Abstract
The capsid (CA) protein, the major structural component of retroviruses, forms a shell that encases the ribonucleoprotein complex in the virion core. The most conserved region of CA, ∼20 amino acids of the major homology region (MHR), lies within the carboxy-terminal domain of the protein. Structural and sequence similarities among CA proteins of retroviruses and the CA-like proteins of hepatitis B virus and various retrotransposons suggest that the MHR is involved in an aspect of replication common to these reverse-transcribing elements. Conservative substitutions in this region of the Rous sarcoma virus protein were lethal due to a severe deficiency in reverse transcription, in spite of the presence of an intact genome and active reverse transcriptase in the particles. This finding suggests that the mutations interfered with normal interactions among these constituents. A total of four genetic suppressors of three lethal MHR mutations have now been identified. All four map to the sequence encoding the CA-spacer peptide (SP) region of Gag. The F167Y mutation in the MHR was fully suppressed by a single amino acid change in the alpha helix immediately downstream of the MHR, a region that forms the major dimer interface in human immunodeficiency virus CA. This finding suggests that the F167Y mutation indirectly interfered with dimerization. The F167Y defect could also be repaired by a second, independent suppressor in the C-terminal SP that was removed from CA during maturation. This single residue change, which increased the rate of SP cleavage, apparently corrected the F167Y defect by modifying the maturation pathway. More surprising was the isolation of suppressors of the R170Q and L171V MHR mutations, which mapped to the N-terminal domain of the CA protein. This finding suggests that the two domains, which in the monomeric protein are separated by a flexible linker, must communicate with each other at some unidentified point in the viral replication cycle.
Rous sarcoma virus (RSV) capsid (CA) sequences exist in multiple forms during the viral replication cycle. Initially they are synthesized as part of the larger Gag polyprotein (Fig. 1). During or shortly after budding, the virus-encoded protease (PR) becomes activated, initiating the cleavage of Gag and the release of CA and the adjacent spacer peptide (SP; previously referred to as CA1 [28]). Further trimming at the C terminus by PR, removing either 9 or 12 amino acids, yields a mixture of two mature CA molecules of 240 and 237 amino acids (CA3 and CA2, respectively). These CA species, together with the viral replication machinery, form the electron-dense core of mature virions (9, 14, 36).
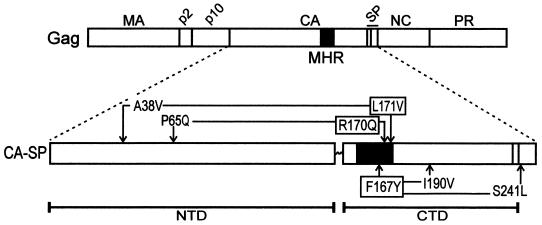
FIG. 1.
Positions of MHR and suppressor mutations in Gag. The wild-type RSV Gag protein is illustrated with the locations of the major cleavage products indicated along the top. Expanded below Gag is the CA-SP protein with the two domains of CA (NTD and CTD) separated by the flexible linker and the MHR (black box). The three MHR alleles are indicated by boxes and are connected by arrows to their respective suppressors.
The contributions that these CA sequences make to the viral replication cycle have been investigated extensively with several viruses by site-directed mutagenesis. The variety of phenotypes found in these studies present a complex picture of CA function. For example, deletions throughout RSV CA and SP sequences lead to the production of exceedingly large particles (23). Although some human immunodeficiency virus type 1 (HIV-1) CA mutants also show increased heterogeneity of virion size (8, 22), this region does not appear to be the major factor constraining HIV-1 particle dimensions (13). Rather, the N-terminal half of CA appears to be involved in maintaining the shape of the conical HIV-1 core (8, 16). In spite of these apparent functions, all of the RSV CA sequences (7, 34a) and most of the CA domain of HIV-1 Gag (1, 3) can be deleted without prevention of budding. Taken together, these results suggest that CA sequences within Gag are required for the assembly of particles normal in size, shape, appearance, and infectivity but are not absolutely essential for particle release.
In contrast to mutations within the CA coding sequence that perturb normal particle assembly and structure, others have been isolated that only appear to block infectivity. The most well studied of these mutants have substitutions in a highly conserved part of CA named the major homology region (MHR) (7, 25, 32, 34a). For RSV, biochemical analyses of these mutants have shown that the released particles are normal in size and density and contain normal amounts of the envelope glycoproteins, genomic RNA, and tRNA primer but exhibit a defect in reverse transcription (4, 7). These data suggest that CA has an important postassembly function that is needed for the establishment of infection once the particle enters a new cell. However, none of these findings has provided an explanation for what that critical activity might be.
At least two scenarios could explain the role of CA early in infection. In one, the arrangement of the mature CA proteins to form the protein shell of the viral core may provide a protective environment essential for the operation of the reverse transcription machinery. Another possibility is that CA may have a nonstructural function, perhaps involving a direct interaction with another viral component that is required for reverse transcription. Techniques such as X-ray crystallography and nuclear magnetic resonance spectroscopy are beginning to provide a framework for CA assembly that may assist in the interpretation of the biochemical and genetic data. Structures of the CA monomer from a variety of retroviruses are very similar and indicate that CA folds into two domains separated by a flexible linker (2, 5,11, 15, 18, 20, 21, 27) (Fig. 1). The MHR is located in the C-terminal domain (CTD) immediately following the interdomain linker, and many of the conserved MHR residues appear to be involved in a hydrogen-bonding network that is critical for the maintenance of the overall CTD structure (11). Genetic data for RSV CA support these biophysical models, since mutations predicted to destroy critical hydrogen bonding or disrupt the hydrophobic core of the CTD destroy particle assembly and budding (4, 21). However, neither the structural nor the genetic studies provide any insights to explain the loss of infectivity in MHR mutants with normal assembly and budding. Furthermore, all of the forms of CA examined to date exist only in the mature core, and it is impossible for these individual static structure determinations to provide comprehensive information about rearrangements or alternate conformations adopted by the multiple, dynamic CA sequences during particle assembly, PR-dependent maturation, and core disassembly.
