Abstract
Termination of mRNA synthesis in vesicular stomatitis virus (VSV), the prototypic rhabdovirus, is controlled by a 13-nucleotide gene end sequence which comprises the conserved tetranucleotide 3′-AUAC-5′, the U7 tract and the intergenic dinucleotide. mRNAs terminated at this sequence possess 100- to 300-nucleotide-long 3′ poly(A) tails which are thought to result from polymerase slippage (reiterative transcription) by the VSV polymerase on the U7 tract. Previously we determined that in addition to the AUAC tetranucleotide, the U7 tract was an essential signal in the termination process. Shortening or interrupting the U7 tract abolished termination. These altered U tracts also prevented the polymerase from performing reiterative transcription necessary for generation of the mRNA poly(A) tail and thus established seven residues as the minimum length of U tract that allowed reiterative transcription to occur. In this study we investigated whether sequences other than the essential U7 tract are involved in controlling polymerase slippage. We investigated whether the AUAC tetranucleotide affected the process of reiterative transcription by analyzing the nucleotide sequence of RNAs transcribed from altered subgenomic templates and infectious VSV variants. The tetranucleotide was found to regulate reiterative transcription on the U7 tract. The extent of polymerase slippage was governed not by specific tetranucleotide sequences but rather by nucleotide composition such that slippage occurred when the tetranucleotide was composed of A or U residues but not when it was composed of G or C residues. This suggested that polymerase slippage was controlled, at least in part, by the strength of base pairing between the template and nascent strands. Further data presented here indicate that the tetranucleotide contains both a signal that directs the VSV polymerase to slip on the downstream U7 tract and also a signal that directs a slipping polymerase to terminate mRNA synthesis.
Vesicular stomatitis virus (VSV) is one of the best-studied and best-characterized members of the order Mononegavirales, a group which comprises RNA viruses having single-stranded genomes of negative-sense polarity. The 11,161-nucleotide VSV genome is composed of five genes flanked by leader and trailer sequences arranged in the order 3′ (leader)-N-P-M-G-L-(trailer) 5′. The VSV RNA-dependent RNA polymerase (RdRp), comprised of the catalytic L protein component along with the P protein cofactor (10), binds to the nucleocapsid (N) protein encapsidated RNA template at or near the genomic 3′ terminus (9). The RdRp traverses the VSV template in the 3′-to-5′ direction, synthesizing a 47-nucleotide leader RNA, which is neither 5′ capped nor 3′ polyadenylated (6, 7, 19), and five capped and polyadenylated mRNAs from each of the VSV genes in a sequential fashion (1, 2, 38). The five VSV mRNAs are not transcribed with equal abundance; instead, due to a poorly understood process known as attenuation, genes nearer to the template 3′ end are transcribed more frequently than those more 5′ (18). There is much experimental evidence to indicate that access to any downstream gene is possible only when mRNA from the gene directly upstream has been transcribed and terminated by the same polymerase, and for this reason the model which describes VSV mRNA synthesis is often referred to as stop-start transcription (1–4, 17).
Transcription by the VSV RdRp is controlled by sequences within the leader region, sequences at the leader-N gene junction, and sequences of the gene junctions (3, 4, 20, 24, 33, 35, 41). The gene junctions comprise 23 highly conserved nucleotides made up of short sequences located at the end of an upstream gene and the beginning of a downstream gene. These sequences direct the VSV polymerase to polyadenylate and terminate the upstream mRNA, to initiate and cap the downstream mRNA, and are also responsible, at least in part, for the process of attenuation (3, 4, 17, 27, 30, 31, 34–36).
The signal within the gene junction which specifies termination has been well studied and has been localized to 13 nucleotides comprising a conserved tetranucleotide 3′-AUAC-5′, a tract of seven uridylates found at the end of each gene (the U7 tract), and a dinucleotide 3′-GA-5′ or 3′-CA-5′ known as the intergenic sequence (3, 4, 17) which is not observed in either up- or downstream mRNA products. This wild-type termination signal is highly efficient, and so readthrough RNAs which arise when a termination signal is ignored are generated with a frequency of only 1 to 3% (18). Gene junction sequences downstream of the termination signal have little effect on termination and are involved primarily in signaling initiation and correct processing of the nascent downstream mRNA 5′ end (3, 35, 36).
Because of its sequence and its position at the end of each gene, the U7 tract is thought to provide the template used to generate the 100- to 300-nucleotide long 3′ poly(A) tails that are characteristic of VSV mRNAs (13–15, 23, 31). To account for the size of these tails relative to the short U7 tract, the RdRp is believed to perform a cycle which involves realignment of the template and polymerase such that the nascent strand moves backward relative to the RdRp polymerization site, followed by correctly templated RNA synthesis. Repeated cycles of these two steps allow synthesis of RNAs containing nontemplated nucleotides; this process is known as reiterative transcription or polymerase slippage. Supporting the suggestion that reiterative transcription is responsible for the generation of VSV poly(A) tails, the VSV RdRp is known to perform this mode of transcription during other aspects of RNA synthesis. Examples include synthesis of large poly(A) tracts in readthrough RNAs synthesized in vitro and in vivo (13, 14, 16, 32), generation of homopolymeric (U) sequences from a subgenomic template having an A7 tract (3), and extensive copying of the sequence 3′-UUUUUAA-5′ during G mRNA transcription (5).
We previously investigated how changes within the conserved gene junction sequence affected transcription termination using VSV subgenomic templates in a baby hamster kidney (BHK) cell-based assay system (3). We found that termination of VSV mRNA synthesis depended implicitly on the presence of the conserved U7 tract and that shortening the tract to U6 or interrupting it with a non-U nucleotide abolished termination, which led to exclusive synthesis of a readthrough RNA (3). To determine why these changes abolished termination, we investigated whether the altered U tract templates affected the ability of the VSV RdRp to perform reiterative transcription; we found that slippage by the VSV RdRp was abrogated by these changes and concluded that slippage to form the mRNA poly(A) tail was a crucial step in the termination process. Furthermore, we demonstrated that slippage alone was not sufficient to cause termination, leading us to suggest that the VSV gene junction contained a signal which was required to allow a slipping polymerase to terminate synthesis of a corresponding mRNA.
In work described here, we have extended our previous studies and investigated whether sequences outside the U7 tract can affect polymerase slippage. Our results show that the upstream 3′-AUAC-5′ tetranucleotide of the gene junction exerts a major influence on whether the VSV polymerase can slip on the downstream U7 tract. Furthermore, our results suggest that the tetranucleotide sequence contains two or more signals that are required in combination for efficient termination: an element that allows initial polymerase slippage on the U7 tract, and an element that allows termination of an RNA which is being reiteratively transcribed.
MATERIALS AND METHODS
Plasmid constructions.
Plasmid p8(+)NP, described previously (3, 4), directed bacteriophage T7 RNA polymerase to transcribe a positive-sense VSV subgenomic template containing the wild-type leader and trailer regions flanking two transcription units separated by a wild-type N-P gene junction (Fig. 1A). This plasmid was constructed by positioning nucleotides 1 to 215, 1236 to 1686, and 10897 to 11161 of the VSV genome between a T7 RNA polymerase promoter and the cDNA encoding the self-cleaving ribozyme from the antigenomic strand of hepatitis delta virus. Following ribozyme cleavage, the T7 RNA polymerase-transcribed RNA had a 3′ end which corresponded exactly with the 3′ VSV genomic terminus but a 5′ end that had two non-VSV nucleotides (GG). As described previously (3, 4), alterations were engineered into the gene junction sequence of p8(+)NP by exchanging a restriction fragment spanning the gene junction sequence for a PCR-generated fragment into which specific changes had been incorporated. All alterations were confirmed by sequence analysis.
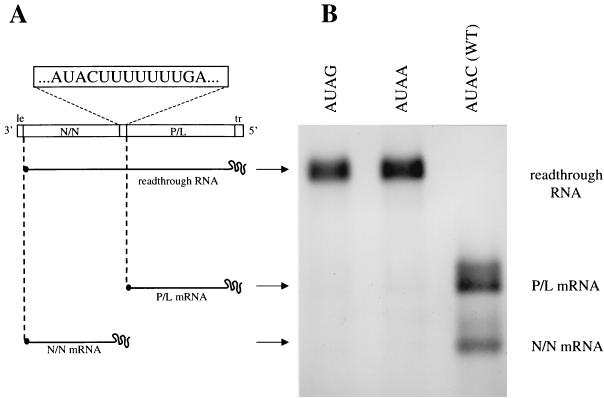
FIG. 1.
(A) Diagrammatic representation of the subgenomic template transcribed from plasmid p8(+)NP and the RNA products that it encodes. (A) The RNA template transcribed from p8(+)NP contains wild-type leader (le) and trailer (tr) regions flanking two transcriptional units, N/N and P/L, separated by the wild-type N-P gene junction. Plasmids encoding this and related subgenomic templates were transfected into vTF7-3-infected BHK cells, where they yielded positive-strand RNAs. These RNAs formed active templates by associating with supporting N, P, and L proteins encoded by transfected plasmids. Replication of the antigenomic RNA generated progeny negative-strand genomes which could act as templates for transcription or for generation of additional antigenomes by a further round of replication. All nucleotide alterations described in the text were made within the termination signal, shown 3′-5′ in negative sense. (B) The subgenomic template transcribed from plasmid p8(+)NP transcribes positive-strand RNA transcripts; these are shown as actinomycin D-resistant, metabolically labeled RNAs harvested from BHK cells. The wild-type template (WT) transcribes a very low abundance of readthrough RNA; to indicate its position, RNAs transcribed from templates with tetranucleotide sequences 3′-AUAA-5′ and 3′-AUAG-5′ are shown alongside.
