Abstract
The expression of the Kaposi's sarcoma-associated herpesvirus (KSHV) open reading frame 50 (ORF50) protein, Lyta (lytic transactivator), marks the switch from latent KSHV infection to the lytic phase. ORF50/Lyta upregulates several target KSHV genes, such as K8 (K-bZip), K9 (vIRF1), and ORF57, finally leading to the production of mature viruses. The auto-upregulation of ORF50/Lyta is thought to be an important mechanism for efficient lytic viral replication. In this study, we focused on this autoregulation and identified the promoter element required for it. An electrophoretic mobility shift assay indicated that the octamer-binding protein 1 (Oct-1) bound to this element. Mutations in the octamer-binding motif resulted in refractoriness of the ORF50/Lyta promoter to transactivation by ORF50/Lyta, and Oct-1 expression enhanced this transactivation. These results suggest that the autoregulation of ORF50/Lyta is mediated by Oct-1.
Kaposi's sarcoma-associated herpesvirus (KSHV), also called human herpesvirus 8, was discovered in Kaposi's sarcoma lesions of human immunodeficiency virus-infected patients by representational difference analysis (5). Sequence analysis revealed that KSHV was related to gammaherpesviruses, such as Epstein-Barr virus (EBV), and suggested that it was an oncogenic DNA virus. KSHV infection is tightly linked to primary effusion lymphoma and multicentric Castleman's disease (2, 32). It is likely that viral infection with KSHV is necessary for development of Kaposi's sarcoma (14).
Lytic replication of herpesviruses is usually initiated by immediate-early (IE) gene expression. Several IE genes have been reported in KSHV (32). Among these, the ORF50/Lyta gene is highly conserved in gammaherpesviruses and plays a key role in viral lytic replication (16, 23, 24, 36, 37, 38, 39). Some chemical reagents, such as 12-O-tetradecanoylphorbol-13-acetate (TPA), n-butyrate, and the calcium ionophore A23187, can induce viral lytic replication in cell lines that are latently infected with KSHV (4, 21, 27, 33). ORF50/Lyta can transactivate the expression of its target genes, including those corresponding to ORF6 (single-stranded-DNA-binding protein), ORF9 (DNA polymerase), ORF21 (thymidine kinase), ORF57 (Mta), ORF59 (PF8), K8 (K-bZip), K9 (vIRF1), K12 (Kaposin), and nut-1/PAN (6, 17, 18, 27). The gene product of ORF9 and ORF59 has been reported to be involved in the lytic replication (3). Therefore, ORF50/Lyta is an initiator for lytic viral replication. Lukac et al. reported that the KSHV ORF50/Lyta mutant, ΔSTAD, which lacks a serine/threonine-rich domain and an acidic domain, can act as a dominant-negative mutant for ORF50/Lyta (18). This mutant inhibits the transactivation activity of ORF50/Lyta by heterodimerizing with the ORF50/Lyta protein.
In the case of EBV, autoregulation of BRLF1/Rta, which is a homologue of ORF50/Lyta, is also observed in EBV lytic replication, and this regulation occurs through a non-DNA-binding mechanism of Rta in certain cell lines (24, 39). Recently it was reported that KSHV ORF50/Lyta can upregulate its own expression (10, 12). However, little is known about the molecular mechanisms of how ORF50/Lyta autoregulates its transcription.
Here we focused on the autoregulation of ORF50/Lyta and analyzed its mechanism. We identified a critical element for the responsiveness of the ORF50/Lyta promoter to ORF50/Lyta transactivation. An electrophoretic mobility shift assay (EMSA) indicated that this regulation might be through a non-DNA-binding mechanism involving ORF50/Lyta and the octamer-binding protein (Oct-1), which could bind with an element in the ORF50/Lyta promoter in vitro. Mutations in the octamer-binding motif resulted in refractoriness of the ORF50/Lyta promoter to transactivation by ORF50/Lyta. Cotransfection of ORF50/Lyta with an Oct-1 expression vector enhanced the transactivation mediated by the Oct-1-binding element. We propose that Oct-1 plays a key role in the autoregulation of ORF50/Lyta and participates in the transactivation of other promoters by ORF50/Lyta.
MATERIALS AND METHODS
Cell lines.
The KSHV-infected cell line BCBL-1 was cultured in RPMI-1640 (Nissui, Tokyo, Japan) supplemented with 100 IU of penicillin G (PEN)/ml, 0.1 mg of streptomycin (STR) (Meiji Seika, Tokyo, Japan)/ml, and 20% heat-inactivated fetal bovine serum (FBS) (Gibco, Rockville, Md.). Ramos cells (EBV negative) were cultured in RPMI-1640 containing PEN, STR, and 10% heat-inactivated FBS. A human embryonal kidney epithelial cell line, 293L, was cultured in Dulbecco's modified Eagle medium (Nissui) with PEN, STR, and 10% heat-inactivated FBS. All cell lines were cultured at 37°C in a humidified atmosphere with 5% CO2.
Plasmids.
The cDNA of ORF50/Lyta was inserted into pcDNA3.1(-)/Myc-His-B (Invitrogen, San Diego, Calif.). In the resultant construct, all 691 amino acids of ORF50/Lyta were expressed and a Myc-histidine tag was fused in frame at the C terminus (6). For the reporter construct designated pGL3-FL (−914), the region between nucleotides 70646 and 71593 of the KSHV genome (GenBank accession no. U75698) was amplified with the primers 50p.F (5′-CTGCCCATGGGCGGGTGGGTGACAGTCCGC-3′) and 50p.R (5′-TGCGCCATGGTTGTGGCTGCCTGGACAGTA-3′) and then inserted into the NcoI site of pGL3-Basic (Promega, Madison, Wisc.). The 5′ deletion mutants were constructed from pGL3-FL (−914) by digesting pGL3-FL (−914) with ExoIII exonuclease (Takara Shuzo, Kyoto, Japan) from the HindIII site in the multicloning site of the pGL3-vector (6).
