Abstract
Immunopeptides are cell surface-located protein fragments that aid our immune system to recognise and respond to pathogenic insult and malignant transformation. In this two-part communication, we firstly summarise and reflect on our recent discovery documenting that MHC-II-bound immunopeptides from immortalised cell lines prevalently carry N-glycans that differ from the cellular glycoproteome (Goodson, Front Immunol, 2023). These findings are important as immunopeptide glycosylation remains poorly understood in immunosurveillance. The study also opened up new technical and biological questions that we address in the second part of this communication. Our study highlighted that the performance of the search engines used to detect glycosylated immunopeptides from LC–MS/MS data remains untested and, importantly, that little biochemical in vivo evidence is available to document the nature of glycopeptide antigens in tumour tissues. To this end, we compared the N-glycosylated MHC-II-bound immunopeptides that were reported from tumour tissues of 14 meningioma patients in the MSFragger-HLA-Glyco database (Bedran, Nat Commun, 2023) to those we identified with the commercial Byonic software. Encouragingly, the search engines produced similar outputs supporting that N-glycosylated MHC-II-bound immunopeptides are prevalent in meningioma tumour tissues. Consistent also with in vitro findings, the tissue-derived MHC-II-bound immunopeptides were found to predominantly carry hyper-processed (paucimannosidic- and chitobiose core-type) and hypo-processed (oligomannosidic-type) N-glycans that varied in prevalence and distribution between patients. Taken together, evidence is emerging suggesting that α-mannosidic glycoepitopes abundantly decorate MHC-II-bound immunopeptides presented in both immortalised cells and tumour tissues warranting further research into their functional roles in immunosurveillance.
Keywords: glycosylation, HLA-II, immunopeptide, meningioma, peptide antigen
Introduction
Immunosurveillance mechanisms are governed by immunopeptides (or peptide antigens), fragments of exogeneous and endogenous proteins, presented on the cell surface by major histocompatibility complex (MHC), also known as human leukocyte antigen (HLA), class I (MHC-I, or HLA-I) and class II (MHC-II, or HLA-II) molecules. The MHC molecules form a highly diverse family of proteins known to present peptide antigens derived from the cellular proteome in an HLA allotype-dependent manner. The diverse population of immunopeptides, the immunopeptidome, is constantly scanned and, if danger is impending, perturbations in the immunopeptidome are recognised by T-cell receptor molecules that then signal a T-cell-mediated immune response (Pagliuca et al. 2022; Santambrogio and Franco 2022; Pinho et al. 2023).
Given their critical role in immunosurveillance, immunopeptides have been studied for decades, with mass spectrometry (MS) being the current gold-standard for deep immunopeptidome profiling (Illing et al. 2022). Sensitive immunopeptidomics methods now enable confident identification of thousands of unmodified immunopeptides from complex biological samples within a single experiment (Purcell et al. 2019). However, modified immunopeptides, such as those carrying glycosylation, remain less well studied. Building on early observations of functionally relevant immunopeptide-conjugated glycans (Chicz et al. 1993; Housseau et al. 2001; Backlund et al. 2002) and the now wide-spread recognition that protein glycosylation is critically important for a wide range of immunomodulatory processes (Ugonotti et al. 2021; Gagneux et al. 2022), the field has experienced a growing interest in studying glycosylated peptide antigens (Kalka-Moll et al. 2002; Cobb et al. 2004; Dengjel et al. 2005; Malaker et al. 2017; Mei et al. 2020; Hoek et al. 2021; Mukherjee et al. 2021; Parker et al. 2021; Bedran et al. 2023; Goodson et al. 2023). Owing to their non-templated enzymatic biosynthesis (Stanley et al. 2022), polypeptide-linked glycans are inherently heterogeneous and their structures and expression patterns unpredictable from the genetic blueprint. Intuitively, glycans therefore have the potential to dramatically expand the structural and functional diversity of immunopeptides, but our knowledge of the glycosylated immunopeptidome remains in its infancy.
Hindering rapid progress in this burgeoning field, the large-scale identification of glycosylated immunopeptides is impeded by profound challenges in computational proteomics. Firstly, the nonspecific proteolytic processing of immunopeptides requires open nonenzymatic searches against a set of target proteins resulting in an impractically large peptide search space. Secondly, the fact that glycosylation is a highly heterogenous often sub-stoichiometric modification, calls for the use of a large glycan search space. Even without considering other challenging variables i.e. presence of other post-translational modifications (oxidation, deamidation) and the localisation of glycosylation site(s), the wide peptide and glycan search space dramatically expand the search time and increase the false discovery rates (FDRs). Compounding further their identification, glycosylated peptides are known to ionise and fragment poorly relative to their unmodified counterparts (Stavenhagen et al. 2013) leaving only a subset of the generated LC–MS/MS spectra suitable for interpretation. The large-scale profiling of glycopeptide antigens therefore poses unique informatics challenges not encountered in other proteomic-type data analysis approaches. Despite the advent of numerous glycoproteomics search engines capable of identifying tryptic and other protease-restricted glycopeptides (Kawahara et al. 2021; Polasky and Nesvizhskii 2023), few search engines have, to the best of our knowledge, shown the ability to identify glycosylated peptide antigens from immunopeptidomics data including the open access MSFragger-HLA-Glyco (Bedran et al. 2023) and the commercial Byonic (Goodson et al. 2023) software.