Plasmid pVSV-AUAC directed T7 RNA polymerase to generate a positive-sense antigenomic strand of an infectious recombinant VSV variant in which chloramphenicol acetyltransferase (CAT) and green fluorescent protein (GFP) genes were positioned immediately following the leader region and separated by a wild-type gene junction sequence (see Fig. 7A). This plasmid was constructed by the stepwise insertion of two PCR-generated fragments containing the CAT and GFP coding sequences, respectively, into the previously described plasmid pVSV(+)41 (40). Briefly, the CAT cDNA represented in plasmid pcDNA3CAT was amplified by PCR using oligonucleotide primers CAT1 (5′-GTCACTCGAGTAAATGGAGAAGAAAATCACTGG-3′) and CAT2 (5′-ACGTACTAGTGCAAAAATTACGCCCCGCCCTGCCACTC-3′), and the resulting fragment was digested and inserted at the MseI and XhoI sites of pVSV(+)41 to give plasmid pVSV(+)CAT. The GFP sequence represented in the previously described plasmid pVSV(+)1098 (40) was amplified by PCR using oligonucleotide primer GFP-1 (5′-TGACTCGAGCTACTAGTTATGAAAAAAACTAACAGATATCACCATGAGCAAGGGCGAGGAAC-3′), which annealed to the 3′ end of the GFP gene, and oligonucleotide BstZ17I-n/c (5′-GTAAGAGATGTATACTCAATGTCATCAGGC-3′), which annealed to VSV N gene nucleotides 1017 to 1046. The amplified fragment was digested and inserted into the SpeI and BstZ17I sites of plasmid pVSV(+)CAT. Plasmids pVSV(+)GCGG and pVSV(+)AUAA, designed to express infectious recombinant VSV variants, were derived from plasmid pVSV(+)AUAC by exchanging the SpeI- Bstz17I fragment described above for a fragment in which the wild-type gene junction 3′-AUAC-5′ tetranucleotide sequence was replaced with 3′-AUAA-5′, or 3′-GCGG-5′ using an appropriately designed oligonucleotide primer.
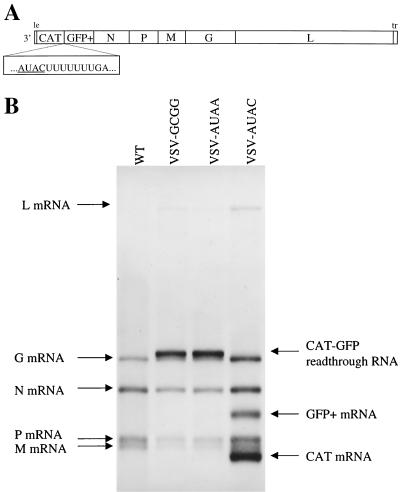
FIG. 7.
Diagrammatic representation of recombinant VSV variants containing two additional transcriptional units, and visualization of the RNAs that these viruses encode. (A) CAT and GFP sequences were inserted into plasmid pVSV(+) to generate the recombinant virus VSV-AUAC, which contained two additional transcriptional units separated by a wild-type gene junction. Altered tetranucleotides 3′-AUAA-5′ and 3′-GCGG-5′ were engineered into the CAT-GFP gene junction of this cDNA (underlined) to generate the VSV recombinants VSV-AUAA and VSV-GCGG. (B) Metabolically labeled, actinomycin D-resistant RNAs transcribed from recombinant VSV variants in infected BHK cells were visualized by agarose-urea gel electrophoresis followed by fluorography. The VSV N, P, M, G, and L mRNAs are marked, along with the CAT, GFP, and bicistronic CAT-GFP readthrough RNAs synthesized from the additional transcriptional units. For reference, the RNAs transcribed by wild-type VSV (WT) are shown alongside.
Transfections.
Both single-round and budded-particle transfections were performed as described previously (26). Briefly, for generating RNAs from single-round transfections, transcription plasmids encoding the VSV N, P, and L proteins along with a specialized plasmid expressing either a subgenomic or infectious virus template were transfected into BHK cells expressing T7 RNA polymerase due to prior infection with vaccinia virus recombinant vTF7-3. For budded-particle transfections, support plasmids for N, P, M, G, and L proteins were transfected into vTF7-3-infected BHK cells along with a subgenomic template-expressing plasmid. Budded particles released into the cell culture supernatant were used to infect a second vTF7-3-infected BHK monolayer previously transfected with N, P, and L support plasmids. This approach was taken to eliminate the generation of RNAs by T7 RNA polymerase and thus ensure that all RNAs which could act as templates for reverse transcriptase-mediated first-strand synthesis were made by the VSV polymerase. Recombinant viruses were recovered from corresponding cDNAs as described previously (42), and the nucleotide sequences of the resulting virus genomes were confirmed by performing sequence analysis on purified virus preparations.
Visualization of VSV specific RNAs.
VSV-specific RNAs generated from subgenomic templates were labeled metabolically 16 h posttransfection by incubating transfected and vTF7-3-infected BHK cells with [3H]uridine (33 μCi/ml) and actinomycin D (10 μl/ml) for 6 h as described previously (26). RNAs generated from infectious recombinant VSV variants were labeled using the same approach except that labeling was performed at 2 h postinfection, for a total of 3 h. Total cell RNA was harvested from monolayers using the RNeasy purification system (Qiagen Inc., Valencia, Calif.), and resulting RNAs were subjected to agarose-urea gel electrophoresis followed by fluorography and autoradiography (26).
Polymerase slippage assays.
RNAs were analyzed for evidence of polymerase slippage using three approaches. The first assay used RNase H-directed cleavage in the presence of oligo(dT) to identify RNAs having additional A residues that arise due to polymerase slippage on a template U tract. This assay is based on the ability of RNase H to recognize and cleave a short RNA-DNA hybrid. Briefly, 3H-labeled RNAs transcribed from subgenomic templates in first-round transfections were heated at 95°C for 1 min with 1 μg of oligo(dT) and then rapidly cooled on ice. Duplicate RNA samples without oligo(dT) were also prepared. After the addition of an equal volume of 2× RNase H buffer (16) and 1 U of RNase H (Gibco-BRL), samples were incubated at 37°C for 20 min, after which they were purified and any resulting cleavage products were visualized by electrophoresis and fluorography as previously described (26).
A second approach detected evidence of polymerase slippage by determining the nucleotide sequence of individually cloned reverse transcriptase PCR (RT-PCR)-derived cDNAs. Briefly, first-strand cDNA synthesis was performed on magnetic bead-selected poly(A)+ readthrough RNAs (Miltenyi Biotech Inc., Auburn, Calif.), using a modified Moloney murine leukemia virus reverse transcriptase (Superscript II; Gibco-BRL) and oligonucleotide pext-xba (5′-GCTCTAGACGAGAATAGGACTTGAGATACTCACG-3′), which annealed downstream of the VSV gene junction, at nucleotides 1417 to 1450. Following purification, the cDNA population was used as template for PCR amplification using oligonucleotides pext-xba and 9121-EcoR1 (5′-AACCGAATTCTGATATGATGCAGTATGCG-3′), which annealed upstream of the VSV gene junction, at VSV nucleotides 1226 to 1254. PCR-amplified fragments were digested with restriction enzymes XbaI and EcoRI and ligated into a similarly digested plasmid pGEM3. Single-nucleotide sequence analysis (using ddA chain terminator) was performed to detect whether slippage had occurred on the U7 tract. This same approach was used to sequence individual readthrough RNAs synthesized by infectious recombinant VSV variants except that first-strand cDNA synthesis was primed using oligonucleotide eco-cloner (5′-ACGAGAATTCGGACCACGCCAGTGAACAGTTCC-5′), which annealed within the 3′ end of the GFP gene. Subsequent PCR amplification utilized oligonucleotides eco-cloner and xba-cloner (5′-TGATCTAGACGTGGCCAATATGACAACTTCTTCGCCCCCG-3′), which annealed within the 5′ end of the CAT gene.
The third slippage assay, termed the RT/Klenow assay, detected evidence of polymerase slippage by determining the spacing of two defined points on either side of the U7 gene junction slippage site (see Figure 4). Briefly, oligonucleotide pext-xba annealed to positive-sense readthrough RNAs downstream of the VSV gene junction to prime for [35S]dATP-labeled first-strand cDNA synthesis using modified Moloney murine leukemia virus reverse transcriptase (Superscript II; Gibco-BRL). After purification (PCR purification column; Qiagen), the labeled cDNA was primed for unlabeled second-strand cDNA synthesis using oligonucleotide 9121 -eco and the Klenow fragment of DNA polymerase I. After repeat purification, cDNAs were digested using restriction enzymes EcoRV and StuI; the resulting DNA fragments were subjected to polyacrylamide gel electrophoresis (PAGE) using a standard sequencing gel and visualized by autoradiography. RNAs derived from infectious recombinant VSV variants were analyzed using the same assay except that first-strand cDNA synthesis was primed using oligonucleotide eco-cloner and second-strand synthesis was primed using oligonucleotide xba-cloner, both described above. The resulting cDNAs were digested with EcoRV and NcoI.