Putative ORF50/Lyta responsive elements (LREs) and the mutant reporter (LRE2.1-mt) were chemically synthesized with an NheI restriction enzyme recognition sequence at the 5′ end (see Table 1). After annealing, the double-stranded DNAs were phosphorylated at the 5′ end with T4 polynucleotide kinase (Takara Shuzo). They were then inserted in tandem into the NheI site of pe1b-TATA-luc, a plasmid that was based on the pGL3-Basic vector, with an adenovirus E1b minimal TATA box upstream of the luciferase gene (6). These reporter plasmids (pe1b-LRE) contained three copies of each fragment in tandem (see Fig. 2A).
TABLE 1.
ORF50/Lyta promoter fragment sequence used for reporter plasmid
| Name | Sequence | Start/end positions |
|---|---|---|
| LRE1 | ctagcGATGTGGTACCGAATGCCACAATCTGTGCCCTCCAGCTCg | −264/−226 |
| gCTACACCATGGCTTACGGTGTTAGACACGGGAGGTCGAGcgatc | ||
| LRE2 | ctagcTCTCACAATTTTCATCTCCAATACCCGGAATTGGGATAg | −227/−190 |
| gAGAGTGTTAAAAGTAGAGGTTATGGGCCTTAACCCTATcgatc | ||
| LRE3 | ctagcGAATTGGGATACACACCTCCATGTTCAGTCACATGTACGCTg | −200/−160 |
| gCTTAACCCTATGTGTGGAGGTACAAGTCAGTGTACATGCGAcgatc | ||
| LRE2.1 | ctagcTCTCACAATTTTCATCTCCAg | −227/−208 |
| gAGAGTGTTAAAAGTAGAGGTcgatc | ||
| LRE2.2 | ctagcTTTTCATCTCCAATACCCGGg | −219/−200 |
| gAAAAGTAGAGGTTATGGGCCcgatc | ||
| LRE2.3 | ctagcCAATACCCGGAATTGGGATAg | −209/−190 |
| gGTTATGGGCCTTAACCCTATcgatc |
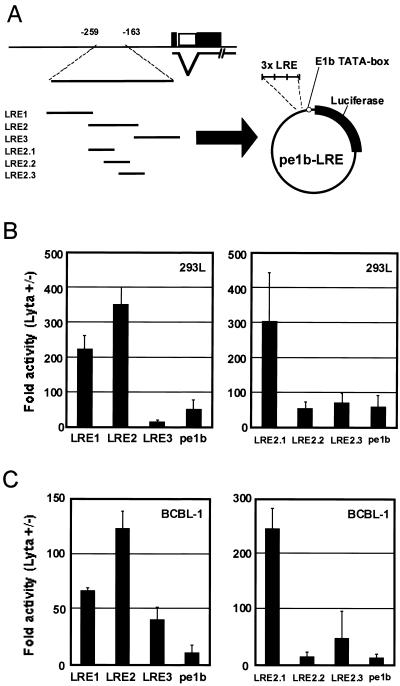
FIG. 2.
Determination of the critical element required for transactivation by ORF50/Lyta. (A) Schematic representation of the ORF50/Lyta promoter and 3×LRE-TATA-luc reporter constructs (pe1b-LREs) for the putative responsive element. (B and C) Transient cotransfection assays with 293L (B) and BCBL-1 (C) cells. Transfection was performed with Superfect reagent (Qiagen), and after a 48-h cultivation, the cells were harvested and assayed. pCMV-β-gal was cotransfected as described (see the legend for Fig. 1), and these experiments were performed at least three times. The results are shown as fold activation of the reporter construct with pcDNA3.1 as the control vector.
To construct the octamer reporter plasmid (p4×oct-e1b-luc), the consensus octamer-binding sequence (Promega), which was also used as a cold competitor in an EMSA, was inserted into the upstream of pe1b-TATA-luc. p4Xoct-e1b-luc contained four copies of 22-bp fragment (5′-TGTCGAATGCAAATCACTAGAA-3′) derived from the immunoglobulin promoter.
pGL3-FLmtoct, which had the same sequence of LRE2.1mt (see Fig. 4A) in the corresponding site of pGL3-FL (−914), was constructed. The method to introduce a directed mutation by PCR-mediated mutagenesis was described elsewhere (9) and was done using the primers mt-Oct-S (5′-CCAGCTCTCACAATTTTCGCCTCCAATACCCGGAATTGG-3′) and mt-Oct-AS (a complementary sequence of mt-Oct-S), which had two mutations in the octamer motif (bold). The mutagenesis was confirmed by an ABI PRISM 310 Genetic Analyzer (Applied Biosystems, Foster City, Calif.).
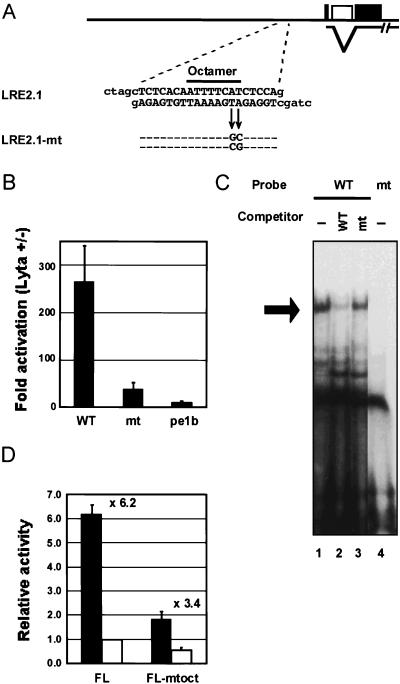
FIG. 4.
