Abstract
Fungal phytotoxins cause significant harm to agricultural production or lead to plant diseases. Discovering new phytotoxins, dissecting their formation mechanism and understanding their action mode are important for controlling the harmful effects of fungal phytopathogens. In this study, a long-term unsolved cluster (polyketide synthase 16, PKS16 cluster) from Fusarium species was thoroughly investigated and a series of new metabolites including both complex α-pyrone-polyketide glycosides and simple polyketide carboxylates were identified from F. proliferatum. The whole pathway reveals an unusual assembly and inactivation process for phytotoxin biosynthesis, with key points as follows: (1) a flavin dependent monooxygenase catalyzes Baeyer–Villiger oxidation on the linear polyketide side chain of α-pyrone-polyketide glycoside 8 to form ester bond compound 1; (2) a β-glucosidase unexpectedly mediates the ester bond hydrolysis of 1 to generate polyketide carboxylate phytotoxin 2; (3) oxidation occurring on the terminal inert carbons of 2 by intracellular oxidase(s) eliminates its phytotoxicity. Our work identifies the chemical basis of the PKS16 cluster in phytotoxicity, shows that polyketide carboxylate is a new structural type of phytotoxin in Fusarium and importantly uncovers a rare ester bond hydrolysis function of β-glucosidase family enzymes.
Investigation of the PKS16 cluster in Fusarium reveals an unusual assembly and inactivation process for polyketide carboxylate phytotoxin biosynthesis from polyketide glycosides and uncovers a rare ester bond hydrolysis function of β-glucosidases.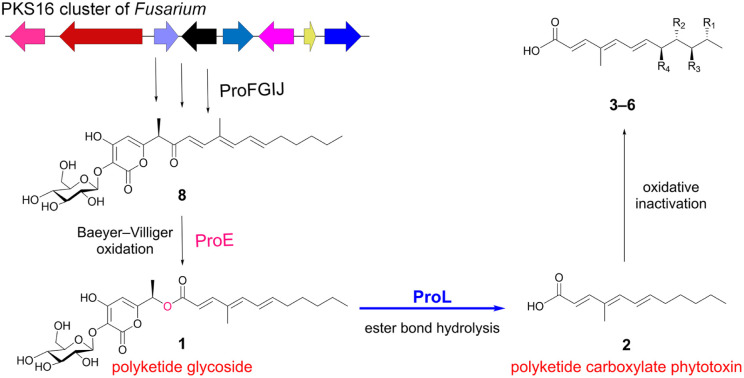
Introduction
Fungal phytotoxins are a large group of specialized secondary metabolites mainly produced and secreted by fungal phytopathogens, which lead to significant losses in crop yields or plant diseases.1–3 They usually serve as the virulence factors that subvert the host defense system and promote the invasion of fungal phytopathogens at different growth stages of plants.4–7 Throughout the entire infection process, fungal phytopathogens typically produce different phytotoxins, utilizing their different chemical spaces and structural types to cope with complex interactions with plants.8,9 Therefore, a deep understanding of the biosynthetic pathway, regulation mechanism, maturation mechanism, and action mode of phytotoxins is of great significance for effectively controlling the harmful effects of fungal phytopathogens.10
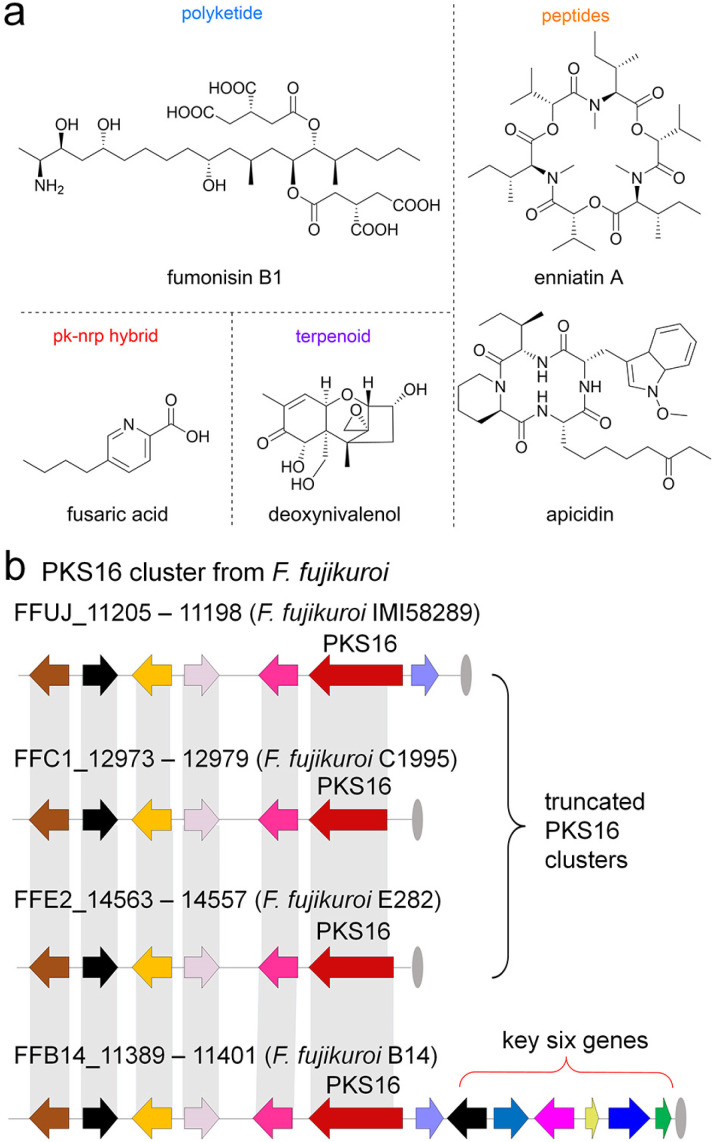
So far, a series of phytotoxins, for instance, polyketides, terpenoids, polyketide–nonribosomal peptide (pk–nrp) hybrids and peptide alkaloids have been identified from many famous fungal phytopathogens like Alternaria, Botrytis, Colletotrichum, Helminthosporium, and Phoma.2 In addition to the above-mentioned phytopathogenic fungal species, we mainly focused on the Fusarium genus, which is one of the top ten fungal phytopathogens worldwide,11 comprising more than 70 species with a wide range of plant hosts,12 and possessing a good ability to produce various phytotoxins13 such as fumonisins,14 deoxynivalenol,15 fusaric acid,16 apicidin,17 enniatins18 and so on (Fig. 1a and S2†).
Fig. 1. Fungal phytotoxins and PKS16 clusters from Fusarium species. (a) The well-known phytotoxins produced by Fusarium sp. (b) PKS16 and its homologue clusters in F. fujikuroi species. The PKS16 cluster in F. fujikuroi IMI 58289 lacks the key tailoring-step genes, while the core hrPKS is truncated in stains C1995 and E282.