In this two-part communication, we firstly summarise and reflect on our recent discovery that MHC-II-bound immunopeptides from cultured breast cancer-derived (MDA-MB-231) and monocytic-like (THP-1) cells are richly modified with N-glycans that structurally differ from those in the cellular glycoproteome (Goodson et al. 2023). Guided by pertinent technical and biological questions that arose from this study, the second part of this communication sets out to compare MSFragger-HLA-Glyco and Byonic in terms of their identification efficiency of glycosylated immunopeptides and, importantly, to establish, using available MHC-II immunopeptidomics data from meningioma patients (Racle et al. 2019), in vivoevidence of glycopeptide antigens in tumour tissues.
Results and discussion
Hyper- and hypo-processed N-glycans decorate MHC-II immunopeptides of immortalised cells
In our original report (Goodson et al. 2023), we comprehensively analysed paired multi-omics datasets generated from two cell systems, the breast cancer-derived (MDA-MB-231) and the monocytic-like (THP-1) cell line, enabling us to map the N-glycosylation of MHC-II-bound immunopeptides relative to the cellular N-glycoproteome. Interestingly, for both MDA-MB-231 and THP-1 we observed considerable levels (~5-10% based on diagnostic ions) of N-glycosylation modifying the heterogeneous populations of MHC-II-bound immunopeptides. The N-glycans were dominated by both truncated chitobiose core- (GlcNAc1-2Fuc0-1, 8.0%–39.1%) and paucimannosidic-type (Man1-3GlcNAc1-2Fuc0-1, 47.8%–56.6%) as well as oligomannosidic-type N-glycans (Man5-9GlcNAc2, 13.0%–31.0%). The immunopeptides were strikingly lacking (or were near-devoid of) complex-type N-glycans including structures carrying sialylation, galactosylation and other glycans typically associated with the glycocalyx. Importantly, the prevalence of biosynthetically hyper-processed (truncated, chitobiose core- and paucimannosidic-type) and hypo-processed (immature, oligomannosidic-type) N-glycan structures on the peptide antigens was orthogonally confirmed by LC–MS/MS-based glycomics of the same population of isolated immunopeptides.
In contrast, glycomics and glycoproteomics applied to lysates of MDA-MB-231 and THP-1 revealed that the cellular N-glycoproteome, as expected, was dominated by complex- and oligomannosidic-type N-glycans. In line with previous studies, truncated N-glycans formed a relatively small component of the N-glycome of MDA-MB-231 cells (Chatterjee et al. 2019) whereas these hyper-processed N-glycans formed a larger proportion in THP-1 (Hare et al. 2017). The stark differences in glycan type distribution between the peptide antigens and cell lysates indicated extensive N-glycan remodelling before, during or after MHC-II antigen processing and presentation. We have previously observed similar N-glycan remodelling on MHC-II-bound immunopeptides of recombinant SARS-CoV-2 spike glycoprotein fed to monocyte-derived dendritic cells (Parker et al. 2021). In the MDA-MB-231 and THP-1 cells, the prominent glycophenotypic differences were corroborated by comparative glycoprofiling of several cell surface glycoproteins identified in both the lysates and immunopeptidome including integrin alpha-2, transferrin receptor-1, and protocadherin FAT1.
Catalysed by N-acetyl-β-hexosaminidase (Hex) and possibly other glycoside hydrolases, we have recently reported on a non-canonical truncation pathway that in human neutrophils operates in parallel to the canonical N-glycoprotein biosynthetic route (referred to as the “elongation pathway”) (Ugonotti et al. 2022). Drawing parallels to this truncation pathway that is now recognised to exist across most eukaryotic kingdoms (Tjondro et al. 2019; West et al. 2021), we indeed identified the Hex enzyme and other glycoside hydrolases (MAN2B1, MANBA) putatively involved in the N-glycan remodelling of immunopeptides in the MDA-MB-231 and THP-1 lysates. By overlaying our findings of remodelled immunopeptide N-glycans observed across two biological systems on the otherwise well-established MHC-II processing and presentation pathway, we proposed that the dramatic glycophenotypic transformation of endogenous cell surface glycoproteins occurs after their endocytosis and exposure to the hydrolytic endo/lysosomal environment. We are currently following up on these leads using glycoside hydrolase inhibitors and genetic ablation of truncating lysosomal glyco-enzymes.