Effect of mutating the octamer-binding sequence on ORF50/Lyta transactivation. (A) Sequences of LRE2.1 and LRE2.1-mt. (B) The fold activation of wild-type LRE2.1 (WT) and its mutant (mt) reporter by ORF50/Lyta in 293L cells. The transfection assay is described in the legend for Fig. 2. (C) EMSA with LRE2.1 (WT) and its mutant (mt). Nuclear extract from BCBL-1 cells treated with TPA was used. The [α-32P]dCTP-labeled LRE2.1 was used as a probe, and an unlabeled LRE2.1 mutant was added as a competitor at a 50-fold excess (lane 3). The labeled LRE2.1-mt was used as a probe in lane 4. (D) Effect of site-directed mutagenesis on ORF50/Lyta autoregulation. The mutant octamer-binding sequence (mtoct) shown in panel A was induced in pGL3-FL (FL) (−914). The solid and open bars indicate the relative activities with pcDNA3.1-ORF50/Lyta and the pcDNA3.1 vector, respectively. The procedure of the transfection assay is described in the legend for Fig. 2.
The octamer-binding protein expression vectors pCGNoct-1 and pCGNoct-2 were gifts from W. Herr (Cold Spring Harbor Laboratory) (34). The plasmids contained a small epitope derived from the influenza virus hemagglutinin at the N terminus of each oct gene. The XbaI-BamHI fragment of pCGNoct-1 was deleted to construct an empty vector.
Transfection.
Electroporation of BCBL-1 and Ramos cells was performed as follows. Cells (5.0 × 106) suspended in 250 μl of serum- and antibiotic-free medium containing each plasmid DNA were electroporated at 950 μF and 250 mV in a cuvette (0.4 cm; Bio-Rad Laboratories, Hercules, Calif.). The cells were then incubated in 10 ml of medium with 10% FBS. For the autoregulation assay by ORF50/Lyta, 5 μg of pcDNA3.1-ORF50/Lyta and 2 μg of the reporter plasmid were cotransfected.
Superfect transfection reagent (Qiagen) was used to transfect the 293L and BCBL-1 cells, according to the manufacturer's protocol. In the case of 293L, cells were plated at a concentration of 2.0 × 105 cells/well in 6-well plates (Iwaki, Chiba, Japan), 1 day prior to transfection. For identification of the element required for ORF50/Lyta autoregulation, 1.0 μg of pcDNA3.1-ORF50/Lyta/well and 0.1 μg of the reporter plasmid/well were cotransfected. For BCBL-1 cells, 106 cells/well in six-well plates were cotransfected with 2 μg of pcDNA3.1-ORF50/well and 0.2 μg of the reporter plasmid/well.
To examine the effects of the overexpression of octamer-binding proteins on the autoregulation, pCGNoct-1 and -2 (none, 0.25, 0.5, and 1.0 μg/well) were cotransfected with 0.1 μg of pe1b-LRE2.1/well and 1.0 μg of pcDNA3.1-ORF50/Lyta or the pcDNA3.1 control vector/well, respectively. To normalize the total amount of DNA, pCGN was used.
In all transfection assays, after 48 h in culture, cells were harvested and the luciferase activity was assayed. pCMV-β-gal (Clonetech, Palo Alto, Calif.) expression was used for normalization.
Luciferase and β-galactosidase assays.
Cell lysate was prepared in 50 μl of the reporter lysis buffer (Promega). Ten microliters of the lysate and 50 μl of luciferase substrate buffer were mixed in a measuring tube, and the luciferase activity expressed in relative luciferase units was measured immediately with a luminometer (LUMAT LB 9507: EG & G Berthold, Bad Wildbad, Germany).
The β-galactosidase activity was measured with a β-galactosidase assay system (Clonetech), according to the manufacturer's protocol.
EMSA.
Synthetic oligonucleotides were annealed at room temperature after incubation at 80°C in the annealing buffer containing 2 mM Tris-HCl (pH 7.5), 10 mM MgCl2, and 50 mM NaCl. Single-stranded regions of the annealed double-stranded DNA were filled in with [α-32P]dCTP (∼3,000 Ci/mmol; Amersham Pharmacia Biotech, Buckinghamshire, United Kingdom) using the Klenow fragment (Takara Shuzo). Nuclear extracts were prepared from BCBL-1 cells that were untreated or treated with 25 ng of TPA (Sigma, St. Louis, Mo.)/ml for 48 h. The Oct-1 consensus oligonucleotide was purchased from Promega to be used in competition analysis. Rabbit anti-Oct-1 polyclonal antibodies (Santa Cruz Biotechnology, Inc., Santa Cruz, Calif.) and a mouse anti-ORF50/Lyta monoclonal antibody (generated in our laboratory) were used for the supershift analysis.
RESULTS
Identification of cis-acting elements required for ORF50/Lyta autoregulation.