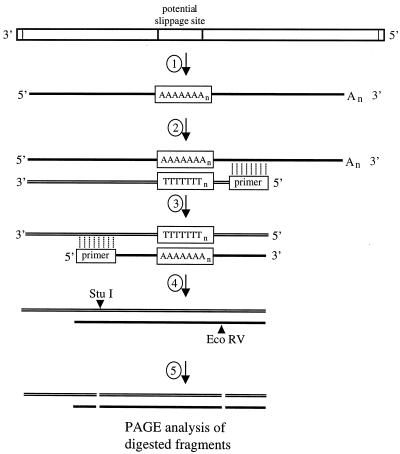
FIG. 4.
Schematic representation of the RT/Klenow slippage assay used to analyze RNAs for evidence of slippage by the VSV polymerase. 1, VSV genomes or subgenomic templates that included a potential slippage sequence (boxed) were used as templates for VSV polymerase-directed RNA synthesis. The slippage sequence comprised a U7 tract preceded by a tetranucleotide sequence which was the site of all specific template alterations. 2, first-strand cDNA synthesis by reverse transcriptase in the presence of [35S]dATP was primed using an oligonucleotide that annealed downstream of the slippage sequence. 3, labeled cDNAs acted as templates for unlabeled second-strand synthesis, using the Klenow fragment of E. coli DNA polymerase I to extend an oligonucleotide primer that annealed to a site corresponding to the upstream side of the slippage site. 4, resulting cDNAs were digested with restriction enzymes EcoRV and StuI. 5, digested cDNAs were denatured and subjected to PAGE and autoradiography to visualize the labeled cDNA fragments.
In each case, total cell RNAs were harvested as described above, treated with RQ1 DNase (Promega Corp., Madison, Wis.), and repurified on an RNeasy column (Qiagen). All RNA samples were confirmed to be free of DNA by PCR (results not shown).
RESULTS
Generation of subgenomic templates for investigation of polymerase slippage.
As described above, we previously established that polymerase slippage on the essential U7 tract of the VSV gene junction was a crucial part of the termination process. We also determined that templates in which the tetranucleotide (3′-AUAC-5′) was changed to AUAA, AUAU, and AUAG were able to signal slippage but were unable to signal termination, indicating that slippage alone was not sufficient to cause termination. To further investigate the process of transcription termination, we wanted to determine whether changes to the tetranucleotide sequence could affect slippage on the downstream U7 tract, and if so, determine the role of the tetranucleotide in regulation of polymerase slippage. To achieve this, we generated a panel of subgenomic VSV templates derived from the previously described plasmid p8(+)NP (3). This plasmid, shown diagrammatically in Fig. 1A, expressed a subgenomic VSV template which contained two cistrons separated by the wild-type VSV N-P gene junction. This template directed the transcription of two major mRNAs generated from the upstream (N/N) and downstream (P/L) cistrons, and also a minor readthrough RNA species synthesized when the termination signals of the gene junction were ignored (Fig. 1B, AUAC). This readthrough RNA is the only transcription product generated from templates unable to terminate transcription, for example, those with the altered tetranucleotide sequences AUAA and AUAG (3), shown in Fig. 1B. To investigate whether alterations within the conserved tetranucleotide sequence of the VSV gene junction affected polymerase slippage, we constructed subgenomic templates which contained the essential wild-type U7 slippage sequence (3) but contained altered tetranucleotide sequences located immediately upstream. We chose nucleotide alterations previously determined to lead to exclusive readthrough RNA synthesis; this was achieved by ensuring that the 5′ C residue of the wild-type 3′-AUAC-5′ tetranucleotide was altered, thus abrogating termination but still allowing polymerase slippage (3).
Our decision to choose this design of template, and thus analyze the process of VSV polymerase slippage by analyzing readthrough RNAs, is based on the finding that VSV polymerase slippage is known to occur during synthesis of readthrough RNAs in response to template sequences (3). Therefore, by determining the extent to which reiterative transcription had occurred as the polymerase encountered the essential U7 slippage template in combination with a specific tetranucleotide sequence, we could assess the ability of the corresponding templates to signal slippage. This method of analysis is similar to that used to determine the sequence requirements of paramyxovirus P gene editing, a related process that is also governed by sequence signals within the template (12).
At this time, we did not address the question of how tetranucleotide alterations affected the degree of polymerase slippage that occurred on a mRNA that was eventually terminated at the altered gene junction. Such studies are complicated by the possibility that the 3′ end of these terminated mRNAs will be modified by either cellular processes or activities of recombinant vaccinia virus.
Rapid analysis of polymerase slippage using an RNase H digestion assay.
To rapidly analyze a large number of subgenomic templates with altered tetranucleotide sequences for their ability to promote polymerase slippage, we developed an assay based on a previous procedure (22) which used RNase H in the presence of oligo(dT) to identify long stretches of poly(A) sequences within transcribed readthrough RNAs. Although qualitative, this assay procedure was attractive for two reasons. First, the assay analyzed all labeled RNAs in an RNA preparation, and thus the sample size was very large. Second, the assay did not depend on additional polymerases commonly used to copy and amplify RNA sequences before their analysis.
By exposing [3H]uridine-labeled RNAs transcribed by T7 RNA polymerase in vTF7-3-infected BHK cells to RNase H in the presence of oligo(dT), we determined that RNAs having A9 or longer tracts were cleaved within this A tract to yield two faster-migrating RNAs (Fig. 2A). In contrast, RNAs with an A6 or A7 tract did not lead to the significant production of faster-mobility bands, indicating that these shorter A tracts were resistant to cleavage. The consequence of this was that a readthrough RNA transcribed by the VSV polymerase from a subgenomic replicon that was a precise copy of the template would not be cleaved by RNase H since the RNA would have precisely seven A residues at the position corresponding to the U7 tract. Conversely, if the VSV polymerase had slipped during transcription of the readthrough RNA, the resulting species would contain additional A residues and thus could be cleaved by RNase H, provided that sufficient A residues were inserted. The replication products generated from these templates are not visible in this system and so do not contribute to the population of cleaved RNAs.
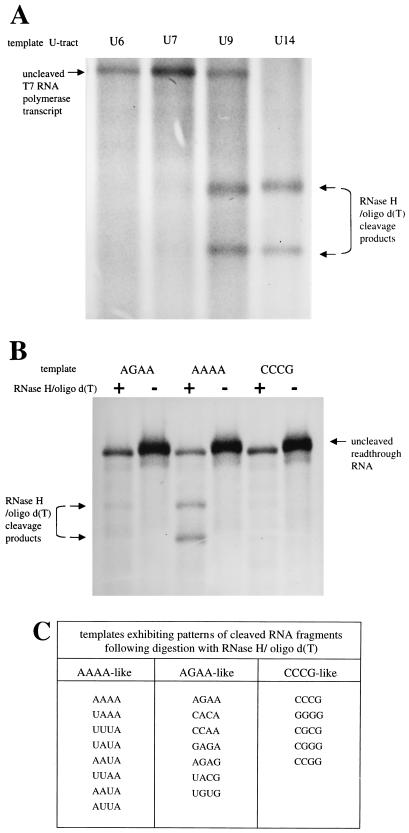
FIG. 2.
Analysis of VSV polymerase slippage by an RNase H-oligo(dT) digestion assay. (A) To determine what length of A tract was cleaved by RNase H in the presence of oligo(dT), positive-strand RNAs were transcribed in BHK cells by T7 RNA polymerase from four plasmids encoding subgenomic templates with gene junction U tracts of 6, 7, 9, and 14 nucleotides. These templates were transcribed to yield positive-stranded RNAs having corresponding A tracts of 6, 7, 9, and 14 nucleotides. Metabolically labeled RNAs were annealed to oligo(dT), digested with RNase H, and then visualized by agarose-urea gel electrophoresis and fluorography. Digestion of the labeled T7 RNA polymerase transcripts with an A6 or A7 tract was insignificant, whereas an expanded tract of nine A's was subjected to extensive digestion, as shown by the presence of two faster-migrating cleavage products. RNAs having A14 tracts were digested to completion. (B) Metabolically labeled RNAs transcribed by the VSV polymerase from three subgenomic templates were subjected to the RNase H-oligo(dT) cleavage assay and visualized by agarose-urea gel electrophoresis and fluorography. The tetranucleotide sequences of gene junctions of these templates are shown above the corresponding lanes, written 3′-5′; + and − denote presence and absence of oligo(dT). As described above, cleavage products following RNase H/oligo(dT) digestion were released only when the A tract contained more than seven residues. This assay demonstrated that VSV polymerase slippage was more prevalent on RNAs transcribed from the subgenomic template with the tetranucleotide sequence 3′-AAAA-5′ than on templates with tetranucleotide sequence 3′-AGAA-5′ or 3′-CCCG-5′. (C). RNAs transcribed from a total of 20 templates with altered tetranucleotide sequences were analyzed by the RNase H/oligo(dT) slippage assay, and the results were tabulated according to a qualitative assessment of whether digestion of the corresponding RNAs resembled the digestion pattern of RNAs transcribed from the three templates shown in panel 2B. AAAA-like corresponds to abundant digestion, CCCG-like corresponds to no detectable digestion, and AGAA-like corresponds to an intermediate level of digestion.