The MHC-II preference for presenting glycopeptide antigens exhibiting both hyper- and hypo-processed N-glycans in the two investigated cell systems raised important questions about the in vivopresence of glycosylated immunopeptides. Moreover, while our study employed multiple lines of -omics evidence to support conclusions, the work also prompted us to scrutinise the search engines used to identify glycosylated peptide antigens due to the technical challenges associated with these experiments. These topics are explored below.
In vivo support for prevalent N-glycosylation on MHC-II immunopeptides of meningioma tumours
Congruence in MSFragger-HLA-Glyco and Byonic outputs elevates confidence in findings
In recognising that MSFragger-Glyco is a strong search engine in glycoproteomics (Polasky et al. 2020) and that the HLA-Glyco database is a valuable library containing over 3,400 N-glycosylated MHC-II-bound immunopeptides (Bedran et al. 2023), we decided to use this resource to benchmark the Byonic software used in our previous study (Goodson et al. 2023).
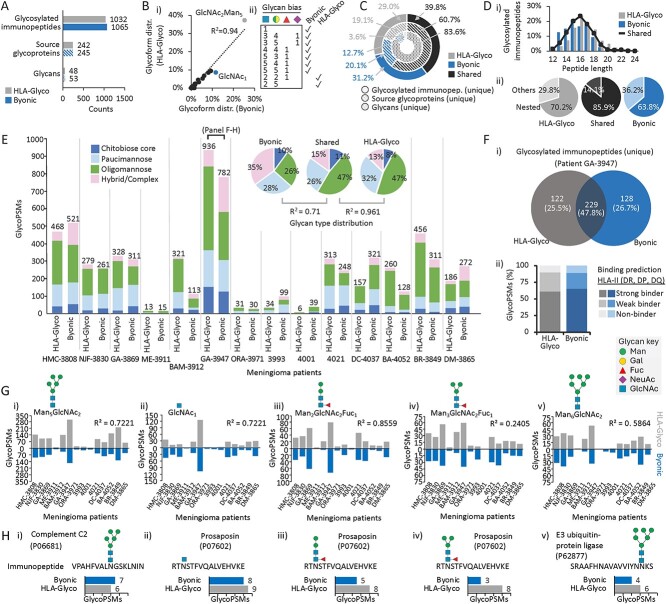
To this end, we searched MHC-II immunopeptidomics LC–MS/MS datasets obtained from resected tumours of 14 meningioma patients using Byonic (Supplementary Table 1) and compared the resulting data (“output”) to the HLA-Glyco database (Supplementary Table 2). HLA-Glyco and Byonic confidently identified a total of 3,788 and 3,451 glycoPSMs across the meningioma tumour tissues, respectively, corresponding to ~1,050 unique (non-redundant) N-glycosylated immunopeptides, ~240 source glycoproteins and ~50 glycan compositions for each search engine (Fig. 1A). Encouragingly, similar glycoform distributions (R2 = 0.94) were reported by the two approaches (Fig. 1Bi) with only relatively few glycans enriched in the output from either software (Fig. 1Bii). While a minor over-reporting of paucimannosidic- and oligomannosidic-type N-glycans was observed for HLA-Glyco, a slight over-reporting of complex-type N-glycans was observed for Byonic. Such differences in search output may be explained by the fact that Byonic lacks a glycan-specific scoring mechanism to distinguish between glycan candidates of relatively low (often complex-type N-glycans) and high (often oligo- and paucimannosidic-type N-glycans) confidence.
Fig. 1.
Congruence of HLA-Glyco and Byonic outputs documents mannose-rich HLA-II glycopeptide antigens in meningioma tumours. A) Glycosylated immunopeptides, their source glycoproteins and glycans identified across 14 MHC-II immunopeptidomics datasets from meningioma tumour tissues by HLA-Glyco (grey) or using Byonic (blue). B. i) Glycoform distribution of glycosylated immunopeptides (average of 14 biological replicates) identified by HLA-Glyco or using Byonic. ii) Glycoforms enriched in either HLA-Glyco or Byonic (n = 14, paired t-test, P < 0.05). C) Glycosylated immunopeptides, their source glycoproteins and glycans reported by HLA-Glyco and/or Byonic. D. i) Peptide length and ii) proportion of nested sets within the glycosylated immunopeptides identified by HLA-glyco and/or Byonic. E) Glycan type distribution across all patients. Insert: The glycan type distribution of glycosylated immunopeptides identified by HLA-Glyco and/or Byonic were compared using Pearson correlation. F. i) Sequence overlap and ii) binding prediction of glycosylated immunopeptides (unique) identified from patient GA-3947 by HLA-Glyco and/or Byonic. G. i–v) The five most common glycoforms identified across all patients by HLA-Glyco and/or Byonic. H. i–v) The most common immunopeptide sequences identified from patient GA-3947 and their source glycoproteins from the five abundant glycoforms as well as their spectral identification rate by HLA-Glyco and Byonic.