As a continuation of our recent research on genome mining for new enzymes and the biosynthesis of mycotoxin natural products from Fusarium sp.,19–23 an important but long-term unsolved cluster (polyketide synthase 16, PKS16 cluster, Fig. 1b) from the main phytopathogenic fungus causing the “bakanae” disease of rice, F. fujikuroi IMI 58289,24–27 has attracted our attention. Previous studies revealed the following information: (1) the PKS16 cluster is located at the left end of chromosome 11 of F. fujikuroi IMI 58289;24 (2) similar to other phytotoxins like fumonisins and enniatins, the expression of the PKS gene in the PKS16 cluster is also co-induced by low-nitrogen conditions;25 most importantly, (3) the wide distribution of the PKS16 cluster among all sequenced F. fujikuroi species isolates strongly imply that the products derived from this cluster may act as phytotoxins in the F. fujikuroi species,26 although their structures and mode of actions are still unclear to date.
In this work, a complete and homologue gene cluster (namely pro cluster) of PKS16 was discovered from F. proliferatum by genome mining. A series of new products were successfully identified from the pro pathway, which include both complex α-pyrone-polyketide glycosides and simple polyketide carboxylate compounds. The whole process reveals an unusual assembly and inactivation process for phytotoxin biosynthesis, with key points including (1) a flavin monooxygenase (FMO) catalyzing Baeyer–Villiger (B–V) oxidation on the linear polyketide side chain of α-pyrone-polyketide glycoside 8 to form ester bond compound 1; (2) a β-glucosidase unexpectedly mediating the ester bond hydrolysis of 1 to generate a new polyketide carboxylate phytotoxin 2 from Fusarium sp., representing a rare function of β-glucosidase family enzymes; and furthermore, (3) the oxidation occurring on the terminal inert carbons of 2 by intracellular oxidase(s), which eliminates the phytotoxicity of 2.
Results and discussion
Genome mining discovers the complete and homologue pro cluster of PKS16 in F. proliferatum
Since the production of phytotoxins is always transient and sometimes requires interactions between fungal phytopathogens and plants to stimulate their generation,8 we initially inferred that the PKS16 products may be produced in a very small amount, making them difficult to identify in F. fujikuroi IMI 58289. However, the position of PKS16 at the left end of chromosome 11 highlights another possibility that, the absence of some key genes in the PKS16 cluster leads to F. fujikuroi IMI 58289 losing its ability to produce the corresponding compounds.
To test this hypothesis, we first carefully investigated the homologue gene clusters from publicly available genome sequences of F. fujikuroi species. As shown in Fig. 1b, (1) compared to PKS16, the two clusters from F. fujikuroi C1995 and F. fujikuroi E282 are also incomplete; moreover, the truncated core PKS genes in these two clusters indicate that they have no function; however, (2) the cluster in F. fujikuroi B14 is considered to be complete, with additional six genes located downstream of the core highly reducing PKS (hrPKS) gene. We further compared this cluster with other publicly available Fusarium genomes and our lab databases using local BLAST with the sequence of the core hrPKS gene. Indeed, the complete and homologue gene clusters of PKS16 have also been found in many Fusarium strains, such as F. denticulatum, F. mexicanum, F. subglutinans, F. sphaerosporum and F. proliferatum (Fig. S3†), which indicates the wide distribution of PKS16 homologue clusters in Fusarium species. Interestingly, in addition to Fusarium species fungi, the PKS16 homologue clusters are also found in other phytopathogenic fungi like Neonectria ditissima and Xylona heveae (Fig. S3†).
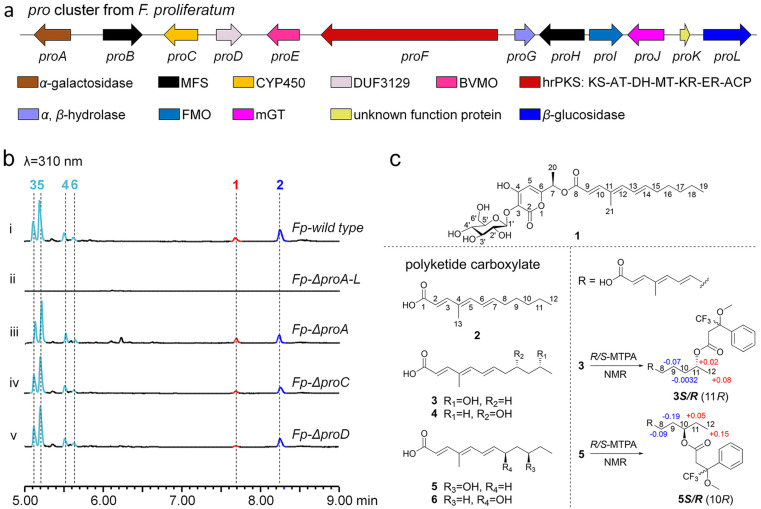
The representative pro cluster (NCBI accession number PQ271635) from F. proliferatum CGMCC 3.4710 is shown in Fig. 2a. Aside from the core hrPKS gene (proF), the pro cluster contains other genes encoding α-galactosidase (proA), transporters (proB and proH), cytochrome P450 (CYP450, proC), DUF3129 family protein (proD), B–V monooxygenase (BVMO, proE), α,β-hydrolase (proG), FMO (proI), membrane-bound glycosyltransferase (mGT, proJ, Fig. S4†), a protein (proK) of unknown function and β-glucosidase (proL). It is worth noting that the discovery of a glycosyltransferase in the pro cluster indicates that the products of PKS16 and its homologue clusters are possible new polyketide glycoside hybrid compounds in Fusarium species;28 the only previously identified polyketide glycoside from Fusarium is fusapyrone.29–31 Moreover, proA, proC and proD are not conserved genes in other homologue clusters (Fig. 2a and S3†), indicating that they may not be involved in the biosynthesis of pro pathway products.
Fig. 2. Identification of the homologue pro cluster and confirmation of the corresponding products in F. proliferatum. (a) Organization and proposed gene functions of the pro cluster. (b) LC-MS analysis of the extracts from F. proliferatum and its knockout mutants. (c) Chemical structures of compounds 1–6.