To overcome these limitations and to provide an additional approach to understanding the puzzle posed by these assembly-competent, noninfectious mutants, we sought second-site suppressors of deleterious MHR mutations. This method takes advantage of the powerful selection for the restoration of viral infectivity to determine which genetic changes are sufficient to repair these severely compromised mutants. In addition, this method is devoid of the investigator bias that is inherent in site-directed mutagenesis studies, and suppressors can theoretically be isolated for mutants blocked at any step during replication. Our findings suggest that the structures of CA and the capsid shell are critical for the assembly and/or function of the reverse transcription machinery. In addition, this study yielded the unanticipated conclusion that the proper formation of the mature core likely depends both on communication between the two domains of CA at some point during the replication cycle and on the rate of CA maturation.
MATERIALS AND METHODS
Mutant proviral genomes.
The following mutant proviral genomes used to initiate suppressor searches have all been described previously: pRC.F167Y, pRC.L171I, pRC.L171F, pRC.L171A, pRC.Q158E, pRC.Q158N, pJD.R170Q, and pJD.L171V (4, 7). Each contains the gag gene from the RSV Prague C genome (31) cloned into either pJD100 (Prague A) or pBH-RCAN (Schmidt-Ruppin A). The infectivity defect caused by these various gag alleles has never been found to be influenced by either the vector used or the choice of avian cells.
Outgrowth of infectious viruses.
Infections were begun by transfection of mutant proviral genomes into either turkey embryo fibroblasts (TEFs) or transformed quail cells (QT6 cells). The use of TEFs appeared to slightly favor the outgrowth of infectious viruses, and so these cells were used for most experiments. Transfected cells were serially passaged over a period of 1 month. Culture supernatants were monitored for increases in cell-free reverse transcriptase (RT) activity, which would have indicated that a reversion event had occurred in the culture. To isolate revertant viruses, RT-positive culture supernatants were placed onto uninfected TEFs (for JD100 proviral genomes) or QT6 cells (RCAN genomes). Infected cell clones were grown either under an agar overlay to identify v-src-transformed TEFs or under selection with 300 μg of hygromycin/ml for RCAN-infected cells. After 2 weeks, colonies were picked and expanded into cell lines. Viruses from the resulting producer lines were tested as described below to confirm their infectivity.
Preparation, PCR amplification, and sequencing of genomic DNA.
Genomic DNA from producer lines was isolated by washing cells twice with Tris-buffered saline and incubating them for 2 h in lysis solution (50 mM Tris [pH 8.3], 100 mM EDTA, 200 mM NaCl, 0.5% sodium dodecyl sulfate, 333 μg of proteinase K/ml). DNA in cell lysates was then subjected to phenol-chloroform extraction, chloroform extraction, and isopropanol precipitation. Viral sequences in the genomic DNA were amplified by PCR using primers complementary to gag. Duplicate PCRs generated from each genomic DNA preparation were then sequenced by standard protocols.
Construction of single and double mutants by use of RCAN vectors.
Suspected suppressors identified by sequencing were recreated in RCAN by oligonucleotide-directed mutagenesis (35) both with and without the original MHR mutations. The following oligonucleotides were used to create mutants not described elsewhere: I190V, 5′-CCGGTGATCGTTGACTGCTTT-3′; A38V, 5′-CGATTACTATGGTAGAAGTGGAAGC-3′; V40M, 5′-CTATGGCAGAAATGGAAGCGCTTATG-3′; P65Q, 5′-GCCTGCCCAATATGCCTTATG-3′; A38V/V40 M, 5′-CGATTACTATGGTAGAAATGGAAGCGCTTATG-3′; and S241L 5′-GCGGCCATGTTGTCTGCTATCC-3′. RCAN vectors containing these mutations were constructed by exchanging the small SstI/HpaI fragment of RCAN with the corresponding fragment of mutant M13 DNA.
Transfections and infectivity assays.
The infectivity of the reconstructed proviral plasmids was measured with TEFs. The cells were transfected by the calcium phosphate method as previously described (6, 7), and the resulting culture medium was collected and filtered through a 0.45-μm-pore-size filter. A portion of the filtered medium was spun (126,000 × g for 40 min) through a 25% sucrose (in phosphate-buffered saline) cushion and analyzed for RT activity (7). The remaining portions of the filtered medium were adjusted to equal amounts of RT activity and added to fresh plates of TEFs to initiate infections. The cells were serially passaged. A small portion of medium was collected prior to each passage and frozen. At the end of the experiment, the amount of virus released into the medium of infected cells at each time point was determined by an RT assay.
Kinetics of CA processing.
QT6 cells were transfected with RCAN proviral plasmids in quadruplicate. At 48 h posttransfection, the cells were labeled for 15 min with [35S]methionine (200 μCi per ml of labeling medium), after which the labeling medium was replaced with medium containing 15 mg of cold methionine/ml. Cells on one plate from each set were lysed immediately; cells on the remaining plates were lysed 1, 2, and 4 h after the labeling was started. Gag proteins were analyzed by immunoprecipitation with anti-RSV serum followed by electrophoresis and autoradiography (35).
Detergent sensitivity assays.
