Abstract
Background
Lung cancer is a heterogeneous disease and the primary cause of cancer-related mortality worldwide. Somatic mutations, including large structural variants, are important biomarkers in lung cancer for selecting targeted therapy. Genomic studies in lung cancer have been conducted using short-read sequencing. Emerging long-read sequencing technologies are a promising alternative to study somatic structural variants, however there is no current consensus on how to process data and call somatic events. In this study, we preformed whole genome sequencing of lung cancer and matched non-tumour samples using long and short read sequencing to comprehensively benchmark three sequence aligners and seven structural variant callers comprised of generic callers (SVIM, Sniffles2, DELLY in generic mode and cuteSV) and somatic callers (Severus, SAVANA, nanomonsv and DELLY in somatic modes).
Results
Different combinations of aligners and variant callers influenced somatic structural variant detection. The choice of caller had a significant influence on somatic structural variant detection in terms of variant type, size, sensitivity, and accuracy. The performance of each variant caller was assessed by comparing to somatic structural variants identified by short-read sequencing. When compared to somatic structural variants detected with short-read sequencing, more events were detected with long-read sequencing. The mean recall of somatic variant events identified by long-read sequencing was higher for the somatic callers (72%) than generic callers (53%). Among the somatic callers when using the minimap2 aligner, SAVANA and Severus achieved the highest recall at 79.5% and 79.25% respectively, followed by nanomonsv with a recall of 72.5%.
Conclusion
Long-read sequencing can identify somatic structural variants in clincal samples. The longer reads have the potential to improve our understanding of cancer development and inform personalized cancer treatment.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12864-024-10792-3.
Keywords: Long read sequencing, Somatic structural variants detection, Benchmarking long read approaches, Small cell lung cancer
Background
Lung cancers are the most lethal type of cancer worldwide[1], with an overall five-year patient survival of 18.5% with certain subtypes like small cell lung cancer (SCLC) being as low as 5%[2]. The poor prognosis is linked to often late diagnosis and limited efficacy of current treatment modalities[3, 4]. Lung cancers exhibit a wide range of somatic genomic alterations, including single-nucleotide variants (SNV) as well as chromosomal structural variants (SV) [5–7]. Somatic SVs in lung cancer encompass large deletions, duplications, inversions and translocations, with implications for disease progression and treatment [8]. This is exemplified by the ALK gene fusions in non-small cell lung cancer (NSCLC), which generate abnormal proteins targetable with ALK inhibitors [9, 10]. The genomics of SCLC has been less explored compared to other subtypes. However somatic events such as bi-allelic inactivation of TP53 or RB1 [11–14], and complex genomic rearrangements including chromothripsis which was associated with CCND1 overexpression and deregulation of Rb1 have been reported as contributing to tumour development [14]. Further work to resolve somatic SV events may reveal mechanisms of lung tumorigenesis and could help develop more targeted therapies.
Long-read sequencing (LRS), such as the PromethION from Oxford Nanopore Technologies (ONT) or the Revio from PacBio, offers advantages for somatic SV detection compared to short-read sequencing (SRS) [15, 16]. This is due to the ability of LRS to produce sequence reads spanning thousands of bases, enabling comprehensive characterization of large (> 10 kb) and complex SVs [17]. Nanopore sequencing has been used to detect a novel type of SV occurring in cancer previously not identified using SRS [18], was used to resolve complex structural rearrangements and intra-tumour genomic heterogeneity [19, 20] and was proposed as a diagnostic approach for brain cancer [21]. Recent advancements in LRS have also enhanced sequencing accuracy, exceeding 99.9% [22], establishing LRS as a feasible method for cancer research and clinical genomics.
The potential of LRS for SV analysis has prompted the development of numerous computational tools [23–27]. However, independent benchmarking to assess the performance of each approach is limited, absent in clinical tumour samples, with previous studies focusing on the detection of germline SVs [28–31]. In our study, we benchmarked the performance of three long-read sequence aligners in conjunction with seven SV callers to identify somatic SVs in clinical samples from SCLC patients. For benchmarking, we compared the results of different LRS sequence aligners and variant callers against somatic SV events detected by short read sequencing. This work provides a comprehensive guide for clinical genomic studies for somatic SV identification.
Results
Overiew of benchmarking study
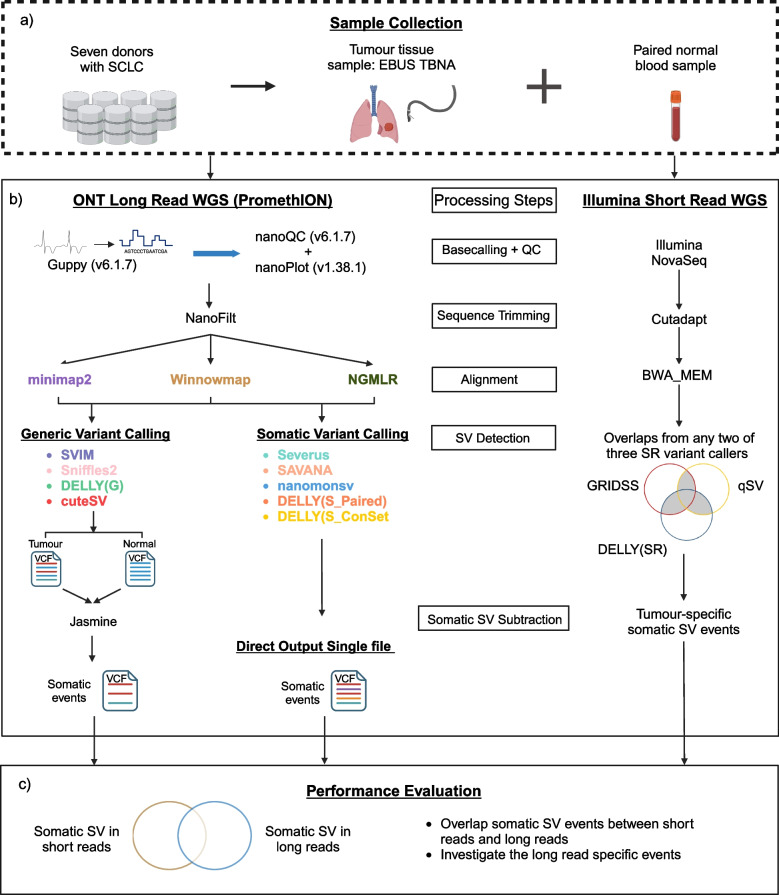
To assess the performance of somatic SV detection using LRS we sequenced DNA from a panel of seven small cell lung carcinoma (SCLC) tissue and paired non-tumour (normal) samples using the Oxford Nanopore PromethION (R9.4) (Fig. 1a). We compared the performance of three long-read sequence aligners (NGMLR, minimap2, and Winnowmap) with different SV calling approaches including four generic approaches (SVIM, Sniffles2, DELLY in generic mode and cuteSV) and five somatic approaches (Severus, SAVANA, nanomonsv and DELLY in two different somatic modes) (Fig. 1b). The performance of each SV caller was assessed by comparing to SV events identified from Illumina short read sequencing (Fig. 1c).