A group of 20 plasmids encoding subgenomic VSV templates with altered tetranucleotide sequences were constructed and individually transfected along with support plasmids expressing VSV N, P, and L proteins into BHK cells previously infected with recombinant vaccinia virus vTF7-3. After labeling of the VSV polymerase-generated RNAs with [3H]uridine in the presence of actinomycin D, these RNAs were subjected to the RNase H cleavage assay and agarose-urea gel analysis described above. Figure 2B shows the results of RNase H-oligo(dT) treatment on RNA populations transcribed from three templates selected from this group having tetranucleotide sequences 3′-AGAA-5′, 3′-AAAA-5′ and 3′-CCCG-5′. The RNase H-oligo(dT) cleavage results of templates AAAA and CCCG were selected because they represented the greatest and least cleavage, respectively, observed from the 20 templates. Template AGAA was selected because it represented an intermediate phenotype. These three templates directed exclusive transcription of the readthrough RNA, which is the only RNA species in lanes loaded with undigested RNA (Fig. 2B, lanes -). RNase H digestion of readthrough RNAs transcribed from template 3′-CCCG-5′ did not result in the production of the two characteristic cleaved RNAs; as described above, this indicated that slippage had not occurred during readthrough RNA synthesis. In contrast, RNase H digestion products derived from readthrough RNAs transcribed from the template 3′-AAAA-5′ are clearly present, indicating that slippage occurred during synthesis of these RNAs. RNase H cleavage of readthrough RNAs transcribed from the template 3′-AGAA-5′ are faintly visible, indicating that a low level of slippage may have occurred during their synthesis.
The primary results of RNase H-oligo(dT)-directed cleavage on readthrough RNAs transcribed from the other 17 templates of this group are not shown but are summarized in Fig. 2C, where these templates are categorized into three groups according to which of the three templates shown in Fig. 2B best resembled their corresponding pattern of RNAs following digestion. In this way, templates that transcribed RNAs that were subjected to a high degree of cleavage were categorized as AAAA-like, templates transcribing RNAs that were not cleaved were grouped as CCCG-like, and templates that transcribed RNAs subjected to an intermediate degree of digestion were grouped as AGAA-like. This classification indicated a trend in which readthrough RNAs transcribed from templates with A/U-rich tetranucleotides were cleaved, whereas RNAs transcribed from templates with G or C containing tetranucleotides were not cleaved. Templates with tetranucleotides having a mixture of A/U and G/C nucleotides transcribed RNAs exhibiting an intermediate degree of cleavage. These findings indicated that the tetranucleotide sequence was able to affect the degree of cleavage of transcribed RNAs, which in turn suggested that this sequence was able to affect the degree of slippage that occurred on the downstream U7 tract.
Construction of the G/C and A/U groups of subgenomic templates.
The results of the RNase H-oligo(dT) analysis described above suggested that the tetranucleotide sequence was able to affect the occurrence of polymerase slippage on the U7 tract. To further investigate the relationship between tetranucleotide sequence and polymerase slippage, we generated a more extensive panel of subgenomic templates which had tetranucleotide sequences composed entirely of either A or U residues (the A/U group) or of G or C residues (the G/C group).
Within the A/U group of templates, we restricted the 5′-most position of the A- and U-containing tetranucleotides to an A residue and in this way maintained the U tract as U7 rather than U8 or longer. The three remaining positions of the A/U-containing tetranucleotide were varied to every possible permutation, generating eight different subgenomic templates. For reasons discussed above, the 5′ position of the 3′-AUAC-5′ tetranucleotide in the G/C tetranucleotide template group was restricted to a G residue to ensure that termination did not occur, and by using every possible permutation of G or C residues within the remaining three positions of this sequence, a further eight subgenomic templates were constructed. The precise sequences of the 16 altered tetranucleotides that were constructed are shown in Fig. 3. We assessed the extent of polymerase slippage directed by these templates by using two different approaches as described below.
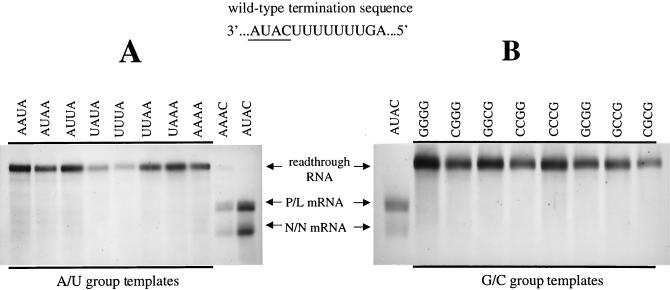
FIG. 3.
Subgenomic templates designed to exclusively transcribe readthrough RNAs. Subgenomic templates having tetranucleotide sequences composed of A and U residues (the A/U group) (A) and templates having tetranucleotide sequences composed of G and C residues (the G/C group) (B) were constructed and used to generate metabolically labeled and actinomycin D-resistant RNAs in BHK cells as described in Materials and Methods. RNAs were visualized by agarose-urea gel electrophoresis and fluorography. The altered tetranucleotide sequences (negative sense, 3′-5′) of templates are shown above the lanes in which the corresponding RNAs products were electrophoresed. For reference, the wild-type termination sequence is shown, with the tetranucleotide sequence underlined. RNAs transcribed from subgenomic templates with tetranucleotide sequences AUAC (wild type) and AAAC, are shown alongside to indicate the mobility of terminated and readthrough RNAs. All tetranucleotide alterations within the A/U and G/C groups resulted in templates that directed exclusive synthesis of readthrough RNAs. In contrast to RNAs electrophoresed in Fig. 2B, the RNAs were not digested with RNase H.
Prior to assessing the extent of polymerase slippage, we determined that these subgenomic templates were unable to terminate mRNA synthesis at their altered gene junctions by visualizing the actinomycin D-resistant, [3H]uridine-labeled RNAs they transcribed, using agarose-urea gel electrophoresis followed by fluorography (Fig. 3). All of the templates within the G/C or A/U groups exclusively synthesized the previously characterized readthrough RNA, which indicated that the tetranucleotide alterations we had chosen had indeed prevented termination at the N/N gene end.
Further analysis of readthrough RNAs for evidence of VSV polymerase slippage.
To further investigate the control of VSV polymerase slippage by the tetranucleotide sequence, we designed an assay (RT/Klenow assay [Fig. 4]) that precisely determined the length of the adenylate tract contained in readthrough RNAs. By analyzing the lengths of poly(A) tracts within a bulk preparation of RNAs, this assay allowed us to assess the relative abundance that each A-tract length represented out of the total population.
The RT/Klenow assay analyzed RNAs for the presence of additional nucleotides inserted at the U7 slippage site by determining the spacing of two defined points within the readthrough RNA that corresponded to either sides of the gene junction in the template. This assay utilized a single polymerase (reverse transcriptase) to synthesize the labeled cDNA and thus reduced the potential for polymerase error to a minimum. Oligonucleotide primer pext-1 annealed downstream of the gene junction and was extended using reverse transcriptase in the presence of [35S]dATP. After purification, the labeled first-strand cDNA was primed for unlabeled second-strand synthesis with oligonucleotide 9121, using the Klenow fragment of DNA polymerase I. Purified duplex cDNAs were digested with restriction enzymes StuI and EcoRV, which recognized sites on either side of the potential slippage sequence, and cleaved cDNA fragments were analyzed by PAGE and autoradiography. The RNAs analyzed using this assay were generated from subgenomic templates supplied as budded particles produced in a previous transfection rather than as T7 RNA polymerase transcripts synthesized from plasmid cDNAs. This approach ensured that DNA-templated RNAs produced by T7 RNA polymerase could not contribute to the pool of RNAs under analysis.
We also carried out control experiments to determine whether slippage by T7 RNA polymerase had occurred during transcription of the subgenomic DNA templates. T7 RNA polymerase primary transcripts were generated by transfecting template encoding cDNAs into vTF7-3-infected BHK cells but without adding the VSV N, P, and L protein support plasmids required for VSV-specific RNA synthesis. RNAs transcribed either from budded-particle templates by the VSV polymerase or from plasmids by T7 RNA polymerase were concurrently analyzed using the slippage assay. By this means, differences detected between the two sets of resulting cDNA fragments could be attributed to the activity of the VSV polymerase, and in this way we were able to rule out the possibility that slippage by T7 RNA polymerase had contributed to the degree of slippage attributed to the VSV polymerase.
Analysis of RNAs transcribed from the G/C group of templates.
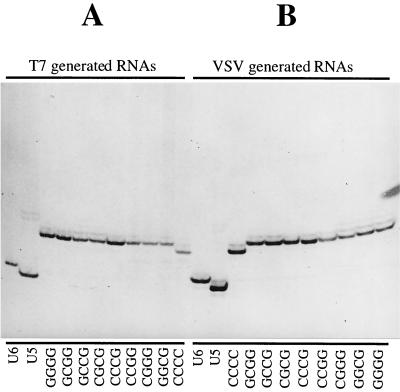
When subjected to the slippage assay described above, RNA transcripts generated from the eight subgenomic templates in the G/C group by both T7 and VSV RNA polymerases gave rise to a single major species of labeled cDNA product (Fig. 5). The two sets of cDNA bands exactly comigrated, indicating that the major RNAs produced by both the T7 RNA and VSV polymerases were identical in length (cDNA fragments generated from additional template CCCC are one nucleotide shorter due to the construction of this template). An additional cDNA band having a mobility indicating that it was one nucleotide longer was also detected in all of the lanes, and this band had an abundance between 5 and 10% of that of the major species. This minor band most likely resulted from infrequent addition of a single extra nucleotide at the slippage site by either or both of the RNA polymerases or by reverse transcriptase. Because abundance of this minor band did not noticeably increase in the lanes specific for VSV polymerase-generated RNAs (Fig. 5B) compared to the lanes specific for T7 RNA polymerase-generated RNAs (Fig. 5A), we suggest that the addition is not likely caused by the VSV RNA polymerase. Whether T7 RNA polymerase or reverse transcriptase is responsible is unknown, although T7 RNA polymerase must be a strong candidate, as instances of nontemplated nucleotide addition by this polymerase have been previously reported in a variety of systems (21, 39).
