Evan W. Miller is associate professor of chemistry and molecular and cell biology at UC Berkeley. Evan earned his PhD in Berkeley in 2009 and then did postdoctoral research at UC San Diego with Roger Tsien, the scientific innovator and Nobel laureate whose work led to many of the chemical and genetically encoded fluorescent sensors in use today. Evan took a unique approach to developing small molecule voltage sensors by employing compounds that exhibited photo-induced electron transfer (PeT). Evan has extended this work since establishing his own laboratory in Berkeley, developing PeT sensors across the visible spectrum and into the infrared, and developing hybrid PeT with molecules that can be targeted to label particular cells expressing a genetically expressed membrane anchoring protein.
Ahmed Abdelfattah is assistant professor of neuroscience at Brown University, Providence, RI. Ahmed earned his PhD in 2016 from the University of Alberta, and moved to the Janelia research campus of the Howard Hughes Medical Institute where he carried out postdoctoral work with Eric Schreiter. Together with an international team of scientists from several institutions, Ahmed developed a state-of-art genetically encoded voltage indicator named Voltron, and used it in mice, zebrafish, and fruit flies to reveal action potentials and subthreshold events in vivo from dozens of neurons simultaneously. In his newly established laboratory at Brown, Ahmed is continuing to use bioengineering and chemical approaches to develop and refine new molecular tools to visualize and study the brain.
Bradley Baker is principal scientist at the Brain Science Institute, Korea Institute of Science and Technology (KIST), located in Seoul, Korea. After earning a PhD in biochemistry from Ohio State University in 1998, Brad moved to Yale University where he spent many years collaborating with Larry Cohen, Dejan Zecevic, and others, carrying out pioneering work on fluorescent proteins for optical recording of electrical activity in vertebrate nervous systems. Brad moved to KIST in 2011, where he has continued to develop genetically encoded voltage indicators.
Recent advances made possible by these tools include the discovery of fast electrical interactions between the plasma membrane and endoplasmic reticulum (ER), a previously hidden process that may coordinate biochemical events on the surface and interior of cells (https://doi.org/10.1038/s41598-018-25083-7).
Richard H. Kramer (moderator) is professor of molecular and cell biology at UC Berkeley. Richard earned his PhD in Berkeley in 1985 before postdoctoral studies with Irwin Levitan at Brandeis University. He then moved to Columbia University as an HHMI research associate with Steve Siegelbaum, where he studied ion channels underlying sensory transduction in vision and olfaction. Richard established his own laboratory at University of Miami in 1993 where he developed novel techniques for sensing intracellular messengers in neurons. He moved back to UC Berkeley in 2000 where his laboratory maintains a variety of interests, from mechanisms of ion channel and synaptic transmission, understanding information processing in the retina, and developing light-sensitive compounds for restoring vision in degenerative blinding disease.
Richard H. Kramer: Ahmed, Brad, and Evan, I want to thank all of you for being here.
Today's discussion is an exploration of fluorescent voltage sensors for monitoring electrophysiological events in tissues and organisms. Our discussion is not limited to the nervous system and not limited to excitable cells, but it will more broadly consider the role of ionic currents and electrical fields in tissue and organismal development and function. Fluorescent reporters of voltage seem to have been underutilized for studying bioelectric phenomenon in these contexts. I am wondering whether there are any design principles that might optimize voltage sensors for these.
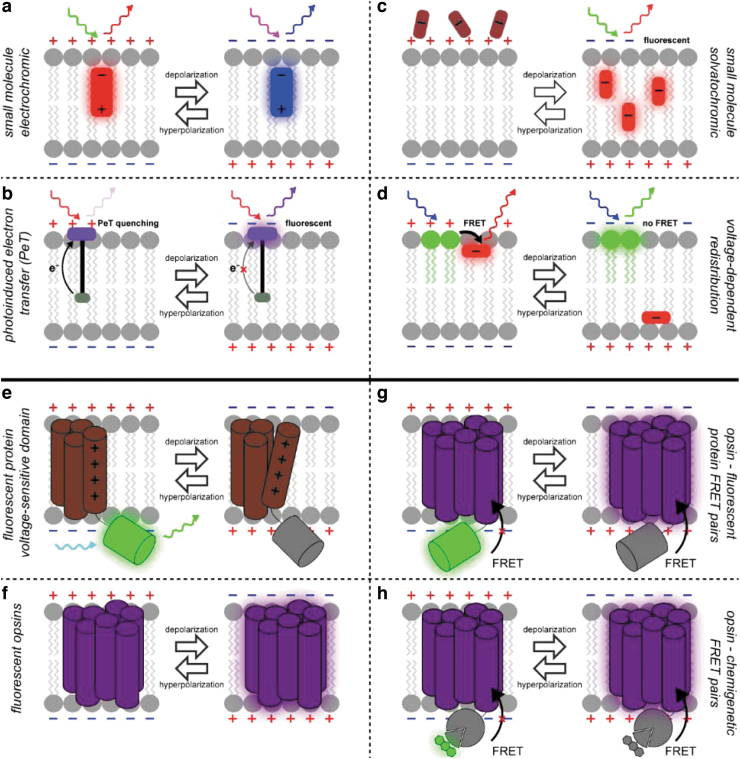
All of you wear two hats—you are both inventors and users of different kinds of fluorescent voltage sensors, and as such you represent a cross section of the field. One place to start our discussion is by listing the types of tools that are out there (Fig. 1) . Evan, you have developed things at the small molecule end of this spectrum, do I have this right?
FIG. 1.