Fig. 1.
Overview of study design to benchmark alignment and somatic SV calling. The workflow includes multiple steps. a Sample collection. DNA was extracted from seven tumour lung samples collected from EBUS-TBNA and seven matching blood samples. b The normal and tumour DNA underwent whole genome sequencing (WGS) using ONT PromethION for long-read sequencing (LRS) and Illumina NovaSeq for short-read sequencing (SRS). The processing steps for identifying somatic SV events in both long-read and short-read data used different tools but followed a similar process: sequence base calling and alignment, followed by the application of various variant calling methods for SV detection. In LRS, three aligners (minimap2, Winnowmap, and NGMLR) were evaluated, in combination with two approaches (generic and somatic calling) for detecting somatic SVs. The four generic SV callers were cuteSV, DELLY (G), Sniffles2, and SVIM, which required manual subtraction to determine somatic SVs with Jasmine. The somatic callers were DELLY (S_Paired), DELLY (S_ConSet), nanomonsv, SAVANA, and Severus. c The performance of each approach in LRS was evaluated by comparing the somatic SVs identified to high-confidence somatic SV events (obtained from SRS of the same samples and called by two or more of these approaches: qSV, DELLY, and GRIDSS)
A comparison of the performance of three long-read sequence aligners
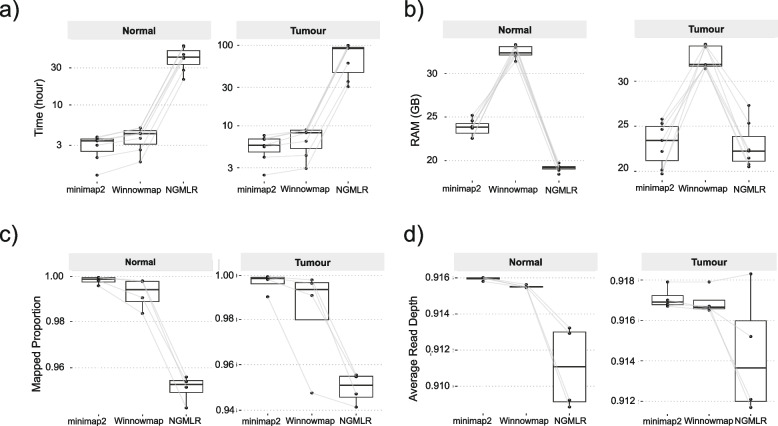
We compared the computational performance of three long-read aligners to process seven tumour and non-tumour patient samples by assessing runtime, RAM and CPU usage. The runtime for NGMLR was 11 times higher compared to minimap2 or Winnowmap for tumour and normal BAM files (Fig. 2a). The RAM used by Winnowmap was approximately 40% higher than that of NGMLR and minimap2 (Fig. 2b), and Winnowmap did not have consistent CPU usage even with thread count explicitly specified (Additional file 1: Fig. S1). RAM usage for each aligner was not associated with SV number (Additional file 1: Fig. S2 and S3).
Fig. 2.
Assessment of three sequence aligners for LRS. a Processing time in hours (y-axis) for three aligners (x-axis). Each box represents seven normal samples (shown on left) and seven tumour samples (shown on right). All data points are shown. The lines between boxes indicate the same samples. b RAM usage in Gb (y-axis) for three aligners (x-axis). Each box represents seven normal samples (shown on left) and seven tumour samples (shown on right). All data points are shown. c Mapping rate (y-axis) for three aligners (x-axis). Four tumour samples (shown on the right) and their paired normal samples (shown on the left) which passed sample quality control are included in the plot. d Genome Coverage (y-axis) for three aligners (x-axis). Four tumour samples (shown on the right) and their paired normal samples (shown on the left) which passed sample quality control are included in the plot
Sequence alignment metrics (proportion of mapped reads, average read depth and N50) were used to assess the alignments for four tumour and normal paired samples that passed quality control (average read depth of 20 × in normal and 50 × in tumour). The mapping results were similar across the three aligners, although minimap2 consistently exhibited a higher proportion of mapped reads (Fig. 2c) and a higher average read depth (Fig. 2d) with the lowest variance, while NGMLR had lower values with higher variance. The N50 values for the reads mapped by the three aligners were similar, however minimap2 generally produced shorter N50 compared to other aligners (Additional file 2: Table S1). Since minimap2 showed a good balance of computational performance and alignment quality, we selected it as the preferred aligner and subsequent results in the main figures are based on minimap2, with other aligners shown in the supplementary figures.
Somatic approaches called less events which were larger in size compared to generic approaches
Four of the seven patients obtained sufficient read depth (average read depth of > 50 in tumour samples and > 20 in non-tumour samples) and were included in this benchmarking study. The DNA Integrity (DIN) score for all samples with sufficient read depth were > 6 (Additional file 2: Table S1). Nine SV calling approaches were used to call SV events including four generic callers and four somatic callers (run in five calling approaches), with Jasmine used to distinguish germline from somatic events for the generic callers (Fig. 1b).
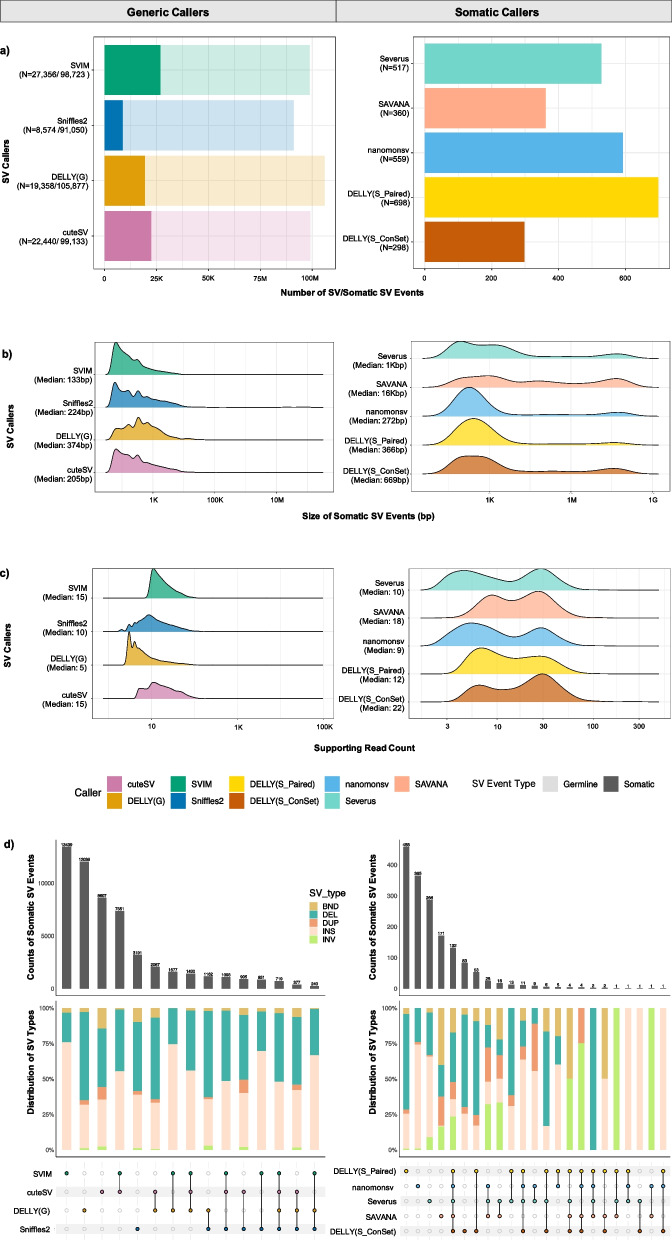
The generic callers collectively reported more than 20-fold as many somatic events as the somatic-specific callers did (Fig. 3a, Additional file 1: Fig. S4). SV events called by generic callers before somatic subtraction were similar. DELLY (G) exhibited the highest count of SV events (N = 105,877) with 19,358 of the events predicted as somatic. cuteSV (N = 99,133) and SVIM (N = 98,723) reported similar number of events with 21,440 and 27,356 predicted as somatic respectively, while Sniffles2 detected N = 98,723 events with 27,356 predicted as somatic. The SV events identified by cuteSV contained a higher proportion of translocation (BND) events compared to other generic approaches (Additional file 1: Fig. S4). Within the somatic-specific callers, DELLY (S_Paired) identified a greater number of somatic SV events (N = 698) with over half of the events were deletion (DEL). Compared to DELLY (S_Paired), DELLY (S_ConSet) detected the fewest somatic events (N = 298). nanomonsv reported the second-highest number of events (N = 559) with over half of the events were insertion (INS) (Fig. 3a, Additional file 1: Fig. 4).