FIG. 5.
RT/Klenow slippage assay analysis of RNAs transcribed from subgenomic templates with G/C tetranucleotides. (A) To control for slippage by either T7 RNA polymerase or reverse transcriptase, positive-stranded RNAs harvested from vTF7-3-infected BHK cells transfected with subgenomic template expressing cDNAs alone were subjected to the RT/Klenow slippage assay (Fig. 4). (B) To detect VSV polymerase slippage, RNAs transcribed from subgenomic templates supplied as budded particles were analyzed by the RT/Klenow slippage assay. The tetranucleotide sequences of altered subgenomic templates are shown beneath the corresponding lanes. For reference and as a control, templates unable to signal polymerase slippage due to their shortened U5 and U6 tracts were also included in the assay. The additional template CCCC produced RNAs one nucleotide shorter than others due to a deletion during construction of the corresponding cDNA.
As VSV polymerase slippage was not detected during transcription of these templates, we can conclude that the U7 tract is not sufficient to allow slippage on its own. This finding indicates that sequences outside the U7 tract are involved in the regulation of polymerase slippage. Furthermore, when RNAs transcribed by the VSV polymerase were analyzed using the slippage assay, the pattern of labeled bands did not noticeably vary between any of the eight templates, indicating that all of the corresponding tetranucleotide sequences were able to suppress VSV polymerase slippage on the U7 tract. Prevention of polymerase slippage was thus a feature common to each of these altered tetranucleotide sequences, and we suggest this feature is the overall G/C content of this sequence.
Analysis of RNAs transcribed from the A/U group of templates.
Using the slippage assay described above, analysis of T7 RNA polymerase transcripts generated from the A/U group of templates gave rise to a single major species of labeled product (Fig. 6A) similar to that seen with the G/C group of templates. Similar to the G/C group of RNAs described above, a minor cDNA band one nucleotide longer with an abundance of approximately 10% of that of the major species was also present. As described above, this species may have resulted from a low level of slippage by T7 RNA polymerase. If T7 RNA polymerase is the cause of this faint band, then slippage must be restricted to allow only a single nucleotide addition and also must be a rare event.
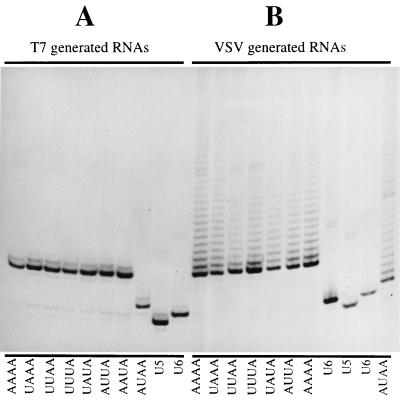
FIG. 6.
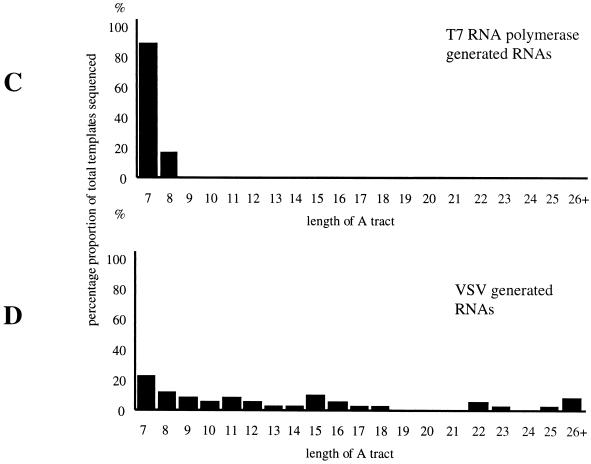
RT/Klenow slippage assay analysis of RNAs transcribed from subgenomic templates with A/U tetranucleotides. To detect VSV polymerase slippage, RNAs were analyzed by the RT/Klenow slippage assay (Fig. 4) and also by determining the nucleotide sequence of RT-PCR-generated cDNA products. (A) To control for slippage by either T7 RNA polymerase or reverse transcriptase, RNAs harvested from vTF7-3-infected cells BHK cells transfected with only subgenomic template expressing cDNAs were subjected to the RT/Klenow slippage assay. (B). To detect slippage by the VSV polymerase, RNAs transcribed from templates supplied as budded particles were subjected to the RT/Klenow slippage assay. In both panels, the tetranucleotide sequences of the templates are shown beneath the corresponding lanes. RNAs transcribed from template AUAA were of faster mobility than those transcribed from other templates due to a four-nucleotide deletion in the cDNA. For reference and as a control, templates unable to signal polymerase slippage (U5 and U6) were included in the assay. To determine the number of A residues present in the A tracts of individual RNAs, RNAs synthesized by both VSV and T7 RNA polymerases from the template with the tetranucleotide 3′-AUAA-5′ were used to generate RT-PCR fragments, which were cloned into pGEM3 and subjected to nucleotide sequence analysis. A total of 46 cDNAs generated by RT-PCR from T7 RNA polymerase transcripts (C) and 46 cDNAs generated by RT-PCR from VSV polymerase transcripts (D) were sequenced, and the results are shown in the form of a bar chart.
In marked contrast to the results of the G/C group, slippage analysis of the VSV polymerase transcribed RNAs from the A/U group of templates gave rise to a series of labeled cDNA bands that gave the appearance of a ladder, extending upward in the gel (Fig. 6B). The mobility of these bands indicated that they contained additional nucleotides, suggesting that the A/U-rich tetranucleotide templates had allowed slippage to occur on the U7 tract during synthesis of the corresponding RNAs.
The activities of the VSV polymerase on all eight of the A/U group of templates under assay appeared similar, despite these templates having different tetranucleotide sequences. This finding indicated that promotion of VSV polymerase slippage was controlled by a feature common to all of the eight different templates, and we suggest that this was the A/U content of the tetranucleotide.
Sequence analysis of cDNAs synthesized from T7 and VSV RNA polymerase-generated RNAs.
Using the RT/Klenow assay described above, we analyzed large populations of RNA transcribed from altered subgenomic templates in order to investigate how the tetranucleotide sequence was able to control VSV polymerase slippage. While this assay allowed us to assess the relative extent to which polymerase slippage had occurred during transcription of a population of RNAs, it did not allow us to precisely determine the exact number of nucleotides added during individual slippage events. To determine the extent to which polymerase slippage had occurred in individual instances, we determined the nucleotide sequences of 46 cDNA clones generated by RT-PCR from populations of VSV-specific RNAs.
RT-PCR was performed on RNAs transcribed by the VSV polymerase in budded-particle transfections, and the sequences of individually cloned cDNAs were determined. Because the RNA population transcribed from all members of the A/U group gave very similar patterns of labeled cDNAs in the slippage assay, we performed sequence analysis on only one template of this group, the one having the tetranucleotide sequence AUAA. As a control for polymerase slippage by T7 RNA polymerase, RNA transcripts of this template generated by this polymerase were analyzed by the same procedure. We did not individually analyze RNAs transcribed from members of the G/C group because these RNAs previously showed no signs of slippage in the RT/Klenow slippage assay.
The data in Fig. 6C show that T7 RNA polymerase synthesized a great majority (86%) of correctly sized RNAs with a tract of seven U residues, with the remaining 14% having U tracts of eight nucleotides. The slightly higher proportion of U8 to U7 templates than previously estimated by the RT/Klenow slippage assay on this template (Fig. 6A) may be due to either the relatively small number of cDNA clones that were sequenced (46 clones) or errors introduced during PCR amplification. In contrast, the RNAs generated by VSV polymerase displayed greater heterogeneity of A-tract size, with only 20% having the exact templated A-tract length of 7 (Fig. 6D). The remaining 80% of the RNAs all had additional A residues present in their A tracts, with the longest A-tract length being 40. The relative abundance of the RNAs that showed evidence of polymerase slippage by direct sequencing correlated with the results generated by the RT/Klenow slippage assay (Fig. 6A). Nucleotide sequence analysis of these cDNAs indicated that all additional nucleotides detected in the readthrough RNAs were A residues.
Construction of recombinant VSV variants possessing an additional gene junction.
In the previous sections we investigated how alterations of the tetranucleotide sequence affected the ability of the VSV polymerase to slip on the adjacent U7 tract. The assays were performed using RNAs generated in the well-established subgenomic template system because it offered a combination of ease of template manipulation and biological authenticity (3, 4). However, to further strengthen our findings, we also chose to analyze the occurrence of VSV polymerase slippage during RNA synthesis by infectious recombinant VSV variants. This approach would allow us to investigate polymerase slippage in a system that did not rely on coinfection with a vaccinia virus recombinant or the transient expression of supporting proteins. In particular, the ability of vaccinia virus-encoded enzymatic activities to modify both 3′ and 5′ ends of RNAs were of potential concern, as the consequences of such modifications on VSV polymerase activity are unknown.