QT6 cells were transfected and labeled with [35S]methionine as described previously (6). Medium samples were collected and spun for 30 s at 15,000 × g to remove cellular debris. Step gradients were prepared with 0.5 ml of either 5% sucrose or 5% sucrose plus 1% Triton X-100 layered on top of 2.0 ml of 10% sucrose. Each cleared medium sample was split in half and layered onto a sucrose gradient and a sucrose–Triton X-100 gradient. Both sets of gradients were spun at 126,000 × g for 40 min. The supernatant was recovered from each tube, and the pellet was resuspended in 3 ml of immunoprecipitation buffer B (35). Gag proteins were immunoprecipitated from the supernatant and pellet fractions and resolved on sodium dodecyl sulfate-polyacrylamide gels as described previously (35). The [35S]methionine counts associated with the triplet of CA proteins were determined by phosphorimage analysis. For each half of the experiment (i.e., with detergent or without), the amount of pelletable CA was expressed as a percentage of total CA (pellet plus supernatant).
RESULTS
Isolation of revertants of deleterious MHR mutants.
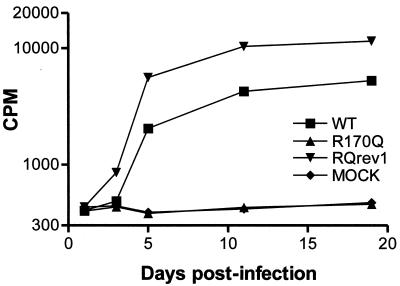
The extensive conservation of the MHR, not only among retroviruses (34a) but also among various retrotransposons and hepatitis B virus (26, 38), suggests that it is of central importance in the viral replication cycle. For this reason, we sought suppressors for MHR point mutants that assemble normally but exhibit severe replication defects. Proviral DNA from each of these particle-producing, noninfectious MHR mutants (Table 1) was transfected into susceptible avian cells as described above. Variable numbers of transfections were done with each mutant, but the combined total exceeded 50 independent transfections. After 1 month, approximately 30% of the cultures exhibited elevated levels of RT activity. To isolate individual infectious viruses, fresh cells were incubated with cell-free medium from the RT-positive cultures, and clones of infected cells were selected as described in Materials and Methods. The replication kinetics of particles from each of these producer lines were compared to the growth kinetics of the parental mutant and wild-type virus. A typical growth curve from these experiments, for a revertant of the R170Q MHR mutant (R170Qrev1), is shown in Fig. 2.
TABLE 1.
Nucleotide changes present in gag genes of revertant viruses
| MHR mutation | Revertant isolatea | Additional nucleotide change(s)b | Resulting amino acid substitution(s)c |
|---|---|---|---|
| F167Y | F167Yrev1 | A1664G | L190V |
| F167Y | F167Yrev2 | C1818T | S241L |
| R170Q | R170Qrev1 | C1300A | P65Q |
| L171V | L171Vrev1 | C1209T and G1214A | A38V and V40M |
| Q158E | None | NA | NA |
| Q158N | None | NA | NA |
| L171A | None | NA | NA |
| L171L | None | NA | NA |
| L171F | None | NA | NA |
Revertant viruses were isolated by transfection of proviral genomes of the indicated MHR mutants and repeated passage of the transfected cells as described in the text.
Sequencing of the gag gene of integrated revertant viruses revealed nucleotide changes in addition to those present in the original MHR mutants. The first letter represents the wild-type nucleotide, and the second letter is the nucleotide present in the revertant virus. The number separating the two letters is the position of the change, with the numbering beginning at the first nucleotide of the RSV genome. NA, not applicable.
The position of the change is numbered starting with the first amino acid of CA.
FIG. 2.
Growth kinetics of infectious R170Qrev1 (RQrev1) virus. A cell line releasing RQrev1 virus was created by passage of RT-positive culture medium from a pJD.R170Q-transfected TEF culture on fresh TEFs and selection of individual transformed cell clones by focus formation (see Materials and Methods). To confirm the infectivity of the RQrev1 virus released by the cells, culture medium was collected and used to infect fresh TEFs. Virus particles bearing the original R170Q mutation but which had not had the opportunity to undergo reversion were included for comparison. These, as well as the wild-type (WT) control particles, were produced by transfection of QT6 cells and normalized for RT activity against the RQrev1 virus preparation prior to initiation of infection in TEFs.
Genomic DNA was prepared from each of the cell lines producing infectious particles, and the MHR of the integrated proviruses was amplified and sequenced. Four of the integrated proviruses retained their respective parental MHR mutation, suggesting that they also contained second-site-suppressing mutations. One was derived from a primary culture transfected with the L171V mutant, one was from an R170Q primary culture, and two were from independent cultures of F167Y-transfected cells (Table 1).
Identification and characterization of second-site-suppressing mutations.
To identify the compensatory mutations, we began by sequencing the gag gene of the integrated proviruses. All four gag alleles contained at least one nucleotide change (in addition to the MHR mutation) that would result in an amino acid substitution (Table 1), and all substitutions mapped to the CA or CA-SP region of Gag. Using M13-based site-directed mutagenesis, we recreated each potential suppressing mutation in an otherwise wild-type RCAN plasmid, both alone and in combination with the parental MHR mutation. Growth kinetics of virus produced from each construct were tested with TEFs and compared to that of wild-type virus and that of virus with the appropriate MHR mutation (Fig. 3).