Fig. 3.
Comparison of somatic structural variant (SV) detection approaches. a Bar charts display the counts of SVs (x-axis) with the four generic callers on the left (y-axis) and the five somatic SV callers on the right (y-axis). Denser colours on the chart signify somatic SV event counts, while lighter colours correspond to the subtracted germline SV events detected in the four tumour samples. Ridgeline plots showing distributions of b SV size and c SV supporting read counts on the x-axis for four generic and five somatic SV callers (y-axis). d The UpSet plot of SV events among different variant callers (y-axis) within each approach. The top section shows the counts of shared and unique somatic SV events among SV callers. The middle section displays the percentage distribution of detected SV types among unique and shared events, categorised and coloured into five distinct types: BND: translocations (dark green); DEL: deletions (green); DUP: duplicates (pale pink); INS: insertions (red); and INV: inversions (dark red). The bottom panel illustrates the matching variant callers. Unique caller events are represented by single dots, while overlaps are indicated by linked dots. Variant callers are assigned colours—green: SVIM; dark blue: Sniffles2; dark yellow: DELLY (G); and pink: cuteSV, for generic callers; gold for DELLY (S_Paired); light brown for DELLY (S_ConSet); light blue for nanomonsv; light salmon for SAVANA; and light aqua for Severus. The N values represent the count of somatic SV events from the total events discovered in four tumour samples. Median values are shown with Median under the name of each SV callers
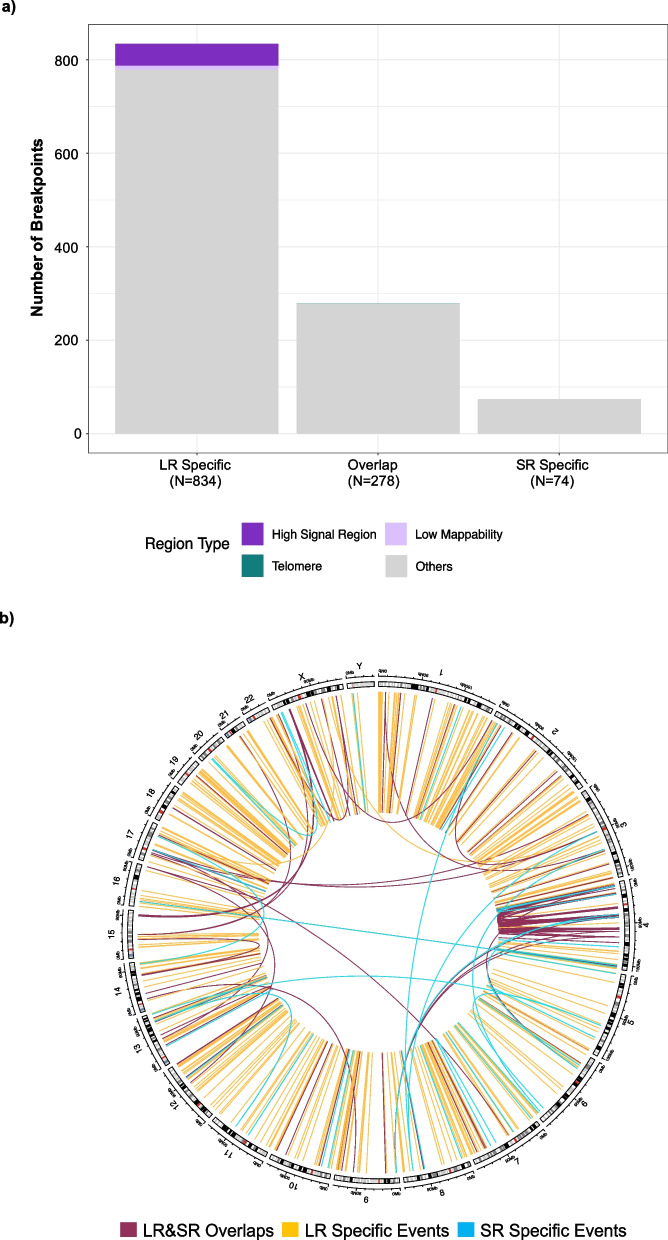
Fig. 4.
Characterization of concordant and unique somatic SV events identified by long and short read sequencing in four lung cancer patients. For long-read sequencing, the data were aligned with minimap2 and somatic SV events identified with nanomonsv. a The counts of SV breakpoints that overlap with various genomic region types, represented by distinct colors: dark purple indicates high signal regions, light purple represents low mappability regions, dark green denotes telomere regions, light green corresponds to centromere regions, and other regions (regions outside of the problematic regions, which short reads can align with high confidence) are shown in light grey. b Circos plot showing the somatic SV events from four patients. The outer ring shows chromosomes (GRCh38), while the inner track shows somatic SV events that are categorized into three groups: LR and SR overlaps (red), LR-specific (yellow), and SR-specific (blue)
A similar number of SV events were observed for the generic approaches using the NGMLR and Winnowmap aligners (Additional file 1: Fig. S5, S6). However, DELLY running in both generic and somatic modes called over 25% fewer events using reads aligned with NGMLR compared to the other two aligners (Fig. 3a, Additional file 1: Fig. S5, S6). The somatic callers nanomonsv and Severus identified more than one-third of the somatic SVs using NGMLR alignments (908 for nanomonsv and 921 for Severus), compared to those identified using minimap2 and Winnowmap (Additional file 1: Fig. S5). Severus primarily identified additional events such as INV and INS, whereas nanomonsv primarily detected DUP (Additional file 1: Fig. S7). In general, a higher proportion of duplications was noted across all variant callers when using sequence data aligned with NGMLR (Additional file 1: Fig. S7 and S8).
