Abstract
Background:
HCC is a highly vascular tumor, and many effective drug regimens target the tumor blood vessels. Prior bulk HCC subtyping data used bulk transcriptomes, which contained a mixture of parenchymal and stromal contributions.
Methods:
We utilized computational deconvolution and cell-cell interaction analyses to cell type-specific (tumor-enriched and vessel-enriched) spatial transcriptomic data collected from 41 resected HCC tissue specimens.
Results:
We report that the prior Hoshida bulk transcriptional subtyping schema is driven largely by an endothelial fraction, show an alternative tumor-specific schema has potential prognostic value, and use spatially paired ligand-receptor analyses to identify known and novel (LGALS9 tumor-HAVCR2 vessel) signaling relationships that drive HCC biology in a subtype-specific and potentially targetable manner.
Conclusions:
Our study leverages spatial gene expression profiling technologies to dissect HCC heterogeneity and identify heterogeneous signaling relationships between cancer cells and their endothelial cells. Future validation and expansion of these findings may validate novel cancer-endothelial cell interactions and related drug targets.
Keywords: hcc, liver cancer, tumor microenvironment, spatial profiling
INTRODUCTION
HCC is the sixth leading cause of cancer death in the United States and the fourth leading cause of cancer death worldwide. 1 HCC incidence is increasing due to the rising rates of obesity and associated metabolic steatotic liver disease, and it still has a significant contribution from the continued impacts of viral hepatitis, heavy alcohol use, and additional etiologies. 2 Although significant genomic profiling efforts in HCC have been described, 3 most common genomic variants are not therapeutically targetable with existing drugs. Rather, many active systemic therapy regimens empirically target the tumor vasculature, either alone or in combination with immune checkpoint inhibitors, 4 and this fact underscores the privileged role of tumor-resident endothelial cells (ECs) in HCC pathogenesis.
The understanding of EC roles in tumor pathophysiology has evolved considerably over the past few decades. Folkman’s original paradigm of tumor-perfusing vasculature 5 in the 1970s gave way to Jain’s biophysical paradigm—whereby the abnormal structure of tumor vessels (in the setting of a local imbalance of proangiogenic and antiangiogenic growth factors) leads to heterogeneous hypoxia and barriers to drug delivery 6 —in the 2000s. The “angiocrine” model proposed by Rafii and others in the 2010s added another layer, in which local bidirectional paracrine cancer-EC interactions ultimately stimulate tumor growth and spread. 7 Further, tumor-resident ECs are well positioned as critical immune regulatory cells, 8 implying the possibility of multipartner interactions between cancer cells, ECs, and leukocytes. Indeed, many of cancer’s hallmarks 9 are directly impacted by local, stromal ECs.
Although HCC natural history 10 and modern systemic therapy landscape 4 are both clearly impacted by EC presence and phenotype, established bulk tissue classification schemas 11 are unable to resolve potential contributions by local ECs or other stromal cell types. High-throughput single-cell transcriptomic profiling can identify multiple EC subtypes in the HCC tumor microenvironment (TME),12,13 a finding concordant with similar studies in other cancers14,15 and other noncancerous organs. 16 Spatial transcriptomics technologies 17 aim to characterize the molecular phenotypes of different cell types in the TME.
In the current study, we used multiple spatial transcriptomics platforms to dissect linked gene expression programs in the tumor vasculature—mainly comprised of ECs—and cancer cell–enriched areas. We demonstrate that: (1) this technique accurately enriches for regions containing blood vessels separately from tumor-enriched areas, (2) the prior Hoshida bulk tumor classification system included a significant contribution of vascular EC signal in the aggressive S1 subtype, (3) transcriptional phenotypes of cancer cell–enriched areas are correlated with survival, and (4) the heterogeneous subtypes contain distinct cancer-EC interactions. Taken together, our work harmonizes prior bulk tissue transcriptomic profiling work with single-cell data for which spatial context is destroyed, demonstrates a significant contribution of the EC compartment to previous canonical bulk gene profiles, and provides further evidence that there is a fundamental link between our redefined tumor-enriched subtyping system and survival using internal and public data sets.
METHODS
Specimen collection and use
All patients in this study consented without compensation to excess tissue biobank protocol 2013P001854, which was reviewed and approved by the Massachusetts General Hospital Institutional Review Board. This study is compliant with all relevant ethical regulations, including the Declarations of Helsinki and Istanbul. For inclusion in this study, patients had nonmetastatic HCC and went to surgical resection without prior neoadjuvant treatment (Table 1). For DSP (Nanostring) analysis, formalin-fixed, paraffin-embedded tissue specimens from the Massachusetts General Hospital Pathology archives were assessed for tumor content by a board-certified pathologist (Vikram Deshpande) and then 2-mm cores were obtained for tissue microarray generation. After tissue microarray generation, cut slides were stored at −80°C until use.
TABLE 1.
Patient clinical characteristics
| ID | Sex | Status | Time | Serum_afp | Tbili | Alb | Inr | Etiology | Tumor_Number | Tumor_Size | Differentiation | Tstage | Nstage | Lvi | Pni | Fibrosis | Steatosis |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | F | 1 | 240 | 7574 | 0.3 | 3.1 | 1.2 | NA | 1 | 13.0 | Moderate | 2 | x | Yes | x | NA | NA |
| 2 | M | 0 | 4241 | 3.8 | 0.3 | 4.4 | 1.3 | Viral | 1 | 1.5 | Moderate | 1 | x | x | x | NA | NA |
| 3 | M | 1 | 2260 | 4.5 | 1.1 | 4 | 1.3 | Nash | NQ | 3.5 | Moderate | 1 | x | x | x | 6 | Macrovesicular |
| 4 | M | 1 | 2587 | 54.1 | 1.8 | 3.3 | 1.5 | Viral | 2 | 2.5 | Moderate | 2 | x | x | x | 6 | Microvesicular |