The α-pyrone-polyketide glycoside 1 and polyketide carboxylate 2 are products of the pro cluster
We first screened various low-nitrogen incubation conditions and found that all pro genes were under transcription when F. proliferatum was incubated in glycerol medium (Fig. S5†), which strongly implies that the corresponding products of the pro cluster were produced. To probe the products of the pro pathway, we knocked out (KO) the whole pro cluster (Fp-ΔproA–L, Fig. S6†) and compared the metabolite profiles between the F. proliferatum wild type and the Fp-ΔproA–L strain, and it was found that the production of six compounds (1–6) was eliminated in Fp-ΔproA–L (Fig. 2b(i and ii)). Subsequent large-scale fermentation of the F. proliferatum wild type, purification, and confirmation of the structures of 1–6via HR-MS and NMR analysis showed that (1) compound 1, namely, proliferapyrone A, is an α-pyrone-polyketide glycoside (Fig. 2c, Table S6 and Fig. S36–S42†), and its linear polyketide side chain is linked to α-pyrone via an unusual ester bond, and O-glucosylation occurs at the C3–OH of the α-pyrone moiety via a β-glycosidic bond (C3–O–C1′ ether bond), which was further confirmed to be d-glucose by comparison with the standard after hydrolysis with 4 M trifluoroacetic acid (Fig. S7†); (2) unlike the complex structure of 1, compounds 2–6, namely, proliferic acids A–E, are all polyketide carboxylate compounds, where 3–6 are likely hydroxyl-derivatives of 2via oxidation of the inert carbon atoms at C8–C11 of 2, respectively (Fig. 2c, S43–S77 and Tables S7–S11†); (3) using 3 and 5 as examples, based on the Mosher derivation32 results (Fig. 2c, S78–S81, Tables S12 and S13†), the stereochemistry of the hydroxyl group in 3–6 was assigned as R configuration, respectively.
Next, we further investigated the relationship between 1–6 and three non-conserved genes (proA, proC and proD) of the pro cluster. Individual KO mutants of proA, proC and proD were first generated (Fig. S6†), and subsequent analysis of the metabolites from each KO mutant showed that these three non-conserved genes are indeed not involved in 1–6 formation; the Fp-ΔproA, Fp-ΔproC and Fp-ΔproD mutants retain the ability to produce compounds 1–6, respectively (Fig. 2b(iii–v)). The exclusion of CYP450 ProC acting as the radical-type oxidase in the hydroxylation of the inert C8–C11 of 2 to form 3–6 strongly suggests that other oxidase(s) besides the pro cluster is(are) responsible for these conversions. Indeed, when 2 was fed into the Fp-ΔproA–L strain, the formation of 3–6 was observed (Fig. S8†). These results clearly show that compounds 3–6 are over-oxidized off-pathway derivatives of 2 by F. proliferatum; thus, 1 and 2 are the final on-pathway products corresponding to the pro cluster.
hrPKS and α,β-hydrolase collaboratively produce α-pyrone-polyketide precursor 7
Structural analysis of 1 and 2 indicates that these two compounds feature significantly different skeletons, suggesting that an unusual biosynthetic process possibly occurred during their biosynthesis. We initially inferred that the generation of 2 results from the ester bond hydrolysis of 1; therefore, according to the proposed function of pro cluster enzymes, the preferred candidate responsible for this step should be α,β-hydrolase ProG. However, when 1 was incubated with purified ProG (Fig. S9†), the conversion of 1 to 2 was not observed (Fig. S10†). We also considered that the hydrolysis of 1 to 2 could be an off-pathway conversion in F. proliferatum; however, when 1 was fed into the Fp-ΔproA–L strain, only trace amounts of 2 were observed (Fig. S11†). Moreover, the chemical conversion of 1 to 2 was investigated under various pH buffer conditions, and the formation of 2 was only detected under strongly basic conditions (pH > 11, Fig. S12†). These conversion investigations indicate that the formation of 2 should be an enzyme-catalyzed route, with the corresponding enzyme(s) located in the pro cluster.
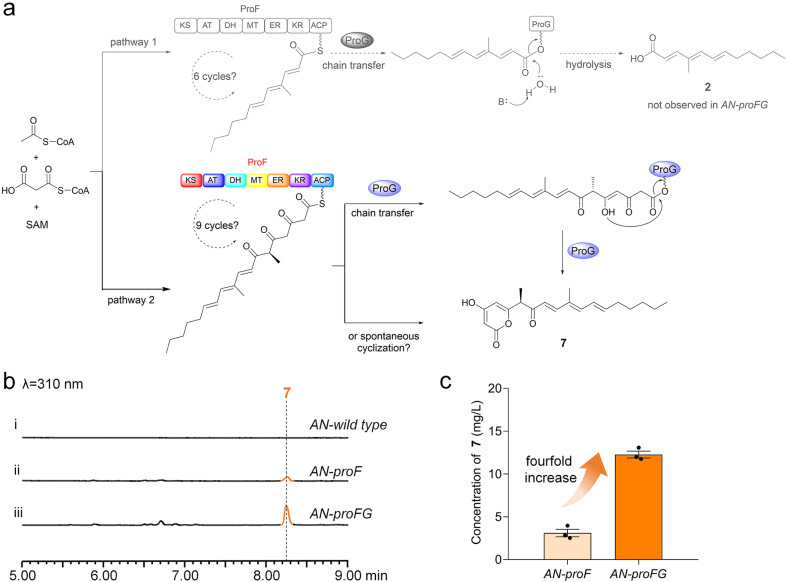
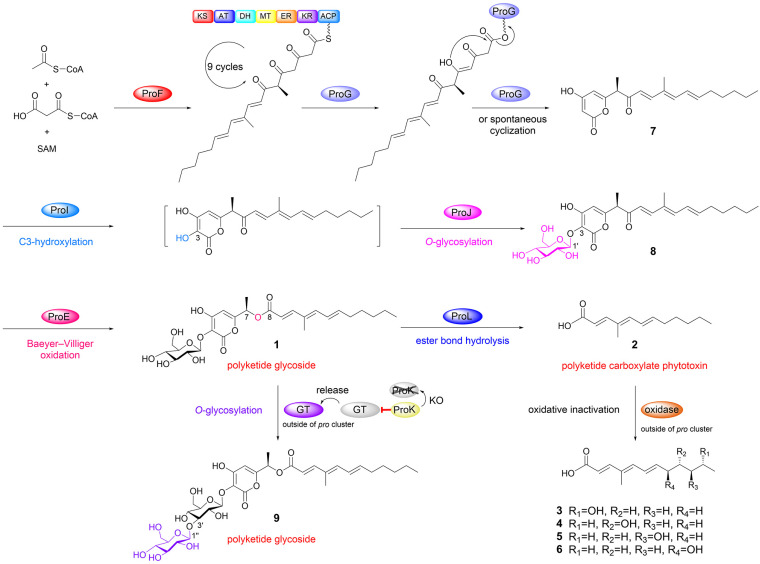
Based on these observations, we alternatively reasoned that the skeletal differences between 1 and 2 may be due to differences in the collaboration of hrPKS ProF and α,β-hydrolase ProG. As shown in Fig. 3a, (1) during six cycles of catalysis by ProF, ProG may catalyze the hydrolysis of the ACP-bound polyketide intermediate to form 2; (2) after nine cycles of catalysis by ProF, an α-pyrone polyketide is ultimately generated through the spontaneous cyclization of the ACP-bound terminal triketone polyketide intermediate.33 This cyclization may also be catalyzed by ProG.