The majority of somatic SV events detected by the generic SV callers tended to be shorter in size (over 200 bp with a range from 133 to 374 bp), compared to the events detected by somatic callers (over 3Kbp with a range from 272 bp to 16Kbp) (Fig. 3b). The somatic SVs detected by SAVANA contained the highest proportion of events that were > 10Mbp in size. The number of reads supporting an SV event was higher in the somatic approaches (~ 14 reads with a range from 9 to 22 reads) compared to the generic approaches (~ 10 reads with a range from 5 to 15 reads) (Fig. 3c), suggesting that the somatic approaches required a greater number of supporting reads from tumour samples to classify somatic SV events. Similar results were seen in sequence data aligned with NGMLR (Additional file 1: Fig. S5) or Winnowmap (Additional file 1: Fig. S6).
A comparison of the SV events detected by the generic and somatic SV approaches
There is little overlap among somatic SVs called by the generic callers, with a large number of SVs called uniquely by each tool (ranging from 3,191 in Sniffles2 to 13,439 in SVIM) (Fig. 3d). The number of concordant events, detected by any combination of two generic variant callers, is 13,913. For combinations of three generic callers, it is 3,140, and the number of SVs called by all four generic callers was only 719. The largest proportion of SVs called by each caller were deletions or insertions (Fig. 3d). The somatic callers exhibited greater concordance with each other. DELLY (S_Paired) was the somatic caller that identified the highest number of caller-specific events (n = 458). The overlap among all the somatic callers was n = 132, with a larger proportion of inversion and duplication events compared to SRS (Fig. 3d).
Somatic approaches detect more high-confidence SV events compared to generic approaches
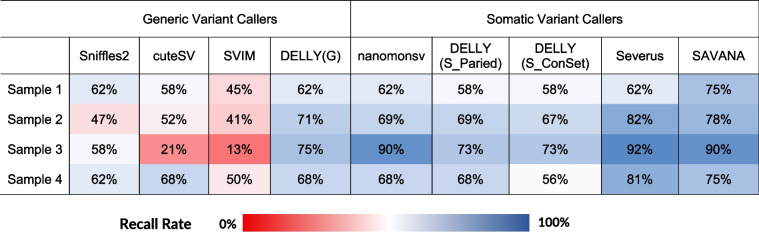
We assessed the performance of each SV tool by determining how many of the high confidence somatic SVs identified by SRS were called by each LRS approach (recall rate). The somatic variant callers outperformed generic variant callers, consistently demonstrating higher recall rates in the four samples (patients 1 to 4) that passed sequencing QC metrics (Table 1). Among the generic variant callers, DELLY(G) achieved the highest recall rate across all samples. Even though SVIM identified the most somatic SV events after subtraction of somatic events from normal samples (Fig. 3a), it had the lowest recall rate in three out of the four samples. Compared to the generic callers, the somatic callers showed an improved performance. Severus and SAVANA called fewer somatic SV events (Fig. 3a) and displayed the highest recall rate in all the samples with the highest recall of 92% (Table 1). Using minimap2, on average SAVANA and Severus achieved the highest recall at 79.5% and 79.25% respectively, followed by nanomonsv with a recall of 72.5% (Table 1). Similar results were observed using sequence data aligned with Winnowmap (Additional file 3: Table S3). Interestingly, the performance of nanomonsv improved using data aligned with NGMLR, with an average recall of 80.75% for the four samples that passed sequencing QC metrics (Additional file 3: Table S2).
Table 1.
Recall rate for somatic SV events in long-read sequence data aligned using minimap2 for each variant caller in four lung cancer patients
We also evaluated the recall rate in the three samples which did not meet the sequencing quality metrics for all somatic SV calling methods across the three aligners (Additional file 3: Table S2-S4). Our findings indicate that sample 6, which was characterized by the lowest read depth and yield, as well as the second smallest N50 and DIN values (Additional file 2: Table S1), exhibited the poorest recall rate among all samples when considering the combined performance of all somatic SV calling methods across the three aligners. In addition, the performance of the generic callers was adversely impacted by lower quality samples with three of four callers showing recall rates below 30%. The DELLY (G) approach was the only generic caller that demonstrated a similar recall rate to somatic callers in the lower quality samples.
A comparison of the total number of events detected by nanomonsv and SRS in the four lung samples revealed a total of 139 SV events (or N = 278 breakpoints) detected in SRS and LRS (Fig. 4a). Many of these SV events map to chromosome 4 (Fig. 4b) and were from one sample that contained a complex genomic rearrangement on chromosome 4 (Additional file 1: Fig. S9). A small number of SV events (37 events, or N = 74 breakpoints) were unique to SRS, whereas LRS identified more unique somatic events (417 events, N = 834 breakpoints) (Fig. 4a). To determine if the events unique to LRS could be potential artefacts, we annotated each break with known ‘problematic’ regions [32]. The number of breakpoints mapped to the problematic regions in the LRS data (49 of 834) was comparable to the SRS data (0) (Fig. 4a). We performed a similar analysis to determine the number of breakpoints within ‘problematic’ regions for the other SV callers. DELLY (G) produced the most events annotated in the 'problematic' regions (740 events, or N = 1,480 breakpoints), whereas SAVANA called the fewest events in that region (13 events, or N = 26 breakpoints) (Additional file 1: Fig. S10).
Discussion
In this study, we show that LRS can reliably identify somatic SVs in clinical cancer samples. We highlight the key differences when using three long-read aligners in combination with generic and somatic structural variant calling approaches. The somatic SV callers outperformed the generic SV tools showing high recall rates against events identified by short-read sequencing data.
We found that the choice of aligner had minimal impact on the identification of SV events, but differed in computational performance. Minimap2 was the fastest tool and demonstrated the least demand for computational resources with the highest mapping rate. Winnowmap was previously shown to enable robust SV calling with low false positive rate and a short computing time by providing improved alignment accuracy in highly repetitive regions [33, 34]. Here, we show the alignments from Winnowmap identified similar SV calls to minimap2 with slightly longer compute time for Winnowmap. This reflects the effective improvements in recent versions of minimap2 [35]. Notably, we also found that Winnowmap has inconsistent CPU usage even when specifying the number of threads, which makes it less suitable for shared High-Performance Computing (HPC) clusters. NGMLR required the longest computing time and had the lowest mapping rate. NGMLR was designed specifically to improve alignments for reads spanning structural variation [25], interestingly this aligner improved the recall of nanomonsv. However, in general, when SV tools used the alignments from NGMLR they called a higher proportion of duplications and a high number of very large variants (> 100,000 bp). These events were not observed with short-read sequencing, therefore future work using targeted approaches such as PCR with Sanger sequencing or spectral karyotyping [36] should be used to verify these NGMLR-unique SV events. Together, minimap2 stands as the current state-of-the-art aligner, offering precise whole-genome alignment and ongoing improvements. However, in cases where sensitivity of SV detection is critical and the computational cost and environment is less constrained, there is potential value in the generation of alignments with both minimap2 and NGMLR. In support of this, the Vulcan aligner which is a dual-mode aligner has been implemented to use both minimap2 and NGMLR [37].Therefore, multiple perspectives on the same genome may be needed to account for inherent differences in downstream SV tools and the algorithms they use to handle read alignment [38].