| 5 | M | 1 | 311 | 28.6 | 0.5 | 3.9 | 1.2 | Viral | NQ | 6.5 | Moderate_poor | 4 | x | Yes | x | 3 | Moderate |
| 6 | M | 1 | 1416 | 3.5 | 0.8 | 4 | 1.2 | NA | NQ | 5.0 | Moderate | 1 | 0 | x | x | 3 | NA |
| 7 | M | 0 | 5338 | 90 | 0.6 | 4.5 | 1.1 | Viral | 2 | 4.0 | Poor | 2 | x | Yes | x | 4 | NA |
| 8 | F | 1 | 11 | 3.6 | 0.4 | 4.2 | NA | Nash | 1 | 13.0 | Well | 1 | x | x | x | 0 | Moderate |
| 9 | M | 0 | 1272 | 31 | 0.8 | 4.3 | 1 | Viral | 1 | 3.9 | Moderate | 1 | x | x | x | 5 | NA |
| 10 | M | 1 | 746 | 3979 | 0.5 | 3.4 | 0.9 | NA | 1 | 10.1 | Moderate | 1 | x | x | x | 2 | NA |
| 11 | F | 0 | 3769 | 7.6 | 1.1 | 4.6 | 1.3 | Viral | 2 | 2.2 | Well | 2 | x | Yes | x | NA | Moderate |
| 12 | M | 0 | NA | 168 | 2 | 3.3 | 1.3 | Viral | 2 | 2.5 | Moderate_poor | 2 | x | x | x | NA | NA |
| 13 | M | 1 | 592 | 80.4 | 0.8 | 3.1 | 1.2 | Viral | 2 | 7.5 | Moderate_poor | 3 | x | Yes | x | 4 | NA |
| 14 | M | 1 | 856 | 1037 | 0.5 | 3.3 | 1.1 | Hemochromatosis | NQ | 3.8 | Moderate | 2 | x | Yes | x | 5 | Moderate |
| 15 | M | 1 | 3496 | 2.6 | 0.8 | 3.8 | 0.95 | NA | NQ | 4.6 | Moderate | 1 | x | x | x | 5 | Moderate |
| 16 | M | 1 | 479 | 17292 | 0.9 | 3.4 | 1 | Viral | NQ | 10.0 | Poor | 2 | x | Yes | x | 2 | NA |
| 17 | F | 1 | 4049 | 6.1 | 0.8 | 4.6 | 1.2 | etoh | NQ | 1.5 | Moderate | 1 | x | x | x | 6 | Mild |
| 18 | M | 1 | 1540 | 45.8 | 0.5 | 3.9 | 1.1 | dm | 1 | 5.7 | Moderate | 2 | x | Yes | x | 2 | Mild |
| 19 | M | 1 | 324 | 9415 | 1.2 | 3.6 | 1.2 | Viral | NQ | 3.8 | Poor | 2 | 0 | Yes | x | NA | NA |
| 20 | M | 1 | 306 | 37510 | 0.6 | 4.1 | 1.1 | Viral | NQ | 5.5 | Moderate | 1 | x | x | x | 6 | NA |
| 21 | M | 0 | 5030 | 1743 | 1.1 | 3.4 | 1.6 | NA | NQ | 3.5 | Moderate_poor | 1 | x | x | x | 5 | Moderate |
| 22 | M | 0 | 2197 | 9.4 | 1.1 | 5.5 | 1.2 | Viral | NQ | 4.5 | Poor | 2 | 0 | Yes | x | 6 | NA |
| 23 | M | 1 | 2884 | 76.7 | 2.5 | 2.7 | 1.6 | Viral | 2 | 5.0 | NA | 2 | 0 | Yes | x | NA | NA |
| 24 | M | 0 | 4611 | 10 | 0.4 | 4.6 | 1 | Viral | 2 | 5.0 | Moderate | 2 | x | x | x | 5 | NA |
| 25 | M | 1 | 627 | 313.2 | 0.5 | 4.2 | 1.1 | Viral | >50 | 3.0 | Moderate | 3 | x | Yes | x | 6 | Minimal |
| 26 | M | 1 | 2245 | 9635 | 1.2 | 3.6 | 1.3 | Viral | 4 | 1.2 | Well | 2 | x | x | x | 5 | Mild |
| 27 | M | 1 | 40 | 30.2 | 1.6 | 3.4 | 1.3 | Viral | 1 | 4.5 | Moderate | 1 | x | x | x | 6 | Mixed |
| 28 | M | 0 | 1480 | 2 | 0.3 | 3.6 | 1.1 | etoh | 1 | 7.1 | Moderate | 2 | x | Yes | x | NA | NA |
| 29 | M | 0 | 2955 | 64.9 | 0.3 | 3.9 | 1.2 | Viral | 1 | 10.5 | Moderate | 2 | x | Yes | x | 5 | NA |
| 30 | F | 1 | 475 | 5.6 | 0.8 | 3.3 | 2.7 | NA | NQ | 4.0 | Moderate | 1 | 0 | x | x | NA | NA |
| 31 | F | 1 | 15 | 3909 | 0.9 | 3.5 | 1.1 | Viral | NQ | 5.0 | Moderate | 2 | x | Yes | x | NA | NA |
| 32 | F | 1 | 696 | NA | 0.4 | 3.7 | 1.1 | dm | 1 | 4.3 | Moderate | 2 | x | Yes | x | NA | NA |
| 33 | M | 0 | 2886 | 21 | 0.7 | 4.6 | 1.1 | Viral | 1 | 4.8 | Moderate | 1 | x | x | x | 5 | NA |
| 34 | M | 1 | 11 | 9.5 | 1.8 | 3.8 | 1.5 | Viral | NQ | 7.2 | Moderate | 3 | x | Yes | x | 6 | NA |
| 35 | M | 1 | 957 | 105.3 | 0.8 | 3.6 | 1.2 | Viral | NQ | 11.0 | Well | 1 | x | x | x | 6 | NA |
| 36 | M | 1 | 2679 | NA | 0.8 | 4.8 | 1.4 | NA | NQ | 3.1 | Moderate | 2 | x | Yes | x | 6 | Moderate |
| 37 | F | 0 | 3466 | 39.3 | 0.6 | 3.9 | 1 | NA | 1 | 15.0 | Moderate | 2 | 0 | Yes | x | 0 | NA |
| 38 | M | 1 | 0 | 801.5 | 1.4 | 3.2 | 1.2 | Viral | 1 | 2.8 | Moderate | 2 | x | Yes | x | 5 | NA |
| 39 | M | 0 | 3519 | 1.66 | 1.3 | 2.9 | 1 | Viral | 1 | 4.2 | Moderate_poor | 2 | x | Yes | Yes | 6 | NA |
| 40 | F | 1 | 1097 | 2 | 0.5 | 5 | 1.1 | dm | 1 | 6.5 | Moderate | 2 | x | Yes | x | 0 | Moderate |
| 41 | M | 1 | 1034 | 119 | 0.4 | 4.4 | 1.1 | NA | 2 | 6.2 | Moderate | 3 | x | x | x | 0 | NA |
Abbreviations: NA, none; NQ, not quantified.
Hierarchical clustering
Counts were Q3 normalized as described in Supplemental Methods, http://links.lww.com/HC9/B46. We then performed log2 transformation of the Q3 normalized counts and mean-centered all the genes such that every gene to be clustered has a mean of 0 while keeping the variance. We further performed agglomerative hierarchical clustering with the hclust package and used average linkage as the distance metric.
Differential expression analyses
For any paired group comparisons, we obtained the log2 fold change between the mean of the 2 groups. To obtain p values for tumor-vessel area of interest (AOI) comparison, we used linear mixed-effects models with the lmerTest package where AOI type was modeled as the fixed effect and resection sample ID as the random effect. To compare the 3 tumor niches in GeoMx data and between cancer and tumor-associated endothelial cells (TECs) in pseudo-bulk scAtlasLC data, we used the Wilcoxon rank sum test. For visualization of the results, we plotted volcano plots with log2 fold change on the x-axis and false discovery rate-adjusted p values on the y-axis.
Vascularization and Hoshida subtype correlation
We defined the extent of vascularization as the percentage of vessel AOI area in a region of interest (ROI). Example images of ROIs of high and low vascularization are shown in Supplemental Figures S3B and C, http://links.lww.com/HC9/B46. To assign a Hoshida subtype for each ROI, we aggregated raw counts from tumor and vessel AOIs to represent bulk transcriptomics data, similar to what was analyzed by Hoshida et al in 2009. We then assigned a subtype per ROI based on the relative expression levels of S1, S2, and S3 genes.
Tumor AOI subtype analysis and ligand-receptor analysis
To identify distinct tumor-vessel niches, we performed hierarchical clustering on the top 100 most variable genes in the tumor AOIs, where we found 3 clusters, termed T1, T2, and T3 AOIs. We performed pairwise differential expression analyses as described above and found genes enriched in the 3 tumor clusters, which we call T1, T2, and T3 genes. To understand interactions in a spatial context, we used CellChat, a curated database of known ligand-receptor pairs. 18 For each ligand-receptor pair, we calculated its correlation and effect size between the ligand expression in the tumor AOIs and the receptor expression in the matched vessel AOIs. This analysis was performed across all ROIs as well as in each of the T1, T2, and T3 subtypes.