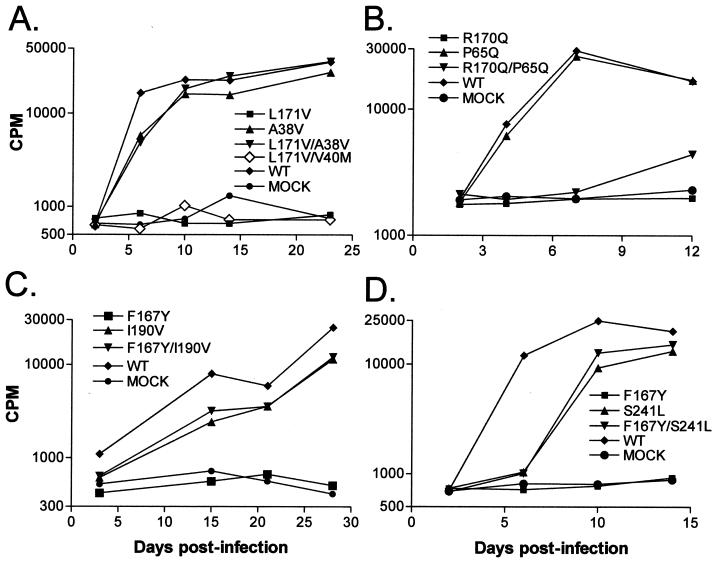
FIG. 3.
Assay of potential MHR suppressors. Virus particles bearing the indicated single or double mutations were produced in QT6 cells by transfection with the corresponding proviral plasmids. At 48 h posttransfection, medium was collected from the plates, normalized for RT activity, and used to initiate infections of TEFs (see Materials and Methods). Potential suppressors tested were A38V and V40M (A), P65Q (B), I190V (C), and S241L (D). WT, wild type.
Since L171Vrev1 contained two mutations in gag, both were tested for suppression activity. Only the A38V substitution was able to restore efficient replication to the L171V parent (Fig. 1 and 3A). No other changes outside of Gag were necessary. Interestingly, viruses containing either the A38V or the V40M change alone or both together replicated with wild-type kinetics (Fig. 3A and data not shown), as did a mutant containing all three substitutions (L171V/A38V/V40 M) (data not shown).
The R170Q substitution is adjacent to the leucine at position 171 in the MHR, and its only identified potential suppressing mutation (P65Q) lies close to the L171V suppressor in the amino terminus of CA (Fig. 1). The P65Q substitution had no detrimental effect on the replication of an otherwise wild-type genome (Fig. 3B). However, P65Q provided only a low level of rescue to the R170Q mutant. The possibility has not been ruled out that the weakness of the rescue is due to the choice of an RCAN vector rather than a JD100 vector, although this possibility seems unlikely. Another, more interesting possibility is that the particles from the R170Qrev1 cell line, which exhibited replication kinetics similar to those of wild-type virus (Fig. 2), contain an additional suppressor of R170Q that remains to be identified. Since the entire gag gene from the R170Qrev1 cell line has been sequenced and no additional mutations have been found, it appears that a second suppressor would have to lie within the pol, env, or noncoding sequences. A search for any such contributing mutations is currently under way.
Two independently isolated suppressors were identified for the F167Y mutation and, unlike the L171V and R170Q suppressors, both were located downstream of the MHR (Fig. 1). One change, just 23 amino acids away from the original substitution, is I190V in the CTD of CA. This change alone was not detrimental to viral replication and was able to restore wild-type growth kinetics, as seen with the F167Y/I190V double mutant (Fig. 3C). The other potential suppressor was not contained within the coding sequence of the 240-amino-acid mature CA protein but, instead, mapped to the first residue of the adjacent spacer region, where serine was changed to leucine (S241L). Even though SP (and this altered amino acid) is proteolytically removed from CA during maturation, its presence in the Gag precursor is able to restore infectivity to the F167Y mutant virus (Fig. 3D). Since the first residue of SP constitutes the P1′ position of a PR recognition sequence between CA and the nucleocapsid protein (NC), we examined the proteolytic cleavage profile of Gag proteins containing the S241L change. Extensive analyses of PR cleavage events indicated that substitution of a bulky hydrophobic residue at this position should lead to an increase in the rate of cleavage (35a). Indeed, when examined by pulse-chase analysis, Gag proteins with the S241L change alone or in combination with F167Y showed more rapid CA processing than did the wild-type protein (Fig. 4), as indicated by the more rapid disappearance of the fastest-migrating CA band (CA-SP). Whether the faster CA maturation exhibited by this mutant is required for the suppression of the MHR mutation or is simply a secondary effect of the amino acid substitution in the PR cleavage region remains unclear.
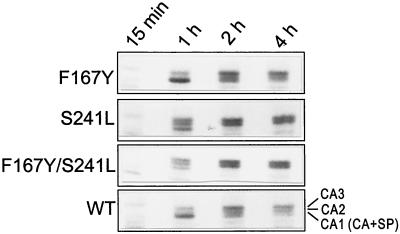
FIG. 4.
Kinetics of maturation of S241L CA protein. QT6 cells transfected with RCAN plasmids bearing each of the indicated mutations or pair of mutations were labeled as described in Materials and Methods. At 15 min, most of the CA protein remained in the Gag precursor (data not shown). CA-SP, the precursor to the CA2 and CA3 proteins, ran more rapidly on the gel than predicted from its molecular mass (28). Unusually rapid processing in the S241L and F167Y/S241L mutants, compared to that in the wild type (WT), is evident from the appearance of abundant CA2 and CA3 proteins by 1 h.
Detergent sensitivity of viral cores.