Our results underscore the necessity for developing somatic-specific SV tools as joint calling with matched tumour and normal data is known to exhibit superior accuracy compared to generic approaches [39]. This advantage stems from the inherent design of somatic callers, which leverage both tumour and paired normal BAM files for evidence of a variant, allowing for a more robust identification of somatic events. This allows somatic callers to better cope with intra-tumour genomic heterogeneity and tumour purity. In contrast, generic SV callers are designed for germline cells and assume that variants are homozygous (present in all reads) or heterozygous (present in approximately half of the reads) in a homogeneous sample. This makes it difficult for generic SV callers to sensitively and accurately capture the intra-tumour heterogeneity of somatic events in clinical samples, which can lead to increased false positive somatic calls through miscalling germline events as somatic as well as lower recall rates. To increase sensitivity of SV detection, our results support the use of multiple tools, and combining the output of multiple somatic approaches may allow more events to be detected.
In this study, we set out to test the utility of LRS for SV detection in clinical tissue samples. The inherent difficulty in controlling DNA quantity and quality in clinical samples, coupled with the influence of tumour purity on sequencing outcomes, poses significant hurdles. The samples were collected during Endobronchial Ultrasound Guided Transbronchial Needle Aspiration (EBUS TBNA), a common procedure for diagnosing suspected lung cancer [40]. During the diagnosis of lung cancer, there is benefit to targeted molecular genomic testing, this is particularly true for non-small cell lung cancer (NSCLC) samples where there are approved therapies for recurrent actionable mutations [41, 42]. In contrast, sequence analysis of SCLC has identified fewer approved treatment targets [14]. Therefore more comprehensive WGS approaches may be beneficial. A small study showed that short-read WGS from samples collected by EBUS TBNA was feasible [43]. However the DNA integrity of EBUS TBNA samples is frequently below 6 [44], meaning the DNA is somewhat degraded, which may be an issue for LRS. In this study, we observed that the quality of DNA and the amount of the data sequenced significantly influenced somatic SV detection, with lower quality samples associated with reduced recall rates compared to short-read sequencing. This emphasizes the critical role of sample quality in achieving accurate somatic SV detection, with implications for both research and clinical applications.
LRS has the potential to provide a more complete WGS analysis and to identify novel SV events. This is evident by a study that used LRS to identify causal germline SV events in paediatric rare diseases [44]. In our study, using LRS of matched tumour and non-tumour samples, we were able to detect somatic SVs events identified by short-read sequencing and additionally identified numerous events unique to LRS. To determine if the events unique to LRS are likely artefacts, we annotated the genome position for each break point with known regions that are problematic to call SV events (encompassing high signal, low mappability areas as well as telomeric and centromeric regions) [32, 45]. The majority of breakpoints from SV events that were identified in both LRS by nanomonsv and SRS, or those unique to one approach mapped to regions outside of the problematic regions, suggesting that these events may be real. In terms of the other somatic and generic SV approaches, for the SV events unique to LRS there is an increase in breakpoints mapping to high signal regions and centromeric regions. Whether these events are false positives or real as enabled by the ability by the ability of LRS to align to problematic regions is uncertain. Similar to what has occurred in short- read sequencing [46], future work to create and share standard long-read datasets with confirmed variants or simulated data containing known events will allow further benchmarking of analytical approaches for variant detection.
A key limitation of our study is the use of an older version of pores on flow cells (R9.4) and the Guppy basecaller. The frequent updates in platforms and softwares add a recognised complexity to benchmarking analyses [47]. Furthermore, the demanding computational requirements for analysing and storing extensive long-read datasets will present logistical challenges. Despite these obstacles, ongoing advancements in basecalling models, updates to ONT pores, and improvements in chemistries collectively signal a promising trajectory for enhancing the performance of LRS. As the accuracy and efficiency of LRS continues to improve, so should our ability to detect somatic SVs in clinical applications.
Conclusions
A critical factor of tumour development and treatment resistance is intra-tumour heterogeneity. We performed benchmarking of sequence aligners and SV tools to identify somatic SV events in clinical SCLC biopsy samples that were sequenced using a PromethION LRS. Among the aligners investigated, minimap2 emerges as particularly noteworthy, while the somatic SV callers outperformed other approaches. Our study highlights the challenges of using clinical samples for SV detection due to differences in tumour purity, DNA quantity and quality that are inherent in clinical samples. However, LRS of clinical samples to identify and resolve complex somatic SVs is likely to advance our understanding of cancer genomics, with the longer reads enabling phasing of cancer genomes to unravel intra-tumour heterogeneity.
Methods
Overview of study design
To assess the performance of somatic SV detection using LRS, we sequenced seven small cell lung carcinoma (SCLC) and paired normal samples using the Oxford Nanopore PromethION (R9.4). Basecalling was conducted with Guppy (v6.1.7) using a high-accuracy model and data were trimmed with NanoFlit (v2.8.0). We compared the performance of three long-read sequence aligners: minimap2 (v2.24), Winnowmap (v2.01), and NGMLR (v0.2.7) to identify somatic SVs using nine SV callers (Fig. 1, Additional file 1: Table S5). The SV approaches comprised of four generic SV callers: cuteSV (v1.0.13), DELLY (v1.1.6) in generic mode (DELLY (G)), Sniffles2 (v2.3.3), SVIM (v2.0.0), and four somatic SV callers: DELLY in somatic modes (using two approaches: DELLY (S_Paired) or a set of control samples (DELLY (S_ConSet)), nanomonsv (v0.3.6), Severus (v1.0), and SAVANA (v1.0.5). The generic approaches call SV events independently in tumour and normal samples, requiring subsequent subtraction of the germline events to determine somatic events, while the somatic approaches perform joint calling to process tumour and normal sequence files together and report somatic events. To assess the performance of the long-read SV callers, we compared SV events to a set of high-confidence events detected in SRS.
Sample collection and preparation
The lung cancer samples sequenced in this study were obtained from seven patients with SCLC. The research was conducted in accordance with the Declaration of Helsinki and the Australian National Statement on Ethical Conduct in Human Research. Human research ethics was approved by the Royal Brisbane and Women’s Hospital human research ethics committee (HREC, HREC/17/QRBW/301) and ratified by the HRECs of QIMR Berghofer (P2404) and the University of Queensland (2018/HE001615). Tumour tissue samples were obtained by Endobronchial Ultrasound Guided Transbronchial Needle Aspiration (EBUS TBNA) and blood collected from each patient. DNA was extracted from tissue and blood samples using a Qiagen DNA ALL prep kit. DNA samples ranged in quality from a DIN of 5.5 to 8.8 (Additional file 2: Table S1).