Fig. 3. The hrPKS and α,β-hydrolase collaboratively produce α-pyrone-polyketide precursor 7. (a) Two proposed pathways for the collaboration of hrPKS ProF and α,β-hydrolase ProG. (b) LC-MS analysis of the culture extracts from AN-proF and AN-proFG. (c) The yields of 7 increased nearly fourfold in AN-proFG indicating that ProG plays a supportive role with ProF in the biosynthesis of 7.
To validate this hypothesis, we cloned and transferred these two genes into the typical heterologous host A. nidulans.34–37 In contrast to the A. nidulans wild type control, (1) a compound (7, proliferapyrone B) was produced by AN-proF (Fig. 3b(i and ii)), and its structure was confirmed to be α-pyrone polyketide through HR-MS and NMR analysis (Fig. 3a, Table S14 and Fig. S82–S88†). The C7-methyl stereochemistry of 7 has been confirmed to be the R configuration by comparing the experimental and calculated ECD spectra38 (Fig. 3 and S13†); (2) the expected compound 2 was not observed in AN-proFG; instead, the yields of 7 increased nearly fourfold (Fig. 3b(iii) and 3c), indicating that ProG plays a supportive role to ProF in the biosynthesis of 7. Therefore, although these results do not support the hypothesis that 2 originates from the ProF branch pathway (pathway 1, Fig. 3a), they clearly elucidate the collaborative mechanism of ProF and ProG in the production of 7, where ProF undergoes complete nine cycles to generate an on-pathway intermediate with the terminal triketone moiety; ProG then mediates its transfer and cyclization to yield the α-pyrone polyketide precursor 7 (pathway 2, Fig. 3a).33
Confirmation of the biosynthetic genes and the catalytic order from 7 to 1
We next investigated the biosynthetic order from 7 to 1. A structural comparison between 7 and 1 indicates that some tailoring steps, such as sequential hydroxylation and O-glucosylation at the C3 of the α-pyrone ring and subsequent B–V oxidation at the C7–C8 of 7, could accomplish the conversion of 7 to 1.
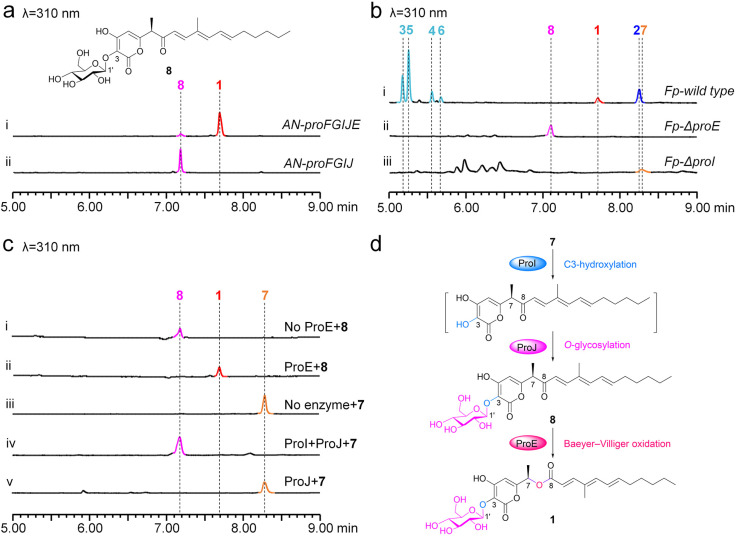
Based on this assumption, three enzymes ProI, ProJ and ProE are ideal candidates for catalyzing these steps. Indeed, when proI, proJ and proE were co-expressed with proFG in A. nidulans, (1) compound 1 and one minor compound (8, proliferapyrone C) were produced by AN-proFGIJE (Fig. 4a(i)); (2) removal of proE eliminates the production of 1; however, 8 was accumulated in AN-proFGIJ (Fig. 4a(ii)). Subsequent purification, and HR-MS and NMR analysis confirmed 8 to be an α-pyrone-polyketide glycoside lacking the C7–C8 ester bond (Fig. 4a, Table S16 and Fig. S89–S95†). Therefore, ProE is responsible for the conversion of 8 to 1 by catalyzing the BV oxidation between C7 and C8; this observation is further confirmed by the in vivo KO proE in F. proliferatum (Fp-ΔproE. Fig. S6†) to yield 8 (Fig. 4b(ii)) and the in vitro incubation of 8 with ProE to form 1 (Fig. 4c(i and ii)). Moreover, (3) when 7 was incubated with purified FMO ProI (Fig. S9†), along with microsome fractions of mGT ProJ and cofactors (FAD, NADPH and UDP-glucose), the formation of 8 was clearly observed (Fig. 4c(iii and iv)), confirming that these two enzymes are indeed responsible for converting 7 to 8; (4) when 7 was incubated with ProJ and UDP-glucose, no products were observed (Fig. 4c(v)), indicating that ProJ does not recognize C4–OH of 7 to perform O-glucosylation, showing a particular preference towards the C3–OH; (5) when 7 was incubated with ProI, a time-dependent decrease in 7 was observed; however, the generation of the proposed C3-hydroxyl derivative of 7 was not observed (Fig. S14†), implying that the hydroxylation occurring at the C3 position of the pyrone ring of 7 possibly makes it unstable.33,39 We further confirmed that 7 is the substrate of ProI, as the in vivo KO proI in F. proliferatum (Fp-ΔproI. Fig. S6†) yields 7 (Fig. 4b(iii)). Therefore, based on these results, the route from 7 to 1 by ProIJE was established (Fig. 4d).
Fig. 4. Confirmation of the function of ProE, ProI and ProJ. (a) LC-MS analysis of the culture extracts from AN-proFGIJE and AN-proFGIJ. (b) LC-MS analysis of the culture extracts from proE and proI knockout mutants in F. proliferatum. (c) The in vitro biochemical assays of ProE, ProI and ProJ. (d) The proposed pathway from 7 to 1.
β-Glucosidase is involved in the formation of 2 from 1
It is worth noting that, clarification of the biosynthesis process from 7 to 1 confirms that 2 is not a product of this conversion stage, thus it should have been originated from 1. Previous results excluded the involvement of ProG and other enzymes outside the pro cluster in this conversion step (Fig. S8 and S11†); thus ProK and ProL seem to be the last candidates for the coversion of 1 to 2.
Careful bioinformatic analysis showed that (1) ProL belongs to the glycoside hydrolase family 3 (GH3) of β-glucosidase proteins (Fig. S15†);40–44 members of this family usually employ an acid/base mechanism to hydrolyze the glycosidic bond (ether bond) of glycosides and oligosaccharides (Fig. S16†), and the corresponding catalytic residues43 (aspartate and glutamate) are indeed conserved in ProL (Fig. S15†); (2) ProK is a very small protein (142 aa); unlike ProL, it is difficult to predict its function through sequence alignment or structural simulation. Therefore, from a bioinformatic analysis perspective, it seems that neither of these enzymes should catalyze the ester bond hydrolysis process from 1 to 2.
