Abstract
Colorectal cancer (CRC) has become a significant global health concern and ranks among the leading causes of morbidity and mortality worldwide. Due to its malignant nature, current immunotherapeutic treatments are used to tackle this issue. However, not all patients respond positively to treatment, thereby limiting clinical effectiveness and requiring the identification of novel therapeutic targets to optimise current strategies. The putative ligand of Siglec-15, Sialyl-Tn (STn), is associated with tumour progression and is synthesised by the sialyltransferases ST6GALNAC1 and ST6GALNAC2. However, the deregulation of both sialyltransferases within the literature remain limited, and the involvement of microRNAs (miRNAs) in STn production require further elucidation. Here, we identified miRNAs involved in the regulation of ST6GALNAC1 via a computational approach and further analysis of miRNA binding sites were determined. In silico tools predicted miR-21, miR-30e and miR-26b to regulate the ST6GALNAC1 gene, all of which had shown significant upregulated expression in the tumour cohort. Moreover, each miRNA displayed a high binding affinity towards the seed region of ST6GALNAC1. Additionally, enrichment analysis outlined pathways associated with several cancer hallmarks, including epithelial to mesenchymal transition (EMT) and MYC targets associated with tumour progression. Furthermore, our in silico findings demonstrated that the ST6GALNAC1 expression profile was significantly downregulated in CRC tumours, and its low expression correlated with poor survival outcomes when compared with patient survival data. In comparison to its counterpart, there were no significant differences in the expression of ST6GALNAC2 between normal and malignant tissues, which was further evidenced in our immunohistochemistry analysis. Immunohistochemistry staining highlighted significantly higher expression was more prevalent in normal human tissues with regard to ST6GALNAC1. In conclusion, the integrated in silico analysis highlighted that STn production is not reliant on deregulated sialyltransferase expression in CRC, and ST6GALNAC1 expression is regulated by several oncomirs. We proposed the involvement of other sialyltransferases in the production of the STn antigen and CRC progression via the Siglec-15/Sia axis.
Introduction
Colorectal cancer (CRC) is one of the most prevalent and lethal malignancies worldwide, with patient diagnoses increasing each consecutive year [1]. Due to its malignant nature, CRC treatment is heavily reliant on early screening and detection to improve patient survival [2]. Current immune checkpoint blockade therapies have shown positive patient responses against programmed cell death protein-1 (PD-1), with blocking antibodies such as pembrolizumab and nivolumab being food and drug administration (FDA) approved for treatment in metastatic CRC patients with significant microsatellite instability (MSI) mutations [3]. However, not all patients respond positively to immunotherapeutic treatments, limiting clinical effectiveness and resulting in a poor prognosis. There is an urgent need to further identify therapeutic targets to optimise current treatment strategies for CRC.
The sialic acid-binding immunoglobulin-type lectin (Siglec)/sialic acid (Sia) axis is an immunoregulatory pathway that establishes immune tolerance to self-cells. More recently, this has been implicated in several cancer types, with Siglecs being focused upon as emerging therapeutic cancer targets [4]. Siglec-15 is one such promising immune checkpoint protein that exhibits distinct expression on cancer subpopulations compared to other immune checkpoints, such as programmed death-ligand 1 (PD-L1) [5]. The interaction of sialoglycan ligands upon Siglec-15 engagement prevents effective activation of T-lymphocytes [6]. The implications of Siglec-15 binding to sialoglycan ligands are those of inducing hypersialylation and promoting immunosuppression, consequently resulting in the immune evasion of tumour cells [7]. Several treatment strategies are being employed in the disruption of the Siglec-15/Sia axis. Current therapeutic strategies involve blocking antibodies (NC318) that are in clinical trials [8]. Similarly, another study highlighted the development of a monoclonal antibody (1-15D1) that had a high binding affinity to the Siglec-15 protein and was capable of stimulating T-cell response in vitro [9]. Moreover, other approaches for Siglec-15 disruption have utilised protein aptamers to assist in checkpoint blockade and have high affinity against the Siglec-15 protein [10]. Further to this, our previous study also highlighted the development of a small molecule inhibitor binding to the V-set binding domain to prevent sialoglycan binding [11]. Interestingly, upregulation of these Sia glycans has also been correlated with cancer-specific glycosylation and serves as a unique driver in cancer onset and progression [12]. More specifically, Siglec-15/Sia binding is dependent on Sialyl-Tn (STn), likely synthesised by glycosyltransferases such as ST6GALNAC1, which have shown aberrant expression in cancer [13]. ST6GALNAC1’s involvement in STn antigen presentation is frequently upregulated in multiple cancer types and has shown poor prognosis in CRC and prostate cancer (PCa) [14,15]. Similarly, ST6GALNAC2 participates in STn antigen generation to a lesser extent and demonstrates high expression in CRC, and exhibits advanced cancer progression in follicular thyroid carcinomas (FTC) [16,17]. Targeting sialyltransferases such as ST6GALNAC1 and ST6GALNAC2 may provide anti-tumour immunity and a greater response in current immune checkpoint blockade therapies. However, evidence regarding miRNAs involved in their regulation and STn production in the literature remain limited.
MiRNAs are small single-stranded RNA molecules capable of modulating gene expression via 3’ untranslated region (UTR) binding post-transcriptionally [18]. Furthermore, the interplay of miRNAs enhances cancer hallmarks and its regulation, and correlates with targeting key genes involved in promoting angiogenesis, cancer proliferation, and metastasis [19,20]. Similarly, the Siglec/Sia axis can also be modulated via miRNA expression. The upregulation of miR-135b and miR-182 directly targets ST6GALNAC2 via the phosphoinositide 3-kinase/protein kinase B (PI3K)/AKT signalling pathway, enhancing chemoresistance in CRC [21]. The crosstalk between miRNA expression patterns and the Siglec-15/Sia axis could underline certain dysregulated mechanisms.
In the current study, a multiomics approach was adopted to investigate the roles of ST6GALNAC1 and ST6GALNAC2 in mediating the Siglec/Sia axis and its clinical relevance to CRC tumorigenesis at the gene and protein levels. We further highlighted enriched pathways associated with cancer hallmarks and identified possible signalling pathways related to tumour onset. Moreover, we addressed the impact of each corresponding gene on the infiltration of myeloid cells and their association with prominent immune checkpoints and immune function. This may provide insights into highlighting their roles in cancer progression.
Methodology
Search Tool for the Retrieval of Interacting Genes/Proteins (STRING)
The STRING database identifies known and predicted protein-protein interactions (PPI) and displays direct associations via computational data mining. The STRING 11.0 (https://version-11-0b.string-db.org/cgi/input?sessionId=brxKfZgAG9Au&input_page_show_search=on; accessed 2 August 2023) software highlighted PPI relationships in ST6GALNAC1 and ST6GALNAC2. The development of the full PPI network included both functional and physical protein associations with ≤ 10 protein interactors. Additionally, each individual protein queried and ≤ 20 protein interactors were included in the second expansion of the framework with a high confidence interval, with ≥ 0.700 considered significant.
GeneMANIA
The GeneMANIA database (https://genemania.org/; accessed 26 July 2023) predicts functional information based on corresponding genes and gene datasets. The gene framework highlights functional communication between genes via criterion comprising of physical interactions, co-expression, predicted datamining, co-localisation, genetic interactions, and common pathway interactions with respect to ST6GALNAC1 and ST6GALNAC2. Correlating genes with a high degree of association were also represented [22].
UALCAN
The UALCAN database (http://ualcan.path.uab.edu; accessed 26 July 2023) is used as a tool for cancer transcriptomics. Utilising TCGA genomic data for analysis, the mRNA expression of ST6GALNAC1 (ENSG00000070526) and ST6GALNAC2 (ENSG00000070731) were compared for normal and colon adenocarcinoma (COAD) cohorts. Similarly, gene expression of identified protein targets obtained from the STRING analysis were determined and compared between normal and COAD subgroups. Additionally, predicted miRNA candidates involved in gene regulation were also compared for normal and COAD cohorts. Statistical significance of gene expression data (transcript per million; TPM) generated by the UALCAN database for box plot construction was assessed via a Welch’s t-test PERL script encoded to identify significant differences between cohorts based on clinicopathological features [23]. P < 0.05 was considered statistically significant.
In silico miRNA datamining
In silico analysis tools were utilised to determine common miRNA targets predicted to modulate ST6GALNAC1 expression and possible clinical relevance to CRC across several databases. Venn diagrams (Venny 2.1.0) (https://bioinfogp.cnb.csic.es/tools/venny/; accessed 28 July 2023) were constructed using three separate miRNA prediction databases including TargetScan (https://www.targetscan.org/vert_80/; accessed 28 July 2023), MiRSystem (http://mirsystem.cgm.ntu.edu.tw/index.php; accessed 28 July 2023), and MiRWalk (http://mirwalk.umm.uni-heidelberg.de/; accessed 28 July 2023), and were further cross-referenced. Furthermore, common miRNA candidates were also determined for the identified in silico targets that were shown to interact with ST6GALNAC1 and ST6GALNAC2. The most frequent miRNA hits associated with ST6GALNAC1 were further explored for predicted binding sites via statistical folding of nucleic acids and studies of regulatory RNAs (Sfold) software (https://sfold.wadsworth.org/cgi-bin/starmir.pl; accessed 13 September 2023).
Gene Set Enrichment Analysis (GSEA)
GSEA of ST6GALNAC1 and ST6GALNAC2 were obtained via the TCGA dataset (TCGA, PanCancer Atlas) from cBioPortal for Cancer Genomics (http://cbioportal.org/ accessed on 19 September 2023). CBioPortal is offered as an open access repository for interactive omics patient data. The dataset for COAD tumours was selected, comprising of a total of 524 patient samples. Each of the corresponding genes were submitted as the queried gene and modified to include the differential mRNA expression relative to the normal cohort with a z-score threshold of ± 2.0. Following this, the mRNA comparative data between the altered (differentially expressed group) and unaltered group (unchanged expression group) was separated. Following this, the altered group containing only the significant differentially expressed genes between the normal and COAD subgroups were selected, which also included the query genes ST6GALNAC1 and ST6GALNAC2, respectively. The number of differentially expressed genes for each dataset were recorded as 6667 genes for the ST6GALNAC1 dataset and 4331 genes for the ST6GALNAC2 dataset and exported as TSV files. The datasetswere uploaded into the GSEA v4.3.2 software as rnk files (https://www.gsea-msigdb.org/ accessed on 19 January 2023) and enriched pathways and cancer hallmarks were identified (FDR < 0.25 and p < 0.05).
TISIDB
The TISIDB database (http://cis.hku.hk/TISIDB/index.php; accessed on 26 July 2023) provides an integrated repository for identifying interactions between tumours and the immune system. The TISIDB database collates information via several datasets including TCGA transcriptomics and clinical data pertaining to multiple cancer types. The Spearman’s rank correlation between the abundance of macrophages, activated CD4+ T-cells, activated CD8+ T-cells, regulatory T-cells (Tregs), and monocytes were assessed in conjunction with the expression of both sialyltransferases, ST6GALNAC1 and ST6GALNAC2, in COAD tumours [24].