To examine whether ORF50/Lyta upregulates its own expression, we performed a transient transfection assay with pGL3-FL (−914), which contained a 950-bp fragment upstream of the ORF50/Lyta coding region, including the transcription start site (18), and with either pcDNA3.1-ORF50/Lyta or pcDNA3.1, as the effector plasmids. Both in Ramos cells and in BCBL-1 cells, ORF50/Lyta transactivated its own promoter (Fig. 1B and C). The increase in reporter activity was about four- and eightfold in Ramos and BCBL-1 cells, respectively. The moderate change in value was probably due to the high background activity of the ORF50/Lyta promoter region in these cell lines in the absence of TPA. And these data were always consistent and independent of the transfection method: electroporation and Superfect transfection reagent (Qiagen). Actual transfection efficiency was 1 to 5% in electroporation and about 1% in Superfect.
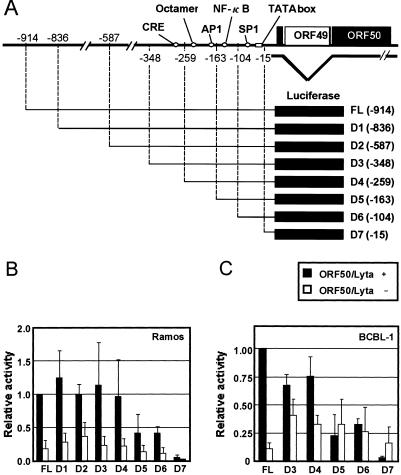
FIG. 1.
Autoregulation of ORF50/Lyta. (A) Schematic representation of the ORF50/Lyta promoter region and its 5′ deletion mutants. (B and C) Relative luciferase activity of 5′ deletion mutants. For Ramos (B) and BCBL-1 (C) cells, a cotransfection assay was performed with the full-length pGL3-FL (FL) or its 5′ deletion mutant constructs (D1 to D7) along with pcDNA3.1-ORF50/Lyta or pcDNA3.1 vector. After 48 h in culture, cells were harvested and assayed. The solid and open bars indicate the relative activities with pcDNA3.1-ORF50/Lyta and pcDNA3.1 vector, respectively. pCMV-β-gal was cotransfected for normalization of transfection efficiency. These experiments were performed at least three times, and the relative mean values with the standard deviations are depicted in comparison with results for FL.
To identify the critical element for the autoregulation, a series of 5′ deletion mutants of pGL3-FL (−914) was constructed. The transcriptional start site of ORF50/Lyta was designated +1, as described elsewhere (18). We tested seven mutants (D1 to D7, shown in Fig. 1A). The activity of the mutants was compared with that of the full-length promoter in the presence of pcDNA3.1-ORF50/Lyta (Fig. 1B and C). Deletion of bp −914 to −259 had little effect, but further deletion up to −163 bp resulted in a remarkable decrease in the luciferase activity in all cell lines (Fig. 1B and C). Deletion mutant D7 (up to −15 bp) showed almost no activity, probably because the putative TATA box was lacking in this promoter (nt 71527 to 71535; GenBank accession no. U75698). In this respect, the site-directed mutagenesis of the TATA box caused the elimination of the luciferase activity (data not shown).
For further determination of the cis-acting element in the region between −259 and −163 bp, we constructed reporter plasmids with three tandemly arranged copies of 40-bp fragments (LRE1 to LRE3) that spanned this interval (as shown in Table 1). These reporters contained the adenovirus E1b minimal TATA box (TATATAA) followed by a luciferase gene (Fig. 2A). In 293L and BCBL-1 cells, although LRE1 also had significant levels of activation, LRE2 resulted in the highest increase in activity of the three fragments tested (Fig. 2B and C, left), suggesting that LRE2 contained the most important element for the ORF50/Lyta transactivation. That was why we focused on LRE2 in this study.
Thus, we constructed three reporters (pe1b-LRE2.1, -2.2, and -2.3) to determine the responsible region in detail (Table 1 and Fig. 2A). The transfection assays indicated that LRE2.1 had a much higher level of activity than did the other reporter plasmids, both in 293L and BCBL-1 cells (Fig. 2B and C, right).
ORF50/Lyta activates its own promoter through a non-DNA-binding mechanism.
The results of the transfection assays indicated that the region from position −227 to −208 contributed greatly to the transactivation by ORF50/Lyta. To determine whether ORF50/Lyta recognized and bound to this specific sequence, we performed an EMSA using an α-32P-labeled double-stranded oligonucleotide of LRE2.1. Nuclear extract was prepared from BCBL-1 cells with or without TPA treatment, which induced ORF50/Lyta expression. One specific DNA-protein complex was detected with extracts from uninduced cells (Fig. 3, lane 1). The competition assay demonstrated that this was a specific shifted band (Fig. 3, lanes 2 and 5). Unexpectedly, however, no increase in expression was observed in TPA-treated extract over that of the nontreated extract (Fig. 3, lanes 1 and 4). The addition of anti-ORF50/Lyta antibody had no effect on the mobility of the band (Fig. 3, lane 8). These results showed that this complex was not involved in the direct binding of ORF50/Lyta.
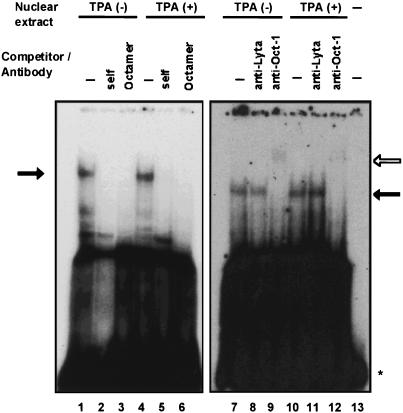
FIG. 3.