Molecular tools for voltage sensing. Fluorescent methods to image membrane potential include (a) electrochromic small molecules, (b) PeT-based voltage-sensitive fluorophores, (c) solvatochromic accumulation type dyes, (d) FRET or quenching-based redistribution, (e) fluorescent protein/voltage sensing domain fusions, (f) fluorescent opsins, (g) fluorescent protein–opsin electrochromic FRET pairs, and (h) chemigenetic opsin electrochromic FRET pairs. PeT, photo-induced electron transfer. Adapted from Current Opinion in Chemical Biology;37 and Biochemistry.38
Evan W. Miller: Yeah, first are small molecules, fluorophores, either a single fluorophore or pairs of fluorophores that interact through Förster Resonance Energy Transfer (FRET) through quenching. So, in the first case you have molecules that respond in some way to the electric field, the changing voltage across the membrane. And in the second case, you have molecules that move through the membrane, which gives you a fluorescent readout either change in FRET efficiency or a change in quenching. And so those are the two styles or flavors of small molecule voltage-sensitive dyes. And those go back quite some time, I think into the early ‘70s. Larry Cohen and others were looking at merocyanines1 and going through catalogs of published compounds and adding them to membranes and then testing for voltage-dependent changes in fluorescence.2 So those have been in use for quite some time.
Richard H. Kramer: And the small molecule FRET probes?
Evan W. Miller: And FRET probes, I think those emerged in the mid ‘90s. The first to appear were combinations of oxonols and coumarins that were added to a cell.3,4 Coumarin anchored in the cell membrane, and then other dyes that could drift from one leaflet to the other and give a nice FRET change. You could also use a molecule like dipicrylamine that is a quencher, and that could be used to FRET with small molecule fluorophore. Or you could use it with a fluorescent protein. I guess that would fall into the next category—hybrid voltage sensors, or hVOS, where dipicrylamine moves through the membrane in a voltage-dependent manner and quenches the fluorescence of a membrane-anchored GFP.5
Richard H. Kramer: And then I guess we move on to fluorescent protein-based chimeric probes, which I think, both of you guys, Ahmed and Brad, have been working with, right?
Bradley Baker: Well, we focus mainly on fluorescent proteins, and we try to limit the number of variables during our probe development. But Evan, Larry Cohen would be very happy you mentioned merocyanine dyes. And I would also like to say that this is the 25th anniversary of the first fluorescent protein voltage sensor that was published by Ehud Isacoff's Group at UC Berkeley.6 This has caused many laboratories a lot of pain and suffering, but we are very happy that he did this because it has given us some really cool research projects to explore. It is hard to believe that was ‘97.
Evan W. Miller: Yeah, 1997.
Ahmed Abdelfattah: Fun fact, it was the same year that the first genetically encoded calcium sensor was published.7 [Note, “GCaMP” emerged later, in 20018].
Evan W. Miller: The first genetically encoded GFP-based Ca2+ indicator was “cameleon,” named as such because it was a FRET-based probe that changed colors upon binding Ca2+, and that was the one that was published in 1997.7
Bradley Baker: Oh really?
Evan W. Miller: Yeah.
Ahmed Abdelfattah: One can appreciate how much harder it has been to evolve and improve the response of genetically encoded voltage sensors so that they are useful for in vivo experiments, versus genetically encoded calcium sensors. And it shows how many people are using each of them. Both were first reported in the same year, 1997.
We briefly talked about the fluorescent protein-based genetically encoded sensor, but really, we did not introduce opsin-based sensors.
Richard H. Kramer: Right. That is next on this list.
Ahmed Abdelfattah: Genetically encoded voltage sensors in general fall in two groups. One is using a conformational change of a voltage-sensitive domain (e.g., from an ion channel6,9,10 or voltage-sensing phosphatase11,12) that relays that conformational change to a fluorophore, whatever that fluorophore is. It can be a fluorescent protein, a small molecule, and so on and so forth. The other class is based on microbial opsin proteins. An opsin is a transmembrane protein that sits in the membrane, and binds the vitamin A derivative, retinaldehyde. And retinaldehyde can change its absorption and fluorescence properties when the membrane potential changes.
I think really the groundwork for exploiting this has been done in Adam Cohen's laboratory about a decade ago now. The first article described an opsin-based voltage sensor named PROPS, which was published in Science in 2011.13 They used it to image voltage in bacteria. Shortly thereafter in a 2011 nature methods article, they introduced a similar tool named Arch, which worked in mammalian neurons to report action potentials.14 The really cool thing about those proteins is that you do not rely on a conformational change at all. It is really protonation/deprotonation of the retinaldehyde molecule. It is inherently much faster than relying on a conformational change in the protein.
Both of them have advantages and disadvantages of course. Opsin fluorescence is inherently very dim. We and others have worked a lot to address their brightness, and we can go into details about that later, but the quantum yield is ∼1%, so it is not as bright as fluorescent proteins or small molecule dyes. But the absorption changes a lot when the membrane potential changes. People have been trying to use that change to record voltage.
Richard H. Kramer: Continuing down the list, the next are opsin-based probes that are chimeric with a fluorescent protein. So, you can make a chimeric combination between an opsin and a fluorescent protein to amplify the voltage sensor effect. Did I get that right?
Ahmed Abdelfattah: A chimera with a fluorescent protein will increase the brightness of your probe and will rely on FRET between the fluorescent protein and the opsin for reporting the voltage signal. In this design you can think of the opsin as a dark quencher, and the fluorescent protein as both the donor and the molecule you are actually imaging.15,16 You can also fuse the opsin to a HaloTag that covalently couples to a dye, like we did with the Voltron work,17 or you can fuse the opsin directly to a dye.18,19 This way you are enhancing the brightness of the voltage sensor, which is very important when you are using it in live tissue to record voltage.