The overall similar outputs produced by the two search engines were also illustrated by the considerable overlap in glycosylated immunopeptide sequences (39.8%), and their source glycoproteins (60.7%) and glycans (83.6%) between HLA-Glyco and Byonic (Fig. 1C). Elevating further the confidence in the outputs, the length distribution of glycosylated immunopeptides identified by HLA-Glyco and/or Byonic matched the expected characteristics of MHC-II immunopeptides averaging 15-16 amino acid residues (Fig. 1Di) (Marino et al. 2020; Gfeller et al. 2023) and formed, as expected, mostly nested sets (Fig. 1Dii).
In line with our in vitro findings (Goodson et al. 2023), the glycopeptide antigens were predominantly modified by oligomannosidic (27.3%–66.7%) and paucimannosidic (10.3%–47.5%) N-glycans while fewer complex- and chitobiose core-type N-glycans were identified by both search engines (Fig. 1E). Dramatic inter-patient variation in glycoPSMs was observed spanning up to two orders of magnitude between tissues with highest (e.g. GA-3947, HMC-3808) and lowest (e.g. ME-3911, ORA-3971, 4001) expression of glycosylated immunopeptides. By factoring in also the non-glycosylated immunopeptides that were detected, the glycosylation frequency (glycoPSMs/all PSMs) remained highly varied across samples (range: 0.4%–33.4%, mean ± SD: 11.4% ± 11.1%), suggesting dramatic biological variation in glycopeptide antigen presentation across tumours. Attempting to explore this inter-patient variation, we focused on the HLA-II typing of the 14 meningioma patients available in the original study (summarised in Supplementary Table 3). While we found a handful of shared allotypes across the investigated patients, we did not identify any apparent associations between neither the immunopeptide glycosylation type nor frequency of glycosylation events and the HLA-II type.
The glycan type distributions of immunopeptides identified exclusively by HLA-Glyco matched the shared output (R2 = 0.96) supporting the accuracy of those identifications (Fig. 1E, insert). The lower correlation between immunopeptides identified exclusively by Byonic and the shared output (R2 = 0.71), suggests, as pointed out above, that Byonic (relative to HLA-Glyco) under-reports on oligomannosidic-type (e.g. Man5GlcNAc2) glycopeptides while some complex−/hybrid-type glycopeptides appear favoured, although the accuracy of these slightly biased identifications remains to be determined.
The outputs from the two search engines were further scrutinised in a focused analysis of data from patient GA-3947 featuring a particularly high immunopeptidome coverage. A considerable overlap in glycosylated immunopeptide sequences (47.8%, 229 counts) were observed between HLA-Glyco and Byonic (Fig. 1Fi). The glycosylated immunopeptides identified by the two search engines were further validated using NetMHCIIpan 4.1 (Reynisson et al. 2020) by showing that most of these were predicted MHC-II-binders (Fig. 1Fii).
While the congruent outputs suggest that MSFragger-HLA-Glyco and Byonic represent search engines that are effective for the identification of N-glycosylated MHC-II immunopeptides, we note that the two programs feature different strengths and weaknesses. For example, the Byonic searches (134.9 min/file) were considerably slower than the MSFragger-HLA-Glyco searches (2.7 min/file), a practical limitation we mitigated in this study by reducing the number of variable modifications and by adding constraints to peptide search space used for the non-specific search strategy. Sequence analysis of the reported immunopeptides revealed a similar proportion of non-tryptic sequences identified by HLA-Glyco (77.0%) and Byonic (70.2%, data not shown). This suggests that the constrained search strategy in Byonic does not favour peptides with tryptic cleavage pattern supporting a similar conclusion obtained from our previous study (Goodson et al. 2023). The comparatively higher search speed makes MSFragger-Glyco an attractive software for analysis of immunopeptidomics data enabling more comprehensive search strategies involving multiple variable modifications such as N- and O-glycosylation as well as other post-translational modifications. In addition, HLA-Glyco features an option for “group FDR” assessment to boost further the glycopeptide confidence in attempts to drive down the high mis-identification rate that remains a well-recognised bottleneck in the field (Chau et al. 2023). On the other hand, Byonic is a well-established commercial software with recognised strengths in N-glycopeptide identification (Kawahara et al. 2021) and feature an intuitive interface, regular updates, and incorporation of user feedback for improved search strategies. Following our 1st HGI study (Kawahara et al. 2021), another inter-laboratory data comparison study of the Human Glycoproteomics Initiative is currently underway, which aims to comprehensively compare the performance of MSFragger-Glyco and Byonic relative to the many other glycopeptide identification software solutions that have emerged in the past decade (Polasky and Nesvizhskii 2023).