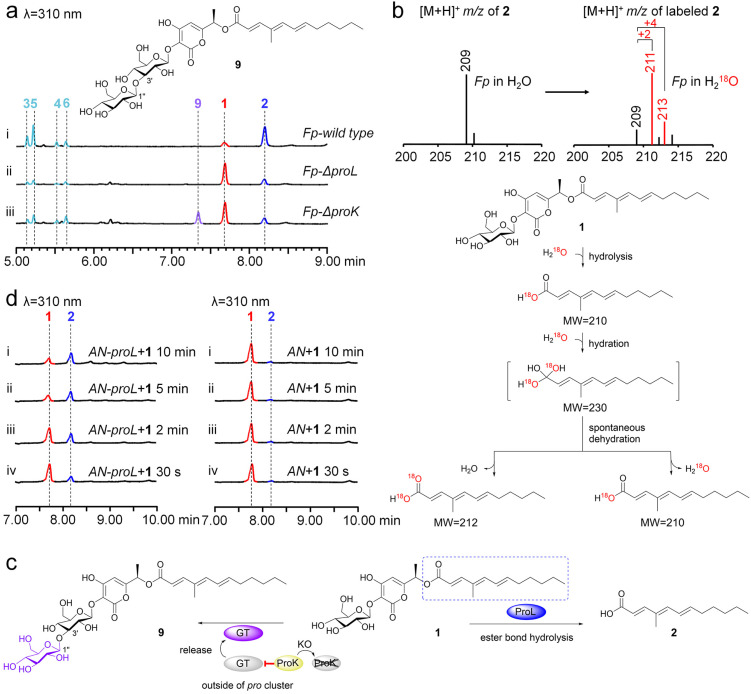
To probe the function of ProK and ProL, in vivo KO of these two genes was performed (Fig. S6†). Analysis of the metabolic profiles of Fp-ΔproK and Fp-ΔproL showed that, compared to the F. proliferatum wild type (Fig. 5a(i)), (1) the production of 2 and its oxidized derivatives (3–6) was almost eliminated, while 1 was accumulated in Fp-ΔproL (Fig. 5a(ii)); (2) unexpectedly, the conversion efficiency of 1 to 2 was greatly decreased; meanwhile a new compound (9, proliferapyrone D) was produced by Fp-ΔproK (Fig. 5a(iii)). Subsequent structural determination through HR-MS and NMR analysis confirmed that 9 features a di-glucose moiety at the C3–OH position; the key HMBC correlation between H1′′ and C3′ shows that the linkage of di-glucose is through an unusual β-1,3 glycosidic bond (Tables S17, S18 and Fig. S96–S109†).45 Thus, the di-glucose moiety of 9 is laminaribiose,46 which is further confirmed by comparison with the standard after hydrolysis (Fig. S17†); moreover, (3) when F. proliferatum was incubated in H218O-labeled medium, the incorporation of 18O from H218O into 2 was successfully observed (Fig. 5b), which confirmed that the hydrolysis reaction indeed occurs during the formation of 2.
Fig. 5. Investigation of the function of ProL and ProK. (a) LC-MS analysis of the culture extracts from proL and proK knockout mutants in F. proliferatum. (b) LC-MS analysis of the incorporation of H218O into 2. (c) The proposed pathway from 1 to 2 and 9, respectively. β-Glucosidase ProL is involved in the hydrolysis of 1 to form 2. (d) The bioconversion of 1 to 2 by the crude enzymes of AN-proL.
These results revealed the following important information (Fig. 5c): (1) the conversion of 1 to 2 is mediated by ProL, where the β-glucosidase-mediated hydrolysis of ester bonds is rarely reported;47,48 (2) the production of 9 seems to be the result of an outside pro cluster enzyme catalyzing the glucosylation of 1, as the formation of 9 was not observed when 1 was incubated with UDP-glucose and the in-cluster mGT ProJ (Fig. S18†); (3) the function of ProK is likely to act as an inhibitor of glycosyltransferase(s) outside the pro cluster; its deletion may eliminate its inhibitory effect, thereby allowing unknown glycosyltransferase(s) to catalyze the glycosylation of 1 to form 9in vivo. We further searched for homologue proteins of ProK using sequence similarity network (SSN) analysis49 (Fig. S19†) and found that they are a very small family clade in fungi (n = 47), with most of them belonging to the Fusarium species, and some ProK homologs are indeed associated with glycosyltransferases in natural product gene clusters (Fig. S19†). However, the actual function and the relationship between ProK family proteins and glycosyltransferases need further in-depth investigation.
We initially attempted to elucidate the unusual function of ProL in conversion of 1 to 2 through in vitro biochemical assays; however, it was insoluble when expressed in E. coli (Fig. S20†). Alternatively, we decided to confirm this conversion via a fungal system. As shown in Fig. 5d, compared to the AN wild type, when 1 was incubated with the crude enzymes from AN-proL, the production of 2 was observably detected within 10 min, where only trace amounts of 2 were observed in AN, suggesting that A. nidulans has endogenous enzyme(s) capable of weakly hydrolyzing 1 to 2. Moreover, when 1 was incubated with the crude enzymes of AN-proK, it was indeed inactive (Fig. S21†). We further performed mutagenesis experiments on the conventional active sites D266 and E493 of β-glucosidases in ProL and found that both AN-proL D266A and AN-proL E493A lost their ability to convert 1 to 2 (Fig. S22†), respectively. Therefore, these results importantly show that the conversion of 1 to 2 is mediated by ProL; unlike traditional β-glucosidases that hydrolyze glycosidic bonds (ether bond),40,43 ProL harbors the classical catalytic amino acid residues (D266 and E493) of β-glucosidases; however it has an unexpected ability to mediate the hydrolysis of the ester bond of 1 to form 2.
Polyketide carboxylate 2 is a phytotoxin
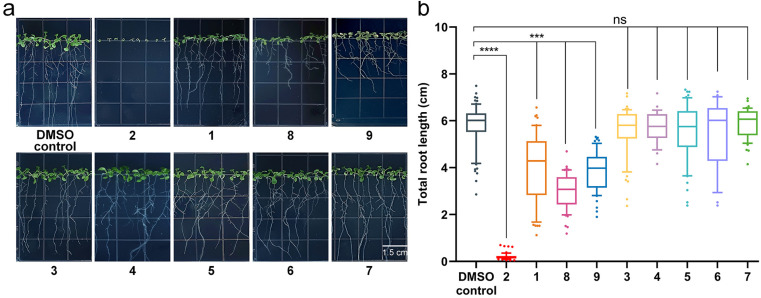
Considering that the core PKS gene of the PKS16 cluster is transcribed in the early stages of F. fujikuroi IMI 58289 infection,27 we proposed that the compounds 1–9 identified from the pro cluster may be new phytotoxins in Fusarium species. As shown in Fig. 6, their phytotoxicity evaluation conducted through the root growth test on Arabidopsis thaliana50 showed that (1) the polyketide glycosides (1, 8 and 9) have moderate inhibition effects on the growth of roots; however, (2) polyketide carboxylate 2 is a phytotoxin, and it has significant inhibitory activity on the growth of roots; (3) the polyketide precursor 7 and compounds 3–6 are not phytotoxins.
