of MLL1 (ca. 16%) and the relative content of FAs in the oil are in the range reported for MLL1 across different environmental conditions [50].
Olive pits, unlike fruits, exhibit a conservative morphology that is not influenced by environmental factors, thus allowing the investigation of genetic variations [18]. Results of the PCA summarizing the analysis of 16 morphometric characteristics of NGOs and reference cultivars (Fig. 8 and Supplementary Fig. S4), indicated that image processing is useful in creating morphometric passport data for cultivar identification [51]. PCA summarizing the total analyzed traits showed a clear divergence between the NGOs and cultivated varieties (Figs. 8 and 9). Pits of NGOs possessed large morphometric variation and were considerably smaller than those of cultivated varieties. However, feral olives also exhibit large variations in morphological traits and distinct shapes and sizes of fruits and pits that are different from cultivated olives [52, 53]. Nevertheless, altogether, the genetic analysis, in combination with phenotypic and morphometric analyses, provides support to the identification of a large NGOs population of O. europaea subsp. europaea var. sylvestris in Atlit.
Conservation recommendations
In situ genetic reserves ensure the protection of populations of plant genetic resources in their natural habitats. Protected genetic reservoirs allow evolutionary processes to continue and thus are considered a complementary approach to ex situ conservation (i.e., in live germplasms, botanical gardens and gene banks). Increasing concern about the loss of valuable genetic diversity have accelerated local conservation programs in Israel, with special emphasis on ex situ seed conservation of crop wild relatives and endangered species [54, 55]. Local plant genetic resources in Israel are protected in 230 terrestrial nature reserves, but only the Amiad reserve was declared as an in situ genetic reserve of emmer wheat (Triticum turgidum var. dicoccoides) and several other cereal wild relatives [56–58]. The NGOs in Idmit and those growing in polygons #2 and #10 in Atlit are found within natural reserves (Fig. 1 and Supplementary File #1). However, GST values, genetic distances (FST) and the admix genetic structure in several polygons (#2, 4, 5 and #9) at Atlit possibly reflect the influence of nearby cultivated trees, remnants of groves in old settlements or those planted in public and private gardens. Nevertheless, the majority of polygons at Atlit, and polygon #7 specifically, remained a sanctuary for O. europaea subsp. europaea var. sylvestris. In Idmit, grazing by rock hyrax and goats shape the appearance of olives (Fig. 1) and also hinders flowering, which so far may be responsible for the limited possible gene-flow from cultivated olive trees planted in a nearby recreational park (as reflected in the GST values, Fig. 3).
Development and urbanization processes necessary to meet the demands of an ever-increasing human population in the southern Levant, and specifically in Israel, rapidly increase the risk of genetic resource loss (https://chm.cbd.int/database/record?documentID=252812, in Hebrew). In the town of Atlit itself, an on-going municipal development program threatens the existence of the NGOs population, both by clearing areas for housing and planting cultivated trees in public and private gardens. Our results suggest the inclusion of specific NGOs in the municipal programs to ensure their protection. In addition, in order to preserve the genetic integrity of NGOs in Idmit and Atlit in the future, park and municipal gardening should avoid planting olive trees and transplant those already planted in the Atlit and Idmit recreational parks elsewhere.
Conclusion
More than 30 years of monitoring the genetic diversity of emmer wheat populations in the Amiad genetic reserve (eastern Galilee) has demonstrated that the establishment of a small, protected site is enough for effective in situ conservation [58]. Thus, the protection of NGOs within the nature reserves in Atlit and Idmit, with appropriate management, should ensure the maintenance of the last two remaining populations of O. europaea subsp. europaea var. sylvestris in the southern Levant. This unique germplasm possesses valuable potential for academic and agronomical studies, especially in global climate change hotspot [59, 60]. Finally, the close proximity of these populations to the archaeological sites with the earliest evidence of olive exploitation in the ancient world [11, 12] emphasizes the importance of designating these populations as heritage resources and conducting the necessary actions to ensure their protection and preservation for future generations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We would like to thank Y. Subbotin, T. Huashla and Y. Ron (Gilat Research Center), D. Farhi and Dr. H. Wasserstrom (Volcani Insitute) and R. Winter Livneh (Open Lanscape Institute, The Steinhardt Museum of Natural History, Tel Aviv University) for their valuable assistance. Many thanks to Prof. Jean-Frédéric Terral (University of Montpellier, France) for guiding us at ealry stages of the morphometric analysis, and Dr. Zachary Dunseth for his valuable assistance in editing and preparing the manuscript for publication.