EMSA for the LRE2.1 fragment of the ORF50/Lyta promoter. Nuclear extracts derived from BCBL-1 with or without TPA (25 ng/ml) were used. [α-32P]dCTP-labeled LRE2.1 was used as a probe. Unlabeled oligonucleotides were added in lanes 2 and 5 (cold LRE2.1) and 3 and 6 (consensus octamer-binding sequence) as a competitor (left panel) at a 100-fold excess. For the right panel, a mouse monoclonal antibody against ORF50/Lyta (lanes 8 and 11) and rabbit polyclonal antibodies against Oct-1 (lanes 9 and 12) were added. The solid arrow shows a specific Oct-1–DNA complex, and the open arrow shows the supershifted band with the anti-Oct-1 antibodies. The asterisk shows free probe (lane 13).
Because the same shifted band appeared with the TPA-treated and untreated nuclear extract of BCBL-1 cells, it seemed possible that the autoregulation of ORF50/Lyta might be mediated through a cellular factor whose expression levels were not affected by TPA stimulation. Computer analysis revealed a putative octamer-binding site in LRE2.1. In competition studies, the specific band disappeared when an oligonucleotide with the octamer-binding consensus (ATGCAAAT) was added to the mixture (Fig. 3, lanes 3 and 6). In addition, incubation with rabbit anti-Oct-1 polyclonal antibodies supershifted the band (Fig. 3, lanes 9 and 12). Therefore, the complex seen in the EMSA contained the Oct-1 protein, which suggested that this factor was involved in the transactivation by ORF50/Lyta.
Mutations in the octamer-binding sequence of LRE2.1 disabled the responsiveness of the reporter to ORF50/Lyta transactivation.
If the octamer-binding sequence were really required for ORF50/Lyta transactivation, the mutant octamer would not be responsible. The mutant reporter construct LRE2.1-mt, in which AT was changed to CG at the end of the motif (Fig. 4A), and pcDNA3.1-ORF50/Lyta were cotransfected into 293L cells, and luciferase assays were performed. As shown in Fig. 4B, the mutant reporter showed a crucial defect in transactivation by ORF50/Lyta.
In an EMSA, the unlabeled LRE2.1-mt fragment could not compete for the binding of Oct-1, and the labeled mutant could not bind with the factor (Fig. 4C). In all likelihood, the mutant reporter was not activated by ORF50/Lyta because Oct-1 could not bind with the mutant sequence. These data suggest that Oct-1 interacted with the responsive element to recruit ORF50/Lyta.
To investigate the responsibility of the octamer-binding sequence for the autoregulation of ORF50/Lyta, a site-directed mutant reporter in FL configuration was constructed. The mutant reporter, designated pGL3-FL-mtoct, contained the same mutation of LRE2.1-mt at the corresponding sites. In 293L cells, a cotransfection assay was carried out, and results were compared with the activation value of the wild type. As shown in Fig. 4D, the pGL3-FLmtoct was activated by the factor of 3.4, while the wild type was 6.2 times as active as in the absence of ORF50/Lyta. Therefore, the autoregulation of ORF50/Lyta was mostly dependent on the octamer-binding sequence, though not completely.
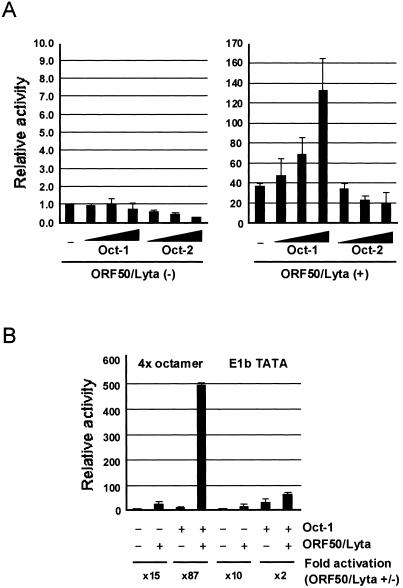
Expression of Oct-1 enhances the transactivation of LRE2.1.
The transfection assay with pe1b-LRE2.1-mt, which was not activated by ORF50/Lyta, suggested that Oct-1 could be involved in transactivation by ORF50/Lyta. To confirm the participation of Oct-1, cotransfection of ORF50/Lyta and an Oct-1 expression vector with pe1b-LRE2.1 was performed. The transactivation by ORF50/Lyta increased with the addition of the Oct-1 expression vector in a dose-dependent manner, whereas the basal activity did not (Fig. 5A, left). In contrast, Oct-2, which is another POU transcription factor that shares the same octamer-binding sequence, did not augment ORF50/Lyta transactivation. These results indicate that ORF50/Lyta interacted specifically with Oct-1.
FIG. 5.
Effects of Oct-1 and -2 overexpression on the transactivation of LRE2.1 (A) and 4× octamer-binding consensus (B) by ORF50/Lyta in 293L cells. (A) The octamer-binding protein expression plasmid, pCGNoct-1 or -2 (none, 0.25, 0.5, and 1.0 μg), and 1.0 μg of pcDNA3.1-ORF50/Lyta or the pcDNA3.1 control vector were cotransfected with 0.1 μg of pe1b-LRE2.1 to 293L cells. The pCGN was used to normalize the total amount of DNA. (B) pCGNoct-1 (1 μg) and pcDNA-ORF50/Lyta (1 μg) were cotransfected with 0.1 μg of p4× oct-e1b-luc or pe1b-TATA-luc. The transfection assay is described in the legend for Fig. 2. The results are shown as fold activation of that in the absence of these effectors.
Furthermore, upregulation of the octamer consensus sequence derived from the immunoglobulin promoter region was tested. Whereas overexpression of Oct-1 resulted in minimal activation, cotransfection of ORF50/Lyta and the Oct-1 expression vector resulted in a significant degree of activation (about 480-fold; Fig. 5B). Both reporter assays indicated that ORF50/Lyta interacted with Oct-1 directly or indirectly, which caused significant and specific activation.