Oxford Nanopore long-read sequencing
LRS DNA libraries for tumour and germline DNA were constructed according to the manufacturer’s instructions using the Ligation Sequencing Kit (SQK-LSK110) and were loaded into flow cell R9.4 (product number: FLO-PRO002) for sequencing with the PromethION. To obtain 60× sequencing depth for tumour samples, each tumour sample was sequenced with two flow cells, while normal samples were sequenced with one flow cell to obtain 30× sequencing depth.
Base-calling, alignment and quality control of long-read sequencing data
The electronic raw signals (in FAST5 format) were translated into bases (A, T, C, or G), using Guppy (v6.1.7), only available to ONT customers via their community site (https://community.nanoporetech.com). Guppy supports running in both GPU and CPU modes, therefore, to reduce the computational resources/time required, GPU-accelerated Guppy base-calling was performed. A configuration file was selected (dna_r9.4.1_450bps_hac_prom.cfg) based on the Flow Cell (FLO-PRO002) and Ligation Sequencing Kit used (SQK-LSK110) to generate the FASTQ files. Trimming was performed based on the QC reports generated by NanoQC. NanoFlit (v2.8.0) was used to remove sequence reads with an average read quality score < 10 and to trim 40 nucleotides from the start and 20 nucleotides from the end of each read, respectively. Trimmed reads were aligned to the human reference genome GRCh38 with minimap2 (v2.24), Winnowmap (v2.01), and NGMLR (v0.2.7). Aligned files were sorted and indexed by SAMtools (v1.17). NanoPlot (v1.38.1) and SAMtools were used to check the quality of the sequencing alignment. Four out of seven samples were included in benchmarking of tools, with three cases excluded since they failed to reach the following quality minimums: an average read depth of 20 × in normal and 50 × in tumour, DNA integrity (DIN) > = 6 and N50 value of > = 5kbp (Additional file 2: Table S2).
Selection of approaches for somatic SV detection from LRS
Tools were selected from a review of the literature and were required to meet multiple criteria: (1) utilised by others as demonstrated by citations; (2) a comprehensive user manual; (3) command-line interface; (4) impartiality for aligners (note: we made an exception with SAVANA, as this tool was included even though it is incompatible with the NGMLR aligner); (5) persistent updating and community interaction on GitHub; and (6) minimal conflicting parameters or dependencies. Additionally, for the selection of somatic variant callers, we only selected tools that use a joint-calling approach to analyse both tumour and paired normal BAMs simultaneously to detect somatic SV events. Consequently, Sniffles2 in somatic mode was excluded due to its use of an un-paired sequenced tumour sample BAM.
Detection of somatic SV events in LRS
All SV approaches were run using default settings where possible, with specific parameters detailed in Supplementary Table 1. Generic calling methods (cuteSV, DELLY (G), Sniffles2, and SVIM) identified SV events in tumour and normal BAM files separately, and required an additional step to filter germline SV events. Jasmine (v1.1.5) was used to merge the SV events from the tumour and normal samples to distinguish somatic from germline events with a 200 bp window. The strand direction was not considered for event subtraction.
DELLY in somatic mode was run in two ways: 1) DELLY(S_Paired): running with paired normal and tumour samples; 2) DELLY (S_ConSet): using all normal samples as a control set to call somatic SV events for each tumour. As nanomonsv provides an optional prebuilt control panel to assist in identifying somatic from germline events, it was used to make precise somatic calling. The provided control panel was previously generated by aligning 30 × Nanopore sequencing data from the Human Pangenome Reference Consortium (HPRC) to the GRCh38 reference genome with minimap2 version 2.24 [48]. In addition, we used an optional step in nanomonsv (as recommended by the tool) to remove indels within simple repeats.
Short read whole genome sequencing and SV identification
Samples were sequenced by Illumina NovaSeq to a targeted average read depth of 30× for normal samples and 60× for tumour samples. Sequence reads were adapter trimmed using Cutadapt (v1.9) [49] and aligned using BWA-MEM (v0.7.15) [50] to the GRCh38 assembly. Duplicate reads were marked with Picard MarkDuplicates (https://broadinstitute.github.io/picard; v1.129). Tumour purity was assessed using ascatNgs [51]. High-confidence somatic SV events were identified using the consensus from any two out of three approaches: qSV [52], DELLY [24], and GRIDSS [53]. To identify somatic SV events reported by multiple short-read SV approaches, breakpoints were compared within a 200-base pair window on both ends. This window was employed during the comparison to accommodate potential variations in event location between different tools. The merging process did not take into account the direction of events, aiming to address potential misclassifications from different tools and to preserve the diversity of SV types identified by each tool. Requiring an SV to be identified by three different SRS SV calling tools should produce a truth set with few false positives but potentially more false negatives.
Assessing the performance of LRS SV detection approaches compared to SRS
The high-confidence somatic SV events obtained from a consensus of any two out of three approaches from SRS data were used as a "ground truth" to evaluate the performance of each LRS SV caller. To determine whether the high-confidence somatic SV events detected in SRS were detected in LRS, we merged the breakpoints detected by each approach using Jasmine with a 200-base pair window. The merging process did not consider the direction of events to account for potential differences in classification between different tools and to preserve the diversity of SV types called by each tool.
Annotating long read calls
We generated a 'blacklist' BED file that included centromere and telomere regions [32, 45] to annotate the events identified using LRS. The blacklist file also contained ENCODE Data Analysis Center (DAC) blacklisted regions, including the high signal and low mappability regions. The high signal regions presumably represent unannotated repeats in the genome. We annotated both long-read and short-read concordant and unique somatic SV events by comparing their breakpoints with the regions in the blacklist file, employing a window of 1000bps for the comparison.
Supplementary Information
Acknowledgements
LL is supported by a University Queensland Graduate School Scholarship and QIMR Berghofer PhD Top-up scholarship. This research was performed on QIMR Berghofer computing infrastructure supported by The Ian Potter Foundation, The John Thomas Wilson Endowment and the Australian Cancer Research Foundation (ACRF).
Authors’ contributions
LL: Conceptualization, Methodology, Software, Formal analysis, Investigation, Writing – Original Draft, Visualisation. JZ: Methodology, Software, Formal analysis, Investigation, Writing - Review and Editing, Visualisation, Supervision. SW: Software, Resources, Writing - Review and Editing. FN: Methodology, Formal analysis, Writing - Review and Editing. CL: Resources, Data Curation, Writing - Review and Editing. LTK: Formal analysis, Writing - Review and Editing. KN: Resources, Writing - Review and Editing, Funding acquisition. AJD: Resources, Writing - Review and Editing. HC: Resources, Writing - Review and Editing. FB: Resources, Writing - Review and Editing. JHS: Resources, Writing - Review and Editing. DS: Resources, Writing - Review and Editing. JPW: Resources, Writing - Review and Editing. MB: Resources, Writing - Review and Editing. CP: Resources, Writing - Review and Editing. PTN: Resources, Writing - Review and Editing. ST: Resources, Writing - Review and Editing. DA: Resources, Writing - Review and Editing. CG: Resources, Writing - Review and Editing. PTS: Resources, Writing - Review and Editing, Funding acquisition. DF: Resources, Writing - Review and Editing, Funding acquisition. NW: Conceptualization, Methodology, Resources, Data Curation, Writing - Review and Editing, Supervision, Project administration, Funding acquisition. JVP: Conceptualization, Methodology, Resources, Data Curation, Writing - Review and Editing, Supervision, Project administration, Funding acquisition.