Tumour IMmune Estimation Resource (TIMER) analysis
The TIMER database (https://cistrome.shinyapps.io/timer/, accessed on 1 August 2023) serves as an extensive platform to systematically analyse immune infiltrates over several cancer types. For this study, specific parameters relating to the correlation of immune checkpoint protein genes, including SIGLEC15, PDCD1, CD274, cytotoxic T-lymphocyte associated protein 4 (CTLA4), T-cell immunoreceptor with Ig and ITIM domains (TIGIT), and Lymphocyte-activation gene 3 (LAG3), were all compared with the abundance of ST6GALNAC1 and ST6GALNAC2. Spearman’s rank correlation coefficient and p < 0.05 for log2 TPM was used to determine the statistical significance of COAD tumours [25].
Kaplan-Meier Plotter (KM PLOTTER)
The Kaplan-Meier plotter (https://kmplot.com/analysis/, accessed on 26 July 2023) database tool was used to identify the prognostic association between the genomic expression data for ST6GALNAC1 and ST6GALNAC2, along with corresponding patient survival outcomes in COAD tumours. The prognostic expression of each highlighted sialyltransferase in CRC patients was analysed over a period of time based on overall survival criterion (OS), relapse-free survival (RFS), post-progression survival (PPS), hazard ratio (HR), 95% confidence interval (CI), and Log-rank P-value [26].
Immunohistochemistry (IHC)
Colorectal cancer tissue array (BC05023a) obtained from Biomax (TissueArray.Com LLC, Maryland, USA), containing 54 cores (18 COAD, 18 cancer adjacent colon tissue (AT), and 18 adjacent normal colon tissue (NAT), was subjected to histological staining. Deparaffinisation was achieved by treating the sections with histoclean/ethanol followed by antigen retrieval via sodium citrate treatment, and subsequently washed in 0.025% triton-x/PBS and incubated in 3% hydrogen peroxide:PBS for a period of 15 min. The slides were washed an additional three times with Triton-X100/PBS for 5 min. Blocking of non-specific binding sites was performed using 5% BSA/PBS, and the sections were then incubated in a humidity chamber for 1h at room temperature (RT) whilst wrapped in parafilm. The slides were incubated overnight at 4°C with the primary ST6GALNAC1 antibody (1:50) (Proteintech Group, Inc., Manchester, UK) and ST6GALNAC2 antibody (1:100) (Life Technologies Limited, Renfrewshire, UK) respectively. The slides were washed with 0.025% Triton X-100/PBS, followed by incubation with a biotin-labelled anti-rabbit secondary antibody. Subsequently, the sections were washed using a 0.025% Triton X-100/PBS wash step and 1h incubation with a streptavidin-HRP conjugate. Visualisation of both ST6GALNAC1 and ST6GALNAC2 antibody staining was achieved via addition of DAB solution (Zytomed Systems GmBH, Berlin, Germany) and haematoxylin as the nuclei stain for a period of 10 min at RT. Quantification of the staining was conducted via an AxioCam Hrc (Zeiss Microscopy, Oberkochen, Germany) microscope at x4 and x10 magnifications. Both normal and malignant tissues were compared via the unpaired student’s t-test.
Results
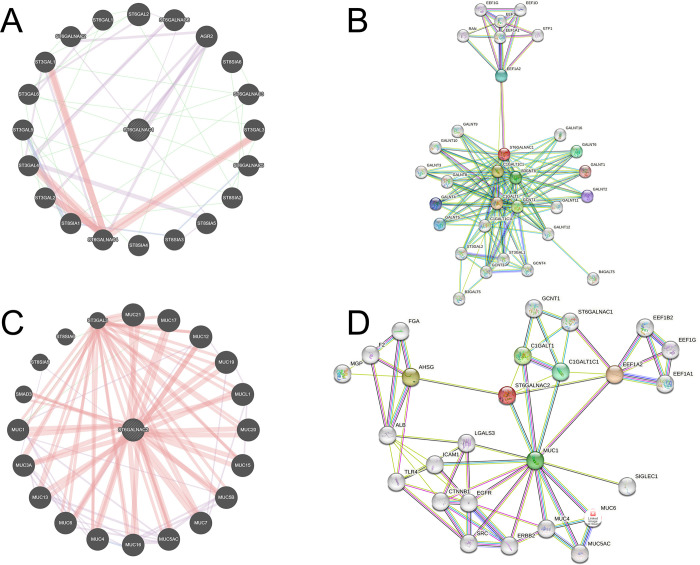
ST6GALNAC1 and ST6GALNAC2 gene and protein-protein molecular networks
The STRING and GeneMANIA databases highlight predicted genes and PPI between interacting queried proteins/genes. The ST6GALNAC1 gene framework, produced by GeneMANIA, shows co-expression (purple) with anterior gradient 2 (AGR2) (Fig 1A) (Fig 1A). The ST6GALNAC1 PPI network (Fig 1B) has shown multiple hits that correspond with N-acetylgalactosaminyltransferase (GALNT) enzymes and other sialyltransferases. Likewise, the average local clustering coefficient (ALCC) (0.83) shows a strong correlation with multiple protein targets, and the PPI enrichment score upon normalization shows significant node interactions at the highest confidence interval (< 1.0 x 10–16) (S1 Table). Predicted gene communication involving ST6GALNAC2 has shown significant physical interactions with the MUC gene family (Fig 1C). The ST6GALNAC2 PPI framework (Fig 1D) has also demonstrated a comprehensive predicted interaction clustering of several protein targets with 26 nodes. The ALCC score (0.708) and PPI enrichment (1.09 x 10−5) (S1 Table) show strong correlation between several protein targets and with primary interactions associated with α-2-HS-glycoprotein (AHSG). Furthermore, both sialyltransferases show interactions with a cluster of EEF1 enzymes, particularly with EEF1A2, and communication with Core-1 synthase-glycoprotein-N-acetylgalactosamine 3-b-galactosyltransferase-1 (C1GALT1) and C1GALT1 Specific Chaperone 1 (C1GALT1C1) (Fig 1B and 1D).
Fig 1. ST6GALNAC1 and ST6GALNAC2 share significant gene and protein-protein interactions and may highlight key novel targets.
(A) ST6GALNAC1 gene framework predicts functional information based on corresponding genes and gene datasets. (B) ST6GALNAC1 STRING framework highlighting possible PPI from interacting proteins and direct associations via computational datamining. The framework is developed with a confidence interval of ≥ 0.700. (C) ST6GALNAC2 gene framework predicts functional information based on corresponding genes and gene datasets. (D) ST6GALNAC2 STRING framework highlighting possible PPI from interacting proteins and direct associations via computational datamining. The framework is developed with a confidence interval of ≥ 0.700.
Candidate miRNAs predicted in gene regulation and possible binding sites
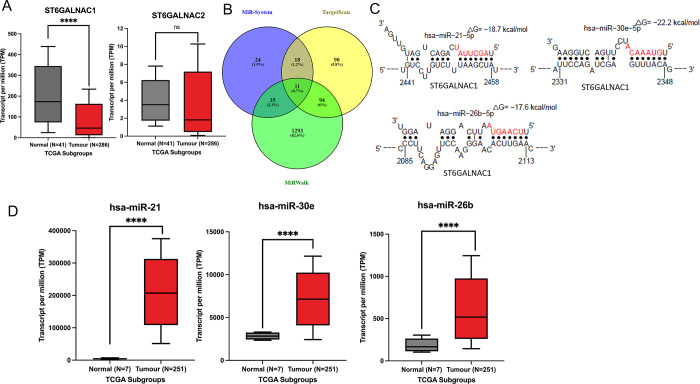
UALCAN TCGA datamining highlighted the sialyltransferase expression profile of ST6GALNAC1 and ST6ALNAC2 in CRC (Fig 2A). ST6GALNAC1 shows significant gene downregulation in the tumour cohort of 46.056 TPM in comparison to the normal subgroup with a mean expression of 173.703 TPM (p < 0.0001). Additionally, UALCAN expression analysis shows no significant change in the expression of ST6GALNAC2 between normal (3.509 TPM) and tumour (1.813 TPM) cohorts. Similarly, the gene expression of the predicted targets AGR2 and AHSG were also determined between normal and CRC tumour groups. UALCAN expression identified significant expression of AHSG in the tumour subgroup (0.032 TPM) in comparison to the normal tissue subgroup (0.00 TPM) (S1B Fig). In contrast, AGR2 had shown no significant difference in the expression of both subgroups. Furthermore, only ST6GALNAC2 displays enhanced promoter methylation in the tumour cohort (S2B Fig). Moreover, the Pearson correlation between both ST6GALNAC1 and ST6GALNAC2 expression in CRC shows a low association between the two genes, but the relationship has been shown to be significant (S3 Fig).
Fig 2. In silico analysis demonstrates that sialyltransferase expression is downregulated in COAD tumours.
(A) TCGA genomic data demonstrating the mRNA expression of ST6GALNAC1 for both normal and colon adenocarcinoma (COAD) cohorts. Welch’s t-test PERL script. **** P < 0.0001 (B) In silico analysis tools were utilised to construct Venn diagrams identifying common miRNA targets predicted to modulate ST6GALNAC1 expression. (C) The most frequent miRNA hits associated with ST6GALNAC1 were further explored for predicted binding sites via the Sfold software. (D) TCGA genomic data demonstrating the mRNA expression of candidate miRNAs with the highest binding affinities for normal and colon adenocarcinoma (COAD) cohorts. Welch’s t-test PERL script; **** P < 0.0001.
To identify the miRNAs involved in regulating the gene expression of ST6GALNAC1, several databases were compared to identify the most frequent hits (Fig 2B). Of the multiple miRNAs cross-referenced, a total of 11 were predicted to be involved in the regulation of ST6GALNAC1 based on targetscan, miRsystem and miRwalk databases (Fig 2B) (S2 Table). We have found that none of the 11 commonly deregulated miRNAs (Fig 2B) were regulating ST6GALNAC1 as their expression levels seem to be downregulated as well. Hence, we focused on the miRNAs found common between Targetscan and miRsystem, and we found that miR-21-5p, miR-30e-5p and miR-26b-5p as potential regulators of ST6GALNAC1 promoting CRC progression when overexpressed (Fig 2C and 2D). All three miRNAs were also shown to regulate ST6GALNAC1 by miRPathDB database as well (data not shown). Moreover, common miRNAs from the TargetScan database were identified between regulating AHSG and AGR2 in association with ST6GALNAC1 and ST6GALNAC2 (S4A Fig). Although no singular miRNA was shown to regulate all four genes, common miRNAs across other predicted protein targets were also determined. The common miRNA element that correlated between the targets GALNT3, GALNT8, B3GNT6 and ST6GALNAC1 identified hsa-miR-30a-5p to be involved in gene regulation (S4B Fig). Similarly, predicted targets C1GALT1, C1GALT1C1, AHSG and ST6GALNAC2 were compared for any common miRNA elements, although none were identified (S4C Fig). Further to this, predicted miRNAs involved in ST6GALNAC1 expression were further investigated, and the binding affinities to key binding sites were identified (Fig 2C). MiRNA activity for gene regulation typically occurs at the 3’ end of the UTR. MiR-21-5p exhibited a high binding affinity (-18.700 kcal/mol) towards the seed region of the target ST6GALNAC1 mRNA strand at position 2441–2458. Similarly, the predicted binding of miR-30e-5p and miR-26b-5p also displayed high binding affinities to the target mRNA strand at -22.200 kcal/mol and -17.600 kcal/mol, respectively. MiR-30e-5p had a predicted binding site at position 2331–2348. In contrast, miR-26b-5p had demonstrated a predicted binding site at position 2085–2113, all of which indicated a strong association between nucleotide bases.