Richard H. Kramer: I guess we covered the list except for the very last one—nanoparticle-based methods. I know people have used graphenes and other nanoparticles as voltage sensors. One advantage is that they do not need to be genetically encoded. You just apply them onto tissue like you would a small molecule, but they are much larger particles. Do you guys know much about these? Evan, maybe?
Evan W. Miller: There is a couple … nanopillars or nanoscale electrodes for action potential monitoring.20 There is nitrogen vacancy diamonds.21 Single molecule voltage imaging with nanoparticles is another approach.22 Similar to the small molecule di-4-ANEPPS types of dyes that are electrochromic. They have a change in their band gap, their Higest Occupied Molecular Orbital and Lowest Unoccupied Molecular Orbital levels, and so you get a spectroscopic shift, like a Stark effect. But they are nanoparticles, they are very bright and so you can get very very localized readings from a single molecule as opposed to having a bunch of dyes.
Richard H. Kramer: So, do these things integrate into the membrane?
Evan W. Miller: That is one of the challenges: integrating these particles into the membrane.23
Richard H. Kramer: The next question is directed at each of you, to talk specifically about your favorite tool in more detail. Just a chance for you to tell us what you have developed recently.
Bradley Baker: I will start. My name is Brad Baker, my laboratory is in Seoul, Republic of Korea. I have been here for 10 years. And for those students who are looking for an international experience and want to travel the world, Korea is an exciting place to come do research. There is good energy here and I have enjoyed it a lot. So, we have been primarily focusing on how a conformational change in a protein voltage sensor results in a fluorescent response, and it has led through a very fascinating sort of journey.
A couple of highlights from what we have done. We have shown that an interaction between different probes is capable of generating a FRET response. You could have intermolecular FRET that could potentially be developed into intersectional expression in distinct cell types. You could have the donor construct expressed by one promoter and the acceptor construct expressed by a different promoter, and where those two intersect, you could use the combinatorial effect, for example, to give you much better resolution of specific cell types in a neuronal circuit.
That is the goal, but the problem is that we do not know how this interaction works, and so we cannot control the association of donor to acceptor. You get a combination of donor–donor and acceptor–acceptor pairs, which also generate a voltage-dependent signal convoluting the voltage-dependent FRET signal. So that was a recent publication we had last year.24
And then, we have also explored what I consider now a new universe. We are actually starting to be able to see charge migration through the fluorescent protein, and start to look at how proton wires and dipole moments are interacting in the protein itself, and how that interaction affects the fluorescent properties of the protein.25 So that has been a remarkable journey, and hopefully we will have a submission on that soon.
As Ahmed mentioned, I was surprised to hear that the first article on GCaMP was published in the same year as the first genetically encoded fluorescent voltage sensor. When people ask me, should they do calcium imaging or voltage imaging, the answer is, well if you can answer your question with calcium imaging, your life will be much easier because since it is a cytoplasmic probe, you can just get way more of it expressed in a cell. I think trying to image membrane potential suffers from photon starvation. We need photons, photons, photons, because we are limited to using a probe reporting exclusively from the plasma membrane.
So, as a result, we have also been looking at protein trafficking to the surface membrane and how to increase it. We have an article in press (https://doi.org/10.1016/j.bpr.2022.100047) that shows that some of these trafficking partners appear to have persistent interactions that affect our conformational change at the plasma membrane, even after it has been trafficked correctly to the plasma membrane. So that is a cool result.
But then we also stumbled upon imaging the ER and internal membrane potentials. Masoud Rad, a very astute scientist in my laboratory, saw not only a signal coming from voltage changes at plasma membrane, but also a signal with opposite polarity coming from internal membranes. And so it was our first indication that you could image the membrane potential of an internal membrane on an intracellular organelle with a fluorescent protein.26 But then the challenge becomes, how do you manipulate the voltage across internal membrane? Do changes happen spontaneously? So that is a new universe, but it is also a fun universe because there has not been a lot of exploration there.
Richard H. Kramer: As long as we are on the subject, can we begin to deliberately target genetically expressed voltage sensors to particular subcellular compartments? I do not know, whether you guys have thoughts about this, targeting it to mitochondria or to specific organelles. And a related question, to what extent do you have to worry about contamination of plasma membrane signals by a probe that is retained in internal membranes?
Bradley Baker: Well, we got very lucky because for whatever reason, the fluorescent signal of our internal membrane was in the opposite direction of the signal on the surface membrane. The internal membrane got brighter as a cell depolarized, whereas the plasma membrane signal got dimmer. It was only in cells with very high internal expression levels where we could distinguish the internal signal from the surface signal. So, there is a lot of optimization that can be done to make the internal signal stronger.
Can we figure out ways to subtract out the plasma membrane signal to observe the membrane potential of internal membranes in isolation? I think so. One idea for the future is to use a probe that interacts with a fluorescent protein expressed on an internal membrane and then measure FRET. Perhaps the environment of plasma membrane and internal membrane is different enough that FRET will differ. That could give a signal that is exclusively from one source or the other.
Another challenge, how do you stimulate a change in the potential across internal membranes? We managed to accidentally target our sensor to internal membranes, and then we saw consistent responses when we altered the plasma membrane voltage, even though there is no direct continuity between the two membrane systems. Surprisingly we see the internal membrane also responds. How exactly does that work? We do not yet know!
Richard H. Kramer: How do you calibrate the voltage response of a probe that is targeted to an intracellular membrane?