Previous studies with MHR mutants suggested that weakened interactions within the core (as measured by increased solubility of CA after detergent treatment) correlated with the loss of infectivity (7) and therefore might be part of the explanation of the lethality of the mutations. The revertants identified here exhibited a range of detergent resistance. Particles produced from cells transfected with each revertant, parental mutant, and wild-type DNA were collected and pelleted through sucrose step gradients in the presence and absence of 1% Triton X-100. Table 2 summarizes the results from three representative experiments done with the F167Y/I190V, L171V/A38V, and F167Y/S241L double mutants. Without detergent present, approximately 90% of the CA from wild-type virus was immunoprecipitated from the pellet fraction in each experiment. Between 30 and 40% of wild-type CA protein remained pelletable in the presence of detergent. In contrast, only 9% of the CA protein from the noninfectious F167Y mutant pelleted through the gradient containing detergent. The CA proteins from both I190V virus (42%) and the F167Y/I190V revertant (43%) behaved in a manner similar to that of the wild type, indicating that full detergent resistance was restored to the F167Y mutant by the I190V suppressor. The correlation between detergent resistance and infectivity did not extend to the other mutants and their suppressors, however. The A38V suppressor, while able to restore full infectivity, only partially restored resistance (∼25% relative to the wild type), and the S241L suppressor did not restore any detectable detergent resistance over the F167Y substitution alone (Table 2). Thus, infectivity can be restored without full repair of the structural defect that is detected by the detergent sensitivity assay.
TABLE 2.
Detergent resistance of mutant viral cores
| Virus | % Gag protein in pellet in the absence (−) or the presence (+) of Triton X-100a
|
|
|---|---|---|
| − | + | |
| F167Y | 88 | 9 |
| L190V | 92 | 42 |
| F167Y/I190V | 93 | 43 |
| Wild type | 89 | 30 |
| L171V | 88 | 1 |
| A38V | 86 | 28 |
| L171V/A38V | 83 | 10 |
| Wild type | 91 | 41 |
| F167Y | 90 | 7 |
| S241L | 95 | 13 |
| F167Y/S241L | 95 | 4 |
| Wild type | 92 | 35 |
Virus particles were spun through gradients containing or lacking Triton X-100 as described in Materials and Methods. Gag proteins were immunoprecipitated from the pellet and supernatant fractions. The percentage of the total Gag protein found in the pellet fraction is shown for each virus and each detergent condition.
DISCUSSION
At the outset, we suspected that the high degree of conservation of the MHR might impose severe restrictions that would make suppressor isolation difficult. Although some MHR mutants yielded no revertants, others spawned suppressors with a frequency sufficient to make their isolation feasible. Precise reversion events that restored the wild-type amino acid sequence occurred in some cases, but four viruses that grew out of the approximately 50 starting cultures contained the original MHR mutations. In each, a secondary substitution that conferred suppression was mapped to the CA-SP region of Gag. None of these suppressors fell within the MHR itself. Rather, all suppressors compensated for the original defects “at a distance” and, in three of the four, actually acted between different protein domains in the CA-SP region (Fig. 5).
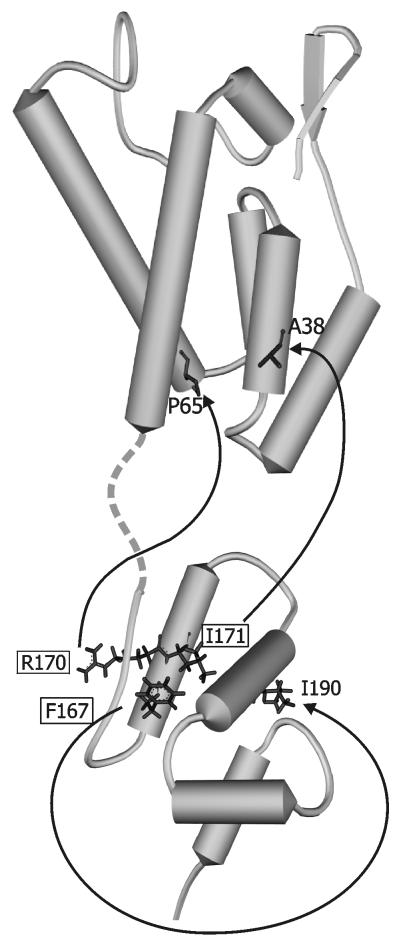
FIG. 5.
Positions of relevant residues on the RSV CA protein structure. Models of the CA NTD (top) and the CA CTD (below) are combined in this diagram using PDB files 1EM9 and 1EOQ for the NTD and the CTD, respectively. The flexible interdomain linker is represented by the broken line. MHR residues F167, R170, and I171 in the CTD are marked with boxes and are connected by arrows to their respective suppressors. The S241L mutation, which suppresses the F167 MHR substitution, is not shown since it occurs in the SP and is not represented in the CA model.
Potential effects of suppressors on the mature CA shell.
Three-dimensional structures (Fig. 5) for the RSV CA monomer have been recently published by two groups (5, 21). The folding of the two protein domains is similar to those in dimers of HIV-1 CA (10, 11, 15) and equine infectious anemia virus (EIAV) CA (18) and monomers of human T-cell leukemia virus CA (20). Although it appears not to be involved in direct CA-CA interactions, the alpha-helical portion of the MHR packs against the second CTD helix, which in turn forms the major dimer interface (11). A model of the HIV-1 core, derived from cryoelectron microscopy of core-like particles assembled from purified CA, indicates that CA monomers are arranged in hexameric rings through interactions between their N-terminal domains (NTDs) (12). The dimerization domain in the CTD of each monomer provides a connection to the CTD in a neighboring hexamer to form a three-dimensional network with the MHR domains arrayed on the interior surface of the shell (24). Although no similar work has been published for RSV, Ganser et al. (12) have made a convincing argument that the same principles are likely to apply to all retroviruses.