To understand the role of these identified candidate miRNAs and their relevance in relation to CRC (Fig 2D), UALCAN data demonstrated significantly upregulated expression of all three miRNAs in the COAD tumour subgroup. MiR-21 displayed a mean expression of 207,192.191 TPM in comparison to normal colon tissues. This was also similarly observed by the mean expression of miR-30e with 7142.537 TPM and a mean expression of 518.048 TPM for miR-26b in comparison to the normal colon tissue cohort.
GSEA analysis of sialyltransferases and cancer hallmarks
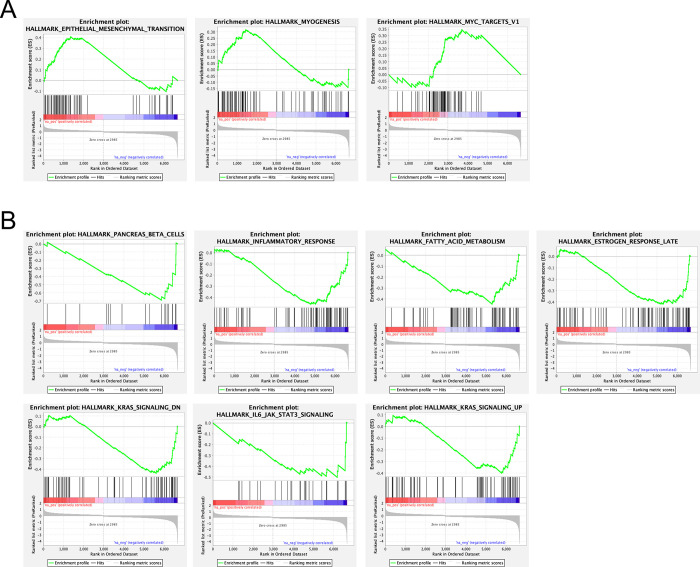
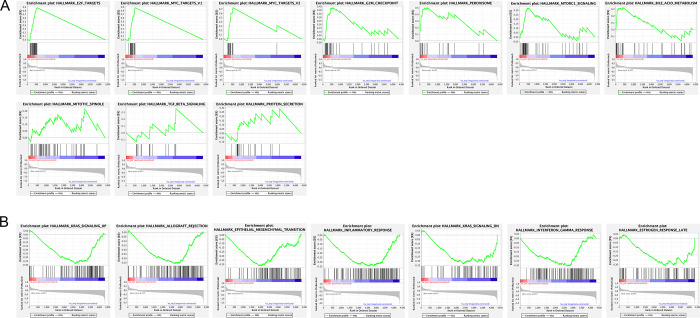
To identify various enriched pathways relating to ST6GALNAC1 and ST6GALNAC2 and cancer hallmarks, the GSEA of the TCGA, PanCancer Atlas dataset with a specific focus on significantly differentially expressed (DE) genes relating to CRC were highlighted with respect to ST6GALNAC1 (Fig 3) and ST6ALNAC2 (Fig 4). Out of a total of 47 gene sets displayed for ST6GALNAC1 and the CRC phenotype, three gene sets were identified as significantly upregulated, whilst seven gene sets were identified as being significantly downregulated with respect to CRC (FDR < 0.25 and p < 0.05) (Fig 3A and 3B, S3 Table). Significant enrichment of EMT, MYC targets, and hallmarks related to myogenesis were identified as being significantly enriched. Additionally, enriched gene sets that had shown downregulation including the inflammatory response, IL-6 mediated JAK/STAT3 signalling, and deregulated KRAS signalling, among others. Similarly, ST6GALNAC2 highlighted 10 significantly enriched gene sets as being upregulated (FDR < 0.25 and p < 0.05) (Fig 4A), and seven gene sets that were significantly enriched as being downregulated out of a total of 42 gene sets (S4 Table). Upregulated gene sets included the enrichment of E2F targets, MYC targets, the G2M checkpoint, mTOR signalling, and other pathways (Fig 4A). Downregulated gene sets that were also enriched included KRAS signalling, allograft rejection, EMT, inflammatory response, and IFNγ response, among other pathways (Fig 4B).
Fig 3. GSEA of ST6GALNAC1 identified significant enrichment between DEG’s and the CRC phenotype.
GSEA of ST6GALNAC1 was obtained via the TCGA dataset (TCGA, PanCancer Atlas) from cBioPortal outlining DEG’s and a total of 47 gene sets associated with cancer hallmarks. (A) The enrichment of three gene sets was identified as being upregulated. FDR < 0.25 and p < 0.05. (B) Enrichment of seven gene sets was identified as being downregulated. FDR < 0.25 and p < 0.05.
Fig 4. GSEA of ST6GALNAC2 identified significant enrichment between DEG’s and the CRC phenotype.
GSEA of ST6GALNAC2 was obtained via the TCGA dataset (TCGA, PanCancer Atlas) from cBioPortal outlining DEG’s and a total of 42 gene sets associated with cancer hallmarks. (A) ST6GALNAC2 highlighted 10 significantly enriched gene sets. FDR < 0.25 and p < 0.05. (B) Significant enrichment of seven gene sets was identified as being significantly downregulated. FDR < 0.25 and p < 0.05.
Sialyltransferase expression correlates to the abundance of myeloid cells
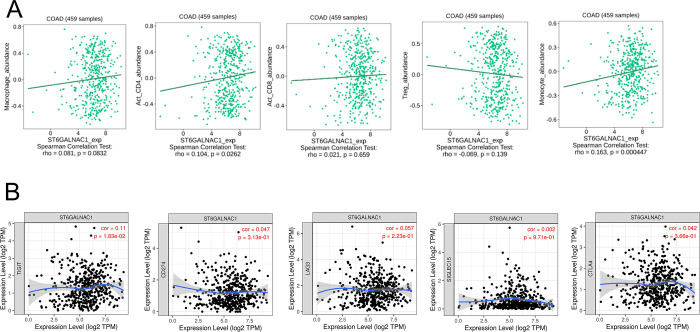
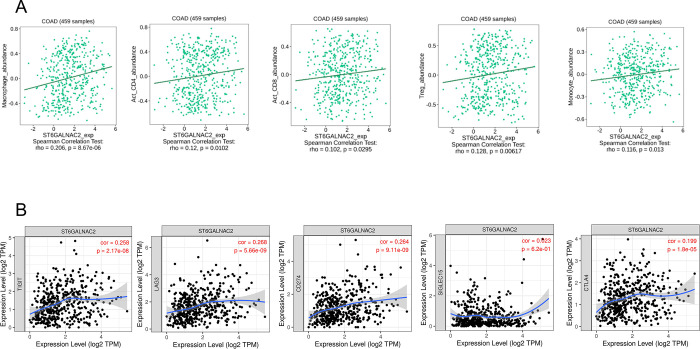
To determine the role of both sialyltransferases and immune infiltration in situ, the abundance of immune infiltrating cell populations correlating with ST6GALNAC1 and ST6GALNAC2 expression in colon adenocarcinomas (COAD) was determined (Figs 5A and 6A). Spearman’s rank correlation coefficient (SRCC) significantly associated the abundance of activated CD4+ T-lymphocytes and monocyte populations with ST6GALNAC1 expression (Fig 5A). In comparison, ST6GALNAC2 expression correlated with the abundance of several infiltrating immune cell populations including activated CD4+ T-lymphocytes, activated CD8+ T-lymphocytes, Tregs, monocytes, and macrophages, with the most significant SRCC correlation with the latter (0.206) (Fig 6A). Additionally, the association between the abundance of both ST6GALNAC1 and ST6GALNAC2 and pro-tumorigenic immune checkpoints were also determined via TIMER analyses (Figs 5B and 6B). ST6GALNAC1 expression showed no significant correlation with other pro-tumorigenic immune checkpoints (Fig 5B). However, ST6GALNAC2 expression displayed an association with most pro-tumorigenic immune checkpoints, excluding Siglec-15 (Fig 6B).
Fig 5. ST6GALNAC1 correlates with CD4+ T-lymphocytes and monocyte populations but shows no significant correlation with other pro-tumorigenic immune checkpoints.
(A) SRCC between the abundance of macrophages, activated CD4+ T-cells, activated CD8+ T-cells, regulatory T-cells (Tregs), and monocytes were assessed in conjunction with the expression of ST6GALNAC1 in COAD tumours. P < 0.05 was considered statistically significant. (B) The association between the abundance of ST6GALNAC1 and pro-tumorigenic immune checkpoints was determined via TIMER analysis.
Fig 6. ST6GALNAC2 correlated with the abundance of several infiltrating immune cell populations and displayed an association with most pro-tumorigenic immune checkpoints, excluding Siglec-15.
(A) SRCC between the abundance of macrophages, activated CD4+ T-cells, activated CD8+ T-cells, regulatory T-cells (Tregs), and monocytes were assessed in conjunction with the expression of ST6GALNAC2 in COAD tumours. P < 0.05 was considered statistically significant. (B) The association between the abundance of ST6GALNAC2 and pro-tumorigenic immune checkpoints was determined via TIMER analysis.
Dysregulated sialyltransferase expression correlated with poor clinical outcomes
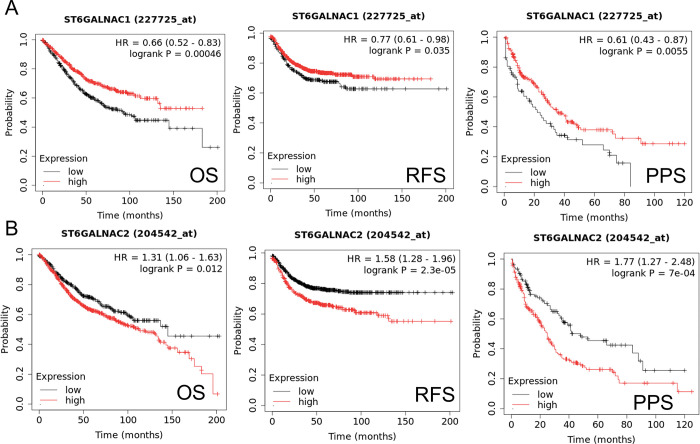
To correlate the expression of ST6GALNAC1 and ST6GALNAC2 to patient survival data, survival curves outlining overall survival (OS), post-progression survival (PPS), and relapse-free survival (RFS) were determined (Fig 7). ST6GALNAC1 shows low expression that is significantly correlated to poor prognosis based on OS criteria (p < 0.05), the RFS criterion (p < 0.05), and PPS (p < 0.05) survival in CRC patients (Fig 7A). Moreover, high ST6GALNAC2 expression was significantly associated with poor prognosis in CRC patients under all survival criteria (p < 0.05) (Fig 7B).