Bradley Baker: How do you even validate whether it gets to the membrane—for example ER? Is it displayed uniformly in the entire ER? Is it on other organelles? The interactome of intercellular organelles is a fascinating topic and there is much more to learn, and I think voltage sensors will be important for understanding how organelles interact.
Ahmed Abdelfattah: I think also maybe Evan can talk on this more, but aren't there cell permeable small molecule voltage sensors that are tuned for imaging voltage changes in mitochondria and the ER? Because I think I used some of those in the early days.
Evan W. Miller: Traditionally, mitochondria have been targeted with a lipophilic cationic type of dye. Rhodamine esters have been used for this since the early ‘80s.27 So back to Richard's original question, we have been working on small molecule voltage-sensitive dyes, and when we turned to rhodamine-based indicators, the first problem was that they are all lipophilic. We made them lipophilic—to localize to the plasma membrane—but went right through the plasma membrane and straight to the mitochondria, and we spent a lot of time trying to make cell impermeant versions that stayed on the outside.
So we have many “failed” dyes that we were pretty certain would be voltage sensitive, but they were all inside the mitochondria. It took some time to figure out how to make them cell impermeant, and then they worked really well as plasma membrane voltage sensors.28
After that, we did go back then and because they are all rhodamine esters, we came up with the idea of making them more hydrolyzable by intracellular esterases so that the charged acid form would become trapped. The idea was that they would then stick in the inner membrane of the mitochondria, and then act as an electron transfer-based indicator.29 And getting back to how you can calibrate—-we spent some time with FCCP (trifluoromethoxy carbonyl cyanide phenylhydrazone), a protonophore, to try and collapse that mitochondrial membrane potential for calibration. Folks typically use micromolar amount of FCCP, but we found that this concentration of the compound affects fluorescent membrane dyes that are not voltage sensitive when you add that much FCCP. So, the message is—do not use micromolar FCCP!
So yes, I think mitochondria are some internal membranes that have been scrutinized for some time. More recently, lysosomes have been targeted with DNA nanostructures coupled to PeT-based indicators30 or with the hVOS system.31
Richard H. Kramer: Changing gears a little bit, for the most part fluorescent voltage sensors have developed specifically for measuring signals in the nervous system, most importantly action potentials. For accurate reporting spikes, you need a probe that is sensitive enough to report changes on the order of tens of millivolts, and fast enough to do this on a millisecond by millisecond timescale. The field has worked hard to develop probes to meet these requirements. But what if you wanted to report voltage signals in nonexcitable cells, maybe slower events.
Or not necessarily in individual cells but in tissues or organisms over a slower longer timeframe? Standing electrical fields, for example. Are there different design principles that you would apply to developing your probes for this purpose? And what might they be? I guess there is a lot in that question.
Bradley Baker: Well when you start to imaging nonexcitable cells, there is a new variable that crops up, and that is your frame rate. At what rate are you going to image? How long are you going to image? People studying circadian events use bioluminescent proteins (luciferin) to monitor these long oscillations over days. But voltage is a different story. How much voltage change is attributable to noise and how much is really signal? We need to know the dynamic range of a voltage probe to be appropriate for the signal we are trying to measure. That is a problem when we know so little about these signals!
Richard H. Kramer: So, do you guys know of anyone who has tried to actually optically monitor standing electrical fields in a purely macroscopic sense? For many years, people have been using voltage-sensitive dyes to deduce events in single excitable cells, or when single cell resolution is impossible in populations of cells, including in the cerebral cortex in vivo.32 But the goal in all these studies is to detect changes in membrane potential, not a steady-state electrical field. Is there on the horizon any optical approach for doing this? This is the opposite of what you are usually trying to do, which is to localize signals to individual cells, but now looking at the broader picture of electrical fields in tissue or even a whole organism?
Bradley Baker: Adam Cohen I think used that as a screening method where they had this high throughput with an electric field.
Ahmed Abdelfattah: I was going to say that we and others stimulate cell lines using electric field potentials in developing voltage sensors. Sometimes we do not start by screening those probes in neurons, but instead we do it in nonexcitable HEK-293 cells or other cell lines. We basically apply an electrical field to generate fluorescent signals in many cells at once. We put two electrodes across a line of HEK cells on a cover slip and apply an electrical field. We induce a transmembrane voltage; one side of the cell would hyperpolarize, the other would depolarize. And this way, we can screen for voltage sensors with a sufficiently large dynamic range.
The other thing I wanted to say is that there is a ton of protein space that you can explore. We have evolved fluorescent protein voltage sensors, but we have probably tested fewer than 1% of the different variations in the amino acids that are theoretically possible. You can change the properties of a voltage sensor dramatically with single point mutations. The genetically encoded probes we have now may be the tip of the iceberg.
For example, with the Voltron, we flipped the signal of the voltage probe from a probe that shows decreased fluorescence to increased fluorescence but with the same dynamic range, using three-point mutations in the protein. So, you can imagine that there is a large headway there. It is also important to keep in mind the use case of the voltage sensor being developed, is it to monitor action potentials in excitable cells like neurons? Or is it meant to record small changes in membrane potential of nonexcitable cells, and you need to magnify a few millivolt change?
The other thing I wanted to say is that proteins may not be the only answer for these problems especially where you do not really need genetically enabled single cell specificity. I think Evan can talk more about this.
Evan W. Miller: Sure. I think to the original question, I think it depends on what you want to measure in a nonexcitable cell. If you want to compare membrane potentials in two regions of a tissue or compare a transformed tumor versus normal tissue, I think that would be challenging. How can you ensure equal distribution and loading of a small molecule dye? How can you control equal expression level and proper trafficking of a protein-based sensor?