Resolve Molecular Cartography data visualization
The Resolve Molecular Cartography technology, with subcellular transcript resolution, is based on combinatorial single-molecule fluorescent in situ hybridization. This technology accommodates tens of transcript-specific probes per target RNA for up to 100 target RNAs for each sample. We designed a custom 100-gene panel and applied it to snap-frozen tumors and matched liver resection samples from an additional 4 patients who had undergone resection of HCC primary tumors. Snap-frozen tissues were run by the Resolve company scientists according to the manufacturer’s instructions (protocol 3.0, available for download from Resolve’s website for registered users) and described elsewhere. 19 We visualized genes from the Hoshida gene sets (S1: ACTA2, CD8A, IL7R, LGALS9, VCAN, CCR7, CXCL1, LGALS1, and PTPRC; S2: FGFR4, GPC3, and HMGCR; S3: ALDH1A1, ARG1, ASGR1, FLT4, and IFIT1) onto all the samples, as well as locations of nuclei (DAPI staining) and canonical endothelial genes (PECAM1, VWF, KDR, CD34, IL33, PLVAP, VCAM1, and LIVE1). To avoid overplotting, we represent endothelial transcripts by their density, with darker pink depicting a region of higher transcript density than lighter pink regions.
scAtlasLC data analysis
We used HCC samples from public liver cancer single-cell RNA sequencing data and associated cell typing 12 and subsetted cancer and TECs for our analyses. Similar to AOI-level GeoMx analyses, we aggregated raw counts from cancer and TECs for each bulk tumor and plotted gene expression in a heatmap and annotated by Hoshida gene sets, shown in Figure 3A. We also performed differential expression analysis to compare genes enriched in tumor cells and TECs, colored by Hoshida gene sets.
FIGURE 3.
Single-cell dissociative and in situ gene expression profiling directly confirms endothelial expression of most “S1” HCC tumor genes. (A) Unsupervised hierarchical clustering of quasi-bulk cancer cells and TECs from the scAtlasLC data set confirm S1 genes are more highly expressed in TECs and S3 genes are in cancer cells. (B) Volcano plot showing differentially expressed genes between quasi-bulk cancer cells and TECs with genes colored by Hoshida gene set membership (S1 black, S2 blue, and S3 green) and labeled if their fold changes exceed 2 and meet FDR-adjusted p value threshold. (C) Molecular Cartography direct transcript visualization demonstrates colocalization of S3 genes (green) with cancer cells and S1 genes (blue) with endothelial gene density (red). Abbreviations: FDR, false discovery rate; TEC, tumor-associated endothelial cell.
Survival analysis
We used survival and survminer packages in R and performed survival analyses on both TCGA and our GeoMx data sets. TCGA data contained bulk transcriptome expression of 367 patients with HCC across 20,518 genes. For each gene, we divided patients into high- and low-expressors based on median split and then used the Cox Proportional Hazard test to compare survival between the 2 groups. We visualized results across all genes on a volcano plot and colored genes based on our T1/T2/T3 designations. In our GeoMx data set, we plotted Kaplan-Meier plots across the tumor AOIs, stratifying by their T1/T2/T3 phenotypes.
Code availability
Analyses were performed in R and Python, and all code used for the paper can be found at https://github.com/clu413/HCC_GeoMx_paper. The data are deposited in the Gene Expression Omnibus (GEO), accession number GSE277104.
RESULTS
GeoMx technology accurately segments vessel regions from tumor-rich regions in HCC tissue
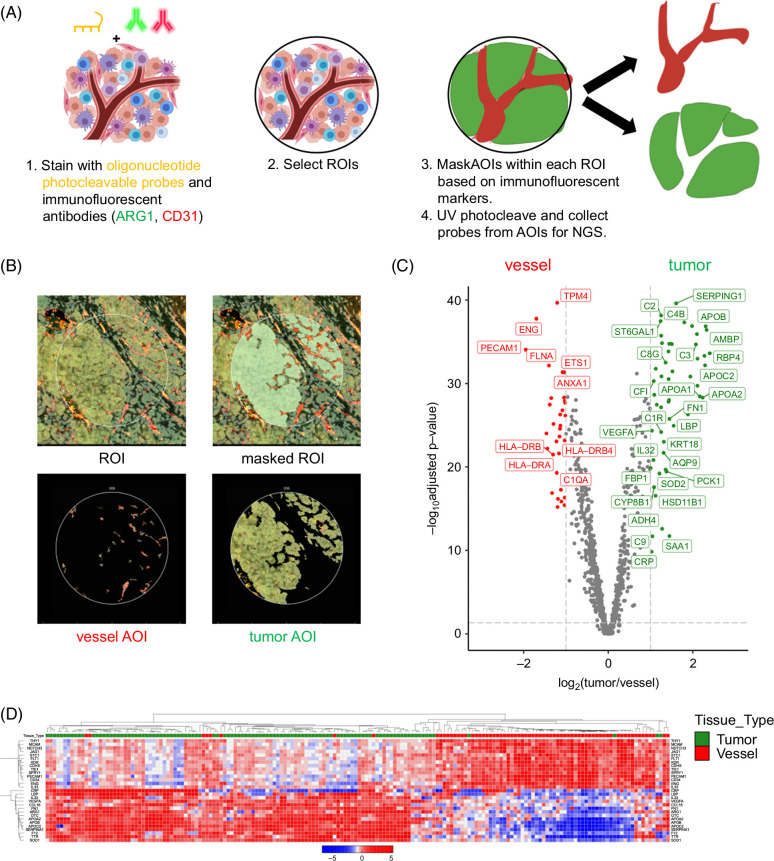
To understand how compartment-specific gene expression programs are spatially organized in situ in HCC tissue, we used the Nanostring GeoMx Digital Spatial Profiling platform and the Cancer Transcriptome Atlas (~1800 genes) oligonucleotide probe set. We analyzed a cohort of formalin-fixed, paraffin-embedded specimens from 41 patients with resected, treatment-naïve HCC (Table 1) for our primary analysis. In this workflow, we hybridized oligonucleotide probes targeting the gene set of interest to tag mRNAs from microscopic ROIs, stained with fluorescent primary antibodies against Arginase 1 (ARG1; the most sensitive and specific marker for hepatocellular differentiation 20 and CD31 to identify areas enriched in HCC cells and blood vessels, respectively, and then used fluorescence-guided digital segmentation to liberate the probe tags for downstream next-generation sequencing processing (Figure 1A). This protocol captures highly purified AOIs (Figure 1B). After data quality control and preprocessing, we obtained 65 ROIs with matched tumor and vessel AOIs from these 41 patients.
FIGURE 1.
GeoMx spatial profiling precisely separates vessels from tumor regions for gene expression profiling. (A) Workflow of sequential tissue staining with photocleavable oligonucleotide probes followed by staining with immunofluorescent tissue subcompartment labels. (B) Example of 1 HCC tumor “region of interest” (ROI), masked ROI to identify CD31-positive (red) and Arg1-positive (green) “areas of interest” (AOIs). (C) Volcano plot of genes differentially expressed across all ROIs and AOIs (false discovery rate–adjusted p value ≥0.05, log2(fold change) ≥1), with genes colored in green for tumor AOIs and red for vessel AOIs. (D) Hierarchically clustered heatmap of all tumor (green column label) and vessel (red column label) AOIs.