Fig 7. ST6GALNAC1 shows low expression to be significantly correlated to poor prognosis based on survival criteria.
High ST6GALNAC2 expression was significantly associated with poor prognosis under all survival criteria. Only miR-940 OS criteria highlighted that low expression was associated with poor prognosis. (A) Survival curves outlining ST6GALNAC1 expression with regard to patient survival data outlining overall survival (OS), post-progression survival (PPS), and relapse-free survival (RFS) criteria. P < 0.05 was considered statistically significant. (B) Survival curves outlining ST6GALNAC2 expression with regard to patient survival data outlining overall survival (OS), post-progression survival (PPS), and relapse-free survival (RFS) criteria. P < 0.05 was considered statistically significant.
ST6GALNAC1 protein expression is more prevalent in normal colon tissues
IHC analysis was performed to investigate the presence of ST6GALNAC1 and ST6GALNAC2 proteins in CRC tissues (Fig 8). Minimal staining for ST6GALNAC1 and ST6GALNAC2 was observed in the malignant tumour tissue cores, with representative images of ST6GALNAC1 and ST6GALNAC2 staining in the tissue sections shown (Fig 8A and 8B). Quantitative analysis of the staining revealed a significant decrease in the expression of ST6GALNAC1 between normal and malignant tumour cores, with non-significant staining found in both subgroups for ST6GALNAC2 staining.
Fig 8. Low staining of ST6GALNAC1 and ST6GALNAC2 was observed in malignant tumour tissues.
(A) IHC analysis was performed to investigate the presence of ST6GALNAC1 in CRC tissues (BC05023a). Representative images of a total of 54 core tissue samples representing normal and malignant tissue cores were stained and visualised. (B) IHC analysis was performed to investigate the presence of ST6GALNAC2 in CRC tissues (BC05023a). Representative images of a total of 54 core tissue samples, representing both normal and malignant tissue cores, were stained and visualised. (C) Quantitative analysis of the IHC staining for both ST6GALNAC1 and ST6GALNAC2 was performed for normal (clear) and malignant (red) tumour tissues. Unpaired student’s t-test. *P < 0.05.
Discussion
The current study has highlighted the multifaceted roles of both ST6GALNAC1 and ST6GALNAC2 and their collective association with the Siglec-15/Sia axis and its clinical relevance to CRC tumorigenesis. However, the regulatory mechanisms underlying their expression profiles remain unclear. Hence, we have identified possible regulatory mechanisms and miRNA candidates that seem to be involved in their regulation and, thereby, have some possible involvement in STn production.
The GeneMANIA database predicted ST6GALNAC1 co-expression with AGR2. AGR2 is localised to the endoplasmic reticulum (ER) and plays a crucial role in maintaining ER homeostasis via the formation, breakage, and isomerisation of the disulphide bonds involved in nascent protein maturation [27]. AGR2 has also been found to be a prominent pro-tumorigenic gene that has consistently been associated with tumour onset and progression in a number of cancer types, including CRC [28,29]. Although AGR2 is predominantly localised in the ER, there is also evidence suggesting the extracellular secretion of AGR2 [30], where ER-resident AGR2 displays disparate O-glycosylation patterns compared to secreted AGR2 [31]. Similarly, ST6GALNAC2 was shown to be co-expressed with and function alongside AHSG, an oncogene that is commonly associated with metabolic processes that have shown abnormal expression in multiple cancer types [32]. Furthermore, PPI interactions have highlighted that both sialyltransferases interact with a cluster of EEF1 proteins, particularly with EEF1A2. EEF1A2 serves an important role in modulating protein translation elongation and can play key roles in several biological processes [33]. To note, little is mentioned in the literature regarding the interactions between sialyltransferases and EEF1A2. However, in vitro studies have demonstrated that EEF1A2 is capable of regulating several signalling pathways, including PI3K/AKT and mTOR, via p53 inactivation [34]. Interaction with EEF1A2 could modulate the activity of ST6GALNAC1/ST6GALNAC2 and sialoglycan synthesis differently in normal and malignant tumour tissues.
Several previous studies have highlighted the aberrant expression of both sialyltransferase enzymes in multiple cancer types and the overexpression of the STn antigen [35,36]. However, TCGA genomic analysis highlighted significantly downregulated expression in the COAD subgroups for ST6GALNAC1. A previous study highlighted that increased ST6GALNAC1 expression was only observed in the presence of M2 tumorigenic macrophages and CRC cells in vitro, which also stimulated the production of the STn antigen [37]. Moreover, another study outlined deficient mismatch repair (dMMR) CRC molecular subtype tumours displayed significantly downregulated expression of GALNT6, a prominent glycosyltransferase in glycan synthesis [38]. Consequently, this enhanced the pro-tumorigenic characteristics of SW480 cells in vitro and increased the expression of the Tn antigen, the precursor of the STn antigen. Additionally, GALNT6 was observed to interact with ST6GALNAC1 via our STRING analysis. This could suggest increased expression of the Tn antigen precursor molecule, and overexpression of STn may be independent of deregulated ST6GALNAC1 expression in CRC and may require intercellular signalling to facilitate STn production in the TME. Further to this point, interactions with TAMs may be necessary for enhanced expression patterns that drive tumour heterogeneity within the TME and CRC progression. Further evidence also highlighted the downregulated expression of ST6GALNAC2 in CRC cells [39]. This was also similarly observed in CRC tumour samples [40]. Possible epigenetic mechanisms and regulatory expression through miRNA activity may indicate dysregulated sialyltransferase activity. In addition, both Tn and STn antigens are more prevalent in CRC tumours, correlating with poor prognosis and reduced clinical survival outcomes in patients [40,41].
UALCAN datamining also indicated enhanced methylation of the promoter region of ST6GALNAC2 in primary COAD tumours, suggesting a possible explanation for silencing gene expression. Additionally, enhanced CpG island methylation in the promoter regions of tumour-specific genes has also been associated with CRC tumorigenesis and further presents as one of the molecular subtypes of CRC [42]. Enhanced methylation and subsequent silencing of ST6GALNAC2 expression could prevent its expression. Similarly, epigenetic modifications of histone proteins, including deacetylation may also repress ST6GALNAC2 transcription [43].
Simultaneously, predicted miRNA candidates involved in sialyltransferase regulation have exhibited high binding affinities for the seed regions on the 3’ UTR of the mRNA target strand, suggesting a high likelihood of gene silencing [44]. Further to this point, full complementarity binding of the miRNA candidate to the target mRNA strand will lead to directed target degradation. In contrast, partial complementary binding can exhibit translational repression [45]. The role of pro-tumorigenic miR-21 has been characterised in the development and progression of multiple tumour malignancies, including CRC, which has also been highlighted as a potential tumour biomarker [46]. Furthermore, miR-21 was shown to induce advanced stage MSI type CRC tumours in conjunction with miR-335 activity, leading to poor prognosis in CRC patients [47]. In addition to this, miR-21 may play a role in tumour associated signalling pathways, including PI3K/AKT and TGF-β signalling [48,49]. This could suggest likely signalling pathways that stimulate CRC progression. Similarly, miR-30e has also been suggested as a potential biomarker for CRC development [50]. Moreover, miR-30e exhibited overexpression in CRC in vitro via stimulating the CXCL12 axis [51]. MiR-30e has also been demonstrated to be consistently deregulated in chemoresistant CRC patients [52]. The overexpression of miR-26b has also been observed in CRC and was identified to correlate with the expression of MMP-9 [53]. However, the role of miR-26b as an oncomir in CRC remains limited and requires further elucidation. Additionally, several miRNAs may facilitate the progression of the TME. Extracellular vesicles containing miR-21 stimulated tumour immune evasion in CRC via upregulated expression of the immune checkpoint PD-L1 in TAMs, thus enhancing tumour migration and invasion [54,55]. It has been understood that changes in miRNA expression patterns drive the onset of malignancies [56]. Moreover, the significance between these miRNA interactions on the seed region of the 3’ UTR and ST6GALNAC1 expression may address its downregulated expression in CRC tumour malignancies. Additionally, it may suggest miRNAs are involved in modulating tumour heterogeneity in CRC and their activity enhanced pro-tumorigenic characteristics including treatment resistance, which inevitably results in poor survival outcomes. However, other epigenetic modifications as were observed with ST6GALNAC2 may downplay the roles of these sialyltransferases in STn production. Therefore, this may provide insights into how ST6GALNAC1 can become deregulated and may suggest the involvement of other sialyltransferase proteins to play a role in STn production.
We have correlated the role of ST6GALNAC1 and ST6GALNAC2 expression and the enrichment of specific gene sets associated with cancer hallmarks.
With respect to ST6GALNAC1, upregulation of EMT and MYC targets provide ample evidence, as reported in the literature, of its role as a possible oncogene, and may also outline possible signalling pathways through which to carry out tumorigenesis [57,58]. Of note, several downregulated mechanisms relating to ST6GALNAC1 also involve IL-6-mediated JAK/STAT3 signalling. Pro-inflammatory stimulation of IL-6 underlines signalling via JAK/STAT3, which promote EMT in multiple cancer types [59]. JAK/STAT3 signalling pathways mediate the activation of EMT through a series of tyrosine and serine/threonine kinases [59]. A previous study associated EMT with CRC metastasis, emphasising crosstalk between CRC tumour cells and TAMs [60] ST6GALNAC1 may facilitate TME heterogeneity following interactions with myeloid cells and EMT pathways. However, the observation of both this pathway and the inflammatory response showing downregulated enrichment supported our findings that EMT is mediated through another mechanism. This may suggest the activity of MYC targets in CRC progression. Disheveled-3 was shown to induce EMT in CRC progression mediated through MYC signalling and Wnt/β-catenin activity [61]. Moreover, syntrophin beta 1 (SNTB1) was shown to mediate EMT progression in CRC through a similar mechanism in knockdown studies [62]. Furthermore, enriched MYC targets may highlight the involvement of Zinc finger protein SNAI1 (SNAIL), a key regulator of the EMT process. Indeed, MYC was shown to induce SNAIL transcription and promote EMT via TGF-β action [63]. Characterisation of ST6GALNAC1 and other gene targets in the MYC/Wnt/β-catenin crosstalk may propose a possible axis in CRC tumorigenesis.