If you wanted to follow changes in membrane potential in tissue in an organism during regeneration, you would need to track signals in the same tissue over time, which presents challenges. For small molecule indicators, you would have to think about internalization over time, for protein-based sensors, you would have issues of changing protein expression and degradation. Maintaining a stable concentration of sensor in the tissue is key, and it is not clear how to do that.
There is going to be some problems with whatever optical modality you use, and the problems are not necessarily the same. It is Rafael Benitez's “small blanket problem”: if you pull the blanket up, you cannot cover the toes. If you pull it down, your head is cold. And hopefully you get multiple blankets to keep you warm. My room is very cold, so I am using that analogy. It is not a one size fits all thing, would be my guess.
Richard H. Kramer: Following up on fluorescence protein-based voltage sensors, how serious a problem is excessive protein expression? I would imagine too much expression could overwhelm protein trafficking machinery, resulting in protein aggregates in places that contaminate the plasma membrane signal. I know this is an issue with GCaMP-calcium sensors; too much expression can actually be toxic or give you fluorescence that no longer represents cytoplasmic free calcium. Is this going to be a similar problem with genetically encoded voltage probes?
Ahmed Abdelfattah: Absolutely. You are expressing an exogenous protein in a cell, so if you are expressing too much, or over a very long time, then probably the cells will die. So, you have to be careful with the titers of virus you use for expressing the sensor and so forth.
And for voltage sensors, you have limited real estate on the cell membrane and that could pose a problem that is not relevant for cytoplasmic sensors such as calcium sensors. So, it is a harder ask for voltage sensors to not affect the physiology. Whenever we develop a new sensor, it is important to monitor the membrane resistance, the membrane capacitance, etc. to see whether or not a cell expressing the sensor is different than a cell that is not expressing. That is the best we can do at least in a neuron culture setting or in a brain slice setting. But for sure, there is an effect, whether that effect is enough to stop you from answering your biological question, that is up to the user to determine.
With the Voltron work, for example, we have recorded from cells that have been expressing it for over a month in a mouse, and we see no changes over that time. But if we inject too much virus and cause too much expression, cells are less healthy. I am sure that is true for other genetically encoded probes as well.
Bradley Baker: You asked about our favorite voltage sensor, and one of my favorite is a chimeric voltage sensor/fluorescent protein (FP) named FlicR.33 It gave a nice signal in cells, it gets brighter with depolarization, I wish it worked better in vivo, but you cannot have everything. The main problem with poorly trafficked probes probably has to do with misfolding. But remarkably, the GFP part of the chimera seems to fold properly. It amazes me how the structural dynamics of that part of the protein remain intact despite the rest of the protein misfolding.
Now as a voltage imager, this is a disaster because it just creates internal fluorescence that does not respond to what you are doing. But from an understanding of protein structure, these sorts of things intrigue me and offer another avenue of research to figure out why. One thing that is a dream of mine is understanding codon usage. Why are some codons more prevalent than others? And does the cell use this as a way to time how a protein folds versus how it is being transcribed and translated?
I would love to see whether there are certain segments of a protein that you can slow down its folding to better accommodate its trafficking or whatever based just on those avenues. And these probes give us a chance to start asking these questions. Unfortunately, I do not know any of the answers yet, but these folding issues are an interesting problem.
Evan W. Miller: Brad, for improved trafficking of FP-based indicators, was it the voltage sensing domain from the phosphatase that really improved trafficking?
Bradley Baker: Yeah, Thomas Knopfel used that one.12 I think Thomas Knopfel, because my first article, one of my most cited articles in this field, had no positive results in mammalian cells! None of the first sensors, FlaSh FlaAre or SPARK, would work in mammalian cells. But, they worked in oocytes. They worked great in frog oocyte and you could see a signal and everyone was happy. And then people would try them in their cell type of interest and got nothing. And so we tried it and got nothing, and it turned out that the indicators did not go to the plasma membrane.34 So one of my most cited contributions to this field is that, if your fluorescent protein voltage indicator is not in the electric field, it cannot report changes in voltage.
Richard H. Kramer: So maybe including a protein domain that ensures timely degradation would be a good idea.
There is another issue Evan and I have talked about over the years. It has to do with how fluorescent voltage sensors affect membrane capacitance. Every voltage sensing requires some type of rearrangement of charges in or near the membrane. And that being the case, every voltage sensor adds to the capacitance of the membrane. In some way, some finite way, the addition of a voltage sensor is going to change the specific capacitance of the membrane. That will change passive membrane properties, such as time constant. How much of that is a concern? Evan, maybe you can say something because we have talked about it a lot, with respect to your voltage sensors.
Evan W. Miller: Yes. I think one of the problems with the FRET probes that we were talking about, dipicrylamine, for example, is a charged molecule, it is moving through the membrane at the same time scales as the biological voltage changes. And so the method that we have been looking at is the electron movement withing the probe itself, which occurs only when it is excited by a photon, which is a very small fraction of time (depending on illumination intensity). And so we have an electron moving from one side of the molecule to the other during the excited state of the fluorophore, and it has to move during that nanosecond time scale.
And then that charge has to move back in order for another round of excitation to happen, otherwise you get some sort of chemical reaction, photo bleaching, redox chemistry, something like that. Both of those processes must be fast. The forward charge transfer has to go in the nanosecond time scale. The charge recombination is usually on a slower time scale but maybe hundreds of nanoseconds, maybe a microsecond.