DISCUSSION
KSHV ORF50/Lyta is believed to be a positional and functional homolog of the EBV BRLF1 (Rta) gene, and it activates KSHV early gene promoters more than 10-fold above the basal expression level (6, 17, 18, 27). This upregulation is thought to lead to the lytic phase of viral reproduction and, ultimately, to the production of mature viral particles (12, 32). Thus, ORF50/Lyta is a key lytic inducer. We previously identified ORF50/Lyta response elements in the ORF K9 promoter and found consensus binding sites for SP-1 in the minimal responsive element (6). In contrast, the data shown here did not indicate that SP-1 was involved in ORF50/Lyta autoregulation.
EMSA revealed that Oct-1 bound with LRE2.1, which was identified as ORF50/Lyta responsive element in the ORF50/Lyta promoter region. Furthermore, transfection assays with a mutant reporter and cotransfection with an Oct-1 expression vector indicated that Oct-1 was required for transactivation of pe1b-LRE2.1 by ORF50/Lyta (Fig. 4B). Disruption of the octamer-binding sequence (LRE2.1) in the ORF50/Lyta promoter did not lose its activity completely, which might suggest that LRE1 is another cis element to be investigated. Previous study has shown that ORF50/Lyta interacted with a cellular transcriptional coactivator, CREB-binding protein, which has histone transferase activity and interacts with some transactivators, such as CREB and c-Jun (13), suggesting that CREB-binding protein might be involved in the autoregulation of ORF50/Lyta.
Here we have shown the interaction of Oct-1 and ORF50/Lyta autoregulation. Oct-1 is a member of the POU family. Both Oct-1 and -2 can specifically interact with the octamer-binding sequence, ATGCAAAT (31). While the Oct-1 protein is ubiquitously expressed, Oct-2 appears to be restricted to B cells (8). Oct-1 itself does not have a strong transactivation activity and is involved in cell cycle regulation of the human histone H2b gene (15) and constitutive expression of small nuclear RNA (35). A B-cell-specific coactivator, OCA-B (OBF-1 or BOB-1) forms a complex with Oct-1 or Oct-2 to upregulate the immunoglobulin promoter (19). Thus, in the absence of OCA-B, Oct-1 should have no effect on the initial expression of ORF50/Lyta.
Some viral proteins, such as herpes simplex virus type 1 (HSV-1) VP16 and varicella-zoster virus (VZV) ORF10, form a complex with Oct-1 to activate expression of their target genes (22, 30). In these cases, the interaction with Oct-1 is so tight that the complexes are easily observed by EMSA (7, 19, 22, 30). An ORF50/Lyta–Oct-1 complex was not observed by EMSA, which suggested that ORF50/Lyta did not associate with octamer sequence strongly.
The cotransfection assay suggested that Oct-1 enhanced the transactivation by ORF50/Lyta (Fig. 5). Furthermore, the reporter containing a 4× octamer-binding consensus was activated by ORF50/Lyta in the presence of excess Oct-1 (Fig. 5B). It suggests that cellular promoters controlled by octamer binding sequence, such as immunoglobulin and histone H2b, might be active in the viral lytic cycle. In this context, ORF50/Lyta may disturb the regulation of cellular genes, affecting the health of the cells.
Previously, it was reported that gamma interferon induces reactivation of KSHV in cell culture (4, 20). However, the induction by gamma interferon in vitro was limited and not as strong as that by chemical reagents such as TPA. Thus, the autoregulation of ORF50/Lyta may be an important event for the virus in vivo. Autoregulation of IE activators is not restricted to KSHV; for example, the BZLF1 (Zta) protein of EBV, which is a key factor for inducing EBV's lytic replication, has an autoregulatory mechanism for amplifying its own expression (11), which may be important for efficient induction of the lytic cycle. The human cytomegalovirus IE1 protein and the simian virus 40 large-T antigen have also been reported to positively autoregulate their own expression (25, 29). On the other hand, some proteins, such as the E1A gene product of adenovirus type 2 and the IE175 (ICP 4) protein of HSV-1, negatively regulate their own expression (1, 26). It is likely that each virus has developed different strategies to produce progeny efficiently. Although negative regulation in KSHV lytic replication has not yet been observed, autoregulation is an important mechanism for controlling the expression of the viral proteins and ultimately of virus production.
In conclusion, we showed that ORF50/Lyta autoregulated its own expression through the octamer-binding sequence, which bound Oct-1. This is the first report that indicates an interaction between KSHV ORF50/Lyta and a cellular transcription factor. Our results provide new information relevant to the reactivation of KSHV and to the molecular mechanisms involved in the transactivation of ORF50/Lyta.
ADDENDUM
We propose referring to the KSHV ORF50 gene product as Lyta, and used ORF50 and Lyta side by side in this article, because the KSHV ORF50 gene product was distinct from EBV BRLF1/Rta in terms of function (a key inducer of lytic replication) and structure (bZip-serine/threonine-rich region–acidic region).
ACKNOWLEDGMENTS
We thank Winship Herr (Cold Spring Harbor Laboratory) for the Oct-1 and Oct-2 expression vectors.
This work was supported by Japan Ministry of Education grants 09CE2007 to K.Y. and 12670282 to K.U.