ST6GALNAC2 enrichment exhibited E2F and MYC targets. E2F target involvement in transcriptional regulation can be correlated with the development of multiple tumour malignancies [64]. Moreover, upregulation of E2F activity was exhibited upon the characterisation of deregulated CRC KRAS mutant tumours and CIN type tumours [65]. E2F expression is also directly associated with clinicopathological features of CRC and correlated with poor clinical outcomes [66]. In addition, synergistic signalling between E2F1/MYC can mediate epigenetic modulation in CRC with targeted inhibition of the axis inducing p-53 independent arrest [67]. Furthermore, a recent study identified the expression of E2F7 activating the transcription of enhancer of zeste homolog 2 (EZH2), thus inducing mTOR signalling in glioblastoma progression [68]. Upon identifying that mTOR signalling is also enriched with regard to ST6GALNAC2, this could possibly indicate that a similar interaction could induce the PI3K/AKT/mTOR signalling pathway, possibly through EEF1A2 and MYC. Similarly, another pro-tumorigenic signalling pathway associated with CRC progression had identified TGF-β signalling as significantly enriched. TGF-β signalling is associated with several characteristics of CRC tumours including EMT, angiogenesis and immunosuppression [69]. Interestingly, the downregulated enrichment of the inflammatory response and IFNγ response were revealed. The downregulation of both pathways may indicate CRC tumours associated with ST6GALNAC2 expression is not dependent on inflammatory stimuli. Stimulated IFNγ response is an immunomodulatory mechanism directed against infection, inflammation and anti-tumour activity [70]. Further to this, several inflammatory mediated diseases, including inflammatory bowel disease and ulcerative colitis, could manifest to colitis-associated CRC as a consequence of chronic inflammation [71]. Therefore, the downregulated enrichment of both pathways in GSEA analysis could suggest ST6GALNAC2 expression is not mediated by inflammatory stimuli and its expression profile may be context-dependent.
The role of sialyltransferases and their involvement with immune infiltration highlighted possible interactions within the TME. Although the exhibited SRCC scores outline a non-correlative relationship > 0.2, there is significance in the expression of ST6GALNAC1 and activated CD4+ T-lymphocytes. A pan-cancer transcriptomics analysis identified monocytes and macrophages accounted for the largest proportion of tumour infiltrating myeloid cells [72]. The heterogeneity of the TME could possibly coincide with the pro-tumorigenic nature of several myeloid cell populations, including CD4+ T-lymphocytes [73]. A pan-cancer analysis of stromal heterogeneity was also able to predict the naïve CD4+ T-lymphocyte response to immunotherapeutic treatment [74]. Furthermore, monocyte depletion is also correlated to an immunosuppressive phenotype [75]. Moreover, a previous study highlighted the differential expression of ST6GALNAC1 and its regulatory miRNAs, corresponding with intra-tumour heterogeneity in CRC metastasis [76]. Immunotherapeutic treatment has shown positive treatment responses in patients. However, further targeting additional myeloid cell populations may drive clinical effectiveness. Monocytes exhibit heterogeneity and plasticity within the TME through differentiation to the polarised M2 immunosuppressive phenotype. Blocking M2 polarisation or stimulating M1 monocyte differentiation may reduce the presence of TAMs, thus improving immunotherapeutic approaches in CRC [77]. Furthermore, stimulating CD4+ T-cells to Th1 cells could enhance CD8+ T-cell activation, promoting anti-tumour activity [78]. Although ST6GALNAC1 shares a weak association with TIGIT, the sialyltransferase also shares significant expression with TIGIT, another immune checkpoint associated with cancer progression [76]. TIGIT has been shown to promote myeloid cell exhaustion, including CD8+ T-lymphocytes and enhanced expression profiles correlated with poor clinical outcomes in CRC [79]. This highlights the possibility of other immune checkpoint-related pathways contributing to tumour onset and tumour heterogeneity.
ST6GALNAC2 had shown significant association with several subsets of myeloid cells and immune infiltration. By directly interacting with cellular components associated with the TME, their interaction could modulate the induction of metastasis and immune tolerance [80]. Additionally, solid tumours could display immunogenicity due to the heterogeneous nature of the TME, largely characterised by an immune-induced inflammatory phenotype [81]. The correlation between ST6GALNAC2 expression and immune cell infiltration may offer insights for improving the efficacy of immunotherapeutic approaches. Further to this, reducing the sialylation of glycans will allow greater antigen recognition and promote anti-tumour immunity [82]. The binding of the STn antigen and Siglec-15 drives TGF-β secretion via monocytes and macrophages, possibly establishing tumoral recruitment and enhancing tumour heterogeneity via ST6GALNAC2 overexpression [83]. Likewise, associations with immune checkpoints excluding Siglec-15 highlight potential combination treatment therapies for immune checkpoint blockade, although the literature associating ST6GALNAC2 and immune checkpoints remains limited.
Several studies have indicated enhanced expression of ST6GALNAC1 in several cancer tissues, including lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC), and clear-cell renal cell carcinoma (ccRCC) samples [84,85]. However, the outlined survival curves obtained via datamining demonstrated low ST6GALNAC1 expression correlated with poor clinical outcomes across all survival criteria (p < 0.05). Similarly, this also corroborated the in silico data and IHC staining, outlining higher expression of ST6GALNAC1 in normal tissues. This possibly highlights the involvement of other sialyltransferases in the production of the STn antigen, and inducing effective binding with Siglec-15.
Interestingly, high levels of ST6GALNAC2 expression were statistically significant in all survival criteria (p < 0.05) and were associated with poor clinical outcomes. However, this was not seen at the gene and protein levels obtained via in silico datamining and IHC staining. One possible explanation could stem from silenced gene expression and modulated expression via miRNA binding. Previous studies have highlighted miR-182 and miR-135b were shown to modulate the expression of ST6GALNAC2 via the PI3K/AKT pathway, promoting chemoresistance and tumour invasiveness [21,86]. This could prove similar as mTOR signalling was significantly enriched with regard to ST6GALNAC2 expression.
To illustrate the role of the STn antigen in the CRC landscape, multiple studies have shown the abnormal expression of the STn antigen and its precursor Tn antigen molecule in CRC tumours [87]. Moreover, the expression of the STn antigen is not directly limited to solid CRC epithelial tumours. Circulating tumour cells of metastatic CRC patients expressed enhanced STn production, indicating its role in CRC metastasis to secondary organ sites [88]. Although the present study has identified possible miRNA candidates involved in the regulation of ST6GALNAC1 and ST6GALNAC2, further sialyltransferase family members could play roles in CRC development. The STn antigen displays high binding affinity towards Siglec-15 [89]; however, other sialylated glycans have also displayed elevated binding affinity for Siglec-15 via ST3GAL4 and ST6GAL1 modulation [90]. Furthermore, GALNT enzymes highlighted by the STRING analysis are also heavily involved in GalNAc type-O glycosylation, this could suggest that members of the GALNT family including GALNT2 and GALNT6 facilitate glycan sialylation, including STn production. In addition, many GALNT genes have displayed dysregulated expression in CRC [91,92]. Therefore, this may suggest that downregulated ST6GALNAC1 expression is independent to STn production. Elevated levels of STn and Tn antigens were also identified in CRC samples. However, their expression was induced by the loss of function of other glycosyltransferases, including C1GALT1 and COSMC, through promoter methylation or mutation [40]. Additionally, sulphation modifications of sialoglycans can also contribute to immune evasion and possible tumorigenesis. The overexpression of carbohydrate sulphotransferases, including CHST1 and CHST2, can greatly induce the occurrence of hypersialylation and Siglec binding [93]. Furthermore, CHST1 exhibited an accentuated effect on sialoglycan ligand binding and greatly impacted Siglec preference for O-glycans [94]. Further elucidation of the Siglec/Sia axis may highlight the role of other sialyltransferases in STn production.
Our findings highlighted a different view in the deregulated expression profiles of ST6GALNAC1 and ST6GALNAC2 in comparison to the literature. We believe that epigenetic modifications in conjunction with miRNA activity greatly impact gene expression and may outline sialyltransferase expression in relation to CRC as tumour specific. Furthermore, we provided insights into possible regulatory pathways and signalling pathways associated with their clinical relevance to CRC. Moreover, we believe that ST6GALNAC1 and ST6GALNAC2 is a minor player in the STn production in the case of CRC. The other sialyltransferase enzymes, such as ST6GAL1, ST3GAL1, and sulphotransferases CHST1 might play a vital role in the production of the STn antigen in CRC.
Whilst a mutliomics approach identified the crosstalk of potential gene targets and regulatory pathways of sialyltransferase expression from the integration of several key datasets. There are significant limitations to consider when utilising multiple databases. Firstly, the well-defined regulatory pathways identified through in silico analysis may not fully highlight the interplay of sialylation and sialyltransferase activity without experimental validation to directly support their potential impact on cellular behaviour [95,96]. Further to this point, variation in methodologies may introduce false discoveries which highlight difficulties for data comparability [97]. Moreover, limited sample size and availability may impact in silico software thus creating challenges that require further sophisticated data mining tools. Additionally, there are also difficulties in histological staining for truncated O-glycan structures. Antibodies are required to have high specificity for antigen staining to prevent non-specific cross-reactive staining of similarly structured glycans [98]. However, addressing these limiting factors in association with experimental validation with further studies will provide more robust data interpretation and elucidation of sialyltransferase regulation.
In conclusion, the present study has predicted possible oncomirs involved in the regulation of ST6GALNAC1 with high binding affinities, all of which displayed significantly upregulated expression in CRC tumours. However, downregulated expression of ST6GALNAC1 in CRC might highlight the involvement of other sialyltransferases in the production of the STn antigen and have suggested the interplay of several key sialyltransferases that could play a role. Moreover, we have identified several regulatory signalling pathways that have highlighted the clinical relevance of both ST6GALNAC1 and ST6GALNAC2 to CRC progression. Further elucidation of this pathway will give significant insights into the regulation of the Siglec-15/Sia axis.
Supporting information
STRING network analysis for predicted protein-protein interactions and direct associations obtained via computational data mining for ST6GALNAC1 and ST6GALNAC2 sialyltransferase enzymes. The obtained framework was developed with a high confidence interval ≥0.700.
(DOCX)
In silico analysis to determine common miRNA targets predicted to modulate ST6GALNAC1 via Venn diagrams.
(DOCX)
GSEA enrichment scores for the association of ST6GALNAC1 with cancer hallmarks (FDR < 0.25 and p < 0.05).
(DOCX)
GSEA enrichment scores for the association of ST6GALNAC2 with cancer hallmarks (FDR < 0.25 and p < 0.05).
(DOCX)
UALCAN genomic data was used to determine the gene expression of STRING protein targets AGR2 (ENSG00000106541) and AHSG (ENSG00000145192) and were compared between normal and tumour cohorts.
(TIF)
UALCAN TCGA genomic data was used to identify promoter methylation of ST6GALNAC1 (ENSG00000070526) and ST6GALNAC2 (ENSG00000070731) and were compared between normal and colon adenocarcinoma (COAD) cohorts.
(TIF)
The Pearson correlation coefficient comparing ST6GALNAC1 and ST6GALNAC2 gene expression was determined.
(TIF)
(A) No common miRNAs were predicted between AGR2, AHSG, ST6GALNAC1 and ST6GALNAC2. MiR-432 was common between AGR2, ST6GALNAC1 and ST6GALNAC2. (B) Common miRNAs were determined between GALNT3, GALNT8, B3GNT6 and ST6GALNAC1. MiR-30a-5p was predicted as the common miRNA between all four genes. (C) Common miRNAs were determined between AHSG, C1GALT1C1, C1GALT1 and ST6GALNAC2. No common miRNAs were determined between each gene.