The way we think about it is that there is no net charge movement on the biological time scale. So rather than moving molecules across the membrane, we are moving those electrons. It is similar to the di-4-ANEPPS, the Stokes shift types of dyes, in the sense that the electric field is interacting with the molecular orbitals of the fluorophore, just that we have decoupled the fluorophore and the sensor part in our case. So that is how we are thinking about the strategy we take.
The other thing we worry about a little bit more is phototoxicity: we have essentially naked fluorophores in the membrane compared with a GFP fluorophore that is surrounded by the beta barrel protein fold. And so it is perhaps a little more protected from interacting with other stuff in the lipid membrane, or interacting with oxygen as much as say, for example, a small-molecule fluorophore. And so I think bleaching and phototoxicity are maybe a bigger problem for small molecules than the capacitance issue. Viral expression of fluorescent protein or genetically encoded indicator will kill cells, and in the same way, you will kill cells with excessive small molecule dye or light that is too intense.
I think that is always the tension as you think about developing these indicators. The trick is finding the right window. And it is just a different set of drawbacks or small molecules, versus hybrids versus proteins, I think. But that is how I think about that particular capacitance or charge issue.
Brad Baker: In the hVOS system, Meyer Jackson's group did a really nice job titrating how much dipicrylamine is effective without being toxic. They have several mice lines that give very nice signals using the hVOS method.35,36 The increased capacitance is an issue, but as long as you are aware of this, it does not prevent you from answering interesting questions. It is like increased calcium buffering with GCaMP. That is always going to be an issue, but GCaMP imaging still gives you information. You do not want to affect the physiology of the cell, but it is unavoidable—any measurement is going to have some effect to the physiology of the cell.
Another accident was Pado, a probe that we made exploiting evolution's high-throughput screening of variable protein structures to guide the development of new genetically encoded voltage indicators (GEVIs). By searching genomes for novel voltage sensing domains, we found that the number of positively charged amino acids in the S4 transmembrane segment is really variable. In voltage-gated potassium or sodium channels, S4 has up to seven positive amino acids, but there can be as few as three in some channels. Most voltage-sensitive phosphatases have three positively charged amino acids in S4. So, we used what nature had done and tried different voltage-sensing domains from various species.
One of these domains, from a voltage-gated proton channel, turned out to be particularly good at generating an optical signal when linked to pH-sensitive fluorescent protein (Super Ecliptic pHluorin). Pado, a fluorescent protein with proton channel activity, can optically monitor membrane potential, intracellular pH, and map gap junctions.25 That GEVI, which we named Pado, maintained its proton channel activity even when the plasma membrane was subjected to strong depolarizing steps. This gave us control over the internal pH while at the same time letting us see a voltage-dependent optical signal.
We could see different changes in pH in different parts of the cell. For example, the cytoplasm became acidified when they fired action potentials, but the pH change was not uniform. Thin processes acidified much more than the soma. Voltage imaging with Pado is complicated, and it is hard to completely avoid altering the physiology of the cell, but at the very least it is important to know what these alterations are.
Richard H. Kramer: I guess this capacitance issue is a bigger issue with the rapid signals, action potential, for example, than it would be with slower signals from nonexcitable cells anyway. So, the final question regards “segmentation,” in other words, how you tell whether a fluorescence signal is coming from one cell from its neighbor when the plasma membranes of the two cells are so closely apposed? If you have sparse labeling, for example, with a genetically encoded indicator, maybe it is not a problem, but if you have dense labeling of cells, how do you tell signals from different cells apart from one another?
It has occurred to me that what is best for calcium imaging is worst for voltage sensing. With calcium imaging, the cell body is a great place to detect a signal because it has got a big volume, and there are many calcium sensors in the cytoplasm, and there is so little cytoplasm in the small processes of the cell.
But with a fluorescent voltage sensor, which is embedded in the plasma membrane instead of the cytoplasm, the cell body is the least favorable place to detect a voltage change because the surface-to-volume ratio is the smallest. In contrast, the dendrites are so small, and they are intertwined that increases the difficulty of segmentation.
Ahmed, this is an issue you have probably thought a lot about.
Ahmed Abdelfattah: Absolutely! One of the advantages of genetically encoded probes is that you can add targeting sequences to enrich trafficking of those proteins to different parts of the cell. A good example is when we tried to image many cells in vivo with Voltron in mice,17 enriching expression on the plasma membrane of a neuron's soma versus processes, we could successfully assign signals from individual cells. Whereas if we did not do that, the contrast would have been too low to record signals at cellular resolution at our labeling density.
Even imaging layer one, which is closest to the surface of the mouse cortex, we were unable to segment any signals. When we added the soma localization signal, which is a 60-amino acid sequence from a voltage-gated ion channel that others have used to localize channelrhodopsin-2 to the soma, you can target Voltron to the somatic plasma membrane, giving you fluorescence in nice donut shapes. If you label sparsely enough, you can see each individual cell.
So, actually, in this particular area, soma localization works really pretty well, at least for opsin/fluorescent protein-based voltage sensors such as Voltron. People have also used the somatic targeting domain with ASAP and ArcLight.
Richard H. Kramer: So, are there equivalent targeting sequences for polarized epithelia, for example, you know of, for apical versus basal lateral?
Ahmed Abdelfattah: No!
Bradley Baker: The Yuste laboratory just published a postsynaptically targeted fluorescent protein voltage indicator where they use a recombinant antibody-like protein (FingR)39 to get it to certain parts of the neuron.40 But I would also stress that new advances in optics and light guides are also helping to localize fluorescent voltage signals. So, in addition to these tricks for getting the voltage sensor where we want, we can also get the light where we want.41
Ahmed Abdelfattah: I should also say that different targeting sequences work differently in different types of cells. So, it depends on what you are looking for.