Abbreviations
- BP
Before Present
- FA
Fatty Acid
- MLG
Multi-Locus Genotypes
- MLL
Multi Locus Analysis
- NGO
Naturally Growing Olives
- PCA
Principal Component Analysis
- PCoA
Principal Coordinate Analysis
- SSR
Simple Sequence Repeats
- TC
Traditional Cultivar
Author contributions
OB, AD, EG and AP conceived the study. AP and DLR conducted the botanical survey. Sampling leaves and fruits for genetic and phenotypic analyses was performed by EBD, TC and DLR. The molecular work was performed by EBD and TC, and they conducted the data analysis together with BH. EBD and YBD developed and conducted the morphometric analysis, and ZT performed the olive oil analysis. EBD and OB wrote the manuscript with the contribution of all authors.
Funding
This research was supported by the Israel Science Foundation (#332/21) and the Israel Land Authority research grant.
Data availability
No datasets were generated or analysed during the current study.
Declarations
Ethical approval
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Melamed Y, Kislev ME, Geffen E, Lev-Yadun S, Goren-Inbar N. The plant component of an Acheulian diet at Gesher Benot Ya ‘aqov, Israel. Proc Natl Acad Sci USA. 2016;113:14674–9. [DOI] [PMC free article] [PubMed]
- 2.Van Zeist W, Bottema S. A palynological study of the Acheulian site of Gesher Benot Ya’aqov, Israel. Veg Hist Archaeobot. 2009;18:105–21. [Google Scholar]
- 3.Finkelstein I, Langgut D. Climate, settlement history, and olive cultivation in the Iron Age Southern Levant. Bull Am Schools Orient Res. 2018;379:153–69. [Google Scholar]
- 4.Langgut D, Almogi-Labin A, Bar-Matthews M, Weinstein-Evron M. Vegetation and climate changes in the South Eastern Mediterranean during the last glacial-interglacial cycle (86 ka): new marine pollen record. Q Sci Rev. 2011;30:3960–72. [Google Scholar]
- 5.Schiebel V, Litt T. Holocene vegetation history of the southern Levant based on a pollen record from Lake Kinneret (Sea of Galilee), Israel. Veg Hist Archaeobot. 2018;27:577–90. [Google Scholar]
- 6.Langgut D, Neumann FH, Stein M, Wagner A, Kagan EJ, Boaretto E, Finkelstein I. Dead sea pollen record and history of human activity in the Judean Highlands (Israel) from the Intermediate Bronze into the Iron Ages (∼ 2500–500 BCE). Palynology. 2014;38:280–302. [Google Scholar]
- 7.Langgut D, Sasi A. The emergence of fruit tree horticulture in Chalcolithic southern Levant. And in Length of Days Understanding(Job 12: 12) Essays on Archaeology in the Eastern Mediterranean and Beyond in Honor of Thomas E Levy 2023:39–58.