Fig. 6. The root growth inhibition activity tests against Arabidopsis thaliana of compounds 1–9. (a) Phenotypic characteristics of seven-day-old Arabidopsis thaliana grown on Murashige and Skoog basal medium containing 50 μg per mL 1–9 (DMSO: dimethyl sulfoxide). (b) Quantification of the total root length of seven-day-old Arabidopsis thaliana grown on medium containing 50 μg per mL 1–9. In box plots, the center line is the median, box edges delineate first and third quartiles and whiskers show the range of values (n = 3 independent experiments), unpaired two-tailed Student's t-test (***p ≤ 0.001, ****p ≤ 0.0001, ns stands for no significant difference).
The above activity tests indicate the assembly and inactivation process of polyketide carboxylate phytotoxin 2 in Fusarium species, where it is generated from polyketide-glycoside 1via β-glucosidase-mediated hydrolysis of ester bonds and is inactivated by the intracellular oxidase-catalyzed oxidation of the terminal inert carbon atoms to form 3–6. It should be noted that, the newly discovered compound 2 is a structural analog of the well-known mitotic progression inhibitor myrmicacin;51,52 thus, it or its structural derivatives are a potential and worth developing herbicide source in the future.
Conclusions
In summary, we discovered the intact PKS16 homologue clusters from various Fusarium strains by genome mining and identified gene functions of the representative pro cluster from F. proliferatum. A plenty of α-pyrone-polyketide glycosides and polyketide carboxylate compounds were characterized from this cluster, importantly revealing an unusual assembly and inactivation process in phytotoxin biosynthesis (Fig. 7). The newly discovered polyketide carboxylate 2, which is generated from the α-pyrone-polyketide glycoside 1via β-glucosidase-mediated hydrolysis of the ester bond, serves as a new phytotoxin that can significantly inhibit the root growth in Arabidopsis thaliana. Our work identifies the chemical substance basis of the long-term unsolved PKS16 cluster in phytotoxicity, shows that polyketide carboxylate is a new structural type of phytotoxins in Fusarium and importantly uncovers a rare ester bond hydrolysis function of β-glucosidase family enzymes.
Fig. 7. Formation of polyketide carboxylate 2 from polyketide glycoside 1 occurs through β-glucosidase mediated ester bond hydrolysis.
Data availability
Materials and methods, additional tables and figures, spectroscopic data and the sequence data of the pro gene cluster from Fusarium proliferatum CGMCC 3.4710 are available in the ESI.† The DNA sequence of the pro cluster has been deposited in the NCBI GenBank with the accession number PQ271635.
Author contributions
X. W. and D-K. K. performed the in vivo and in vitro experiments, as well as compound isolation. X. W. performed bioactive assays. X. W. and H-R. Z. performed compound characterization. All authors analyzed and discussed the results. Y. Z. supervised the research and wrote the manuscript.
Conflicts of interest
There are no conflicts to declare.
Supplementary Material
Acknowledgments
This work was supported by the National Key R&D Program of China (2020YFA0907700), Science and Technology Innovation Key R&D Program of Chongqing (CSTB2022TIAD-STX0015), Special Fund for Youth Team and 2035 Pilot Plan for Innovative Research of Southwest University (SWU-XJLJ202306 and SWU-XDPY22009), and Innovation Research Project for Postgraduate Students of Chongqing (CYB240125).
Electronic supplementary information (ESI) available: Materials and methods, additional tables and figures, and NMR data. See DOI: https://doi.org/10.1039/d4sc05256k
References
- Möbius N. Hertweck C. Fungal phytotoxins as mediators of virulence. Curr. Opin. Plant Biol. 2009;12:390–398. doi: 10.1016/j.pbi.2009.06.004. [DOI] [PubMed] [Google Scholar]
- Xu D. Xue M. Shen Z. Jia X. Hou X. Lai D. Zhou L. Phytotoxic secondary metabolites from fungi. Toxins. 2021;13:261. doi: 10.3390/toxins13040261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohn T. M., in Plant Relationships: Part A, ed. G. C. Carroll and P. Tudzynski, Springer Berlin Heidelberg, Berlin, Heidelberg, 1997, pp. 129–144 [Google Scholar]
- Pontes J. G. d. M. Fernandes L. S. dos Santos R. V. Tasic L. Fill T. P. Virulence factors in the phytopathogen–host interactions: an overview. J. Agric. Food Chem. 2020;68:7555–7570. doi: 10.1021/acs.jafc.0c02389. [DOI] [PubMed] [Google Scholar]
- Huang J.-S., in Plant Pathogenesis and Resistance: Biochemistry and Physiology of Plant-Microbe Interactions, ed. J.-S. Huang, Springer Netherlands, Dordrecht, 2001, pp. 291–411 [Google Scholar]
- Zeilinger S. Gupta V. K. Dahms T. E. S. Silva R. N. Singh H. B. Upadhyay R. S. Gomes E. V. Tsui C. K.-M. Nayak S C. Friends or foes? Emerging insights from fungal interactions with plants. FEMS Microbiol. Rev. 2016;40:182–207. doi: 10.1093/femsre/fuv045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. Hu J. Wei H. Solomon P. S. Vuong D. Lacey E. Stubbs K. A. Piggott A. M. Chooi Y.-H. Chemical ecogenomics-guided discovery of phytotoxic α-pyrones from the fungal wheat pathogen Parastagonospora nodorum. Org. Lett. 2018;20:6148–6152. doi: 10.1021/acs.orglett.8b02617. [DOI] [PubMed] [Google Scholar]
- Keller N. P. Fungal secondary metabolism: regulation, function and drug discovery. Nat. Rev. Microbiol. 2019;17:167–180. doi: 10.1038/s41579-018-0121-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimmino A. Masi M. Evidente M. Superchi S. Evidente A. Fungal phytotoxins with potential herbicidal activity: chemical and biological characterization. Nat. Prod. Rep. 2015;32:1629–1653. doi: 10.1039/C5NP00081E. [DOI] [PubMed] [Google Scholar]