REFERENCES
- 1.Borrelli E R, Hen R, Chambon P. Adenovirus-2 E1A products repress enhancer-induced stimulation of transactivation. Nature. 1984;312:608–612. doi: 10.1038/312608a0. [DOI] [PubMed] [Google Scholar]
- 2.Cesarman E, Chang Y, Moore P S, Said J W, Knowles D M. Kaposi's sarcoma-associated herpesvirus-like DNA sequence in AIDS-related body-cavity-based lymphomas. N Engl J Med. 1995;332:1186–1191. doi: 10.1056/NEJM199505043321802. [DOI] [PubMed] [Google Scholar]
- 3.Chan S R, Chandran B. Characterization of human herpesvirus 8 ORF59 protein (PF8) and mapping of the processivity and viral DNA polymerase-interacting domains. J Virol. 2000;74:10920–10929. doi: 10.1128/jvi.74.23.10920-10929.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang J, Renne R, Ditter D, Ganem D. Inflammatory cytokines and the reactivation of Kaposi's sarcoma-associated herpesvirus lytic replication. Virology. 2000;266:17–25. doi: 10.1006/viro.1999.0077. [DOI] [PubMed] [Google Scholar]
- 5.Chang Y, Cesarman E, Pessin M S, Lee F, Culpeper J, Knowles D M, Moore P S. Identification of herpes-like DNA sequence in AIDS-associated Kaposi's sarcoma. Science. 1994;266:1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- 6.Chen J, Ueda K, Sakakibara S, Okuno T, Yamanishi K. Transcriptional regulation of the Kaposi's sarcoma-associated herpesvirus viral interferon regulatory factor gene. J Virol. 2000;74:8623–8634. doi: 10.1128/jvi.74.18.8623-8634.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cleary A M, Herr W. Mechanisms for flexibility in DNA sequence recognition and VP16-induced complex formation by the Oct-1 POU domain. Mol Cell Biol. 1995;15:2090–2100. doi: 10.1128/mcb.15.4.2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clerc R G, Corcan L M, LeBowitz J H, Baltimore D, Sharp P A. The B-cell-specific Oct-2 protein contains POU box- and homeo box-type domains. Genes Dev. 1988;2:1570–1581. doi: 10.1101/gad.2.12a.1570. [DOI] [PubMed] [Google Scholar]
- 9.Cormack B. Directed mutagenesis using the polymerase chain reaction. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1997. pp. 8.5.1–8.5.9. [Google Scholar]
- 10.Deng H, Young A, Sun R. Auto-activation of the rta gene of human herpesvirus-8/Kaposi's sarcoma-associated herpesvirus. J Gen Virol. 2001;81:3043–3048. doi: 10.1099/0022-1317-81-12-3043. [DOI] [PubMed] [Google Scholar]
- 11.Flemington E, Speck S H. Autoregulation of Epstein-Barr virus putative lytic switch gene BZLF1. J Virol. 1990;64:1227–1232. doi: 10.1128/jvi.64.3.1227-1232.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gradoville L, Gerlach J, Grogan E, Shedd D, Nikiforow S, Metroka C, Miller G. Kaposi's sarcoma-associated herpesvirus open reading frame 50/Rta protein activates the entire viral lytic cycle in the HH-B2 primary effusion lymphoma cell line. J Virol. 2000;74:6207–6212. doi: 10.1128/jvi.74.13.6207-6212.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gwack Y, Byun H, Hwang S, Lim C, Choe J. CREB-binding protein and histone deacetylase regulate the transcriptional activity of Kaposi's sarcoma-associated herpesvirus open reading frame 50. J Virol. 2001;75:1909–1917. doi: 10.1128/JVI.75.4.1909-1917.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kedes D H, Operskalski E, Busch M, Kohn R, Flood J, Ganem D. The seroepidemiology of human herpesvirus 8 (Kaposi's sarcoma-associated herpesvirus): distribution of infection in KS risk groups and evidence for sexual transmission. Nat Med. 1996;2:918–924. doi: 10.1038/nm0896-918. [DOI] [PubMed] [Google Scholar]
- 15.LaBella F, Sive H L, Roeger R G, Heint N. Cell-cycle regulation of a human histone H2b gene is mediated by the H2b subspecific consensus element. Genes Dev. 1988;2:32–39. doi: 10.1101/gad.2.1.32. [DOI] [PubMed] [Google Scholar]
- 16.Liu S, Pavlova I V, Virgin IV H W, Speck S H. Characterization of gammaherpesvirus 68 gene 50 transactivation. J Virol. 2000;74:2029–2037. doi: 10.1128/jvi.74.4.2029-2037.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lukac D M, Renne R, Kirshner J R, Ganem D. Reactivation of Kaposi's sarcoma-associated herpesvirus infection from latency by expression of the ORF50 transactivator, a homolog of the EBV R protein. Virology. 1998;252:304–312. doi: 10.1006/viro.1998.9486. [DOI] [PubMed] [Google Scholar]
- 18.Lukac D M, Kirshner J R, Ganem D. Transcriptional activation by the product of open reading frame 50 of Kaposi's sarcoma-associated herpesvirus is required for lytic viral reactivation in B cells. J Virol. 1999;73:9348–9361. doi: 10.1128/jvi.73.11.9348-9361.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luo Y, Roeder R G. Cloning, functional characterization, and mechanism of action of the B-cell-specific transcriptional coactivator OCA-B. Mol Cell Biol. 1995;15:4115–4124. doi: 10.1128/mcb.15.8.4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mercader M, Taddeo B, Panella J R, Chandran B, Nickoloff B J, Foreman K E. Induction of HHV-8 lytic cycle replication by inflammatory cytokines produced by HIV-1-infected T cells. Am J Pathol. 