- 8.Baruch U. The late Holocene vegetational history of Lake Kinneret (sea of Galilee), Israel. Paléorient. 1986;12:37–48. [Google Scholar]
- 9.Langgut D, Adams MJ, Finkelstein I. Climate, settlement patterns and olive horticulture in the southern Levant during the early bronze and intermediate bronze Ages (c. 3600–1950 BC). Levant. 2016;48:117–34. [Google Scholar]
- 10.Langgut D, Garfinkel Y. 7000-year-old evidence of fruit tree cultivation in the Jordan Valley, Israel. Sci Rep. 2022;12:7463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Galili E, Langgut D, Terral J, Barazani O, Dag A, Kolska Horwitz L, Ogloblin Ramirez I, Rosen B, Weinstein-Evron M, Chaim S. Early production of table olives at a mid-7th millennium BP submerged site off the Carmel coast (Israel). Sci Rep. 2021;11:2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Galili E, Stanley DJ, Sharvit J, Weinstein-Evron M. Evidence for earliest olive-oil production in submerged settlements off the Carmel coast, Israel. J Archaeol Sci. 1997;24:1141–50. [Google Scholar]
- 13.Namdar D, Amrani A, Getzov N, Milevski I. Olive oil storage during the fifth and sixth millennia BC at Ein Zippori, Northern Israel. Isr J Plant Sci. 2015;62:65–74. [Google Scholar]
- 14.Besnard B G, Khadari, Navascués M, Fernández-Mazuecos M, El Bakkali A, Arrigo N, Baali-Cherif D, Brunini-Bronzini de Caraffa V, Santoni S, Vargas P. The complex history of the olive tree: from late quaternary diversification of Mediterranean lineages to primary domestication in the northern Levant. Proc Royal Soc B: Biol Sci. 2013;280:20122833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Julca I, Marcet-Houben M, Cruz F, Gómez-Garrido J, Gaut BS, Díez CM, Gut IG, Alioto TS, Vargas P, Gabaldón T. Genomic evidence for recurrent genetic admixture during the domestication of Mediterranean olive trees (Olea europaea L). BMC Biol. 2020;18:148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Diez CM, Trujillo I, Martinez-Urdiroz N, Barranco D, Rallo L, Marfil P, Gaut BS. Olive domestication and diversification in the Mediterranean Basin. New Phytol. 2015;206:436–47. [DOI] [PubMed] [Google Scholar]
- 17.Kaniewski D, Van Campo E, Boiy T, Terral JF, Khadari B, Besnard G. Primary domestication and early uses of the emblematic olive tree: palaeobotanical, historical and molecular evidence from the Middle East. Biol Rev. 2012;87:885–99. [DOI] [PubMed] [Google Scholar]
- 18.Terral J-F, Bonhomme V, Pagnoux C, Ivorra S, Newton C, Paradis L, Ater M, Kassout J, Limier B, Bouby L. The shape diversity of olive stones resulting from domestication and diversification unveils traits of the oldest known 6500-Years-old table olives from Hishuley Carmel Site (Israel). Agronomy. 2021;11:2187. [Google Scholar]
- 19.Klepo T, De la Rosa R, Satovic Z, León L, Belaj A. Utility of wild germplasm in olive breeding. Sci Hort. 2013;152:92–101. [Google Scholar]
- 20.León L, De la Rosa R, Velasco L, Belaj A. Using wild olives in breeding programs: implications on oil quality composition. Front Plant Sci. 2018;9:232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mariotti R, Belaj A, De La Rosa R, Leòn L, Brizioli F, Baldoni L, Mousavi S. EST–SNP study of Olea europaea L. uncovers functional polymorphisms between cultivated and wild olives. Genes. 2020;11:916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Angiolillo A, Mencuccini M, Baldoni L. Olive genetic diversity assessed using amplified fragment length polymorphisms. Theor Appl Genet. 1999;98:411–21. [Google Scholar]
- 23.Barazani O, Keren-Keiserman A, Westberg E, Hanin N, Dag A, Ben-Ari G, Fragman-Sapir O, Tugendhaft Y, Kerem Z, Kadereit JW. Genetic variation of naturally growing olive trees in Israel: from abandoned groves to feral and wild? BMC Plant Biol. 2016;16:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hannachi H, Breton C, Msallem M, El Hadj SB, El Gazzah M, Bervillé A. Genetic relationships between cultivated and wild olive trees (Olea europaea L. var. europaea and var. sylvestris) based on nuclear and chloroplast SSR markers. Nat Resour. 2010;1:95. [Google Scholar]
- 25.Lumaret R, Ouazzani N, Michaud H, Villemur P. Cultivated olive and oleaster: two very closely connected partners of the same species (Olea europaea). Evidence from enzyme polymorphism. Bocconea. 1997;7:39–42. [Google Scholar]
- 26.Tourvas N, Ganopoulos I, Koubouris G, Kostelenos G, Manthos I, Bazakos C, Stournaras V, Molassiotis A, Aravanopoulos F. Wild and cultivated olive tree genetic diversity in Greece: a diverse resource in danger of erosion. Front Genet. 2023;14. [DOI] [PMC free article] [PubMed]
- 27.Baldoni L, Tosti N, Ricciolini C, Belaj A, Arcioni S, Pannelli G, Germana MA, Mulas M, Porceddu A. Genetic structure of wild and cultivated olives in the central Mediterranean basin. Ann Botany. 2006;98:935–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Turrill WB. Wild and cultivated olives. Kew Bull. 1951;6:437–42. [Google Scholar]
- 29.Hannachi H, Sommerlatte H, Breton C, Msallem M, El Gazzah M, Ben El Hadj S, Bervillé A. Oleaster (var. sylvestris) and subsp. cuspidata are suitable genetic resources for improvement of the olive (Olea europaea subsp. europaea var. europaea). Genet Resour Crop Evol. 2009;56:393–403. [Google Scholar]
- 30.Soto-Berelov M, Fall P, Falconer S. A revised map of plant geographical regions of the Southern Levant. In: GSR: 2012.