Differential expression analysis of the vessel AOIs compared with the tumor AOIs demonstrated 32 genes enriched in the former and 53 genes in the latter (Supplemental Table S1, http://links.lww.com/HC9/B45). As expected, there were many canonical endothelial genes (eg, CD34, CDH5, KDR, and PECAM1/CD31) in the former and many hepatocyte genes (eg, APOA1, ARG1, C2, and CLU) in the latter (Figure 1C). Gene set enrichment analysis supported the significant and consistent enrichment of EC genes in the vessel AOIs (Supplemental Figure S1A, http://links.lww.com/HC9/B46) and liver-related genes in the tumor AOIs (Supplemental Figure S1B, http://links.lww.com/HC9/B46). Unsupervised clustering of both tumor and vessel AOIs annotated with a set of canonical endothelial and liver-related genes confirmed clean separation of vessel and tumor AOI clusters (Figure 1D). Computational mixed cell type deconvolution analysis of the vessel AOIs indicated that the vessel AOIs contain mostly ECs and—to a smaller and more heterogeneous extent—various immune cell subtypes (Supplemental Figure S2, http://links.lww.com/HC9/B46). Taken together, the above results confirm that the GeoMx spatial transcriptomic workflow generates gene expression profiles from highly purified HCC vessel regions and tumor regions. We next sought to discover more about the vessel and tumor compartments using additional unsupervised analyses.
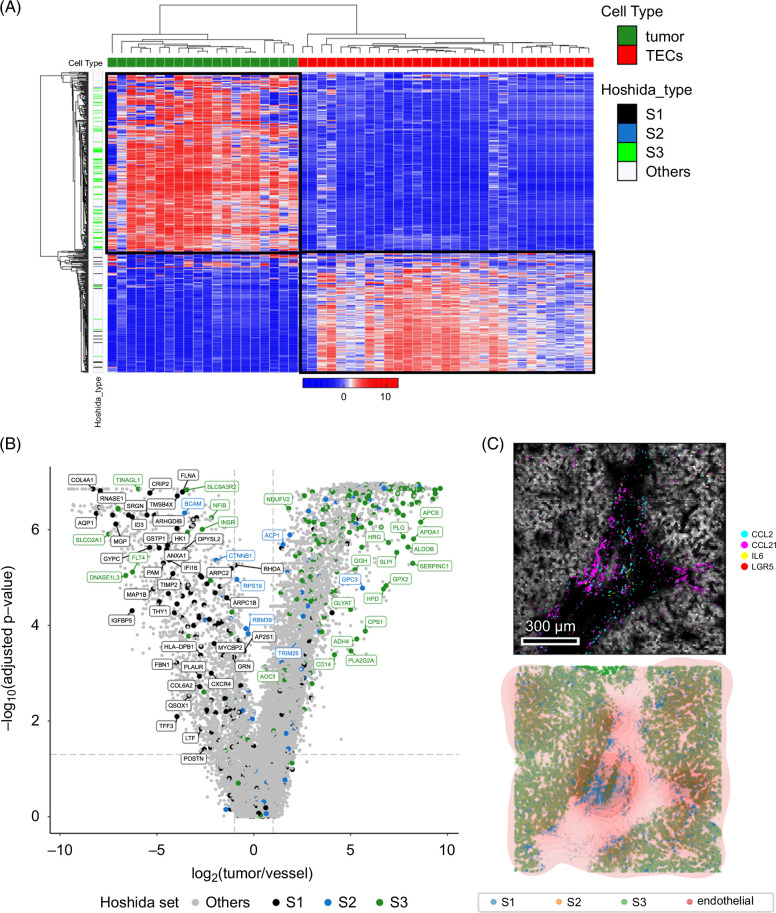
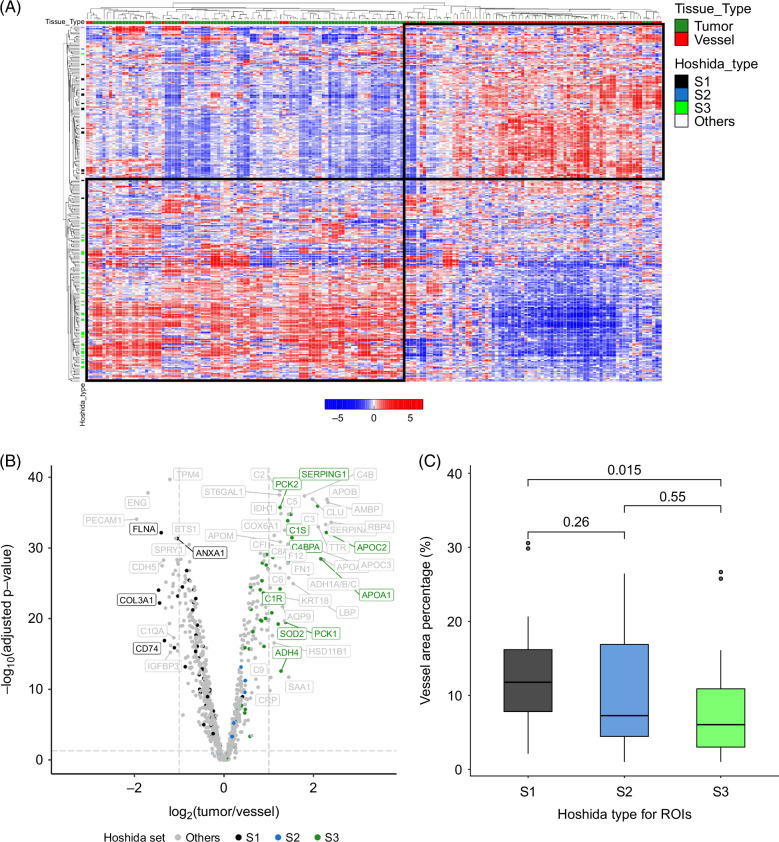
Unsupervised analysis of tumor and vessel AOIs demonstrates a significant enrichment of Hoshida bulk tissue subtype 1 (S1) in the vessel compartment
Unsupervised hierarchical clustering of the entire set of tumor AOIs and vessel AOIs using the 300 most variably expressed genes yielded 2 distinct clusters, with classical endothelial and liver genes associating within one cluster and mostly liver genes associating with the other (Figure 2A). We noticed that those genes present in the canonical “S1” bulk tissue subtype established by Hoshida et al 11 were highly enriched in the vessel AOIs, whereas “S3” and—to a lesser extent due to low gene set representation—“S2” genes were enriched in the tumor AOIs. Specifically, after quality control and preprocessing, the 1800-gene Cancer Transcriptome Atlas (CTA) panel that we used for this study contained 56/235 of the S1 genes, 16/115 of the S2 genes, and 48/266 of the S3 genes. Of the 56 Hoshida S1 genes in the CTA panel, all were more prominent in the vessel AOIs; 43 of the 48 were more prominent in the tumor AOIs. Indeed, when limiting the genes belonging to the S1, S2, and S3 gene sets present in the 1800-gene cancer transcriptome atlas, differential expression analysis between the vessel and tumor AOIs confirmed that the majority of S1 genes were enriched in the vessel AOIs and the majority of S3 genes in the tumor AOIs (Supplemental Figure S3A, http://links.lww.com/HC9/B46 and Supplemental Table S1, http://links.lww.com/HC9/B45). We also noticed a correlation between vessel AOI area and HCC subtype, assigned by relative enrichment of the Hoshida gene sets. ROIs with a larger vessel AOI fraction (Supplemental Figure S3B, http://links.lww.com/HC9/B46) were enriched in S1 genes and thus were assigned S1 phenotype, whereas the ROIs with smaller vessel fractions (Supplemental Figure S3C, http://links.lww.com/HC9/B46) were assigned S3 phenotype (Figure 2C), showing consistent bulk tumor subtype results from the original Hoshida study. 11 Altogether, vascular density was strongly associated with classification as S1 HCC subtype.