Bradley Baker: We have seen tremendous cell-to-cell variability of the expression of these different voltage sensors, even with mammalian cell lines where all the cells should be identical.
Ahmed Abdelfattah: That is true, and that alone can call into question the interpretation of fluorescence signals (or the lack of a signal). Especially as we are developing a probe, consider what we record with a patch electrode as the gold standard. At least at first, this is essential for calibrating the fluorescence signal. So, if we are stepping the membrane potential to different values, and for some reason the seal is leaky or the cell is not healthy enough, the relationship between electrophysiologically and optically recorded voltage can break down.
But the real challenge is when we rely on the fluorescent signal alone. For example, suppose we are stimulating a dish of cultured cells with a field stimulus and we see variable responses in what we thought was a homogeneous population. We can minimize this issue by imaging several hundreds of cells and averaging the results.
Bradley Baker: But in this case, the recordings could be great but perhaps there is a variable degree of expression, even in cultured cells. When I read an article that compares the virtues of several different fluorescent voltage sensors, I just go straight to the supplemental information to see where the real data are, and then I look at the figures to see for myself how well a specific probe can actually do.42,43
Ahmed Abdelfattah: I think we talked a lot about neurons, but the same design principles of those probes apply when you want to use them to other cells. Like, for example, how bacteria communicate to find food, bacterial cultures, for example. Actually, that is what got me interested in making fluorescent voltage sensors to begin with. I was trying to make a probe for Escherichia coli and then somehow learned about neuroscience. I am a biochemist by training, and then somehow I ended up in a neuroscience department.
But really, it is the same design principles. I am sure Evan can say similar things about small molecules, but if you have fluorescent voltage sensing working in excitable cells, it should be even easier in nonexcitable cells. That is because action potentials are so brief (1–2 ms), so the probe not only has to be sensitive and bright, but also very fast.
There is a very strong push now to push the voltage sensors to sense subthreshold changes in membrane potential, Accurately measuring the magnitude and kinetics of subthreshold voltage signals is super important. These are signals that could not otherwise be picked up by fluorescent calcium sensors. This will be directly applicable to other nonexcitable cells or bacterial cultures, or other nonmodel organisms. So, I think the same design principles apply for any type of cell.
Richard H. Kramer: Well, we seem to have run out of time. Pardon the pun but thank you all for an illuminating discussion. People have dreamt about optical detection of changes in membrane potential for a very long time, and it is great to see that thanks to the inventiveness of you and others in this field, tools for high-resolution voltage imaging have become a reality!
Editor's Note
This article is the edited transcript of a round table discussion that was chaired online by R.H.K.. We are grateful to all the participants for their contributions.
References
- 1. Davila HV, Salzberg BM, Cohen LB, et al. A large change in axon fluorescence that provides a promising method for measuring membrane potential. Nat New Biol 1973;241:159–160. [DOI] [PubMed] [Google Scholar]
- 2. Cohen LB, Salzberg BM, Davila HV, et al. Changes in axon fluorescence during activity: Molecular probes of membrane potential. J Membr Biol 1974;19:1–36. [DOI] [PubMed] [Google Scholar]
- 3. González JE, Tsien RY. Voltage sensing by fluorescence resonance energy transfer in single cells. Biophys J 1995;69:1272–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. González JE, Tsien RY. Improved indicators of cell membrane potential that use fluorescence resonance energy transfer. Chem Biol 1997;4:269–277. [DOI] [PubMed] [Google Scholar]
- 5. Chanda B, Blunck R, Faria LC, et al. A hybrid approach to measuring electrical activity in genetically specified neurons. Nat Neurosci 2005;8:1619–1626. [DOI] [PubMed] [Google Scholar]
- 6. Siegel MS, Isacoff EY. A genetically encoded optical probe of membrane voltage. Neuron 1997;19:735–741. [DOI] [PubMed] [Google Scholar]
- 7. Miyawaki A, Llopis J, Heim R, et al. Fluorescent indicators for Ca2+ based on green fluorescent proteins and calmodulin. Nature 1997;388:882–887. [DOI] [PubMed] [Google Scholar]
- 8. Nakai J, Ohkura M, Imoto K. A high signal-to-noise Ca2+ probe composed of a single green fluorescent protein. Nat Biotechnol 2001;19:137–141. [DOI] [PubMed] [Google Scholar]
- 9. Ataka K, Pieribone VA. A genetically targetable fluorescent probe of channel gating with rapid kinetics. Biophys J 2002;82:509–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sakai R, Repunte-Canonigo V, Raj CD, et al. Design and characterization of a DNA-encoded, voltage-sensitive fluorescent protein. Eur J Neurosci 2001;13:2314–2318. [DOI] [PubMed] [Google Scholar]
- 11. Murata Y, Iwasaki H, Sasaki M, et al. Phosphoinositide phosphatase activity coupled to an intrinsic voltage sensor. Nature 2005;435:1239–1243. [DOI] [PubMed] [Google Scholar]
- 12. Dimitrov D, He Y, Mutoh H, et al. Engineering and characterization of an enhanced fluorescent protein voltage sensor. PLoS One 2007;2:e440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kralj JM, Hochbaum DR, Douglass AD, et al. Electrical spiking in Escherichia coli probed with a fluorescent voltage-indicating protein. Science 2011;333:345–348. [DOI] [PubMed] [Google Scholar]