(TIF)
Acknowledgments
We are grateful to the School of Life and Medical Sciences, University of Hertfordshire, United Kingdom, for their support during this work.
Abbreviations
- AGR2
anterior gradient 2
- AHSG
α-2-HS-glycoprotein
- ALCC
average local clustering coefficient
- AT
cancer adjacent colon tissue
- C1GALT1
Core-1 synthase-glycoprotein-N-acetylgalactosamine 3-b-galactosyltransferase- 1
- C1GALT1C1
C1GALT1 Specific Chaperone 1
- ccRCC
clear-cell renal cell carcinoma
- CI
confidence interval
- CIMP
CpG island methylation
- CIN
chromosomal instability
- COAD
colon adenocarcinoma
- CRC
colorectal cancer
- CTLA4
Cytotoxic T-lymphocyte associated protein 4
- EEF
eukaryotic elongation factor
- EEF1A2
Elongation factor 1-a 2
- EMT
epithelial-mesenchymal transition
- ER
endoplasmic reticulum
- EZH2
Enhancer of zeste homolog 2
- FDR
false discovery rate
- FTC
follicular thyroid carcinomas
- GALNT
N-acetylgalactosaminyltransferase
- GALNT
N-acetylgalactosaminyltransferase
- GSEA
Gene Set Enrichment Analysis
- HR
hazard ratio
- IHC
immunohistochemistry
- LAG3
Lymphocyte-activation gene 3
- LUAD
lung adenocarcinoma
- LUSC
lung squamous cell carcinoma
- miRNAs
microRNAs
- MSI
microsatellite instability
- NAT
adjacent normal colon tissue
- OS
overall survival
- PD-L1
programmed death-ligand 1
- PPI
protein-protein interactions
- PPS
post-progression survival
- RECIST
response evaluation criteria in solid tumours
- RFS
relapse-free survival
- RT
room temperature
- Sfold
statistical folding of nucleic acids and studies of regulatory RNAs
- Siglec
Sialic acid-binding immunoglobulin-type lectin
- SNAIL
Zinc finger protein SNAI1SNTB1—syntrophin beta 1
- SRCC
spearman’s rank correlation coefficient
- STn
Sialyl Tn
- STRING
Search Tool for the Retrieval of Interacting Genes/Proteins
- TAMs
tumour-associated macrophages
- TIGIT
T-cell immunoreceptor with Ig and ITIM domains
- TILs
tumour-infiltrating lymphocytes
- TIMER
tumour IMmune Estimation Resource analysis
- TME
tumour microenvironment
- TPM
transcript per million
- Tregs
regulatory T-cells
- UTR
untranslated region
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
The author(s) received no specific funding for this work.
References
- 1.Xie Y-H, Chen Y-X, Fang J-Y. Comprehensive review of targeted therapy for colorectal cancer. Sig Transduct Target Ther. 2020;5: 22. doi: 10.1038/s41392-020-0116-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lam KK, Thean LF, Cheah PY. Advances in colorectal cancer genomics and transcriptomics drive early detection and prevention. The International Journal of Biochemistry & Cell Biology. 2021;137: 106032. doi: 10.1016/j.biocel.2021.106032 [DOI] [PubMed] [Google Scholar]
- 3.Fan A, Wang B, Wang X, Nie Y, Fan D, Zhao X, et al. Immunotherapy in colorectal cancer: current achievements and future perspective. Int J Biol Sci. 2021;17: 3837–3849. doi: 10.7150/ijbs.64077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Läubli H, Nalle SC, Maslyar D. Targeting the Siglec–Sialic Acid Immune Axis in Cancer: Current and Future Approaches. Cancer Immunology Research. 2022;10: 1423–1432. doi: 10.1158/2326-6066.CIR-22-0366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sun J, Lu Q, Sanmamed MF, Wang J. Siglec-15 as an Emerging Target for Next-generation Cancer Immunotherapy. Clinical Cancer Research. 2021;27: 680–688. doi: 10.1158/1078-0432.CCR-19-2925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Y, Xu Z, Wu K-L, Yu L, Wang C, Ding H, et al. Siglec-15/sialic acid axis as a central glyco-immune checkpoint in breast cancer bone metastasis. Proc Natl Acad Sci USA. 2024;121: e2312929121. doi: 10.1073/pnas.2312929121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gianchecchi E, Arena A, Fierabracci A. Sialic Acid-Siglec Axis in Human Immune Regulation, Involvement in Autoimmunity and Cancer and Potential Therapeutic Treatments. IJMS. 2021;22: 5774. doi: 10.3390/ijms22115774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Siglec-15: An Attractive Immunotherapy Target. Cancer Discovery. 2020;10: 7–8. doi: 10.1158/2159-8290.CD-NB2019-136 [DOI] [PubMed] [Google Scholar]
- 9.Ding H, Yao B, Ci L, Feng J, Ouyang P, Chen G, et al. Enhancing the Anti-tumor Potency of a Novel Siglec-15 Antibody by Engineering its Fc-mediated Effector Functions. Journal of Immunotherapy. 2023;46: 161–169. doi: 10.1097/CJI.0000000000000465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu Q, Wei X, Chen F, Huang M, Zhang S, Zhu L, et al. Aptamer‐Assisted Blockade of the Immune Suppressor Sialic Acid‐Binding Immunoglobulin‐Like Lectin‐15 for Cancer Immunotherapy. Angew Chem Int Ed. 2023; e202312609. doi: 10.1002/anie.202312609 [DOI] [PubMed] [Google Scholar]
- 11.Ahmad MS, Braoudaki M, Patel H, Ahmad I, Shagufta, Siddiqui SS. Novel Siglec-15-Sia axis inhibitor leads to colorectal cancer cell death by targeting miR-6715b-3p and oncogenes. Front Immunol. 2023;14: 1254911. doi: 10.3389/fimmu.2023.1254911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stanczak MA, Rodrigues Mantuano N, Kirchhammer N, Sanin DE, Jacob F, Coelho R, et al. Targeting cancer glycosylation repolarizes tumor-associated macrophages allowing effective immune checkpoint blockade. Sci Transl Med. 2022;14: eabj1270. doi: 10.1126/scitranslmed.abj1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Angata T, Tabuchi Y, Nakamura K, Nakamura M. Siglec-15: an immune system Siglec conserved throughout vertebrate evolution. Glycobiology. 2007;17: 838–846. doi: 10.1093/glycob/cwm049 [DOI] [PubMed] [Google Scholar]
- 14.Munkley J, Oltean S, Vodák D, Wilson BT, Livermore KE, Zhou Y, et al. The androgen receptor controls expression of the cancer-associated sTn antigen and cell adhesion through induction of ST6GalNAc1 in prostate cancer. Oncotarget. 2015;6: 34358–34374. doi: 10.18632/oncotarget.6024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ogawa T, Hirohashi Y, Murai A, Nishidate T, Okita K, Wang L, et al. ST6GALNAC1 plays important roles in enhancing cancer stem phenotypes of colorectal cancer via the Akt pathway. Oncotarget. 2017;8: 112550–112564. doi: 10.18632/oncotarget.22545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schneider F, Kemmner W, Haensch W, Franke G, Gretschel S, Karsten U, et al. Overexpression of sialyltransferase CMP-sialic acid:Galbeta1,3GalNAc-R alpha6-Sialyltransferase is related to poor patient survival in human colorectal carcinomas. Cancer Res. 2001;61: 4605–4611. [PubMed] [Google Scholar]
- 17.Miao X, Zhao Y. ST6GalNAcII mediates tumor invasion through PI3K/Akt/NF-κB signaling pathway in follicular thyroid carcinoma. Oncology Reports. 2016;35: 2131–2140. doi: 10.3892/or.2016.4590 [DOI] [PubMed] [Google Scholar]
- 18.Ali Syeda Z, Langden SSS, Munkhzul C, Lee M, Song SJ. Regulatory Mechanism of MicroRNA Expression in Cancer. IJMS. 2020;21: 1723. doi: 10.3390/ijms21051723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ghafouri-Fard S, Hussen BM, Badrlou E, Abak A, Taheri M. MicroRNAs as important contributors in the pathogenesis of colorectal cancer. Biomedicine & Pharmacotherapy. 2021;140: 111759. doi: 10.1016/j.biopha.2021.111759 [DOI] [PubMed] [Google Scholar]
- 20.Shi Y, Liu Z, Lin Q, Luo Q, Cen Y, Li J, et al. MiRNAs and Cancer: Key Link in Diagnosis and Therapy. Genes. 2021;12: 1289. doi: 10.3390/genes12081289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jia L, Luo S, Ren X, Li Y, Hu J, Liu B, et al. miR-182 and miR-135b Mediate the Tumorigenesis and Invasiveness of Colorectal Cancer Cells via Targeting ST6GALNAC2 and PI3K/AKT Pathway. Dig Dis Sci. 2017;62: 3447–3459. doi: 10.1007/s10620-017-4755-z [DOI] [PubMed] [Google Scholar]
- 22.Warde-Farley D, Donaldson SL, Comes O, Zuberi K, Badrawi R, Chao P, et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Research. 2010;38: W214–W220. doi: 10.1093/nar/gkq537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chandrashekar DS, Karthikeyan SK, Korla PK, Patel H, Shovon AR, Athar M, et al. UALCAN: An update to the integrated cancer data analysis platform. Neoplasia. 2022;25: 18–27. doi: 10.1016/j.neo.2022.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ru B, Wong CN, Tong Y, Zhong JY, Zhong SSW, Wu WC, et al. TISIDB: an integrated repository portal for tumor–immune system interactions. Wren J, editor. Bioinformatics. 2019;35: 4200–4202. doi: 10.1093/bioinformatics/btz210 [DOI] [PubMed] [Google Scholar]
- 25.Li T, Fan J, Wang B, Traugh N, Chen Q, Liu JS, et al. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Research. 2017;77: e108–e110. doi: 10.1158/0008-5472.CAN-17-0307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Győrffy B. Survival analysis across the entire transcriptome identifies biomarkers with the highest prognostic power in breast cancer. Computational and Structural Biotechnology Journal. 2021;19: 4101–4109. doi: 10.1016/j.csbj.2021.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moidu NA A Rahman NS, Syafruddin SE, Low TY, Mohtar MA. Secretion of pro-oncogenic AGR2 protein in cancer. Heliyon. 2020;6: e05000. doi: 10.1016/j.heliyon.2020.e05000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xiu B, Chi Y, Liu L, Chi W, Zhang Q, Chen J, et al. LINC02273 drives breast cancer metastasis by epigenetically increasing AGR2 transcription. Mol Cancer. 2019;18: 187. doi: 10.1186/s12943-019-1115-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tian S, Chu Y, Hu J, Ding X, Liu Z, Fu D, et al. Tumour-associated neutrophils secrete AGR2 to promote colorectal cancer metastasis via its receptor CD98hc–xCT. Gut. 2022;71: 2489–2501. doi: 10.1136/gutjnl-2021-325137 [DOI] [PubMed] [Google Scholar]