Hyper- and hypo-processed N-glycans decorate MHC-II immunopeptides of meningioma tumours
Relative quantitation of the N-glycan structures identified on the immunopeptides across the meningioma tumour tissues revealed that both the oligomannosidic-type (Man5-6GlcNAc2) and the truncated paucimannosidic-type (Man2-3GlcNAc2Fuc1) as well as GlcNAc1 structures were the most common glycoforms reported by both HLA-Glyco and Byonic (Fig. 1G). The general agreement between the two search engines for these five glycoforms across the patients (0.24 < R2 < 0.86) provided important in vivo evidence to support our in vitro findings (Goodson et al. 2023) documenting the dominance of both hypo- and hyper-processed N-glycans in meningioma tumour tissues.
By again focusing on patient GA-3947 featuring a high immunopeptidome coverage, we then explored the most common immunopeptide sequences and their source glycoproteins that carried the five abundant peptide antigen glycoforms. Both HLA-Glyco and Byonic identified complement C2 (P06681), E3 ubiquitin-protein ligase DTX3L (Q8TDB6) and the lysosomal prosaposin (P07602) as the main source proteins of the glycopeptide antigens (Fig. 1H). While VPAHFVALNGSKLNIN of complement C2 and SRAAFHNAVAVVIYNNKS of E3 ubiquitin-protein ligase DTX3L carried oligomannosidic-type N-glycans, RTNSTFVQALVEHVKE of prosaposin was the main immunopeptide carrier of the paucimannosidic and chitobiose core glycoforms. All three source proteins are known to play roles in tumorigenic processes (Saleh et al. 2024; Sharma et al. 2024; Zhang et al. 2024). Thus, identifying glycosylated immunopeptides derived from these endogenous glycoproteins in the meningioma tumour tissues is of potential significance as it implicates their involvement in the tumour microenvironment possibly by modulating the immune-tumour synapse of critical importance for tumour growth and dissemination (Vicente et al. 2023), aspects that await future exploration.
Presentation of exposed α-mannosidic residues in a mono- or divalent form through the paucimannosidic-type N-glycans or in a tri-valent form through oligomannosidic-type N-glycans are emerging as a common molecular feature of the abundant immunopeptide-conjugated glycans reported in this communication. Perusing the wider HLA-Glyco database across other biological systems substantiates these trends. Together with our observation of N-glycan remodelling on MHC-II-bound immunopeptides in dendritic cells pulsed with the SARS-CoV-2 spike glycoprotein (Parker et al. 2021), the findings discussed in this communication illustrate that both exogenous and endogenous glycoproteins produce mannose-rich glycopeptide antigens in the MHC-II system. As building literature points to mannosidic glycoepitopes being functional motifs mediating recognition by a range of immunomodulatory glycan-binding (lectin) receptors (Loke et al. 2016; Peters and Peters 2021; Vicente et al. 2023), it may be proposed that the glycoepitopes carried by the MHC-II peptide antigens may play roles in immune cell communication by either engaging directly with such lectin receptors (in cis or trans configuration) or indirectly by altering the peptide antigen recognition by the T-cell receptor of CD4+ T-cells. Regardless of the underlying mechanisms, the sheer prevalence of voluminous and highly flexible N-glycans carrying reactive glyco-epitopes on the MHC-II peptide antigens suggests their potential to impact the downstream immune response.
In conclusion, building evidence points to N-glycosylation being an abundant type of modification of the MHC-II immunopeptidome. As now mapped with different search engines across different immortalised cell lines and in tumour tissues from a cohort of meningioma patients, the immunopeptides consistently display both hyper-processed (paucimannosidic) and hypo-processed (oligomannosidic) N-glycans on the cell surface, structures that differ from the cellular glycoproteome. As both pauci- and oligomannosylation are typically not exposed to the extracellular environment under baseline conditions (Loke et al. 2016; Ugonotti et al. 2021), their prevalence within the MHC-II immunopeptidome suggests yet-to-be-revealed functional roles in immunosurveillance mechanisms.
Materials and methods
This section pertains to the second part of this communication for which new data searches and comparative analyses of outputs were performed. For methods and materials underpinning the findings summarised in the first part, see our original research paper (Goodson et al. 2023).