We constructed plasmids designed to generate recombinant VSV variants having two extra nonviral genes (CAT and GFP) separated by a gene junction into which tetranucleotide alterations could be engineered without affecting the relative expression of the five VSV genes further downstream (Fig. 7A). We selected one tetranucleotide sequence from each of the G/C and A/U groups of subgenomic templates described above, namely, GCGG and AUAA, and inserted these sequences independently at the CAT-GFP gene junction of the relevant cDNAs to create pVSV-GCGG and pVSV-AUAA, respectively. In addition, we constructed a cDNA designed to generate an infectious VSV variant in which the two additional genes were separated by the wild-type VSV N-P gene junction sequence (pVSV-AUAC). Infectious viruses were recovered from these cDNAs and used to infect BHK cells. The VSV-specific RNAs were labeled and visualized as previously described (42). The RNAs synthesized by the three recombinant VSV variants as well as from the wild-type VSV are shown in Fig. 7B. VSV-AUAC-directed the transcription of two additional RNA products compared to the wild type, which corresponded to the individual mRNAs transcribed from the CAT and GFP genes. In addition, a minor RNA of much slower mobility which represented the bicistronic CAT-GFP readthrough RNA was also detected on longer exposure of the gel. Compared to wild-type VSV, the VSV variants VSV-GCGG and VSV-AUAA expressed just one extra RNA of slower mobility which represented the bicistronic CAT-GFP readthrough RNA.
Analysis of RNAs generated by the VSV variants.
Readthrough RNAs generated by these three VSV variants in infected BHK cells were harvested and analyzed for evidence of polymerase slippage. VSV-GCGG and VSV-AUAA synthesized abundant readthrough RNAs (Fig. 7B), and so RNAs expressed from these viruses were analyzed by the RT/Klenow slippage assay described above. Figure 8A shows the labeled cDNA products of the slippage assay. Readthrough RNAs transcribed from VSV-GCGG showed no evidence of polymerase slippage, whereas RNAs transcribed from VSV-AUAA showed abundant evidence of slippage. These results were indistinguishable from those obtained using the plasmid-supported subgenomic template system and thus confirmed our finding that G/C tetranucleotide sequences acted to prevent polymerase slippage, whereas A/U tetranucleotides acted to promote slippage.
FIG. 8.
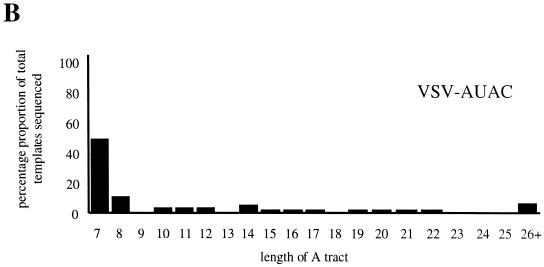
Analysis of RNAs transcribed by the VSV polymerase from recombinant viruses VSV-AUAC, VSV-GCGG, and VSV-AUAA which have altered tetranucleotide sequences within the CAT-GFP gene junction. (A) RNAs transcribed in infected BHK cells from viruses VSV-AUAA and VSV-GCGG were analyzed by the RT/Klenow slippage assay, and the radiolabeled cDNAs were visualized by PAGE analysis and autoradiography. The lanes are marked with the tetranucleotide sequence of the corresponding viruses. (B) Readthrough RNAs transcribed from recombinant virus VSV-AUAC were analyzed not by the slippage assay but by determining the nucleotide sequence of individual RT-PCR generated cDNAs due to the low abundance of the readthrough RNA species. A total of 51 RT-PCR-generated cDNA clones were sequenced, and the results are shown in a bar chart with the numbers on the horizontal axis corresponding to numbers of A residues detected in the A tract. The rounded percentage abundances of each A-tract length are as follows: A7, 52%; A8, 10%; A10, A11, and A12, 4%; A14 6%; A15, A16, A17, A19, A20, A21, and A22, 2%; and A26+, 8%.
VSV-AUAC generated a very low quantity of readthrough RNAs (Fig. 7B), and so these RNAs could not be analyzed by the RT/Klenow slippage assay. Instead, readthrough RNAs were analyzed by nucleotide sequencing of corresponding RT-PCR products. Briefly, polyadenylated RNAs were selected by a magnetic bead poly(A) enrichment procedure, which was necessary to remove the positive-sense antigenome which could also act as template for the initial reverse transcription reaction. The poly(A)+ RNAs were used as the template for RT-PCR amplification, and the resulting PCR products were inserted into pGEM3 and individually sequenced. Of 51 cDNAs analyzed, 25 (49%) were found to have an unaltered A, sequence at the gene junction, indicating faithful copying of the gene junction U7 tract, and a further 7 cDNAs (14%) showed the presence of a single additional A residue (Fig. 8B). The remaining 19 cDNAs contained A tracts that predominantly ranged between 10 and 22 nucleotides in length (3 of the 19 cDNAs had A tracts of 31, 40, and 63 nucleotides), indicating that the tetranucleotide AUAC was able to signal polymerase slippage on the U7 tract. However, by comparing Fig. 8B with Fig. 6D, we noted that the extent and frequency of slippage on the AUAC template is less than that determined for the template with the AUAA tetranucleotide; a possible explanation of this finding is discussed below.
DISCUSSION
The weight of evidence suggests that the long poly(A) tails at the 3′ ends of VSV mRNAs are synthesized by the VSV polymerase performing reiterative transcription, or polymerase slippage, on the relatively short U7 tract located at each of the VSV gene junctions. Since mRNAs lacking poly(A) tails are not observed in wild-type VSV infections, polyadenylation and consequently polymerase slippage are both likely to be crucial steps in the process of VSV mRNA termination. In this study, we investigated whether sequences other than the U7 tract affected the process of polymerase slippage by analyzing the nucleotide sequence of RNAs transcribed from altered subgenomic templates and infectious VSV variants.
Our findings showed that the U7 sequence alone was not sufficient to signal polymerase slippage and that the composition of the upstream tetranucleotide sequence (underlined) of the conserved VSV termination signal (3′-AUACUUUUUUUGA-5′) was a major determinant of the degree of slippage that occurred on the downstream U7 tract. Analysis of readthrough RNAs generated from templates having tetranucleotides composed entirely of various combinations of A and U residues showed extensive and frequent polymerase slippage. The extent to which slippage occurred was approximately the same for all of the eight A/U-containing templates tested, and we interpret this to indicate that the ability of a template to promote polymerase slippage on the U7 tract was controlled by the upstream tetranucleotide in a way that did not depend on a precise sequence signal. Similarly, when we analyzed the readthrough RNAs transcribed from the group of templates with tetranucleotide sequences composed entirely of various combinations of G and C residues, we found that polymerase slippage on the U7 tract was prevented in all cases despite the variety of tetranucleotide sequences. Therefore, for templates within these two groups, promotion or abrogation of polymerase slippage did not depend on precise sequences within the tetranucleotide, which consequently indicated that the signal controlling slippage was a common property of all members of the two individual groups. We suggest that this common element is overall G and C content in the case of slippage abrogation and is overall A and U content in the case of slippage promotion.
How might these various tetranucleotide sequences alter the activity of the VSV polymerase and dictate whether or not it can slip on the U7 tract? There are several possible mechanisms through which this effect may be mediated. These include interaction of the template strand tetranucleotide with the complementary region of the nascent strand or interaction of either of these sequences with the VSV polymerase, with an unidentified component of the host cell, or with a trans-acting nucleic acid molecule.
Our observation that A/U tetranucleotide sequences acted to promote slippage whereas G/C tetranucleotides prevented it makes it tempting to propose that polymerase slippage is controlled, at least in part, by the strength of an interaction between two nucleic acid molecules that are base paired. Because of their proximity to the catalytic site of the VSV polymerase and their ability to form a perfectly base-paired hybrid, we suggest that the most likely candidates for these interacting molecules are the genomic template and the nascent RNA strands. This interpretation can accommodate our observation that tetranucleotide sequences signaled the promotion or abrogation of slippage in a way that was independent of the exact tetranucleotide sequence, so long as the A/U or G/C nucleotide composition of this sequence was maintained. Clearly any alteration of the tetranucleotide would cause the corresponding alteration of the nascent strand, thus allowing maintenance of the base-pairing ability of template and nascent strands. If this hybrid strength is in fact the signal, then several tetranucleotide sequences would likely provide comparable signals, which is in accordance with our findings.
As we stated above, alternative mechanisms by which tetranucleotide alterations could modulate polymerase slippage include interaction with components such as the VSV polymerase, a host cell factor, or an additional nucleic acid molecule. However, in contrast to the template and nascent strands, components such as these will obviously not change in accordance to an alteration in tetranucleotide sequence. Therefore, to permit the type of sequence-independent signaling observed in this study, these components would need to be capable of recognizing, and responding consistently to, a broad range of RNA sequences. We consider this less likely.
If an interaction between the template and nascent strands is important in signaling polymerase slippage, do these findings tell us anything about the nature of such a hybrid? The precise number of nucleotides for which the template and nascent RNA strands of VSV are hybridized during polymerization is not known. However, during the slippage reaction, it seems unlikely that the hybrid could extend for more than the seven residues of the U tract because as soon as even one round of polymerase slippage had occurred, unfavorable base pairing combinations would be required to form between newly inserted (and nontemplated) A residues and non-U (and thus noncomplementary) residues within the template strand tetranucleotide. Why then do changes within the tetranucleotide have such a dramatic effect on polymerase slippage that suggest involvement in a hybrid? A possible answer is that the tetranucleotide may exert its effect only immediately before an initial slippage event takes place. At such a time, the nascent strand and the template will be perfectly complementary, and perhaps a weak hybrid extending across both the U7 tract and an A/U-rich tetranucleotide may provide sufficient instability to allow the initiation of slippage. Once initial slippage has occurred, the length of the hybrid will be reduced to just the U7 tract and its complement, and so potentially unfavorable base pairings are not required to form. In contrast, a G/C-containing tetranucleotide preceding the U7 tract may prevent initial interstrand movement and thus prevent a slippage event.