- 30.Fessart D, Domblides C, Avril T, Eriksson LA, Begueret H, Pineau R, et al. Secretion of protein disulphide isomerase AGR2 confers tumorigenic properties. eLife. 2016;5: e13887. doi: 10.7554/eLife.13887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pirro M, Mohammed Y, De Ru AH, Janssen GMC, Tjokrodirijo RTN, Madunić K, et al. Oxonium Ion Guided Analysis of Quantitative Proteomics Data Reveals Site-Specific O-Glycosylation of Anterior Gradient Protein 2 (AGR2). IJMS. 2021;22: 5369. doi: 10.3390/ijms22105369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xing X, Cao F, Gao L, Song M. AHSG, a Gene Promoting Tumour Proliferation, Migration and Invasion, is an Independent Prognostic Factor for Poor Overall Survival in Lung Adenocarcinoma. Mol Biol Rep. 2023;50: 7659–7666. doi: 10.1007/s11033-023-08623-x [DOI] [PubMed] [Google Scholar]
- 33.Shen Z, Li Y, Fang Y, Lin M, Feng X, Li Z, et al. SNX16 activates c‐Myc signaling by inhibiting ubiquitin‐mediated proteasomal degradation of eEF1A2 in colorectal cancer development. Molecular Oncology. 2020;14: 387–406. doi: 10.1002/1878-0261.12626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pellegrino R, Calvisi DF, Neumann O, Kolluru V, Wesely J, Chen X, et al. EEF1A2 inactivates p53 by way of PI3K/AKT/mTOR-dependent stabilization of MDM4 in hepatocellular carcinoma: Pellegrino et al. Hepatology. 2014;59: 1886–1899. doi: 10.1002/hep.26954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Munkley J. The Role of Sialyl-Tn in Cancer. IJMS. 2016;17: 275. doi: 10.3390/ijms17030275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sanz-Martinez I, Pereira S, Merino P, Corzana F, Hurtado-Guerrero R. Molecular Recognition of GalNAc in Mucin-Type O-Glycosylation. Acc Chem Res. 2023;56: 548–560. doi: 10.1021/acs.accounts.2c00723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kvorjak M, Ahmed Y, Miller ML, Sriram R, Coronnello C, Hashash JG, et al. Cross-talk between Colon Cells and Macrophages Increases ST6GALNAC1 and MUC1-sTn Expression in Ulcerative Colitis and Colitis-Associated Colon Cancer. Cancer Immunology Research. 2020;8: 167–178. doi: 10.1158/2326-6066.CIR-19-0514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Noda M, Okayama H, Tachibana K, Sakamoto W, Saito K, Thar Min AK, et al. Glycosyltransferase Gene Expression Identifies a Poor Prognostic Colorectal Cancer Subtype Associated with Mismatch Repair Deficiency and Incomplete Glycan Synthesis. Clinical Cancer Research. 2018;24: 4468–4481. doi: 10.1158/1078-0432.CCR-17-3533 [DOI] [PubMed] [Google Scholar]
- 39.Fernández-Ponce C, Geribaldi-Doldán N, Sánchez-Gomar I, Navarro Quiroz R, Atencio Ibarra L, Gomez Escorcia L, et al. The Role of Glycosyltransferases in Colorectal Cancer. IJMS. 2021;22: 5822. doi: 10.3390/ijms22115822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sun X, Ju T, Cummings RD. Differential expression of Cosmc, T-synthase and mucins in Tn-positive colorectal cancers. BMC Cancer. 2018;18: 827. doi: 10.1186/s12885-018-4708-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tian H, Yu J-L, Chu X, Guan Q, Liu J, Liu Y. Unraveling the role of C1GALT1 in abnormal glycosylation and colorectal cancer progression. Front Oncol. 2024;14: 1389713. doi: 10.3389/fonc.2024.1389713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nishiyama A, Nakanishi M. Navigating the DNA methylation landscape of cancer. Trends in Genetics. 2021;37: 1012–1027. doi: 10.1016/j.tig.2021.05.002 [DOI] [PubMed] [Google Scholar]
- 43.Erfani M, Zamani M, Mokarram P. Evidence of histone modification affecting ARID1A expression in colorectal cancer cell lines. Gastroenterol Hepatol Bed Bench. 2022;15: 32–38. [PMC free article] [PubMed] [Google Scholar]
- 44.Wang X, He Y, Mackowiak B, Gao B. MicroRNAs as regulators, biomarkers and therapeutic targets in liver diseases. Gut. 2021;70: 784–795. doi: 10.1136/gutjnl-2020-322526 [DOI] [PubMed] [Google Scholar]
- 45.Shang R, Lee S, Senavirathne G, Lai EC. microRNAs in action: biogenesis, function and regulation. Nat Rev Genet. 2023;24: 816–833. doi: 10.1038/s41576-023-00611-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li J, Chen H, Sun G, Zhang X, Ye H, Wang P. Role of miR-21 in the diagnosis of colorectal cancer: Meta-analysis and bioinformatics. Pathology—Research and Practice. 2023;248: 154670. doi: 10.1016/j.prp.2023.154670 [DOI] [PubMed] [Google Scholar]
- 47.Calvo-López T, Paz-Cabezas M, Llovet P, Ibañez MD, Sastre J, Alonso-Orduña V, et al. Association of miR-21 and miR-335 to microsatellite instability and prognosis in stage III colorectal cancer. CBM. 2022;34: 201–210. doi: 10.3233/CBM-210353 [DOI] [PubMed] [Google Scholar]
- 48.Lai C-Y, Yeh K-Y, Liu B-F, Chang T-M, Chang C-H, Liao Y-F, et al. MicroRNA-21 Plays Multiple Oncometabolic Roles in Colitis-Associated Carcinoma and Colorectal Cancer via the PI3K/AKT, STAT3, and PDCD4/TNF-α Signaling Pathways in Zebrafish. Cancers. 2021;13: 5565. doi: 10.3390/cancers13215565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Despotovic J, Dragicevic S, Nikolic A. Effects of Chemotherapy for Metastatic Colorectal Cancer on the TGF-β Signaling and Related miRNAs hsa-miR-17-5p, hsa-miR-21-5p and hsa-miR-93-5p. Cell Biochem Biophys. 2021;79: 757–767. doi: 10.1007/s12013-021-00980-3 [DOI] [PubMed] [Google Scholar]
- 50.Peng X, Wang J, Zhang C, Liu K, Zhao L, Chen X, et al. A three-miRNA panel in serum as a noninvasive biomarker for colorectal cancer detection. Int J Biol Markers. 2020;35: 74–82. doi: 10.1177/1724600820950740 [DOI] [PubMed] [Google Scholar]
- 51.Wei K, Shi J, Xiao Y, Wang W, Yang Q, Chen C. [MiR-30e-5p overexpression promotes proliferation and migration of colorectal cancer cells by activating the CXCL12 axis via downregulating PTEN]. Nan Fang Yi Ke Da Xue Xue Bao. 2023;43: 1081–1092. doi: 10.12122/j.issn.1673-4254.2023.07.04 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Han J, Sun W, Liu R, Zhou Z, Zhang H, Chen X, et al. Plasma Exosomal miRNA Expression Profile as Oxaliplatin-Based Chemoresistant Biomarkers in Colorectal Adenocarcinoma. Front Oncol. 2020;10: 1495. doi: 10.3389/fonc.2020.01495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Farouk S, El-Shenawy R, Khairy AM, Bader El-Din NG. Overexpression of Mirna 26A and 26B with Mmp-9 are Valuable Diagnostic Biomarkers for Colorectal Cancer Patients. Biomark Med. 2023;17: 159–169. doi: 10.2217/bmm-2022-0861 [DOI] [PubMed] [Google Scholar]
- 54.Yin Y, Liu B, Cao Y, Yao S, Liu Y, Jin G, et al. Colorectal Cancer‐Derived Small Extracellular Vesicles Promote Tumor Immune Evasion by Upregulating PD‐L1 Expression in Tumor‐Associated Macrophages. Advanced Science. 2022;9: 2102620. doi: 10.1002/advs.202102620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lan J, Sun L, Xu F, Liu L, Hu F, Song D, et al. M2 Macrophage-Derived Exosomes Promote Cell Migration and Invasion in Colon Cancer. Cancer Research. 2019;79: 146–158. doi: 10.1158/0008-5472.CAN-18-0014 [DOI] [PubMed] [Google Scholar]
- 56.Hussen BM, Hidayat HJ, Salihi A, Sabir DK, Taheri M, Ghafouri-Fard S. MicroRNA: A signature for cancer progression. Biomedicine & Pharmacotherapy. 2021;138: 111528. doi: 10.1016/j.biopha.2021.111528 [DOI] [PubMed] [Google Scholar]
- 57.Gao F, Li X, Xu K, Wang R, Guan X. c-MYC mediates the crosstalk between breast cancer cells and tumor microenvironment. Cell Commun Signal. 2023;21: 28. doi: 10.1186/s12964-023-01043-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shin AE, Giancotti FG, Rustgi AK. Metastatic colorectal cancer: mechanisms and emerging therapeutics. Trends in Pharmacological Sciences. 2023;44: 222–236. doi: 10.1016/j.tips.2023.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Huang B, Lang X, Li X. The role of IL-6/JAK2/STAT3 signaling pathway in cancers. Front Oncol. 2022;12: 1023177. doi: 10.3389/fonc.2022.1023177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wei C, Yang C, Wang S, Shi D, Zhang C, Lin X, et al. Crosstalk between cancer cells and tumor associated macrophages is required for mesenchymal circulating tumor cell-mediated colorectal cancer metastasis. Mol Cancer. 2019;18: 64. doi: 10.1186/s12943-019-0976-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li Z, Yang Z, Liu W, Zhu W, Yin L, Han Z, et al. Disheveled3 enhanced EMT and cancer stem-like cells properties via Wnt/β-catenin/c-Myc/SOX2 pathway in colorectal cancer. J Transl Med. 2023;21: 302. doi: 10.1186/s12967-023-04120-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang H, Li Z, Jiang J, Lei Y, Xie J, Liu Y, et al. SNTB1 regulates colorectal cancer cell proliferation and metastasis through YAP1 and the WNT/β-catenin pathway. Cell Cycle. 2023;22: 1865–1883. doi: 10.1080/15384101.2023.2244778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Meškytė EM, Keskas S, Ciribilli Y. MYC as a Multifaceted Regulator of Tumor Microenvironment Leading to Metastasis. IJMS. 2020;21: 7710. doi: 10.3390/ijms21207710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.He J, Gao R, Yang J, Li F, Fu Y, Cui J, et al. NCAPD2 promotes breast cancer progression through E2F1 transcriptional regulation of CDK1. Cancer Science. 2023;114: 896–907. doi: 10.1111/cas.15347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chong W, Zhu X, Ren H, Ye C, Xu K, Wang Z, et al. Integrated multi-omics characterization of KRAS mutant colorectal cancer. Theranostics. 2022;12: 5138–5154. doi: 10.7150/thno.73089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xu Z, Qu H, Ren Y, Gong Z, Ri HJ, Zhang F, et al. Systematic Analysis of E2 F Expression and Its Relation in Colorectal Cancer Prognosis. IJGM. 2022;Volume 15: 4849–4870. doi: 10.2147/IJGM.S352141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jeon D, Kim N, Um S-J. BET Inhibitors Induce p53-Independent Growth Arrest in HCT116 Cells via Epigenetic Control of the E2F1/c-MYC Axis. Biological & Pharmaceutical Bulletin. 2023;46: 12–18. doi: 10.1248/bpb.b22-00343 [DOI] [PubMed] [Google Scholar]