- Duke S. O. Dayan F. E. Modes of action of microbially-produced phytotoxins. Toxins. 2011;3:1038–1064. doi: 10.3390/toxins3081038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean R. Van Kan J. A. L. Pretorius Z. A. Hammond-Kosack K. E. Di Pietro A. Spanu P. D. Rudd J. J. Dickman M. Kahmann R. Ellis J. Foster G. D. The top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 2012;13:414–430. doi: 10.1111/j.1364-3703.2011.00783.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P. Tedersoo L. Crowther T. W. Wang B. Shi Y. Kuang L. Li T. Wu M. Liu M. Luan L. Liu J. Li D. Li Y. Wang S. Saleem M. Dumbrell A. J. Li Z. Jiang J. Global diversity and biogeography of potential phytopathogenic fungi in a changing world. Nat. Commun. 2023;14:6482. doi: 10.1038/s41467-023-42142-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munkvold G. P. Proctor R. H. Moretti A. Mycotoxin production in Fusarium according to contemporary species concepts. Annu. Rev. Phytopathol. 2021;59:373–402. doi: 10.1146/annurev-phyto-020620-102825. [DOI] [PubMed] [Google Scholar]
- Brandwagt B. F. Mesbah L. A. Takken F. L. W. Laurent P. L. Kneppers T. J. A. Hille J. Nijkamp H. J. J. A longevity assurance gene homolog of tomato mediates resistance to Alternaria alternata f. sp. lycopersici toxins and fumonisin B1. Proc. Natl. Acad. Sci. U. S. A. 2000;97:4961–4966. doi: 10.1073/pnas.97.9.4961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. Ouyang B. Zhang W. Guang C. Xu W. Mu W. Deoxynivalenol: occurrence, toxicity, and degradation. Food Control. 2024;155:110027. doi: 10.1016/j.foodcont.2023.110027. [DOI] [Google Scholar]
- Hai Y. Chen M. Huang A. Tang Y. Biosynthesis of mycotoxin fusaric acid and application of a PLP-dependent enzyme for chemoenzymatic synthesis of substituted L-pipecolic acids. J. Am. Chem. Soc. 2020;142:19668–19677. doi: 10.1021/jacs.0c09352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J. Baek S. R. Lee K. R. Lee J. Yun S. Kang S. Lee Y. Purification and phytotoxicity of apicidins produced by the Fusarium semitectum KCTC16676. Plant Pathol. J. 2008;24:417–422. doi: 10.5423/PPJ.2008.24.4.417. [DOI] [Google Scholar]
- Gautier C. Pinson-Gadais L. Richard-Forget F. Fusarium mycotoxins enniatins: an updated review of their occurrence, the producing Fusarium species, and the abiotic determinants of their accumulation in crop harvests. J. Agric. Food Chem. 2020;68:4788–4798. doi: 10.1021/acs.jafc.0c00411. [DOI] [PubMed] [Google Scholar]
- Chen X.-W. Rao L. Chen J.-L. Zou Y. Unexpected assembly machinery for 4(3H)-quinazolinone scaffold synthesis. Nat. Commun. 2022;13:6522. doi: 10.1038/s41467-022-34340-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan G.-Y. Zhang J.-M. Xu Q.-D. Zhang H.-R. Hu C. Zou Y. Biosynthesis of cosmosporasides reveals the assembly line for fungal hybrid terpenoid saccharides. Angew. Chem., Int. Ed. 2023;62:e202308887. doi: 10.1002/anie.202308887. [DOI] [PubMed] [Google Scholar]
- Zhang H. Huang Y. Tang Y. Kong D. Zou Y. Genome mining of multi-substituted alkylresorcinols from a hybrid highly reducing- and type III- polyketide pathway. Chin. Chem. Lett. 2024;35:108968. doi: 10.1016/j.cclet.2023.108968. [DOI] [Google Scholar]
- Rao L. Yuan G.-Y. Chen X.-Y. Ran J.-L. Zou Y. Reshaping the diversity of oxidized polyquinane sesquiterpenoids by cytochrome P450s. Org. Lett. 2023;25:3276–3280. doi: 10.1021/acs.orglett.3c01024. [DOI] [PubMed] [Google Scholar]
- Zhang H. Zhao H. Huang Y. Zou Y. Genome mining reveals the biosynthesis of sativene and its oxidative conversion to seco-sativene. Org. Lett. 2024;26:338–343. doi: 10.1021/acs.orglett.3c04005. [DOI] [PubMed] [Google Scholar]
- Niehaus E.-M. Kim H.-K. Münsterkötter M. Janevska S. Arndt B. Kalinina S. A. Houterman P. M. Ahn I.-P. Alberti I. Tonti S. Kim D.-W. Sieber C. M. K. Humpf H.-U. Yun S.-H. Güldener U. Tudzynski B. Comparative genomics of geographically distant Fusarium fujikuroi isolates revealed two distinct pathotypes correlating with secondary metabolite profiles. PLoS Pathog. 2017;13:e1006670. doi: 10.1371/journal.ppat.1006670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiemann P. Sieber C. M. K. von Bargen K. W. Studt L. Niehaus E.-M. Espino J. J. Huß K. Michielse C. B. Albermann S. Wagner D. Bergner S. V. Connolly L. R. Fischer A. Reuter G. Kleigrewe K. Bald T. Wingfield B. D. Ophir R. Freeman S. Hippler M. Smith K. M. Brown D. W. Proctor R. H. Münsterkötter M. Freitag M. Humpf H.-U. Güldener U. Tudzynski B. Deciphering the cryptic genome: genome-wide analyses of the rice pathogen Fusarium fujikuroi reveal complex regulation of secondary metabolism and novel metabolites. PLoS Pathog. 2013;9:e1003475. doi: 10.1371/journal.ppat.1003475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiara M. Fanelli F. Mulè G. Logrieco A. F. Pesole G. Leslie J. F. Horner D. S. Toomajian C. Genome sequencing of multiple isolates highlights subtelomeric genomic diversity within Fusarium fujikuroi. Genome Biol. Evol. 2015;7:3062–3069. doi: 10.1093/gbe/evv198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niehaus E.-M. Münsterkötter M. Proctor R. H. Brown D. W. Sharon A. Idan Y. Oren-Young L. Sieber C. M. Novák O. Pěnčík A. Tarkowská D. Hromadová K. Freeman S. Maymon M. Elazar M. Youssef S. A. El-Shabrawy E. S. M. Shalaby A. B. A. Houterman P. Brock N. L. Burkhardt I. Tsavkelova E. A. Dickschat J. S. Galuszka P. Güldener U. Tudzynski B. Comparative “Omics” of the Fusarium fujikuroi species complex highlights differences in genetic potential and metabolite synthesis. Genome Biol. Evol. 2016;8:3574–3599. doi: 10.1093/gbe/evw259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arishi A. A. Shang Z. Lacey E. Crombie A. Vuong D. Li H. Bracegirdle J. Turner P. Lewis W. Flematti G. R. Piggott A. M. Chooi Y.-H. Discovery and heterologous biosynthesis of glycosylated polyketide luteodienoside A reveals unprecedented glucinol-mediated product offloading by a fungal carnitine O-acyltransferase domain. Chem. Sci. 2024;15:3349–3356. doi: 10.1039/D3SC05008D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhen X. Mao M.-J. Wang R.-Z. Chang S.-S. Xiao T.-M. Wu Y.-X. Yu L.-Y. Song Y.-L. Chen M.-H. Si S.-Y. Fusapyrone A, a γ-pyrone derived from a desert Fusarium sp. J. Asian Nat. Prod. Res. 2021;23:504–511. doi: 10.1080/10286020.2020.1794857. [DOI] [PubMed] [Google Scholar]