FIGURE 2.
Established bulk HCC tissue transcriptional subtypes can be refined by separate analysis of the associated vascular and tumor-enriched compartments. (A) Unsupervised hierarchical clustering of the 500 most variable genes yields a clear separation of the vessel (red label on top) and tumor (green label) AOIs, with Hoshida S1 (black label on left) genes showing higher expression in vessel AOIs and S3 (green label) genes showing higher expression in tumor AOIs. (B) Volcano plot showing differentially expressed genes between tumor and vessel AOIs, with genes colored by Hoshida gene set membership (S1 black, S2 blue, S3 green, and others not in Hoshdia gene sets gray) and labeled if their fold changes exceed 2 and meet FDR-adjusted p value threshold. (C) Boxplot of composite ROI gene expression shows that S1 ROI classification is related to high vessel fraction and S3 ROI classification to high tumor fraction (S3 vs. S1 p = 0.015; Kruskal-Wallis test). Abbreviations: AOI, areas of interest; FDR, false discovery rate; ROI, regions of interest.
To confirm our observations that S1 genes tended to be expressed in HCC vessels and S3 genes in tumor regions, we used the single-cell Atlas in Liver Cancer (scAtlasLC), 12 an orthogonal, publicly available data set of treatment-naïve single-cell RNA sequencing data of primary liver cancers. We used the scAtlasLC HCC data subset with associated cell type annotations and combined single-cell RNA sequencing reads in each sample to generate quasi-bulk compartment-level data—with all annotated EC reads combined and all annotated cancer cell reads combined from a given tumor—to compare with our GeoMx analysis. Hierarchical clustering of the data confirmed that most S1 genes were highly expressed in ECs and S3 genes in cancer cells (Figure 3A). Of the genes that were differentially expressed between the ECs and cancer cells in this data set, we found again a significant enrichment (90/228) of S1 genes with higher expression in ECs, whereas the vast majority of S2 (5/111) and S3 genes (18/257) had the opposite pattern, with higher expression in cancer cells (Figure 2C and Supplemental Table S2, http://links.lww.com/HC9/B45). Gene set enrichment analysis of the specific S1 genes that were more highly expressed in ECs implied significant inflammatory and immunomodulatory functions, whereas the S3 genes that were more highly expressed in ECs were more consistent with differentiated, tissue-resident ECs (Supplemental Figure S4, http://links.lww.com/HC9/B46). To provide additional validation of our results, we used the Resolve “Molecular Cartography” technology 19 to visualize 100 genes in situ at single-cell resolution in 4 separate HCC resection specimens. For ease of visualization, we plotted all S1 gene transcripts (ACTA2, CD8A, IL7R, LGALS9, VCAN, CCR7, CXCL1, LGALS1, and PTPRC) in blue, all S2 transcripts (FGFR4, GPC3, and HMGCR) in orange, and all S3 transcripts (ALDH1A1, ARG1, ASGR1, FLT4, and IFIT1) in green. Known markers of ECs (CD34, KDR, PLVAP, VCAM1, VWF, IL33, and PECAM1) density is indicated by the color of pink rings. From direct visualization of transcripts in situ qualitatively, we again noted that S1 genes colocalized with endothelial genes in regions devoid of S3 genes, and S3 genes are highly expressed in regions of tumor parenchyma (Figure 3C; Supplemental Figure S5, http://links.lww.com/HC9/B46).
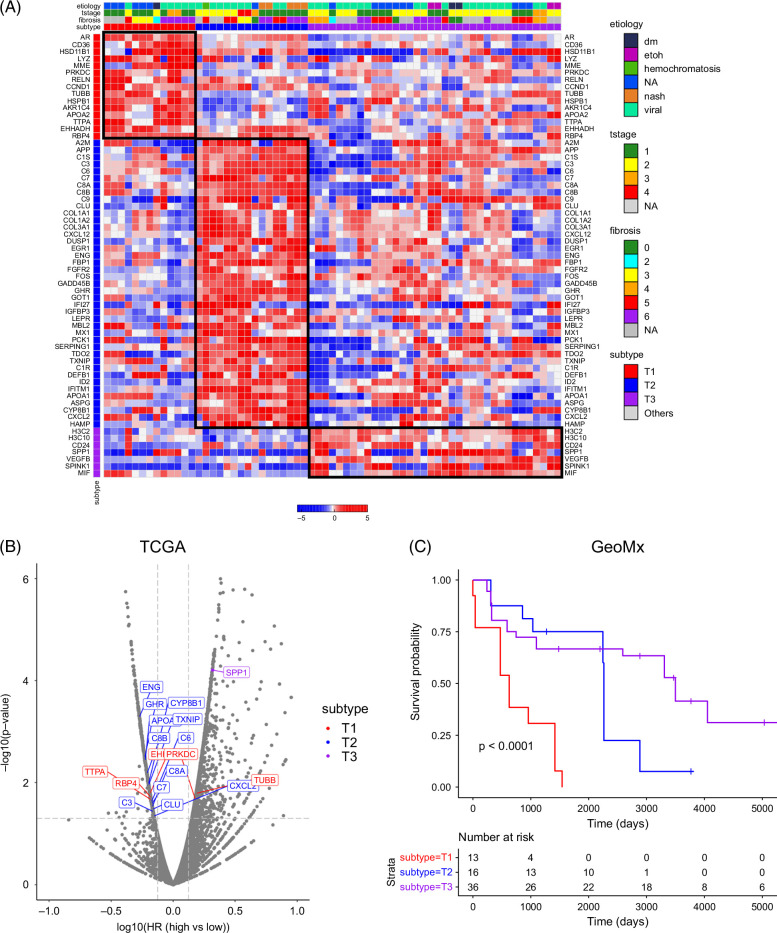
Tumor-enriched clustering demonstrates 3 distinct tumor AOI phenotypes associated with differential survival outcomes
Since previously described HCC bulk tissue signatures contained a significant stromal (EC) contribution, we next sought to define novel clusters that were more specific to the tumor compartment and characterize their microenvironment niches. Hierarchical clustering of tumor AOIs using the 100 most variable genes across the samples yielded 3 clusters, which we denote as T1, T2, and T3 (Figure 4A). Hoshida S3 genes were represented across the groups. Differential expression analysis of the genes across clusters revealed enrichment in inflammatory and oxidative stress–related genes (eg, CD36, EHHADH, and RELN) in cluster T1, well-differentiated and metabolic liver metabolic genes (A2M, APOA1, and HAMP) in cluster T2, and stemness genes (CD24 and SPP1) in cluster T3 (Supplemental Figure S6, http://links.lww.com/HC9/B46; Supplemental Table S3, http://links.lww.com/HC9/B45).
FIGURE 4.
HCC tumor-enriched AOIs cluster into 3 molecular groups with survival correlation. (A) Hierarchical clustering of tumor AOIs demonstrates 3 clusters: T1, T2, and T3. (B) Genes enriched in T1/T2/T3 phenotypes are differentially associated with overall survival in TCGA. (C) T1/T2/T3 phenotypes in our GeoMx data set have different survival outcomes, with “T1” subtype demonstrating inferior survival compared with the “T2” and “T3” subtypes. Abbreviations: AOI, areas of interest; TCGA, The Cancer Genome Atlas.