- 14. Kralj JM, Douglass AD, Hochbaum DR, et al. Optical recording of action potentials in mammalian neurons using a microbial rhodopsin. Nat Methods 2011;9:90–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zou P, Zhao Y, Douglass AD, et al. Bright and fast multicoloured voltage reporters via electrochromic FRET. Nat Commun 2014;5:4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gong Y, Huang C, Li JZ, et al. High-speed recording of neural spikes in awake mice and flies with a fluorescent voltage sensor. Science 2015;350:1361–1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Abdelfattah AS, Kawashima T, Singh A, et al. Bright and photostable chemigenetic indicators for extended in vivo voltage imaging. Science 2019;365:699–704. [DOI] [PubMed] [Google Scholar]
- 18. Xu Y, Peng L, Wang S, et al. Hybrid indicators for fast and sensitive voltage imaging. Angew Chem Int Ed Engl 2018;57:3949–3953. [DOI] [PubMed] [Google Scholar]
- 19. Liu S, Lin C, Xu Y, et al. A far-red hybrid voltage indicator enabled by bioorthogonal engineering of rhodopsin on live neurons. Nat Chem 2021;13:472–479. [DOI] [PubMed] [Google Scholar]
- 20. Xie C, Lin Z, Hanson L, et al. Intracellular recording of action potentials by nanopillar electroporation. Nat Nanotechnol 2012;7:185–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Karaveli S, Gaathon O, Wolcott A, et al. Modulation of nitrogen vacancy charge state and fluorescence in nanodiamonds using electrochemical potential. Proc Natl Acad Sci U S A 2016;113:3938–3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Park J, Kuo Y, Li J, et al. Improved surface functionalization and characterization of membrane-targeted semiconductor voltage nanosensors. J Phys Chem Lett 2019;10:3906–3913. [DOI] [PubMed] [Google Scholar]
- 23. Grupi A, Ashur I, Degani-Katzav N, et al. Interfacing the cell with “biomimetic membrane proteins.” Small 2019;15:e1903006. [DOI] [PubMed] [Google Scholar]
- 24. Leong LM, Kang BE, Baker BJ. Improving the flexibility of genetically encoded voltage indicators via intermolecular FRET. Biophys J 2021;120:1927–1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kang BE, Leong LM, Kim Y, et al. Mechanism of ArcLight derived GEVIs involves electrostatic interactions that can affect proton wires. Biophys J 2021;120:1916–1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sepehri Rad M, Cohen LB, Braubach O, et al. Monitoring voltage fluctuations of intracellular membranes. Sci Rep 2018;8:6911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Johnson LV, Walsh ML, Chen LB. Localization of mitochondria in living cells with rhodamine 123. Proc Natl Acad Sci U S A 1980;77:990–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Deal PE, Kulkarni RU, Al-Abdullatif SH, et al. Isomerically pure tetramethylrhodamine voltage reporters. J Am Chem Soc 2016;138:9085–9088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Klier PEZ, Martin JG, Miller EW. Imaging reversible mitochondrial membrane potential dynamics with a masked rhodamine voltage reporter. J Am Chem Soc 2021;143:4095–4099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Saminathan A, Devany J, Veetil AT, et al. A DNA-based voltmeter for organelles. Nat Nanotechnol 2021;16:96–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Matamala E, Castillo C, Vivar JP, et al. Imaging the electrical activity of organelles in living cells. Commun Biol 2021;4:389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Grinvald A, Hildesheim R. VSDI: a new era in functional imaging of cortical dynamics. Nat Rev Neurosci 2004;5:874–885. [DOI] [PubMed] [Google Scholar]
- 33. Abdelfattah AS, Farhi SL, Zhao Y, et al. A bright and fast red fluorescent protein voltage indicator that reports neuronal activity in organotypic brain slices. J Neurosci 2016;36:2458–2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Baker BJ, Lee H, Pieribone VA, et al. Three fluorescent protein voltage sensors exhibit low plasma membrane expression in mammalian cells. J Neurosci Methods 2007;161:32–38. [DOI] [PubMed] [Google Scholar]
- 35. Wang D, McMahon S, Zhang Z, et al. Hybrid voltage sensor imaging of electrical activity from neurons in hippocampal slices from transgenic mice. J Neurophysiol 2012;108:3147–3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ma Y, Bayguinov PO, Jackson MB. Optical studies of action potential dynamics with hVOS probes. Curr Opin Biomed Eng 2019;12:51–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Miller EW. Small molecule fluorescent voltage indicators for studying membrane potential. Curr Opin Chem Biol 2016;33:74–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kulkarni RU, Miller EW. Voltage Imaging: Pitfalls and Potential. Biochemistry 2017; 56:5171–5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gross GG, Junge JA, Mora RJ, et al. Recombinant probes for visualizing endogenous synaptic proteins in living neurons. Neuron 2013;78:971–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Cornejo VH, Ofer N, Yuste R. Voltage compartmentalization in dendritic spines in vivo. Science 2022;375:82–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Foust AJ, Zampini V, Tanese D, et al. Computer-generated holography enhances voltage dye fluorescence discrimination in adjacent neuronal structures. Neurophotonics 2015;2:021007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bando Y, Sakamoto M, Kim S, et al. Comparative evaluation of genetically encoded voltage indicators. Cell Rep 2019;26:802.e4–813.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Milosevic MM, Jang J, McKimm EJ, et al. In vitro testing of voltage indicators: Archon1, ArcLightD, ASAP1, ASAP2s, ASAP3b, Bongwoori-Pos6, BeRST1, FlicR1, and Chi-VSFP-Butterfly. eNeuro 7:0060–20.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]