Interpretation of the suppressors in light of these structural models raises the possibility that the F167Y, R170Q, and L171V substitutions, all of which exhibit identical phenotypic properties (4), are lethal at least in part because they interfere with normal dimeric interactions between CTDs. The F167Y substitution introduces a bulkier aromatic side chain into the hydrophobic core of the CTD (Fig. 5) and may therefore destabilize this protein domain (21). The suppressor (I190V) cannot directly counteract this effect, since it lies on the surface of the domain. The homologous position in HIV-1 CA lies at the primary interface of interacting CA-CA dimers. Thus, it is likely that in the RSV protein, I190V acts to strengthen CA-CA interactions that have been indirectly weakened by F167Y. This scenario appears consistent with the loss of detergent resistance in the original mutant and its restoration in the double mutant.
The two suppressors found in the NTDs are more difficult to understand using this model. Like the I190V suppressor, the A38V substitution occurred at a surface-exposed position on the RSV monomer (Fig. 5) and would not be expected to alter the secondary and tertiary structures of either domain (5, 21). Both A38V and P65Q are located in positions that could potentially influence CA-CA interactions between the six NTDs in the hexameric rings (24). However, it is difficult to imagine how strengthening these interactions could restore infectivity, since it would leave the defect in the CTD unaltered.
We are left to conclude that there are likely to be other consequences of the suppressors that cannot be deduced from the study of the published models. One possibility is that the suppressors modify the maturation pathway that gives rise to the final core particle (see below). Another intriguing idea is that the existence of NTD suppressors indicates that physical contact between the CTD of one CA monomer and the NTD of a neighboring molecule occurs in the wild-type virus. Such contacts have been found in crystals of HIV-1 CA (2) and EIAV CA (18), but no additional biochemical evidence for their existence in viruses has been documented. The model of Li et al. (24) gives no support for such binding events in an intact CA shell. However, if such direct contact does indeed occur, it may involve a transient intermediate that is not present in the mature core. Alternatively, it could occur in only a minority of the total population of CA proteins or only when interior components of the core are present. In any of these cases, the interaction would not have been identified in the reconstruction of in vitro-assembled CA shells (24).
If the suppressors simply restore dimerization between subunits that has been disrupted by the MHR substitutions, it is not clear why detergent resistance is not restored along with infectivity in all cases. Previous studies from our laboratory concluded that this property reflects the formation of intermolecular interactions involving CA during core maturation. Various mutations that alter the maturation of CA destroy detergent resistance, and until now every CA mutation that causes a loss of detergent resistance has been proven to be lethal (4, 6,7). The exception posed by the L171V/A38V and F167Y/S241L double mutant viruses is perplexing and may indicate that the interactions detected by this assay are actually inconsequential for infectivity. On the other hand, it remains possible that the suppressors truly restore a critical interaction but one with an affinity lower than that present in the wild-type virus and thus undetectable by this crude assay.
Potential effects of suppressors on core maturation.
Suppression of lethality in the F167Y/S241L double mutant clearly cannot be explained from the structural models. The S241 residue is located at the first position in the nine-amino-acid SP separating CA from NC and is not present in mature CA protein. The SP tail appears disordered in purified RSV CA-SP (21), and it is not apparent how the S241L substitution could directly correct any disruption to the core of the CTD caused by the F167Y mutation. Instead, it is more likely that S241L suppression is due to an influence on critical interactions between Gag proteins or cleavage intermediates during assembly or maturation. This notion suggests that the MHR plays a role in controlling the proper reorganization of the particle interior, consistent with previous findings that SP mutants show a replication defect resembling that seen in MHR mutants (4; unpublished data), with published studies suggesting that HIV-1 core formation requires that SP be temporarily attached at the C terminus (6, 22, 29), and with the idea that the removal of SP during maturation can act as a conformational switch to alter interactions between CA subunits (17).
In HIV-1, RSV, and Mason-Pfizer monkey virus, the N-terminal proline of CA and adjacent residues are critical for controlling the shape of assembled core structures (19, 30, 33). It has been suggested that proteolytic cleavage at the N terminus of CA triggers a refolding of the protein, leading to condensation of the entire core structure (33). If so, then proper folding-association of the NTD may be a prerequisite for maturation of the CTD. It is possible, therefore, that perturbations of the NTD folding-association pathway caused by the A38V and P65Q suppressor mutations allow the formation of functional core structures in spite of the downstream MHR mutations.
Given the very strong conservation of the residues in the MHR, both the relative ease with which suppressors were found and the distribution of the confirmed suppressors across the CA protein seem quite surprising. Why these residues are conserved still needs to be answered. The results presented here and elsewhere (4) imply that the MHR has a prominent role in the formation, maintenance, and function of the core structure. MHR suppressors will ultimately provide an important means for evaluating the validity of future models of core structure and assembly, particularly with respect to the definition of important transient intermediates, determination of the influence of PR processing rate on the formation of the normal capsid structure, and/or clarification of the intermolecular interactions that underlie core function in reverse transcription.
ACKNOWLEDGMENTS
We are grateful to Rich Kingston and Michael Rossmann for sharing structures of RSV CA prior to publication and to Volker Vogt and Wes Sundquist for insightful discussions. Special thanks are extended to Carol Wilson for expert technical assistance in the detergent resistance studies and to Tina Cairns for numerous collaborative interactions as well as careful review of the manuscript.
This work was supported in part by National Institutes of Health (NIH) grant CA47482 (to J.W.W.) and by monies from the Four Diamonds Research Fund (to R.C.C.). J.B.B. was supported by NIH training grant CA60395.