Next, we sought to correlate these gene signatures with overall survival, both in our own data set (GeoMx) and in The Cancer Genome Atlas (TCGA). In the TCGA data set, we conducted survival analyses for each gene by comparing survival between high expressors and low expressors of the gene of interest. We found that bulk tumor expression of T2 genes was associated with better survival, as patients with higher expressions of these genes had lower survival hazard ratios than those with lower expression of T2 genes (Figure 4B). In our GeoMx data set, tumor AOIs categorized as T2 and T3 are also associated with better survival than T1 AOIs (Figure 4C). Taken together, our data demonstrate that our tumor-specific signature—with the removal of much of the EC gene expression signals inherent in bulk transcriptomic data—may provide a novel prognostic schema for HCC survival.
Spatial analysis identifies subtype-specific ligand-receptor interactions between microscopic cancer regions and vascular regions
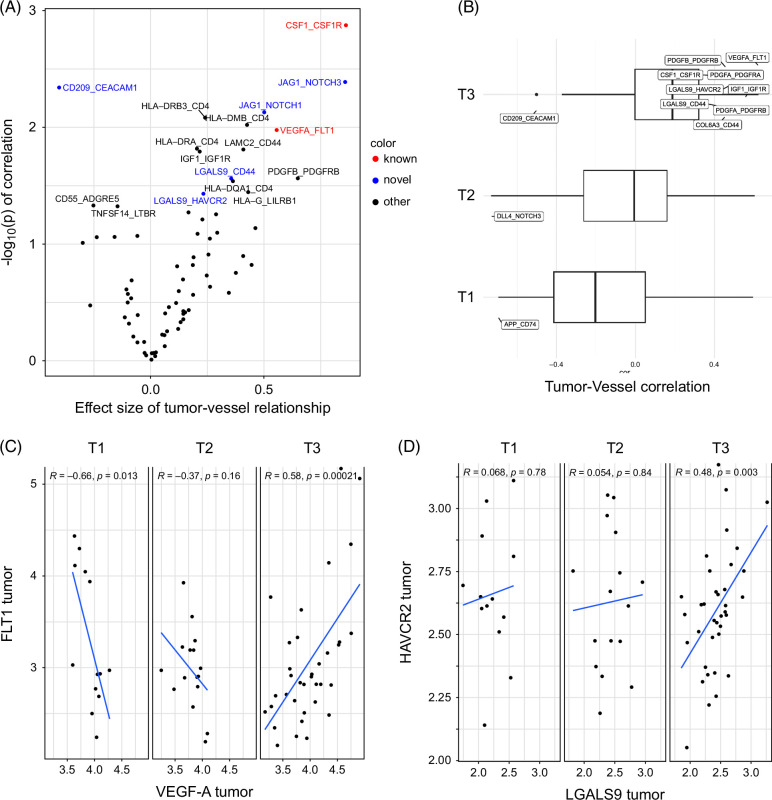
After we identified heterogeneous tumor-enriched regions with clear prognostic differences, we hypothesized that linked tumor-vascular interactions may correlate with these microenvironment subsets. We, therefore, applied a published computational tool designed for inference of cell-cell interactions to our spatial transcriptional data. 18 Using the CellChat curated database, we first defined the correlated tumor-vessel interactions across all samples and found examples of both expected (eg, VEGFA in the tumor and FLT1 a.k.a. VEGFR1 in the vessel) and potentially novel (LGALS9 in tumor and CD44 in vessel) ligand-receptor pairs (Figure 5A). Next, we subdivided the data into T1/T2/T3 subtypes and found differential correlations between tumor-vessel pairs (Figure 5B) in a T-subtype–dependent manner. For example, VEGFA in tumor AOIs correlated strongly with FLT1 in T3 ROIs (Figure 5B), but this correlation was not noted in T2, and there was an anticorrelation of this pair in T1 ROIs (Figure 5C). In addition, LGALS9 in T3 subregions exhibited a strong positive correlation with HAVCR2 (a.k.a. TIM3) in T3-linked vessel regions (Figure 5B) but not T1 or T2 ROIs. Taken together, our data point to heterogeneous tumor-vessel interactions driving local tumor biology and motivating the development of tissue subtyping for future precision medicine approaches.
FIGURE 5.
HCC tumor subtypes contain heterogeneous cancer-EC ligand-receptor interactions. (A) Correlation of tumor-vessel ligand-receptor gene pairs across all samples demonstrates both known therapeutically targeted interactions (red) and potentially novel targets (blue). (B) Ligand-receptor interactions between tumor and vessel AOIs vary depending on T subtypes. (C) Correlation of VEGFA in tumor AOIs and FLT1 in matched vessel AOIs is either positive (T3), negative (T1), or null (T2). (D) Correlation of LGALS9 in tumor AOIs and HAVCR2 (a.k.a. TIM3) in vessel AOIs is either positive (T3) or null (T1, T2). Abbreviations: AOI, areas of interest; EC, endothelial cell.
DISCUSSION
The TME is composed of a dynamic, heterogeneous collection of interacting parenchymal (cancer) and stromal (noncancer) cell types, 21 and many of cancer’s “hallmarks” are fundamentally dependent on these interactions. 9 Spatial transcriptomics technologies aim to bridge the gap between traditional intact tissue characterization techniques such as immunohistochemical staining and RNA in situ hybridization and high-plex but dissociative technologies like single-cell RNA sequencing. 22 ECs are the most common stromal cell type in HCC, and tumor microvascular density correlates with adverse outcomes in HCC. 23 Further, ECs are a direct target for many of the active systemic therapy regimens in advanced HCC. 24
Given the above, we decided to focus our initial HCC spatial transcriptomic profiling effort on ECs. We used the Nanostring GeoMx technology to precisely separate vessel-dense AOIs from tumor-dominant AOIs and found a clear and dominant EC contribution to the Hoshida S1 HCC bulk tissue subgroup. Further, we found that higher vessel density was correlated with an increased likelihood of a tissue microregion being classified as S1 in our cohort of 41 patients with resected HCC. We validated this finding using the publicly available single-cell atlas in the liver cancer (scAtlasLC 12 ) data set and with direct in situ visualization of S1 genes with EC genes using the Resolve “Molecular Cartography” platform.
Using our purified tumor AOIs, we defined 3 novel transcriptional subgroups (T1–T3) of microscopic tumor regions. We found that the T2 subtype was defined by genes like those expressed by normal liver parenchyma and “well-differentiated” HCC tumors, whereas the T1 subtype contains inflammatory and oxidative stress–related genes, and T3 contains inflammatory and stemness genes. Notably, several of the genes from our unbiased clustering and classification scheme have been described to affect HCC outcomes. For example, the highest-risk T1 subgroup contained multiple genes previously shown to contribute to aggressive HCC features, including CD36 (encoding a fatty acid receptor protein that contributes to metabolic derangement in HCC and thereby promotes tumor progression 25 ), LYZ (lysozyme, a secreted enzyme promoting HCC proliferation and migration 26 ), and CCND1 (cyclin D1, a cell cycle proliferation protein 27 ). Conversely, many T2 genes associated with “well-differentiated” HCC—including IGFBP3 (insulin-like growth factor binding protein 3 28 ) and GHR (ghrelin 29 )—have been identified as good prognostic features in prior studies.