- 68.Yang R, Wang M, Zhang G, Bao Y, Wu Y, Li X, et al. E2F7−EZH2 axis regulates PTEN/AKT/mTOR signalling and glioblastoma progression. Br J Cancer. 2020;123: 1445–1455. doi: 10.1038/s41416-020-01032-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li X, Wu Y, Tian T. TGF-β Signaling in Metastatic Colorectal Cancer (mCRC): From Underlying Mechanism to Potential Applications in Clinical Development. IJMS. 2022;23: 14436. doi: 10.3390/ijms232214436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Coelho MA, Cooper S, Strauss ME, Karakoc E, Bhosle S, Gonçalves E, et al. Base editing screens map mutations affecting interferon-γ signaling in cancer. Cancer Cell. 2023;41: 288–303.e6. doi: 10.1016/j.ccell.2022.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhou RW, Harpaz N, Itzkowitz SH, Parsons RE. Molecular mechanisms in colitis-associated colorectal cancer. Oncogenesis. 2023;12: 48. doi: 10.1038/s41389-023-00492-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cheng S, Li Z, Gao R, Xing B, Gao Y, Yang Y, et al. A pan-cancer single-cell transcriptional atlas of tumor infiltrating myeloid cells. Cell. 2021;184: 792–809.e23. doi: 10.1016/j.cell.2021.01.010 [DOI] [PubMed] [Google Scholar]
- 73.Zhao Q, Cheng Y, Xiong Y. LTF Regulates the Immune Microenvironment of Prostate Cancer Through JAK/STAT3 Pathway. Front Oncol. 2021;11: 692117. doi: 10.3389/fonc.2021.692117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Qian J, Olbrecht S, Boeckx B, Vos H, Laoui D, Etlioglu E, et al. A pan-cancer blueprint of the heterogeneous tumor microenvironment revealed by single-cell profiling. Cell Res. 2020;30: 745–762. doi: 10.1038/s41422-020-0355-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ortiz-Muñoz G, Brown M, Carbone CB, Pechuan-Jorge X, Rouilly V, Lindberg H, et al. In situ tumour arrays reveal early environmental control of cancer immunity. Nature. 2023;618: 827–833. doi: 10.1038/s41586-023-06132-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Urh K, Zidar N, Tomažič A, Boštjančič E. Intra‑tumor heterogeneity of cancer stem cell‑related genes and their potential regulatory microRNAs in metastasizing colorectal carcinoma. Oncol Rep. 2022;48: 193. doi: 10.3892/or.2022.8408 [DOI] [PubMed] [Google Scholar]
- 77.Olingy CE, Dinh HQ, Hedrick CC. Monocyte heterogeneity and functions in cancer. Journal of Leukocyte Biology. 2019;106: 309–322. doi: 10.1002/JLB.4RI0818-311R [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Speiser DE, Chijioke O, Schaeuble K, Münz C. CD4+ T cells in cancer. Nat Cancer. 2023;4: 317–329. doi: 10.1038/s43018-023-00521-2 [DOI] [PubMed] [Google Scholar]
- 79.Liang R, Zhu X, Lan T, Ding D, Zheng Z, Chen T, et al. TIGIT promotes CD8+T cells exhaustion and predicts poor prognosis of colorectal cancer. Cancer Immunol Immunother. 2021;70: 2781–2793. doi: 10.1007/s00262-021-02886-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bai Z, Yan C, Chang D. Prediction and therapeutic targeting of the tumor microenvironment-associated gene CTSK in gastric cancer. Discov Onc. 2023;14: 200. doi: 10.1007/s12672-023-00821-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chen DS, Mellman I. Elements of cancer immunity and the cancer–immune set point. Nature. 2017;541: 321–330. doi: 10.1038/nature21349 [DOI] [PubMed] [Google Scholar]
- 82.Munkley J. Aberrant Sialylation in Cancer: Therapeutic Opportunities. Cancers. 2022;14: 4248. doi: 10.3390/cancers14174248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hugonnet M, Singh P, Haas Q, Von Gunten S. The Distinct Roles of Sialyltransferases in Cancer Biology and Onco-Immunology. Front Immunol. 2021;12: 799861. doi: 10.3389/fimmu.2021.799861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bai Q, Liu L, Xi W, Wang J, Xia Y, Qu Y, et al. Prognostic significance of ST6GalNAc-1 expression in patients with non-metastatic clear cell renal cell carcinoma. Oncotarget. 2018;9: 3112–3120. doi: 10.18632/oncotarget.11258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Alabiad M, Harb O, Abozaid M, Embaby A, Mandour D, Hemeda R, et al. The Diagnostic and Prognostic Roles of Combined Expression of Novel Biomarkers in Lung Adenocarcinoma (LUAD) and Lung Squamous Cell Carcinoma (LUSC); An Immunohistochemical Study. Iran J Pathol. 2021;16: 162–173. doi: 10.30699/ijp.2020.130944.2452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Liu B, Liu Y, Zhao L, Pan Y, Shan Y, Li Y, et al. Upregulation of microRNA‐135b and microRNA‐182 promotes chemoresistance of colorectal cancer by targeting ST6GALNAC2 via PI3K/AKT pathway. Molecular Carcinogenesis. 2017;56: 2669–2680. doi: 10.1002/mc.22710 [DOI] [PubMed] [Google Scholar]
- 87.Dombek GE, Ore AS, Cheng J, Matsumoto Y, Glickman JN, Fleishman A, et al. Immunohistochemical analysis of Tn antigen expression in colorectal adenocarcinoma and precursor lesions. BMC Cancer. 2022;22: 1281. doi: 10.1186/s12885-022-10376-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Neves M, Azevedo R, Lima L, Oliveira MI, Peixoto A, Ferreira D, et al. Exploring sialyl-Tn expression in microfluidic-isolated circulating tumour cells: A novel biomarker and an analytical tool for precision oncology applications. New Biotechnology. 2019;49: 77–87. doi: 10.1016/j.nbt.2018.09.004 [DOI] [PubMed] [Google Scholar]
- 89.Murugesan G, Weigle B, Crocker PR. Siglec and anti-Siglec therapies. Current Opinion in Chemical Biology. 2021;62: 34–42. doi: 10.1016/j.cbpa.2021.01.001 [DOI] [PubMed] [Google Scholar]
- 90.Briard JG, Jiang H, Moremen KW, Macauley MS, Wu P. Cell-based glycan arrays for probing glycan–glycan binding protein interactions. Nat Commun. 2018;9: 880. doi: 10.1038/s41467-018-03245-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Liao Y, Chuang Y, Lin H, Lin N, Hsu T, Hsieh S, et al. GALNT2 promotes invasiveness of colorectal cancer cells partly through AXL. Molecular Oncology. 2023;17: 119–133. doi: 10.1002/1878-0261.13347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lavrsen K, Dabelsteen S, Vakhrushev SY, Levann AMR, Haue AD, Dylander A, et al. De novo expression of human polypeptide N-acetylgalactosaminyltransferase 6 (GalNAc-T6) in colon adenocarcinoma inhibits the differentiation of colonic epithelium. Journal of Biological Chemistry. 2018;293: 1298–1314. doi: 10.1074/jbc.M117.812826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jung J, Enterina JR, Bui DT, Mozaneh F, Lin P-H, Nitin, et al. Carbohydrate Sulfation As a Mechanism for Fine-Tuning Siglec Ligands. ACS Chem Biol. 2021;16: 2673–2689. doi: 10.1021/acschembio.1c00501 [DOI] [PubMed] [Google Scholar]
- 94.Büll C, Nason R, Sun L, Van Coillie J, Madriz Sørensen D, Moons SJ, et al. Probing the binding specificities of human Siglecs by cell-based glycan arrays. Proc Natl Acad Sci USA. 2021;118: e2026102118. doi: 10.1073/pnas.2026102118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kaur P, Singh A, Chana I. Computational Techniques and Tools for Omics Data Analysis: State-of-the-Art, Challenges, and Future Directions. Arch Computat Methods Eng. 2021;28: 4595–4631. doi: 10.1007/s11831-021-09547-0 [DOI] [Google Scholar]
- 96.Subramanian I, Verma S, Kumar S, Jere A, Anamika K. Multi-omics Data Integration, Interpretation, and Its Application. Bioinform Biol Insights. 2020;14: 117793221989905. doi: 10.1177/1177932219899051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ye Z, Vakhrushev SY. The Role of Data-Independent Acquisition for Glycoproteomics. Molecular & Cellular Proteomics. 2021;20: 100042. doi: 10.1074/mcp.R120.002204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Loureiro LR, Sousa DP, Ferreira D, Chai W, Lima L, Pereira C, et al. Novel monoclonal antibody L2A5 specifically targeting sialyl-Tn and short glycans terminated by alpha-2–6 sialic acids. Sci Rep. 2018;8: 12196. doi: 10.1038/s41598-018-30421-w [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
STRING network analysis for predicted protein-protein interactions and direct associations obtained via computational data mining for ST6GALNAC1 and ST6GALNAC2 sialyltransferase enzymes. The obtained framework was developed with a high confidence interval ≥0.700.
(DOCX)
In silico analysis to determine common miRNA targets predicted to modulate ST6GALNAC1 via Venn diagrams.
(DOCX)
GSEA enrichment scores for the association of ST6GALNAC1 with cancer hallmarks (FDR < 0.25 and p < 0.05).
(DOCX)
GSEA enrichment scores for the association of ST6GALNAC2 with cancer hallmarks (FDR < 0.25 and p < 0.05).
(DOCX)
UALCAN genomic data was used to determine the gene expression of STRING protein targets AGR2 (ENSG00000106541) and AHSG (ENSG00000145192) and were compared between normal and tumour cohorts.
(TIF)
UALCAN TCGA genomic data was used to identify promoter methylation of ST6GALNAC1 (ENSG00000070526) and ST6GALNAC2 (ENSG00000070731) and were compared between normal and colon adenocarcinoma (COAD) cohorts.
(TIF)
The Pearson correlation coefficient comparing ST6GALNAC1 and ST6GALNAC2 gene expression was determined.
(TIF)
(A) No common miRNAs were predicted between AGR2, AHSG, ST6GALNAC1 and ST6GALNAC2. MiR-432 was common between AGR2, ST6GALNAC1 and ST6GALNAC2. (B) Common miRNAs were determined between GALNT3, GALNT8, B3GNT6 and ST6GALNAC1. MiR-30a-5p was predicted as the common miRNA between all four genes. (C) Common miRNAs were determined between AHSG, C1GALT1C1, C1GALT1 and ST6GALNAC2. No common miRNAs were determined between each gene.
(TIF)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.