Data selection
For the data re-interrogation and cross search engine comparisons, we used MHC-II immunopeptidomics datasets generated from resected tumour tissues of 14 meningioma patients (Racle et al. 2019). These datasets, which were generated without any enrichment for glycosylated immunopeptides, were selected given i) their high quality (high mass accuracy and signal/noise, informative MS/MS, and good LC peak resolution, see original study for acquisition conditions), ii) the considerable number of biological replicates, and iii) the fact that the data were previously interrogated in the HLA-Glyco study (Bedran et al. 2023) enabling us to use their output for comparison and benchmarking purposes against our Byonic-based searches. While both MHC-I and -II immunopeptidomics data were available, we restricted our analysis to the HLA-II datasets (HLA-DR, HLA-DP, HLA-DQ) in attempts to generate in vivo evidence to support our previous observation that N-glycans prevalently decorate MHC-II-bound immunopeptides from immortalised cells in culture (Goodson et al. 2023).
Immunopeptidomics data search and cross search engine comparisons
To enable an informative comparison against the HLA-Glyco search outputs (Bedran et al. 2023), we searched the HCD-MS/MS data from the same LC–MS/MS raw datasets for immunopeptides modified with N-glycans using Byonic v5.5 (Protein Metrics). In doing this, we aimed to mimic how experts operate a specific search engine rather than attempting to use identical search settings across the two search engines. The latter was not practically possible as the two search engines have different requirements and constraints. For the Byonic search, we used 67 N-glycans in the glycan search space; comprising the N-glycans reported by HLA-Glyco and an additional six N-glycans (Supplementary Table 4). In comparison, 198 N-glycans was used by HLA-Glyco (Supplementary Table 5). See Table 1 for overview of key search settings. Both search engines used the entire human proteome as the protein search space and a reverse/decoy and contaminant database to establish FDRs. Mimicking the HLA-Glyco approach that considered any non-specific peptides, the Byonic search permitted a non-specific cleavage pattern of peptide candidates, but included constraints in the peptide search space by allowing only candidate peptides with up to two internal R/K residues to accelerate the otherwise impractically slow search. We have previously demonstrated that this constraint does not bias towards tryptic cleavage patterns when identifying immunopeptides (Goodson et al. 2023). For both search engines, the precursor/product ions were permitted to deviate up to 10–20 or 15–20 ppm from the expected (theoretical) values, respectively. While multiple other variable modifications were included in the HLA-Glyco search (methionine oxidation, N-terminal acetylation, cysteinylation in addition to the N-glycosylation), this approach was impractical for Byonic, which therefore instead restricted the search to a single N-glycan per peptide defined as a variable “common” modification. Similar to HLA-Glyco, the Byonic search used monoisotopic correction (error check equals +/− floor, mass in Da/4000). For HLA-Glyco, glycopeptides (7–25 amino acid residues in length) were identified using 1% FDR, q-value (glycan) < 0.05 and motif deconvolution as confidence thresholds. For Byonic, glycopeptide candidates were filtered to PEP-2D < 0.01 and any candidates from the reverse and contaminant protein lists were manually deleted. The glycosylated MHC-II immunopeptide candidates were additionally filtered by peptide length (12–25 amino acid residues) to improve confidence further. The protein FDR of the filtered glycoPSMs reaching confidence was consistently below 1%. Unique glycosylated immunopeptides (unique peptide, glycosylation site, glycan composition) and all glycosylated peptide-to-spectral matches (glycoPSMs) were quantitatively compared between the two search engines using a commonly used spectral counting approach (Kawahara et al. 2023).
Table 1.
Key search settings used for the HLA-Glyco study (Bedran et al. 2023) and the Byonic-based searches of the same MHC-II immunopeptidomics LC–MS/MS data files.
| MSFragger-HLA-Glyco (Bedran et al. 2023) |
Byonic v5.5 (Protein Metrics) |
|
|---|---|---|
| Protein search space | Human proteome (UniProtKB) | Human proteome (UniProtKB) |
| Decoy database | + | + |
| Contaminant database | + | + |
| Digestion specificity | Non-specific | Non-specific |
| Cleavage site | None | R/K |
| Cleavage side | None | C-terminal |
| Missed cleavages | N/A | 2 |
| Precursor mass tolerance | 20 ppm | 10 ppm |
| Fragmentation type | HCD | HCD |
| Fragment mass tolerance | 15 ppm | 20 ppm |
| Monoisotopic peak picking | Correction enabled | Correction enabled |
| Variable modifications (# variable modifications permitted per peptide) | N-glycosylation, oxidation, N-terminal acetylation, cysteinylation (not stated) | N-glycosylation (1) |
| Glycan database size | 198 | 67 |
| Confidence filtering | 1% FDR, q < 0.05 | 1% FDR, PEP 2D < 0.01 |
| Average search time/file | 2.7 min | 134.9 min |
Supplementary Material
Contributor Information
Hayley Goodson, School of Natural Sciences, Macquarie University, 4 Wally's Walk, NSW-2109, Macquarie Park, Sydney, Australia.