- 31.Zohary D, Hopf M, Weiss E. Domestication of plants in the Old World: the origin and spread of domesticated plants in Southwest Asia. Europe, and the Mediterranean Basin: Oxford University Press; 2012. [Google Scholar]
- 32.Zohary D, Spiegel-Roy P. Beginnings of fruit growing in the old world: Olive, grape, date, and fig emerge as important bronze age additions to grain agriculture in the Near East. Science. 1975;187:319–27. [DOI] [PubMed] [Google Scholar]
- 33.Kutiel H. Climate of Israel. Landscapes and landforms of Israel. Springer; 2024. pp. 39–47.
- 34.Barazani O, Westberg E, Hanin N, Dag A, Kerem Z, Tugendhaft Y, Hmidat M, Hijawi T, Kadereit JW. A comparative analysis of genetic variation in rootstocks and scions of old olive trees–a window into the history of olive cultivation practices and past genetic variation. BMC Plant Biol. 2014;14:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peakall R, Smouse PE. GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics. 2012;28:2537–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smouse PE, Peakall R. Spatial autocorrelation analysis of individual multiallele and multilocus genetic structure. Heredity. 1999;82:561–73. [DOI] [PubMed] [Google Scholar]
- 37.Hubisz MJ, Falush D, Stephens M, Pritchard JK. Inferring weak population structure with the assistance of sample group information. Mol Ecol Resour. 2009;9:1322–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol. 2005;14:2611–20. [DOI] [PubMed] [Google Scholar]
- 40.Naor A, Schneider D, Ben-Gal A, Zipori I, Dag A, Kerem Z, Birger R, Peres M, Gal Y. The effects of crop load and irrigation rate in the oil accumulation stage on oil yield and water relations of ‘Koroneiki’olives. Irrig Sci. 2013;31:781–91. [Google Scholar]
- 41.Tietel Z, Dag A, Yermiyahu U, Zipori I, Beiersdorf I, Krispin S, Ben-Gal A. Irrigation‐induced salinity affects olive oil quality and health‐promoting properties. J Sci Food Agric. 2019;99:1180–9. [DOI] [PubMed] [Google Scholar]
- 42.Zunino L, Cubry P, Sarah G, Mournet P, El Bakkali A, Aqbouch L, Sidibé-Bocs S, Costes E, Khadari B. Genomic evidence of genuine wild versus admixed olive populations evolving in the same natural environments in western Mediterranean Basin. PLoS ONE. 2024;19(1):e0295043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Neumann FH, Kagan EJ, Schwab MJ, Stein M. Palynology, sedimentology and palaeoecology of the late Holocene Dead Sea. Q Sci Rev. 2007;26:1476–98. [Google Scholar]
- 44.Rojo J, Orlandi F, Pérez-Badia R, Aguilera F, Dhiab AB, Bouziane H, de la Guardia CD, Galán C, Gutiérrez-Bustillo A, Moreno-Grau S. Modeling olive pollen intensity in the Mediterranean region through analysis of emission sources. Sci Total Environ. 2016;551:73–82. [DOI] [PubMed] [Google Scholar]
- 45.Raviv O, Zemah-Shamir S, Izhaki I, Lotan A. The effect of wildfire and land-cover changes on the economic value of ecosystem services in Mount Carmel Biosphere Reserve, Israel. Ecosyst Serv. 2021;49:101291. [Google Scholar]
- 46.Johns CN, Pringle D. Pilgrims’ castle (‘Atlit), David’s Tower (Jerusalem) and Qal ‘at ar-Rabad (‘Ajlun): three Middle Eastern castles from the time. of the Crusades: Routledge; 2019. [Google Scholar]
- 47.Kark R, Oppenheim A. Armenian farmers in Palestine/Israel: The hamlet of Sheikh Bureik during the twentieth century In: Cathedra. vol. 162. Jerusalem, Israel: Yad Izhak Ben-Zvi; 2017: 67–94 (in Hebrew).