There have been other recently published HCC spatial transcriptomic profiling efforts that have focused on other components of the tumor milieu, such as carcinoma-associated fibroblast subsets 30 or T-cell subsets. 31 We focused our analyses on potential heterotypic receptor-ligand interactions between purified tumor subregions (as a proxy for cancer cells) and vascular subregions (a proxy for ECs). We found both known interactions and potentially novel clinically relevant interactions (LGALS9 tumor-CD44 or HAVCR2 vessel, JAG1 tumor-NOTCH1/3 vessel) within the HCC TME in a manner that depended on the T subtype. Interestingly, LGALS9 (in tumor) and SLC1A5 (macrophages) have been previously identified as potentially important in the HCC microenvironment. 32 Further, multiple of these heterogeneous signaling interactions represents immune and vascular targets in clinical development, including LGALS9 with HAVCR2,33,34 LGALS9 with CD44, 35 and JAG1 with NOTCH1/3. 36 Our study provides further evidence of the dynamic and potentially targetable interactions between cancer cells and their stromal ECs. 37
This study had some limitations. First, the GeoMx gene probe panel was limited to the 1800 gene “cancer transcriptome atlas” gene panel, which was the highest-plex panel available at the time of the start of the project but did not cover the entire transcriptome. In addition, as with all gene expression analyses, the translated protein concentrations for our gene sets may not fully match. Third, although the GeoMx analyses are limited to microscopic ROIs that have hundreds to thousands of purified cells, each AOI still potentially contains multiple different cell types—including entrained immune cells—and hence the gene expression profile obtained from a given AOI is still a “bulk” profile, for example, with potential fibroblast genes present in vascular AOIs (Supplemental Figure S1, http://links.lww.com/HC9/B46). Fourth, given the imperfect nature of ROI segmentation into AOIs, there is always the possibility of “bleed over” of genes from one AOI into another (eg, the tumor into a vessel). Finally, our tissue specimens were collected from a small, discovery cohort of patients with resected, treatment-naïve HCC, and hence, these samples may not reflect the full breadth of TME heterogeneity that may be reflected within larger data sets or in different background tissue environments (eg, metastatic sites and more severely cirrhotic livers) or after exposure to various treatments.
In summary, we performed spatial transcriptomic analysis of treatment-naïve HCC, focusing on analyzing tumor parenchyma separate from blood vessels. In addition to clarifying and expanding upon prior bulk transcriptional tissue classifiers, our study illustrates the importance of spatial transcriptomics to provide higher precision in characterizing cellular ecosystems and identifying significant heterotypic cellular interactions within the TME, such as the LGALS9 (tumor)-HAVCR2 (vessel) axis. The next-generation of single-cell spatial transcriptomic technologies will provide even higher resolution of cell-cell interactions to identify additional cancer-stroma interactions and thereby nominate potential biomarkers and therapeutic targets for future development. Future functional studies based on these putative interactions using in vitro and in vivo models and perturbations of these TME interactions (eg, with vascular-targeted, immune-targeted, or other treatments) will unlock new therapeutic avenues for HCC.
Supplementary Material
Acknowledgments
FUNDING INFORMATION
David T. Ting received funding support from the NIH (U01CA228963). Yujin Hoshida received funding support from the NIH (CA233794, CA255621), the European Commission (ERC-AdG-2020-101021417), and the Cancer Prevention and Research Institute of Texas (RR180016). Joseph W. Franses received funding support from the NIH (CA263551). Chenyue Lu received fellowship funding support from the Sita Foundation.
CONFLICTS OF INTEREST
Yujin Hoshida advises and owns stock in Alentis and Espervita Therapeutics. He advises Helio Genomics, Roche Diagnostics, and Elevar Therapeutics. David T. Ting consults, advises, and owns stock in Rome Therapeutics and PanTher Therapeutics. He advises and owns stock in TellBio Incorporated. He consults for Moderna, ABRDN, AstraZeneca, Leica Biosystems Imaging, and Sonata Therapeutics and received grants from Sanofi and Incyte. Joseph W. Franses consults for Eisai, Foundation Medicine, Guardant Health, and Genetech. He received grants from Abbvie, Iterion Therapeutics, and Omega Therapeutics. David T. Ting has received an honorarium from NanoString Technologies, whose technology was used in this work. David T. Ting has received consulting fees from ROME Therapeutics and Tekla Capital that are not related to this work. David T. Ting has received honorariums from Moderna, Ikena Oncology, Foundation Medicine, Inc., and Pfizer that are not related to this work. David T. Ting is a founder and has equity in ROME Therapeutics, PanTher Therapeutics, and TellBio, Inc., which is not related to this work. D.T.T. receives research support from ACD-Biotechne, PureTech Health LLC, Ribon Therapeutics, AVA LifeScience GmbH, and Incyte, which was not used in this work. David T. Ting’s interests were reviewed and managed by Massachusetts General Hospital and Mass General Brigham in accordance with their conflict of interest policies. Yujin Hoshida has competing financial interests at Helio Genomics, Alentis Therapeutics, and Espervita Therapeutics, and additionally holds advisory/management and consulting positions at Roche Diagnostics. Joseph W. Franses holds consulting positions at Eisai, Foundation Medicine, Genentech, Guardant Health, and Servier. Martin J. Aryee has financial interests in SeQure Dx, unrelated to this work. The remaining authors have no conflicts to report.
Footnotes
David T. Ting, Martin J. Aryee, and Joseph W. Franses contributed equally to this work.
Abbreviations: AOI, area of interest; CTA, Cancer Transcriptome Atlas; EC, endothelial cell; ROI, region of interest; scAtlasLC, single-cell Atlas in Liver Cancer; TCGA, The Cancer Genome Atlas; TEC, tumor-associated endothelial cell; TME, tumor microenvironment.
Supplemental Digital Content is available for this article. Direct URL citations are provided in the HTML and PDF versions of this article on the journal’s website, www.hepcommjournal.com.
Contributor Information
Chenyue Lu, Email: chenyuel@mit.edu.
Amaya Pankaj, Email: apankaj@mgh.harvard.edu.
Michael Raabe, Email: MRAABE@mgh.harvard.edu.
Cole Nawrocki, Email: CCNAWROCKI@PARTNERS.ORG.
Ann Liu, Email: annliu@caltech.edu.
Nova Xu, Email: nxu1@stanford.edu.
Bidish K. Patel, Email: BPATEL31@mgh.harvard.edu.
Matthew J. Emmett, Email: Matthew_Emmett@DFCI.HARVARD.EDU.
Avril K. Coley, Email: AKCOLEY@mgh.harvard.edu.
Cristina R. Ferrone, Email: Cristina.Ferrone@cshs.org.
Vikram Deshpande, Email: eic.jcp.deshpande@gmail.com.
Irun Bhan, Email: IBHAN1@MGH.HARVARD.EDU.
Yujin Hoshida, Email: yujin.hoshida@utsouthwestern.edu.
David T. Ting, Email: DTING1@mgh.harvard.edu.
Martin J. Aryee, Email: martin.aryee@ds.dfci.harvard.edu.
Joseph W. Franses, Email: joseph.franses@bsd.uchicago.edu.