Funding
LL is supported by a University Queensland Graduate School Scholarship, QIMR Berghofer PhD Top-up scholarship. This work was funded by a Cancer Council Queensland (CCQ) Accelerating Collaborative Cancer Research (AACR) Grant (000000027). NW is funded by the National Health and Medical Research Council of Australia (NHMRC) Senior Research Fellowship (APP1139071) and Investigator Grant (2018244). In addition, we are grateful to research support from the 2017 Priority-driven Collaborative Cancer Research Scheme, funded by Cancer Australia (Grant #1147067); Cancer Council Queensland (Grant #1147067); Australian Genomics (NHMRC grants GNT1113531 and GNT2000001) and the Medical Research Futures Fund Genomics Health Futures Mission (2009160). This research was performed on QIMR Berghofer computing infrastructure supported by The Ian Potter Foundation, The John Thomas Wilson Endowment and the Australian Cancer Research Foundation (ACRF) Centre for Optimised Cancer Therapy.
Availability of data and materials
The sequence data for this project can be accessed in the European Genome-phenome Archive (EGA) under study accession EGAS00001007832 for SR data in dataset EGAD00001015399 and under study accession EGAS00001007819 for LR data in dataset EGAD00001015400.
Declarations
Ethics approval and consent to participate
The lung cancer samples sequenced in this study were obtained from patients with SCLC. Research was conducted in accordance with the Declaration of Helsinki and the Australian National Statement on Ethical Conduct in Human Research. Human research ethics was approved by the Royal Brisbane and Women’s Hospital human research ethics committee (HREC, HREC/17/QRBW/301) and ratified by the HRECs of QIMR Berghofer (P2404) and the University of Queensland (2018/HE001615). All patients provided written informed consent to participate in research.
Consent for publication
Not applicable.
Competing interests
John V. Pearson and Nicola Waddell are co-founders of genomiQa. LL and NW were funded by Oxford Nanopore to present work from this study at meetings. The remaining authors declare that there are no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Nicola Waddell and John V. Pearson jointly contributed to and supervised the work.
References
- 1.WHO. Cancer World Health Organization (Fact sheets). 2022. Available from: https://www.who.int/news-room/fact-sheets/detail/cancer. Cited 2023 23rd Feb.
- 2.Kim K-B, Dunn CT, Park K-S. Recent progress in mapping the emerging landscape of the small-cell lung cancer genome. Exp Mol Med. 2019;51(12):1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kris MG, Johnson BE, Berry LD, Kwiatkowski DJ, Iafrate AJ, Wistuba II, et al. Using multiplexed assays of oncogenic drivers in lung cancers to select targeted drugs. JAMA. 2014;311(19):1998–2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Herbst RS, Morgensztern D, Boshoff C. The biology and management of non-small cell lung cancer. Nature. 2018;553(7689):446–54. [DOI] [PubMed] [Google Scholar]
- 5.Zhang T, Joubert P, Ansari-Pour N, Zhao W, Hoang PH, Lokanga R, et al. Genomic and evolutionary classification of lung cancer in never smokers. Nat Genet. 2021;53(9):1348–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alexandrov LB, Kim J, Haradhvala NJ, Huang MN, Tian Ng AW, Wu Y, et al. The repertoire of mutational signatures in human cancer. Nature. 2020;578(7793):94–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Collisson EA, Campbell JD, Brooks AN, Berger AH, Lee W, Chmielecki J, et al. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511(7511):543–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li Y, Roberts ND, Wala JA, Shapira O, Schumacher SE, Kumar K, et al. Patterns of somatic structural variation in human cancer genomes. Nature. 2020;578(7793):112–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Soda M, Choi YL, Enomoto M, Takada S, Yamashita Y, Ishikawa S, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448(7153):561–6. [DOI] [PubMed] [Google Scholar]
- 10.Shaw AT, Yeap BY, Mino-Kenudson M, Digumarthy SR, Costa DB, Heist RS, et al. Clinical features and outcome of patients with non-small-cell lung cancer who harbor EML4-ALK. J Clin Oncol. 2009;27(26):4247–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rudin CM, Brambilla E, Faivre-Finn C, Sage J. Small-cell lung cancer. Nat Rev Dis Primers. 2021;7(1):3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arakawa S, Yoshida T, Shirasawa M, Takayanagi D, Yagishita S, Motoi N, et al. RB1 loss induced small cell lung cancer transformation as acquired resistance to pembrolizumab in an advanced NSCLC patient. Lung Cancer. 2021;151:101–3. [DOI] [PubMed] [Google Scholar]
- 13.Febres-Aldana CA, Chang JC, Ptashkin R, Wang Y, Gedvilaite E, Baine MK, et al. Rb tumor suppressor in small cell lung cancer: combined genomic and IHC analysis with a description of a distinct rb-proficient subset. Clin Cancer Res. 2022;28(21):4702–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.George J, Lim JS, Jang SJ, Cun Y, Ozretić L, Kong G, et al. Comprehensive genomic profiles of small cell lung cancer. Nature. 2015;524(7563):47–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cretu Stancu M, Van Roosmalen MJ, Renkens I, Nieboer MM, Middelkamp S, De Ligt J, et al. Mapping and phasing of structural variation in patient genomes using nanopore sequencing. Nat Commun. 2017;8(1):1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Merker JD, Wenger AM, Sneddon T, Grove M, Zappala Z, Fresard L, et al. Long-read genome sequencing identifies causal structural variation in a Mendelian disease. Genet Med. 2018;20(1):159–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu L, Wang X, Lu X, Liang F, Liu Z, Zhang H, et al. Long-read sequencing identifies novel structural variations in colorectal cancer. PLoS Genet. 2023;19(2): e1010514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gong L, Wong C-H, Cheng W-C, Tjong H, Menghi F, Ngan CY, et al. Picky comprehensively detects high-resolution structural variants in nanopore long reads. Nat Methods. 2018;15(6):455–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chaisson MJP, Sanders AD, Zhao X, Malhotra A, Porubsky D, Rausch T, et al. Multi-platform discovery of haplotype-resolved structural variation in human genomes. Nat Commun. 2019;10(1):1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spies N, Weng Z, Bishara A, McDaniel J, Catoe D, Zook JM, et al. Genome-wide reconstruction of complex structural variants using read clouds. Nat Methods. 2017;14(9):915–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Euskirchen P, Bielle F, Labreche K, Kloosterman WP, Rosenberg S, Daniau M, et al. Same-day genomic and epigenomic diagnosis of brain tumors using real-time nanopore sequencing. Acta Neuropathol. 2017;134(5):691–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Technologies ON. [Available from: https://nanoporetech.com/accuracy.