REFERENCES
- 1.Accola M A, Strack B, Gottlinger H G. Efficient particle production by minimal Gag constructs which retain the carboxy-terminal domain of human immunodeficiency virus type 1 capsid-p2 and a late assembly domain. J Virol. 2000;74:5395–5402. doi: 10.1128/jvi.74.12.5395-5402.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berthet-Colominas C, Monaco S, Novelli A, Sibai G, Mallet F, Cusack S. Head-to-tail dimers and interdomain flexibility revealed by the crystal structure of HIV-1 capsid protein (p24) complexed with a monoclonal antibody Fab. EMBO J. 1999;18:1124–1136. doi: 10.1093/emboj/18.5.1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Borsetti A, Ohagen A, Gottlinger H G. The C-terminal half of the human immunodeficiency virus type 1 Gag precursor is sufficient for efficient particle assembly. J Virol. 1998;72:9313–9317. doi: 10.1128/jvi.72.11.9313-9317.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cairns T M, Craven R C. Viral DNA synthesis defects in assembly-competent Rous sarcoma virus CA mutants. J Virol. 2001;75:242–250. doi: 10.1128/JVI.75.1.242-250.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Campos-Olivas R, Newman J L, Summers M F. Solution structure and dynamics of the Rous sarcoma virus capsid protein and comparison with capsid proteins of other retroviruses. J Mol Biol. 2000;296:633–649. doi: 10.1006/jmbi.1999.3475. [DOI] [PubMed] [Google Scholar]
- 6.Craven R C, Leure-duPree A E, Erdie C R, Wilson C B, Wills J W. Necessity of the spacer peptide between CA and NC in the Rous sarcoma virus Gag protein. J Virol. 1993;67:6246–6252. doi: 10.1128/jvi.67.10.6246-6252.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Craven R C, Leure-duPree A E, Weldon R A, Wills J W. Genetic analysis of the major homology region of the Rous sarcoma virus Gag protein. J Virol. 1995;69:4213–4227. doi: 10.1128/jvi.69.7.4213-4227.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dorfman T, Bukovsky A, Ohagen A, Hoglund S, Gottlinger H G. Functional domains of the capsid protein of human immunodeficiency virus type 1. J Virol. 1994;68:8180–8187. doi: 10.1128/jvi.68.12.8180-8187.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fuller S D, Wilk T, Gowen B E, Krausslich H G, Vogt V M. Cryo-electron microscopy reveals ordered domains in the immature HIV-1 particle. Curr Biol. 1997;7:729–738. doi: 10.1016/s0960-9822(06)00331-9. [DOI] [PubMed] [Google Scholar]
- 10.Gamble T R, Vajdos F F, Yoo S, Worthylake D K, Houseweart M, Sundquist W I, Hill C P. Crystal structure of human cyclophilin A bound to the amino-terminal domain of HIV-1 capsid. Cell. 1996;87:1285–1294. doi: 10.1016/s0092-8674(00)81823-1. [DOI] [PubMed] [Google Scholar]
- 11.Gamble T R, Yoo S, Vajdos F F, von Schwedler U K, Worthylake D K, Wang H, McCutcheon J P, Sundquist W I, Hill C P. Structure of the carboxyl-terminal dimerization domain of the HIV-1 capsid protein. Science. 1997;278:849–853. doi: 10.1126/science.278.5339.849. [DOI] [PubMed] [Google Scholar]
- 12.Ganser B K, Li S, Klishko V Y, Finch J T, Sundquist W I. Assembly and analysis of conical models for the HIV-1 core. Science. 1999;293:80–83. doi: 10.1126/science.283.5398.80. [DOI] [PubMed] [Google Scholar]
- 13.Garnier L, Ratner L, Rovinski B, Cao S X, Wills J W. Particle size determinants in the human immunodeficiency virus type 1 Gag protein. J Virol. 1998;72:4667–4677. doi: 10.1128/jvi.72.6.4667-4677.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gelderblom H R, Hausmann E H S, Ozel M, Pauli G, Koch M A. Fine structureof human immunodeficiency virus (HIV) and immunolocalization of structural proteins. Virology. 1987;156:171–176. doi: 10.1016/0042-6822(87)90449-1. [DOI] [PubMed] [Google Scholar]
- 15.Gitti R K, Lee B M, Walker J, Summers M F, Yoo S, Sundquist W I. Structure of the amino-terminal core domain of the HIV-1 capsid protein. Science. 1996;273:231–235. doi: 10.1126/science.273.5272.231. [DOI] [PubMed] [Google Scholar]
- 16.Gottlinger H G, Sodroski J G, Haseltine W A. Role of capsid precursor processing and myristoylation in morphogenesis and infectivity of human immunodeficiency virus type 1. Proc Natl Acad Sci USA. 1989;86:5781–5785. doi: 10.1073/pnas.86.15.5781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gross I, Hohenberg H, Wilk T, Wiegers K, Grattinger M, Muller B, Fuller S, Krausslich H G. A conformational switch controlling HIV-1 morphogenesis. EMBO J. 2000;19:103–113. doi: 10.1093/emboj/19.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jin Z, Jin L, Peterson D L, Lawson C L. Model for lentivirus capsid core assembly based on crystal dimers of EIAV p26. J Mol Biol. 1999;286:83–93. doi: 10.1006/jmbi.1998.2443. [DOI] [PubMed] [Google Scholar]
- 19.Joshi S M, Vogt V M. Role of the Rous sarcoma virus p10 domain in shape determination of Gag virus-like particles assembled in vitro and within Escherichia coli. J Virol. 2000;74:10260–10268. doi: 10.1128/jvi.74.21.10260-10268.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khorasanizadeh S, Campos-Olivas R, Summers M F. Solution structure of the capsid protein from the human T-cell leukemia virus type-I. J Mol Biol. 1999;291:491–505. doi: 10.1006/jmbi.1999.2986. [DOI] [PubMed] [Google Scholar]