2000;156:1961–1971. doi: 10.1016/S0002-9440(10)65069-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller G, Heston L, Grogan E, Gradoville L, Rigsby M, Sun R, Shedd D, Kushnaryov V M, Grossberg S, Chang Y. Selective switch between latency and lytic replication of Kaposi's sarcoma-associated herpesvirus and Epstein-Barr virus in dually infected body cavity lymphoma cells. J Virol. 1997;71:314–324. doi: 10.1128/jvi.71.1.314-324.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moriuchi H, Moriuchi M, Cohen J I. Proteins and cis-acting elements associated with transactivation of the varicella-zoster virus (VZV) immediate-early gene 62 promoter by VZV open reading frame 10 protein. J Virol. 1995;69:4693–4701. doi: 10.1128/jvi.69.8.4693-4701.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ragoczy T, Heston L, Miller G. The Epstein-Barr virus Rta protein activates lytic cycle genes and can disrupt latency in B lymphocytes. J Virol. 1998;72:7978–7984. doi: 10.1128/jvi.72.10.7978-7984.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ragoczy T, Miller G. Role of Epstein-Barr virus Rta protein in activation of distinct classes of viral lytic cycle genes. J Virol. 1999;73:9858–9866. doi: 10.1128/jvi.73.12.9858-9866.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reed S T, Stark G R, Alwine J C. Autoregulation of simian virus 40 gene A by T antigen. Proc Natl Acad Sci USA. 1976;73:3083–3087. doi: 10.1073/pnas.73.9.3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roberts M S, Boundy A, O'Hare P, Pizzorno M C, Ciufo D M, Hayward G S. Direct correlation between a negative autoregulatory response element at the cap site of the herpes simplex virus type 1 IE175 (α4) promoter and a specific binding site for the IE175 (ICP4) protein. J Virol. 1988;62:4307–4320. doi: 10.1128/jvi.62.11.4307-4320.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seaman W T, Ye D, Wang R X, Hale E E, Weisse M, Quinlivan E B. Gene expression from the ORF50/K8 region of Kaposi's sarcoma-associated herpesvirus. Virology. 1999;263:436–449. doi: 10.1006/viro.1999.9963. [DOI] [PubMed] [Google Scholar]
- 28.Soulier J, Grollet L, Oksenhendler E, Cacoub P, Cazals-Hatem D, Babinet P, d'Agay M F, Clauvel J P, Raphael M, Degos L. Kaposi's sarcoma-associated herpesvirus-like DNA sequence in multicentric Castleman's disease. Blood. 1995;86:1276–1280. [PubMed] [Google Scholar]
- 29.Stenberg R M, Stinski M F. Autoregulation of the human cytomegalovirus major immediate-early gene. J Virol. 1985;56:676–682. doi: 10.1128/jvi.56.3.676-682.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stern S, Tanaka M, Herr W. The Oct-1 homeodomain directs formation of a multiprotein-DNA complex with the HSV transactivator VP16. Nature. 1989;341:624–630. doi: 10.1038/341624a0. [DOI] [PubMed] [Google Scholar]
- 31.Sturm R A, Das G, Herr W. The ubiquitous octamer-binding protein Oct-1 contains a POU domain with a homeo box subdomain. Genes Dev. 1988;2:1582–1599. doi: 10.1101/gad.2.12a.1582. [DOI] [PubMed] [Google Scholar]
- 32.Sun R, Lin S-F, Gradoville L, Yuan Y, Zhu F, Miller G. A viral gene that activates lytic cycle expression of Kaposi's sarcoma-associated herpesvirus. Proc Natl Acad Sci. 1998;95:10866–10871. doi: 10.1073/pnas.95.18.10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sun R, Lin S-F, Staskus K, Gradoville L, Grogan E, Haase A, Miller G. Kinetics of Kaposi's sarcoma-associated herpesvirus gene expression. J Virol. 1999;73:2232–2242. doi: 10.1128/jvi.73.3.2232-2242.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tanaka M, Herr W. Differential transcriptional activation by Oct-1 and Oct-2: interdependent activation domains induce Oct-2 phosphorylation. Cell. 1990;60:375–386. doi: 10.1016/0092-8674(90)90589-7. [DOI] [PubMed] [Google Scholar]
- 35.Tanaka M, Lai J S, Herr W. Promoter-selective activation domains in Oct-1 and Oct-2 direct differential activation of an snRNA and mRNA promoter. Cell. 1992;68:755–767. doi: 10.1016/0092-8674(92)90150-b. [DOI] [PubMed] [Google Scholar]
- 36.Thurau M, Whitehouse A, Wittemann S, Meredith D, Fickenscher H. Distinct transcriptional and functional properties of the R transactivator gene orf50 of the transforming herpesvirus saimiri strain C488. Virology. 2000;268:167–177. doi: 10.1006/viro.1999.0167. [DOI] [PubMed] [Google Scholar]
- 37.Whitehouse A, Cooper M, Hall K T, Meredith D. The open reading frame (ORF) 50a gene product regulates ORF57 gene expression in herpesvirus saimiri. J Virol. 1998;72:1967–1973. doi: 10.1128/jvi.72.3.1967-1973.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu T, Usherwood E J, Stewart J P, Nash A A, Sun R. Rta of murine gammaherpesvirus 68 reactivates the complete lytic cycle from latency. J Virol. 2000;74:3659–3667. doi: 10.1128/jvi.74.8.3659-3667.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zalani S, Holley-Guthrie E, Kelly S. Epstein-Barr virus latency is disrupted by the immediate-early BRLF1 protein through a cell-specific mechanism. Proc Natl Acad Sci USA. 1996;93:9194–9199. doi: 10.1073/pnas.93.17.9194. [DOI] [PMC free article] [PubMed] [Google Scholar]