- 23.Shiraishi Y, Koya J, Chiba K, Okada A, Arai Y, Saito Y, et al. Precise characterization of somatic complex structural variations from tumor/control paired long-read sequencing data with nanomonsv. Nucleic Acids Res. 2023;51(14):e74-e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rausch T, Zichner T, Schlattl A, Stütz AM, Benes V, Korbel JO. DELLY: structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics. 2012;28(18):i333–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sedlazeck FJ, Rescheneder P, Smolka M, Fang H, Nattestad M, von Haeseler A, et al. Accurate detection of complex structural variations using single-molecule sequencing. Nat Methods. 2018;15(6):461–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiang T, Liu Y, Jiang Y, Li J, Gao Y, Cui Z, et al. Long-read-based human genomic structural variation detection with cuteSV. Genome Biol. 2020;21(1):1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heller D, Vingron M. SVIM: structural variant identification using mapped long reads. Bioinformatics. 2019;35(17):2907–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dierckxsens N, Li T, Vermeesch JR, Xie Z. A benchmark of structural variation detection by long reads through a realistic simulated model. Genome Biol. 2021;22(1):342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin J, Jia P, Wang S, Ye K. Comparison and benchmark of long-read based structural variant detection strategies. bioRxiv. 2022:2022.08.09.503274. 10.1101/2022.08.09.503274.
- 30.Yildiz G, Zanini SF, Afsharyan NP, Obermeier C, Snowdon RJ, Golicz AA. Benchmarking Oxford Nanopore Read Alignment-Based Structural Variant Detection Tools in Crop Plant Genomes. bioRxiv. 2022:2022.09.23.508909. 10.1002/tpg2.20314. [DOI] [PubMed]
- 31.Bolognini D, Magi A. Evaluation of germline structural variant calling methods for nanopore sequencing data. Front Genet. 2021;12: 761791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Amemiya HM, Kundaje A, Boyle AP. The ENCODE Blacklist: identification of problematic regions of the genome. Sci Rep. 2019;9(1):9354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jain C, Rhie A, Hansen NF, Koren S, Phillippy AM. Long-read mapping to repetitive reference sequences using Winnowmap2. Nat Methods. 2022;19(6):705–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.LoTempio J, Delot E, Vilain E. Benchmarking long-read genome sequence alignment tools for human genomics applications. PeerJ. 2023;11:e16515. 10.7717/peerj.16515. [DOI] [PMC free article] [PubMed]
- 35.Li H. New strategies to improve minimap2 alignment accuracy. Bioinformatics. 2021;37(23):4572–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guo B, Han X, Wu Z, Da W, Zhu H. Spectral karyotyping: an unique technique for the detection of complex genomic rearrangements in leukemia. Transl Pediatr. 2014;3(2):135–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fu Y, Mahmoud M, Muraliraman VV, Sedlazeck FJ, Treangen TJ. Vulcan: Improved long-read mapping and structural variant calling via dual-mode alignment. GigaScience. 2021;10(9). 10.1093/gigascience/giab063. [DOI] [PMC free article] [PubMed]
- 38.Jenko Bizjan B, Katsila T, Tesovnik T, Šket R, Debeljak M, Matsoukas MT, et al. Challenges in identifying large germline structural variants for clinical use by long read sequencing. Comput Struct Biotechnol J. 2020;18:83–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ura H, Togi S, Niida Y. Dual deep sequencing improves the accuracy of low-frequency somatic mutation detection in cancer gene panel testing. Int J Mol Sci. 2020;21(10): 3530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Torii A, Oki M, Yamada A, Kogure Y, Kitagawa C, Saka H. EUS-B-FNA enhances the diagnostic yield of EBUS bronchoscope for intrathoracic lesions. Lung. 2022;200(5):643–8. [DOI] [PubMed] [Google Scholar]
- 41.Li T, Kung H-J, Mack PC, Gandara DR. Genotyping and genomic profiling of non–small-cell lung cancer: implications for current and future therapies. J Clin Oncol. 2013;31(8):1039–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ramarao-Milne P, Kondrashova O, Patch AM, Nones K, Koufariotis LT, Newell F, et al. Comparison of actionable events detected in cancer genomes by whole-genome sequencing, in silico whole-exome and mutation panels. ESMO Open. 2022;7(4):100540-. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fielding D, Dalley AJ, Singh M, Nandakumar L, Lakis V, Chittoory H, et al. Whole genome sequencing in advanced lung cancer can be performed using diff-quik cytology smears derived from Endobronchial Ultrasound, Transbronchial Needle Aspiration (EBUS TBNA). Lung. 2023;201(4):407–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fielding D, Dalley AJ, Singh M, Nandakumar L, Nones K, Lakis V, et al. Prospective optimization of endobronchial ultrasound-guided transbronchial needle aspiration lymph node assessment for lung cancer: three needle agitations are noninferior to 10 agitations for adequate tumor cell and DNA yield. JTO Clin Res Rep. 2022;3(10):100403-. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee BT, Barber GP, Benet-Pagès A, Casper J, Clawson H, Diekhans M, et al. The UCSC Genome Browser database: 2022 update. Nucleic Acids Res. 2022;50(D1):D1115–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Craig DW, Nasser S, Corbett R, Chan SK, Murray L, Legendre C, et al. A somatic reference standard for cancer genome sequencing. Sci Rep. 2016;6(1): 24607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dong X, Du MRM, Gouil Q, Tian L, Jabbari JS, Bowden R, et al. Benchmarking long-read RNA-sequencing analysis tools using in silico mixtures. Nat Methods. 2023;20(11):1810–21. [DOI] [PubMed] [Google Scholar]
- 48.Shafin K, Pesout T, Lorig-Roach R, Haukness M, Olsen HE, Bosworth C, et al. Nanopore sequencing and the Shasta toolkit enable efficient de novo assembly of eleven human genomes. Nat Biotechnol. 2020;38(9):1044–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011;17(1):10–2. [Google Scholar]
- 50.Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv: Genomics. 2013;00(00):1–3. 10.48550/arXiv.1303.3997.
- 51.Raine KM, Van Loo P, Wedge DC, Jones D, Menzies A, Butler AP, et al. ascatNgs: identifying somatically acquired copy-number alterations from whole-genome sequencing data. Curr Protoc Bioinform. 2016;56:15.9.1-.9.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hayward NK, Wilmott JS, Waddell N, Johansson PA, Field MA, Nones K, et al. Whole-genome landscapes of major melanoma subtypes. Nature. 2017;545(7653):175–80. [DOI] [PubMed] [Google Scholar]
- 53.Cameron DL, Baber J, Shale C, Valle-Inclan JE, Besselink N, van Hoeck A, et al. GRIDSS2: comprehensive characterisation of somatic structural variation using single breakend variants and structural variant phasing. Genome Biol. 2021;22(1):202. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequence data for this project can be accessed in the European Genome-phenome Archive (EGA) under study accession EGAS00001007832 for SR data in dataset EGAD00001015399 and under study accession EGAS00001007819 for LR data in dataset EGAD00001015400.