- 21.Kingston R L, Fitzon-Ostendorp T, Eisenmesser E Z, Schatz G W, Vogt V M, Post C B, Rossmann M G. Structure and self-association of the Rous sarcoma virus capsid protein. Structure. 2000;8:617–628. doi: 10.1016/s0969-2126(00)00148-9. [DOI] [PubMed] [Google Scholar]
- 22.Krausslich H-G, Facke M, Heuser A-M, Konvalinka J, Zentgraf H. The spacer peptide between human immunodeficiency virus capsid and nucleocapsid proteins is essential for ordered assembly and viral infectivity. J Virol. 1995;69:3407–3419. doi: 10.1128/jvi.69.6.3407-3419.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krishna N K, Campbell S, Vogt V M, Wills J W. Genetic determinants of Rous sarcoma virus particle size. J Virol. 1998;72:564–577. doi: 10.1128/jvi.72.1.564-577.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li S, Hill C P, Sundquist W I, Finch J T. Image reconstructions of helical assemblies of the HIV-1 CA protein. Nature. 2000;407:409–413. doi: 10.1038/35030177. [DOI] [PubMed] [Google Scholar]
- 25.Mammano F, Ohagen A, Hoglund S, Gottlinger H G. Role of the major homology region of human immunodeficiency virus type 1 in virion morphogenesis. J Virol. 1994;68:4927–4936. doi: 10.1128/jvi.68.8.4927-4936.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McClure M A, Johnson M S, Feng D F, Doolittle R F. Sequence comparisons of retroviral proteins: relative rates of change and general phylogeny. Proc Natl Acad Sci USA. 1988;85:2469–2473. doi: 10.1073/pnas.85.8.2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Momany C, Kovari L C, Prongay A J, Keller W, Gitti R K, Lee B M, Gorbalenya A E, Tong L, McClure J, Ehrlich L S, Summers M F, Carter C, Rossmann M G. Crystal structure of dimeric HIV-1 capsid protein. Nat Struct Biol. 1996;3:763–770. doi: 10.1038/nsb0996-763. [DOI] [PubMed] [Google Scholar]
- 28.Pepinsky R B, Papayannopoulos I A, Chow E P, Krishna N K, Craven R C, Vogt V M. Differential proteolytic processing leads to multiple forms of the CA protein in avian sarcoma and leukemia viruses. J Virol. 1995;69:6430–6438. doi: 10.1128/jvi.69.10.6430-6438.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pettit S C, Moody M D, Wehbie R S, Kaplan A H, Nantermet P V, Klein C A, Swanstrom R. The p2 domain of human immunodeficiency virus type 1 Gag regulates sequential proteolytic processing and is required to produce fully infectious virions. J Virol. 1994;68:8017–8027. doi: 10.1128/jvi.68.12.8017-8027.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rumlova-Klikova M, Hunter E, Nermut M V, Pichova I, Ruml T. Analysis of Mason-Pfizer monkey virus Gag domains required for capsid assembly in bacteria: role of the N-terminal proline residue of CA in directing particle shape. J Virol. 2000;74:8452–8459. doi: 10.1128/jvi.74.18.8452-8459.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schwartz D E, Tizard R, Gilbert W. Nucleotide sequence of Rous sarcoma virus. Cell. 1983;32:853–869. doi: 10.1016/0092-8674(83)90071-5. [DOI] [PubMed] [Google Scholar]
- 32.Strambio-de-Castillia C, Hunter E. Mutational analysis of the major homology region of Mason-Pfizer monkey virus by use of saturation mutagenesis. J Virol. 1992;66:7021–7032. doi: 10.1128/jvi.66.12.7021-7032.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.von Schwedler U K, Stemmler T L, Klishko V Y, Li S, Albertine K H, Davis D R, Sundquist W I. Proteolytic refolding of the HIV-1 capsid protein amino-terminus facilitates viral core assembly. EMBO J. 1998;17:1555–1568. doi: 10.1093/emboj/17.6.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weldon R A, Jr, Wills J W. Characterization of a small (25-kilodalton) derivative of the Rous sarcoma virus Gag protein competent for particle release. J Virol. 1993;67:5550–5561. doi: 10.1128/jvi.67.9.5550-5561.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34a.Wills J W, Craven R C. Form, function, and use of retroviral Gag proteins. AIDS. 1991;5:639–654. doi: 10.1097/00002030-199106000-00002. [DOI] [PubMed] [Google Scholar]
- 35.Wills J W, Craven R C, Achacoso J A. Creation and expression of myristylated forms of Rous sarcoma virus Gag protein in mammalian cells. J Virol. 1989;63:4331–4343. doi: 10.1128/jvi.63.10.4331-4343.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35a.Xiang Y, Thorick R, Vana M L, Craven R, Leis J. Proper processing of avian sarcoma/leukosis virus capsid proteins is required for infectivity. J Virol. 2001;75:6016–6021. doi: 10.1128/JVI.75.13.6016-6021.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yeager M, Wilson-Kubalek E M, Weiner S G, Brown P O, Rein A. Supramolecular organization of immature and mature murine leukemia virus revealed by electron cryo-microscopy: implications for retroviral assembly mechanisms. Proc Natl Acad Sci USA. 1998;95:7299–7304. doi: 10.1073/pnas.95.13.7299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zlotnick A, Stahl S J, Wingfield P T, Conway J F, Cheng N, Steven A C. Shared motifs of the capsid proteins of hepadnaviruses and retroviruses suggest a common evolutionary origin. FEBS Lett. 1998;431:301–304. doi: 10.1016/s0014-5793(98)00755-8. [DOI] [PubMed] [Google Scholar]