REFERENCES
- 1.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–249. [DOI] [PubMed] [Google Scholar]
- 2.Vogel A, Meyer T, Sapisochin G, Salem R, Saborowski A. Hepatocellular carcinoma. Lancet. 2022;400:1345–1362. [DOI] [PubMed] [Google Scholar]
- 3.Cancer Genome Atlas Research Network . Electronic address wbe, Cancer Genome Atlas Research N. Comprehensive and integrative genomic characterization of hepatocellular carcinoma. Cell. 2017;169:1327–1341.e1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Llovet JM, Pinyol R, Kelley RK, El-Khoueiry A, Reeves HL, Wang XW, et al. Molecular pathogenesis and systemic therapies for hepatocellular carcinoma. Nat Cancer. 2022;3:386–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Folkman J. Tumor angiogenesis: Therapeutic implications. N Engl J Med. 1971;285:1182–1186. [DOI] [PubMed] [Google Scholar]
- 6.Jain RK. Normalization of tumor vasculature: An emerging concept in antiangiogenic therapy. Science. 2005;307:58–62. [DOI] [PubMed] [Google Scholar]
- 7.Rafii S, Butler JM, Ding BS. Angiocrine functions of organ-specific endothelial cells. Nature. 2016;529:316–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nagl L, Horvath L, Pircher A, Wolf D. Tumor endothelial cells (TECs) as potential immune directors of the tumor microenvironment—New findings and future perspectives. Front Cell Dev Biol. 2020;8:766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hanahan D. Hallmarks of cancer: New dimensions. Cancer Discov. 2022;12:31–46. [DOI] [PubMed] [Google Scholar]
- 10.Ribatti D, Vacca A, Nico B, Sansonno D, Dammacco F. Angiogenesis and anti-angiogenesis in hepatocellular carcinoma. Cancer Treat Rev. 2006;32:437–444. [DOI] [PubMed] [Google Scholar]
- 11.Hoshida Y, Nijman SM, Kobayashi M, Chan JA, Brunet JP, Chiang DY, et al. Integrative transcriptome analysis reveals common molecular subclasses of human hepatocellular carcinoma. Cancer Res. 2009;69:7385–7392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ma L, Hernandez MO, Zhao Y, Mehta M, Tran B, Kelly M, et al. Tumor cell biodiversity drives microenvironmental reprogramming in liver cancer. Cancer Cell. 2019;36:418–430 e416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma L, Wang L, Khatib SA, Chang CW, Heinrich S, Dominguez DA, et al. Single-cell atlas of tumor cell evolution in response to therapy in hepatocellular carcinoma and intrahepatic cholangiocarcinoma. J Hepatol. 2021;75:1397–1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen K, Wang Q, Li M, Guo H, Liu W, Wang F, et al. Single-cell RNA-seq reveals dynamic change in tumor microenvironment during pancreatic ductal adenocarcinoma malignant progression. EBioMedicine. 2021;66:103315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu F, Fan J, He Y, Xiong A, Yu J, Li Y, et al. Single-cell profiling of tumor heterogeneity and the microenvironment in advanced non-small cell lung cancer. Nat Commun. 2021;12:2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kalluri AS, Vellarikkal SK, Edelman ER, Nguyen L, Subramanian A, Ellinor PT, et al. Single-cell analysis of the normal mouse aorta reveals functionally distinct endothelial cell populations. Circulation. 2019;140:147–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rao A, Barkley D, Franca GS, Yanai I. Exploring tissue architecture using spatial transcriptomics. Nature. 2021;596:211–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jin S, Guerrero-Juarez CF, Zhang L, Chang I, Ramos R, Kuan CH, et al. Inference and analysis of cell-cell communication using CellChat. Nat Commun. 2021;12:1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guilliams M, Bonnardel J, Haest B, Vanderborght B, Wagner C, Remmerie A, et al. Spatial proteogenomics reveals distinct and evolutionarily conserved hepatic macrophage niches. Cell. 2022;185:379–396.e338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Umetsu SE, Kakar S. Evaluating liver biopsies with well-differentiated hepatocellular lesions. Surg Pathol Clin. 2023;16:581–598. [DOI] [PubMed] [Google Scholar]
- 21.Liu P, Kong L, Liu Y, Li G, Xie J, Lu X. A key driver to promote HCC: Cellular crosstalk in tumor microenvironment. Front Oncol. 2023;13:1135122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Palla G, Fischer DS, Regev A, Theis FJ. Spatial components of molecular tissue biology. Nat Biotechnol. 2022;40:308–318. [DOI] [PubMed] [Google Scholar]
- 23.Zhang Q, Wu J, Bai X, Liang T. Evaluation of intra-tumoral vascularization in hepatocellular carcinomas. Front Med (Lausanne). 2020;7:584250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang C, Zhang H, Zhang L, Zhu AX, Bernards R, Qin W, et al. Evolving therapeutic landscape of advanced hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol. 2023;20:203–222. [DOI] [PubMed] [Google Scholar]
- 25.Luo X, Zheng E, Wei L, Zeng H, Qin H, Zhang X, et al. The fatty acid receptor CD36 promotes HCC progression through activating Src/PI3K/AKT axis-dependent aerobic glycolysis. Cell Death Dis. 2021;12:328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gu Z, Wang L, Dong Q, Xu K, Ye J, Shao X, et al. Aberrant LYZ expression in tumor cells serves as the potential biomarker and target for HCC and promotes tumor progression via csGRP78. Proc Natl Acad Sci USA. 2023;120:e2215744120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ding H, Wang Y, Zhang H. CCND1 silencing suppresses liver cancer stem cell differentiation and overcomes 5-Fluorouracil resistance in hepatocellular carcinoma. J Pharmacol Sci. 2020;143:219–225. [DOI] [PubMed] [Google Scholar]
- 28.Yan J, Yang X, Li L, Liu P, Wu H, Liu Z, et al. Low expression levels of insulin-like growth factor binding protein-3 are correlated with poor prognosis for patients with hepatocellular carcinoma. Oncol Lett. 2017;13:3395–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Abu El-Makarem MA, Kamel MF, Mohamed AA, Ali HA, Mohamed MR, Mohamed AEM, et al. Down-regulation of hepatic expression of GHR/STAT5/IGF-1 signaling pathway fosters development and aggressiveness of HCV-related hepatocellular carcinoma: Crosstalk with Snail-1 and type 2 transforming growth factor-beta receptor. PLoS One. 2022;17:e0277266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li K, Zhang R, Wen F, Zhao Y, Meng F, Li Q, et al. Single-cell dissection of the multicellular ecosystem and molecular features underlying microvascular invasion in HCC. Hepatology. 2024;79:1293–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu R, Guo W, Qiu X, Wang S, Sui C, Lian Q, et al. Comprehensive analysis of spatial architecture in primary liver cancer. Sci Adv. 2021;7:eabg3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma L, Heinrich S, Wang L, Keggenhoff FL, Khatib S, Forgues M, et al. Multiregional single-cell dissection of tumor and immune cells reveals stable lock-and-key features in liver cancer. Nat Commun. 2022;13:7533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Golden-Mason L, Rosen HR. Galectin-9: Diverse roles in hepatic immune homeostasis and inflammation. Hepatology. 2017;66:271–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sauer N, Janicka N, Szlasa W, Skinderowicz B, Kolodzinska K, Dwernicka W, et al. TIM-3 as a promising target for cancer immunotherapy in a wide range of tumors. Cancer Immunol Immunother. 2023;72:3405–3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Menke-van der Houven van Oordt CW, Gomez-Roca C, van Herpen C, Coveler AL, Mahalingam D, Verheul HM, et al. First-in-human phase I clinical trial of RG7356, an anti-CD44 humanized antibody, in patients with advanced, CD44-expressing solid tumors. Oncotarget. 2016;7:80046–80058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Allen F, Maillard I. Therapeutic targeting of Notch signaling: From cancer to inflammatory disorders. Front Cell Dev Biol. 2021;9:649205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yuan F, Chen Y, Dellian M, Safabakhsh N, Ferrara N, Jain RK. Time-dependent vascular regression and permeability changes in established human tumor xenografts induced by an anti-vascular endothelial growth factor/vascular permeability factor antibody. Proc Natl Acad Sci USA. 1996;93:14765–14770. [DOI] [PMC free article] [PubMed] [Google Scholar]