- 48.Fanelli V, Mascio I, Falek W, Miazzi MM, Montemurro C. Current status of biodiversity assessment and conservation of wild olive (Olea europaea L. subsp. europaea var. sylvestris). Plants. 2022;11:480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dag A, Kerem Z, Yogev N, Zipori I, Lavee S, Ben-David E. Influence of time of harvest and maturity index on olive oil yield and quality. Sci Hort. 2011;127:358–66. [Google Scholar]
- 50.Barazani O, Waitz Y, Tugendhaft Y, Dorman M, Dag A, Hamidat M, Hijawi T, Kerem Z, Westberg E, Kadereit JW. Testing the potential significance of different scion/rootstock genotype combinations on the ecology of old cultivated olive trees in the southeast Mediterranean area. BMC Ecol. 2017;17:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Terral JF, Alonso N, Capdevila RBi, Chatti N, Fabre L, Fiorentino G, Marinval P, Jordá GP, Pradat B, Rovira N. Historical biogeography of olive domestication (Olea europaea L.) as revealed by geometrical morphometry applied to biological and archaeological material. J Biogeogr. 2004;31:63–77. [Google Scholar]
- 52.Belaj A, León L, Satovic Z, de la Rosa R. Variability of wild olives (Olea europaea subsp. europaea var. sylvestris) analyzed by agro-morphological traits and SSR markers. Sci Hortic. 2011;129:561–569.
- 53.Hannachi H, Breton C, Msallem M, El Hadj SB, El Gazzah M, Bervillé A. Differences between native and introduced olive cultivars as revealed by morphology of drupes, oil composition and SSR polymorphisms: a case study in Tunisia. Sci Hort. 2008;116:280–90. [Google Scholar]
- 54.Barazani O, Mayzlish-Gati E, Lifshitz D, Hadas R, Keren-Keiserman A, Golan S, Faraj T, Singer A, Beerman A, Perevolotsky A. Strategies and priorities in field collections for ex situ conservation: the case of the Israel Plant Gene Bank. Genet Resour Crop Evol. 2017;64:1–5. [Google Scholar]
- 55.Barazani O, Perevolotsky A, Hadas R. A problem of the rich: prioritizing local plant genetic resources for ex situ conservation in Israel. Biol Conserv. 2008;141:596–600. [Google Scholar]
- 56.Anikster Y, Feldman M, Horovitz A. The Ammiad experiment. Plant Genetic Conservation: the in situ Approach. Springer; 1997. pp. 239–53.
- 57.Anikster Y, Noy-Meir I. The wild-wheat field laboratory at Ammiad. Isr J Bot. 1991;40:351–62. [Google Scholar]
- 58.Dahan-Meir T, Ellis TJ, Mafessoni F, Sela H, Rudich O, Manisterski J, Avivi-Ragolsky N, Raz A, Feldman M, Anikster Y. 36-year study reveals the effects of microhabitats on a wild wheat population. bioRxiv. 2022;2022(2001):2010–475641. [DOI] [PubMed] [Google Scholar]
- 59.Cockel CP, Guzzon F, Gianella M, Müller JV. The importance of conserving crop wild relatives in preparing agriculture for climate change. CABI Reviews 2022.
- 60.Vincent H, Hole D, Maxted N. Congruence between global crop wild relative hotspots and biodiversity hotspots. Biol Conserv. 2022;265:109432. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No datasets were generated or analysed during the current study.