This explanation would imply that the number of nucleotides involved in the hybrid varies during the termination process. While there is currently no evidence to suggest that the VSV polymerase behaves in this way, the RNA polymerase of Escherichia coli has been shown to move backward in response to weak base-pairing potential between the template and nascent strands, even when these nucleotides extend upstream of an 8- or nine-nucleotide-long hybrid (25). It is conceivable that if such a mechanism existed in VSV, then the weak base pairing potential of an A/U-rich tetranucleotide may allow realignment of the template and polymerase within the U7 tract, and when followed by correctly templated RNA synthesis, cycles of this process could generate the expanded poly(A) tracts that we have observed.
We previously analyzed how changes at the gene junction affected termination ability and established that while all changes within the tetranucleotide decreased termination ability compared to that of the wild-type sequence, certain changes were more deleterious than others (3). We were unable to establish a pattern relating tetranucleotide alteration to corresponding termination ability and thus could not precisely predict how a defined alteration would affect termination, and understanding this relationship has been one of our goals. In particular, we have been intrigued to understand why the single nucleotide alteration of the wild-type gene junction tetranucleotide 3′-AUAC-5′ to one with the sequence 3′-AUAA-5′ decreased termination from 99% to 0%. Also, given the importance of polymerase slippage in the termination process, we were curious to understand why tetranucleotide alterations that, according to the results of this study would not be expected to alter slippage ability, had a major affect on termination. As an example, changing the tetranucleotide 3′-AUAC-5′ to UUAC, AAAC, or AUUC leads to a 33% reduction in termination ability compared to that of wild type (3).
In an attempt to tie our observations together, we suggest that the tetranucleotide comprises at least two signals that are required for termination to occur and that these signals overlap. The first signal is high overall A/U content, which promotes interstrand slippage in the vicinity of the U7 tract, and the second is a signal that allows polymerase molecules engaged in reiterative transcription to terminate synthesis of the corresponding mRNA. We have termed this the commitment signal. This commitment signal likely includes the 5′ C residue of the 3′-AUAC-5′ tetranucleotide, since alteration of this position to any other residue results in the total loss of termination ability (3). However, we suggest that the commitment signal also depends on the identity of the other three positions of the tetranucleotide, and this allows us to explain why seemingly preserving potential slippage ability of a tetranucleotide, as described above, can lead to a loss in termination ability. In these cases, the alterations have maintained slippage potential, but as the two signals overlap, they have perturbed the commitment signal. As any change of the tetranucleotide can alter either or both of these two signals, precisely defining the commitment signal in isolation is problematical. However, we know that the most efficient tetranucleotide sequence for termination of mRNA synthesis is the wild-type sequence 3′-AUAC-5′ (99% termination ability) (3, 4), and we suggest that this is because this sequence provides the most effective balance of the slippage and commitment elements. The lessened ability of the AUAC tetranucleotide to promote polymerase slippage compared to AUAA (compare Fig. 6D and 8B) is perhaps necessary to ensure the presence of the essential C residue that is obligately required for transcription termination (3).
By what mechanism does the C residue of the tetranucleotide provide the essential termination signal? In the same way as described above for signaling slippage, the C residue may be involved in a crucial interaction within the template: nascent strand hybrid. Alternatively, the C residue of the template (or G residue of the nascent strand) may interact with either the VSV polymerase, a host cell component, or an unidentified nucleic acid molecule. However, in contrast to the signal that regulated polymerase slippage, which can be provided by several tetranucleotide sequences, the signaling ability of the essential C residue cannot be provided by any alternative nucleotide (3). This degree of signal specificity does not rule out (nor does it implicate) any of the possible interacting partners listed above. However, because changing the essential C residue to the complementary G residue does not signal termination despite maintaining the potential template-nascent strand association, we favor the notion that the essential C residue provides a signal in a way other than by its base pairing with the nascent RNA strand. A model describing the process of reiterative transcription during editing in the paramyxovirus Sendai virus (see below) has recently been proposed (11). A central theme of this model is that a region of the nascent strand immediately upstream of a slippery sequence provides a signal which modulates reiterative transcription by making sequence-specific contacts with a site on the polymerase. It is possible that the role of the 3′-AUAC-5′ tetranucleotide is to provide a signal within the nascent strand which somehow interacts with the VSV polymerase in a similar way.
Our detection of readthrough RNAs containing additional A residues indicates that a reiteratively transcribing polymerase can resume processive transcription and transcribe the remainder of the template. Since the A/U group of templates exclusively directed synthesis of readthrough RNAs, the polymerase molecules engaged in slippage must always return to the normal mode of transcription. The A tracts found in the readthrough RNAs generated by template AUAA rarely exceeded 26 A residues in length (Fig. 6D) and were thus shorter than typical VSV mRNA poly(A) tails, which are between 100 and 300 nucleotides long. Does this observation suggest that termination of mRNA synthesis can occur only when polymerase slippage makes an A tract that exceeds a certain critical length? We suggest that while A-tract length may be an important factor in the termination mechanism, it is not likely to be the only one, as many previous studies have identified intervening A tracts in readthrough RNAs much larger than those of a typical VSV poly(A) tail (13, 14, 28).
The ability of the VSV polymerase to regain template-dependent transcription after performing reiterative transcription on the U7 tract, as described above, is strongly reminiscent of the editing activity by which many negative-strand viruses, most notably simian virus 5, measles virus, and Sendai virus, have been shown to modulate transcription of their P genes (8, 37). Importantly, however, there are differences between these examples of nontemplated transcription. First, during P gene editing, the addition of nontemplated G residues generally occurs in a controlled manner so that addition of a precise number of nucleotides is favored over all others. This contrasts with the findings of our present study, which revealed that the number of nucleotide additions at the U7 VSV slippage site was distributed over a broad range. Second, unlike our findings reported here, during paramyxovirus P gene editing, the inserted residue (G) is templated by a nucleotide that is not within the slippage sequence itself but is located immediately downstream. Recent work involving Sendai virus has indicated that these two features reflect differences in the template sequence rather than fundamental differences in polymerase activity (11, 12). This result, in combination with the finding that sequences immediately upstream of the U tract can regulate the reiterative transcription activity of the Sendai virus and VSV polymerases, has revealed a strong mechanistic relationship between the process of mRNA editing and the polymerase slippage activity reported here. A parallel may also be drawn between the VSV slippage activity and glycoprotein mRNA editing in Ebola virus, as both activities involve insertion of A residues via slippage on a polyuridylate tract (29). Furthermore, the sequence responsible for the nucleotide additions at the Ebola virus editing site strongly resembles an Ebola virus gene end signal. Perhaps Ebola virus and other viruses which edit their mRNAs via a slippage mechanism have modified what once were termination signals in order to either increase the coding flexibility of their genomes or reduce the attenuating effect of a gene junction.
Comparing the conserved gene end sequence of VSV to those of several other nonsegmented negative-stranded RNA viruses reveals that sequences which precede the universal U tract share an A/U-rich element followed by a C residue that directly abuts the U tract. Because of this conservation, we suggest that many members of the family of nonsegmented negative-strand RNA viruses may utilize a common mechanism of not only polyadenylation but also the way in which the termination process is signaled.
ACKNOWLEDGMENTS
We thank members of the G. W. Wertz and L. A. Ball laboratories for helpful discussions during preparation of the manuscript.
This work was supported by PHS grant R37-A112464 from NIAID to G.W.W.