Rebeca Kawahara, Institute for Glyco-core Research (iGCORE), Nagoya University, Furocho, Chikusa Ward, Nagoya, 464-8601, Aichi, Japan.
Joshua Fehring, Department of Biochemistry and Molecular Biology & Biomedicine Discovery Institute, Monash University, Innovation Walk, VIC-3800, Clayton, Melbourne, Australia.
Anthony W Purcell, Department of Biochemistry and Molecular Biology & Biomedicine Discovery Institute, Monash University, Innovation Walk, VIC-3800, Clayton, Melbourne, Australia.
Nathan P Croft, Department of Biochemistry and Molecular Biology & Biomedicine Discovery Institute, Monash University, Innovation Walk, VIC-3800, Clayton, Melbourne, Australia.
Morten Thaysen-Andersen, School of Natural Sciences, Macquarie University, 4 Wally's Walk, NSW-2109, Macquarie Park, Sydney, Australia; Institute for Glyco-core Research (iGCORE), Nagoya University, Furocho, Chikusa Ward, Nagoya, 464-8601, Aichi, Japan.
Author contributions
Hayley Goodson (Conceptualization [supporting], Data curation [lead], Formal analysis [lead], Investigation [equal], Methodology [lead], Software [lead], Validation [equal], Visualization [lead], Writing—original draft [equal], Writing—review & editing [equal]), Rebeca Kawahara (Conceptualization [supporting], Data curation [supporting], Formal analysis [supporting], Investigation [supporting], Methodology [supporting], Project administration [supporting], Resources [supporting], Software [supporting], Supervision [supporting], Validation [supporting], Visualization [supporting], Writing—original draft [supporting], Writing—review & editing [supporting]), Joshua Fehring (Data curation [supporting], Formal analysis [supporting], Investigation [supporting], Methodology [supporting], Software [supporting], Validation [supporting], Writing—original draft [supporting], Writing—review & editing [supporting]), Anthony W. Purcell (Conceptualization [supporting], Funding acquisition [supporting], Investigation [supporting], Project administration [supporting], Resources [supporting], Supervision [supporting], Writing—original draft [supporting], Writing—review & editing [supporting]), Nathan P. Croft (Conceptualization [supporting], Data curation [supporting], Funding acquisition [supporting], Investigation [supporting], Methodology [supporting], Project administration [supporting], Resources [supporting], Supervision [supporting], Writing—original draft [supporting], Writing—review & editing [supporting]), and Morten Thaysen-Andersen (Conceptualization [lead], Data curation [supporting], Funding acquisition [supporting], Investigation [equal], Methodology [supporting], Project administration [lead], Resources [lead], Software [supporting], Supervision [lead], Validation [supporting], Visualization [supporting], Writing—original draft [equal], Writing—review & editing [equal])
Funding
HG was supported by a Macquarie Research Excellence Scholarship. A.W.P. is a NHMRC Investigator Fellow (APP2016596). M.T.A. is the recipient of an ARC Future Fellowship (FT210100455).
Conflict of interest statement. None declared.
Data availability
Immunopeptidomics LC–MS/MS datasets available through the ProteomeXchange Consortium via the PRIDE repository (PXD012308) in were analysed in this study.