- Kil Y.-S. You J. Wendt K. L. King J. B. Cichewicz R. H. Resolving a natural product cold case: elucidation of fusapyrone structure and absolute configuration and demonstration of their fungal biofilm disrupting properties. J. Org. Chem. 2023;88:9167–9186. doi: 10.1021/acs.joc.3c00765. [DOI] [PubMed] [Google Scholar]
- Atanasoff-Kardjalieff A. K. Lünne F. Kalinina S. Strauss J. Humpf H.-U. Studt L. Biosynthesis of fusapyrone depends on the H3K9 methyltransferase, FmKmt1, in Fusarium mangiferae. Front. Fungal Biol. 2021;2:1–23. doi: 10.3389/ffunb.2021.671796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoye T. R. Jeffrey C. S. Shao F. Mosher ester analysis for the determination of absolute configuration of stereogenic (chiral) carbinol carbons. Nat. Protoc. 2007;2:2451–2458. doi: 10.1038/nprot.2007.354. [DOI] [PubMed] [Google Scholar]
- Mao X.-M. Zhan Z.-J. Grayson M. N. Tang M.-C. Xu W. Li Y.-Q. Yin W.-B. Lin H.-C. Chooi Y.-H. Houk K. N. Tang Y. Efficient biosynthesis of fungal polyketides containing the dioxabicyclo-octane ring system. J. Am. Chem. Soc. 2015;137:11904–11907. doi: 10.1021/jacs.5b07816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang Y.-M. Ahuja M. Oakley C. E. Entwistle R. Asokan A. Zutz C. Wang C. C. C. Oakley B. R. Development of genetic dereplication strains in Aspergillus nidulans results in the discovery of aspercryptin. Angew. Chem., Int. Ed. 2016;55:1662–1665. doi: 10.1002/anie.201507097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yee D. A. and Tang Y., in Engineering Natural Product Biosynthesis: Methods and Protocols, ed. E. Skellam, Springer US, New York, NY, 2022, pp. 41–52 [Google Scholar]
- Chiang Y.-M. Lin T.-S. Wang C. C. C. Total heterologous biosynthesis of fungal natural products in Aspergillus nidulans. J. Nat. Prod. 2022;85:2484–2518. doi: 10.1021/acs.jnatprod.2c00487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang C.-Y. Ohashi M. Tang Y. Deciphering chemical logic of fungal natural product biosynthesis through heterologous expression and genome mining. Nat. Prod. Rep. 2023;40:89–127. doi: 10.1039/D2NP00050D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nugroho A. E. Morita H. Circular dichroism calculation for natural products. J. Nat. Med. 2014;68:1–10. doi: 10.1007/s11418-013-0768-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koryakina I. McArthur J. B. Draelos M. M. Williams G. J. Promiscuity of a modular polyketide synthase towards natural and non-natural extender units. Org. Biomol. Chem. 2013;11:4449–4458. doi: 10.1039/C3OB40633D. [DOI] [PubMed] [Google Scholar]
- Ketudat Cairns J. R. Esen A. β-glucosidases. Cell. Mol. Life Sci. 2010;67:3389–3405. doi: 10.1007/s00018-010-0399-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed A. Nasim F. u.-H. Batool K. Bibi A. Microbial β-glucosidase: sources, production and applications. Appl. Environ. Microbiol. 2017;5:31–46. [Google Scholar]
- Godse R. Bawane H. Tripathi J. Kulkarni R. Unconventional β-Glucosidases: a promising biocatalyst for industrial biotechnology. Appl. Biochem. Biotechnol. 2021;193:2993–3016. doi: 10.1007/s12010-021-03568-y. [DOI] [PubMed] [Google Scholar]
- Erkanli M. E. El-Halabi K. Kim J. R. Exploring the diversity of β-glucosidase: classification, catalytic mechanism, molecular characteristics, kinetic models, and applications. Enzyme Microb. Technol. 2024;173:110363. doi: 10.1016/j.enzmictec.2023.110363. [DOI] [PubMed] [Google Scholar]
- Yang W. Su Y. Wang R. Zhang H. Jing H. Meng J. Zhang G. Huang L. Guo L. Wang J. Gao W. Microbial production and applications of β-glucosidase-a review. Int. J. Biol. Macromol. 2024;256:127915. doi: 10.1016/j.ijbiomac.2023.127915. [DOI] [PubMed] [Google Scholar]
- Fontana C. Widmalm G. Primary structure of glycans by NMR spectroscopy. Chem. Rev. 2023;123:1040–1102. doi: 10.1021/acs.chemrev.2c00580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S. Wei X. You C. The construction of an in vitro synthetic enzymatic biosystem that facilitates laminaribiose biosynthesis from maltodextrin and glucose. Biotechnol. J. 2019;14:1800493. doi: 10.1002/biot.201800493. [DOI] [PubMed] [Google Scholar]
- Kiso T. Kitahata S. Okamoto K. Miyoshi S. Nakano H. Hydrolysis of β-glucosyl ester linkage of p-hydroxybenzoyl β-d-glucose, a chemically synthesized glucoside, by β-glucosidases. J. Biosci. Bioeng. 2000;90:614–618. doi: 10.1016/S1389-1723(00)90005-7. [DOI] [PubMed] [Google Scholar]
- Hua Y. Sansenya S. Saetang C. Wakuta S. Ketudat Cairns J. R. Enzymatic and structural characterization of hydrolysis of gibberellin A4 glucosyl ester by a rice β-d-glucosidase. Arch. Biochem. Biophys. 2013;537:39–48. doi: 10.1016/j.abb.2013.06.005. [DOI] [PubMed] [Google Scholar]
- Oberg N. Zallot R. Gerlt J. A. EFI-EST, EFI-GNT, and EFI-CGFP: enzyme function initiative (EFI) web resource for genomic enzymology tools. J. Mol. Biol. 2023;435:168018. doi: 10.1016/j.jmb.2023.168018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y. Liu Q. Zang X. Yuan S. Bat-Erdene U. Nguyen C. Gan J. Zhou J. Jacobsen S. E. Tang Y. Resistance-gene-directed discovery of a natural-product herbicide with a new mode of action. Nature. 2018;559:415–418. doi: 10.1038/s41586-018-0319-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schildknecht H. Koob K. Myrmicacin, the first insect herbicide. Angew. Chem., Int. Ed. 1971;10:124–125. doi: 10.1002/anie.197101241. [DOI] [PubMed] [Google Scholar]
- Iwanami Y. Myrmicacin, a new inhibitor for mitotic progression after metaphase. Protoplasma. 1978;95:267–271. doi: 10.1007/BF01294455. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Materials and methods, additional tables and figures, spectroscopic data and the sequence data of the pro gene cluster from Fusarium proliferatum CGMCC 3.4710 are available in the ESI.† The DNA sequence of the pro cluster has been deposited in the NCBI GenBank with the accession number PQ271635.