REFERENCES
- 1.Abraham G, Banerjee A K. Sequential transcription of the genes of vesicular stomatitis virus. Proc Natl Acad Sci USA. 1976;73:1504–1508. doi: 10.1073/pnas.73.5.1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ball L A, White C N. Order of transcription of genes of vesicular stomatitis virus. Proc Natl Acad Sci USA. 1976;73:442–446. doi: 10.1073/pnas.73.2.442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barr J N, Whelan S P, Wertz G W. cis-acting signals involved in termination of vesicular stomatitis virus mRNA synthesis include the conserved AUAC and the U7 signal for polyadenylation. J Virol. 1997;71:8718–8725. doi: 10.1128/jvi.71.11.8718-8725.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barr J N, Whelan S P J, Wertz G W. Role of the intergenic dinucleotide in vesicular stomatitis virus RNA transcription. J Virol. 1997;71:1794–1801. doi: 10.1128/jvi.71.3.1794-1801.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bilsel P A, Nichol S T. Polymerase errors accumulating during natural evolution of the glycoprotein gene of vesicular stomatitis virus Indiana serotype isolates. J Virol. 1990;64:4873–4883. doi: 10.1128/jvi.64.10.4873-4883.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Colonno R J, Banerjee A K. Complete nucleotide sequence of the leader RNA synthesized in vitro by vesicular stomatitis virus. Cell. 1978;15:93–101. doi: 10.1016/0092-8674(78)90085-5. [DOI] [PubMed] [Google Scholar]
- 7.Colonno R J, Banerjee A K. A unique RNA species involved in initiation of vesicular stomatitis virus RNA transcription in vitro. Cell. 1976;8:197–204. doi: 10.1016/0092-8674(76)90003-9. [DOI] [PubMed] [Google Scholar]
- 8.Curran J, Boeck R, Kolakofsky D. The Sendai virus P gene expresses both an essential protein and an inhibitor of RNA synthesis by shuffling modules via mRNA editing. EMBO J. 1991;10:3079–3085. doi: 10.1002/j.1460-2075.1991.tb07860.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Emerson S U. Reconstitution studies detect a single polymerase entry site on the vesicular stomatitis virus genome. Cell. 1982;31:635–642. doi: 10.1016/0092-8674(82)90319-1. [DOI] [PubMed] [Google Scholar]
- 10.Emerson S U, Wagner R R. Dissociation and reconstitution of the transcriptase and template activities of vesicular stomatitis virus B and T virions. J Virol. 1972;10:297–309. doi: 10.1128/jvi.10.2.297-309.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hausmann S, Garcin D, Delenda C, Kolakofsky D. The versatility of paramyxovirus RNA polymerase stuttering. J Virol. 1999;73:5568–5576. doi: 10.1128/jvi.73.7.5568-5576.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hausmann S, Garcin D, Morel A S, Kolakofsky D. Two nucleotides immediately upstream of the essential A6G3 slippery sequence modulate the pattern of G insertions during Sendai virus mRNA editing. J Virol. 1999;73:343–351. doi: 10.1128/jvi.73.1.343-351.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herman R C, Adler S, Lazzarini R A, Colonno R J, Banerjee A K, Westphal H. Intervening polyadenylate sequences in RNA transcripts of vesicular stomatitis virus. Cell. 1978;15:587–596. doi: 10.1016/0092-8674(78)90027-2. [DOI] [PubMed] [Google Scholar]
- 14.Herman R C, Schubert M, Keene J D, Lazzarini R A. Polycistronic vesicular stomatitis virus RNA transcripts. Proc Natl Acad Sci USA. 1980;77:4662–4665. doi: 10.1073/pnas.77.8.4662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hunt D M, Smith E F, Buckley D W. Aberrant polyadenylation by a vesicular stomatitis virus mutant is due to an altered L protein. J Virol. 1984;52:515–521. doi: 10.1128/jvi.52.2.515-521.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hutchinson K L, Herman R C, Hunt D M. Increased synthesis of polycistronic mRNA associated with increased polyadenylation by vesicular stomatitis virus. Virology. 1992;189:67–78. doi: 10.1016/0042-6822(92)90682-f. [DOI] [PubMed] [Google Scholar]
- 17.Hwang L N, England N, Pattnaik A K. Polyadenylation of vesicular stomatitis virus mRNA dictates efficient transcription termination at the intercistronic gene junctions. J Virol. 1998;72:1805–1813. doi: 10.1128/jvi.72.3.1805-1813.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iverson L E, Rose J K. Localized attenuation and discontinued synthesis during vesicular stomatitis virus transcription. Cell. 1981;23:477–484. doi: 10.1016/0092-8674(81)90143-4. [DOI] [PubMed] [Google Scholar]
- 19.Leppert M, Rittenhouse L, Perrault J, Summers D F, Kolakofsky D. Plus and minus strand leader RNAs in negative strand virus-infected cells. Cell. 1979;18:735–747. doi: 10.1016/0092-8674(79)90127-2. [DOI] [PubMed] [Google Scholar]
- 20.Li T, Pattnaik A K. Overlapping signals for transcription and replication at the 3′ terminus of the vesicular stomatitis virus genome. J Virol. 1999;73:444–452. doi: 10.1128/jvi.73.1.444-452.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Macdonald L E, Zhou Y, McAllister W T. Termination and slippage by bacteriophage T7 RNA polymerase. J Mol Biol. 1993;232:1030–1047. doi: 10.1006/jmbi.1993.1458. [DOI] [PubMed] [Google Scholar]
- 22.Masters P, Samuel C E. Detection of in vivo synthesis of polycistronic mRNAs of vesicular stomatitis virus. Virology. 1984;134:277–286. doi: 10.1016/0042-6822(84)90297-6. [DOI] [PubMed] [Google Scholar]
- 23.McGeoch D J. Structure of the gene N: gene NS intercistronic junction in the genome of vesicular stomatitis virus. Cell. 1977;17:673–681. doi: 10.1016/0092-8674(79)90274-5. [DOI] [PubMed] [Google Scholar]
- 24.Moyer S A, Smallwood-Kentro S, Haddad A, Prevec L. Assembly and transcription of synthetic vesicular stomatitis virus nucleocapsids. J Virol. 1991;65:2170–2178. doi: 10.1128/jvi.65.5.2170-2178.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nudler E, Mustaev A, Lukhtanov E, Goldfarb A. The RNA-DNA hybrid maintains the register of transcription by preventing backtracking of RNA polymerase. Cell. 1997;89:33–41. doi: 10.1016/s0092-8674(00)80180-4. [DOI] [PubMed] [Google Scholar]
- 26.Pattnaik A K, Ball L A, LeGrone A W, Wertz G W. Infectious defective interfering particles of VSV from transcripts of a cDNA clone. Cell. 1992;69:1011–1020. doi: 10.1016/0092-8674(92)90619-n. [DOI] [PubMed] [Google Scholar]
- 27.Rose J K. Complete intergenic and flanking gene sequences from the genome of vesicular stomatitis virus. Cell. 1980;19:415–421. doi: 10.1016/0092-8674(80)90515-2. [DOI] [PubMed] [Google Scholar]
- 28.Rose J K, Lodish H F, Brock M L. Giant heterogeneous polyadenylic acid on vesicular stomatitis virus mRNA synthesized in vitro in the presence of S-adenosylhomocysteine. J Virol. 1977;21:683–693. doi: 10.1128/jvi.21.2.683-693.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sanchez A, Trappier S G, Mahy B W, Peters C J, Nichol S T. The virion glycoproteins of Ebola viruses are encoded in two reading frames and are expressed through transcriptional editing. Proc Natl Acad Sci USA. 1996;93:3602–3607. doi: 10.1073/pnas.93.8.3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schnell M J, Buonocore L, Whitt M A, Rose J K. The minimal conserved transcriptional stop-start signal promotes stable expression of a foreign gene in vesicular stomatitis virus. J Virol. 1996;70:2318–2323. doi: 10.1128/jvi.70.4.2318-2323.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schubert M, Keene J D, Herman R C, Lazzarini R A. Site on the vesicular stomatitis virus genome specifying polyadenylation and the end of the L gene mRNA. J Virol. 1980;34:550–559. doi: 10.1128/jvi.34.2.550-559.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schubert M, Lazzarini R A. In vivo transcription of the 5′-terminal extracistronic region of vesicular stomatitis virus RNA. J Virol. 1981;38:256–262. doi: 10.1128/jvi.38.1.256-262.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smallwood S, Moyer S A. Promoter analysis of the vesicular stomatitis virus RNA polymerase. Virology. 1993;192:254–263. doi: 10.1006/viro.1993.1028. [DOI] [PubMed] [Google Scholar]
- 34.Stillman E A, Whitt M A. The length and sequence composition of vesicular stomatitis virus intergenic regions affect mRNA levels and the site of transcript initiation. J Virol. 1998;72:5565–5572. doi: 10.1128/jvi.72.7.5565-5572.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stillman E A, Whitt M A. Mutational analyses of the intergenic dinucleotide and the transcriptional start sequence of vesicular stomatitis virus (VSV) define sequences required for efficient termination and initiation of VSV transcripts. J Virol. 1997;71:2127–2137. doi: 10.1128/jvi.71.3.2127-2137.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stillman E A, Whitt M A. Transcript initiation and 5′-end modifications are separable events during vesicular stomatitis virus transcription. J Virol. 1999;73:7199–7209. doi: 10.1128/jvi.73.9.7199-7209.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thomas S M, Lamb R A, Paterson R G. Two mRNAs that differ by two nontemplated nucleotides encode the amino coterminal proteins P and V of the paramyxovirus SV5. Cell. 1988;54:891–902. doi: 10.1016/S0092-8674(88)91285-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Villarreal L P, Breindl M, Holland J J. Determination of molar ratios of vesicular stomatitis virus induced species in BHK-21 cells. Biochemistry. 1976;15:1663–1667. doi: 10.1021/bi00653a012. [DOI] [PubMed] [Google Scholar]
- 39.Volchkov V E, Becker S, Volchkova V A, Ternovoj V A, Kotov A N, Netesov S V, Klenk H D. GP mRNA of Ebola virus is edited by the Ebola virus polymerase and by T7 and vaccinia virus polymerases. Virology. 1995;214:421–430. doi: 10.1006/viro.1995.0052. [DOI] [PubMed] [Google Scholar]
- 40.Whelan S P, Barr J N, Wertz G W. Identification of a minimal size requirement for termination of vesicular stomatitis virus mRNA: implications for the mechanism of transcription. J Virol. 2000;74:8268–8276. doi: 10.1128/jvi.74.18.8268-8276.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Whelan S P J, Wertz G W. Regulation of RNA synthesis by the genomic termini of vesicular stomatitis virus: identification of distinct sequences essential for transcription but not replication. J Virol. 1999;73:297–306. doi: 10.1128/jvi.73.1.297-306.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Whelan S P W, Ball L A, Barr J N, Wertz G W. Efficient recovery of infectious vesicular stomatitis virus entirely from cDNA clones. Proc Natl Acad Sci USA. 1995;92:8388–8392. doi: 10.1073/pnas.92.18.8388. [DOI] [PMC free article] [PubMed] [Google Scholar]