References
- Backlund J, Carlsen S, Hoger T, Holm B, Fugger L, Kihlberg J, Burkhardt H, Holmdahl R. Predominant selection of T cells specific for the glycosylated collagen type II epitope (263-270) in humanized transgenic mice and in rheumatoid arthritis. Proc Natl Acad Sci USA. 2002:99(15):9960–9965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedran G, Polasky DA, Hsiao Y, Yu F, da Veiga LF, Alfaro JA, Cieslik M, Nesvizhskii AI. Unraveling the glycosylated immunopeptidome with HLA-Glyco. Nat Commun. 2023:14(1):3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee S, Lee LY, Kawahara R, Abrahams JL, Adamczyk B, Anugraham M, Ashwood C, Sumer-Bayraktar Z, Briggs MT, Chik JHL, et al. Protein Paucimannosylation is an enriched N-glycosylation signature of human cancers. Proteomics. 2019:19(21-22):e1900010. [DOI] [PubMed] [Google Scholar]
- Chau TH, Chernykh A, Kawahara R, Thaysen-Andersen M. Critical considerations in N-glycoproteomics. Curr Opin Chem Biol. 2023:73:102272. [DOI] [PubMed] [Google Scholar]
- Chicz RM, Urban RG, Gorga JC, Vignali DA, Lane WS, Strominger JL. Specificity and promiscuity among naturally processed peptides bound to HLA-DR alleles. J Exp Med. 1993:178(1):27–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobb BA, Wang Q, Tzianabos AO, Kasper DL. Polysaccharide processing and presentation by the MHCII pathway. Cell. 2004:117(5):677–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dengjel J, Rammensee HG, Stevanovic S. Glycan side chains on naturally presented MHC class II ligands. J Mass Spectrom. 2005:40(1):100–104. [DOI] [PubMed] [Google Scholar]
- Gagneux P, Hennet T, Varki A. Biological functions of Glycans. In: Varki A, Cummings RD, Esko JD, Stanley P, Hart GW, Aebi M, Mohnen D, Kinoshita T, Packer NH, Prestegard JH, et al., editors. Essentials of Glycobiology. 4th ed. Cold Spring Harbor (NY); 2022. pp. 79–92. [PubMed] [Google Scholar]
- Gfeller D, Liu Y, Racle J. Contemplating immunopeptidomes to better predict them. Semin Immunol. 2023:66:101708. [DOI] [PubMed] [Google Scholar]
- Goodson H, Kawahara R, Chatterjee S, Goncalves G, Fehring J, Purcell AW, Croft NP, Thaysen-Andersen M. Profound N-glycan remodelling accompanies MHC-II immunopeptide presentation. Front Immunol. 2023:14:1258518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hare NJ, Lee LY, Loke I, Britton WJ, Saunders BM, Thaysen-Andersen M. Mycobacterium tuberculosis infection manipulates the glycosylation machinery and the N-Glycoproteome of human macrophages and their microparticles. J Proteome Res. 2017:16(1):247–263. [DOI] [PubMed] [Google Scholar]
- Hoek M, Demmers LC, Wu W, Heck AJR. Allotype-specific glycosylation and cellular localization of human leukocyte antigen class I proteins. J Proteome Res. 2021:20(9):4518–4528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Housseau F, Moorthy A, Langer DA, Robbins PF, Gonzales MI, Topalian SL. N-linked carbohydrates in tyrosinase are required for its recognition by human MHC class II-restricted CD4(+) T cells. Eur J Immunol. 2001:31(9):2690–2701. [DOI] [PubMed] [Google Scholar]
- Illing PT, Ramarathinam SH, Purcell AW. New insights and approaches for analyses of immunopeptidomes. Curr Opin Immunol. 2022:77:102216. [DOI] [PubMed] [Google Scholar]
- Kalka-Moll WM, Tzianabos AO, Bryant PW, Niemeyer M, Ploegh HL, Kasper DL. Zwitterionic polysaccharides stimulate T cells by MHC class II-dependent interactions. J Immunol. 2002:169(11):6149–6153. [DOI] [PubMed] [Google Scholar]
- Kawahara R, Chernykh A, Alagesan K, Bern M, Cao W, Chalkley RJ, Cheng K, Choo MS, Edwards N, Goldman R, et al. Community evaluation of glycoproteomics informatics solutions reveals high-performance search strategies for serum glycopeptide analysis. Nat Methods. 2021:18(11):1304–1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara R, Ugonotti J, Chatterjee S, Tjondro HC, Loke I, Parker BL, Venkatakrishnan V, Dieckmann R, Sumer-Bayraktar Z, Karlsson-Bengtsson A, et al. Glycoproteome remodeling and organelle-specific N-glycosylation accompany neutrophil granulopoiesis. Proc Natl Acad Sci USA. 2023:120(36):e2303867120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loke I, Kolarich D, Packer NH, Thaysen-Andersen M. Emerging roles of protein mannosylation in inflammation and infection. Mol Asp Med. 2016:51:31–55. [DOI] [PubMed] [Google Scholar]
- Malaker SA, Ferracane MJ, Depontieu FR, Zarling AL, Shabanowitz J, Bai DL, Topalian SL, Engelhard VH, Hunt DF. Identification and characterization of complex glycosylated peptides presented by the MHC class II processing pathway in melanoma. J Proteome Res. 2017:16(1):228–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marino F, Semilietof A, Michaux J, Pak HS, Coukos G, Muller M, Bassani-Sternberg M. Biogenesis of HLA ligand presentation in immune cells upon activation reveals changes in peptide length preference. Front Immunol. 2020:11:1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei S, Ayala R, Ramarathinam SH, Illing PT, Faridi P, Song J, Purcell AW, Croft NP. Immunopeptidomic analysis reveals that Deamidated HLA-bound peptides Arise predominantly from Deglycosylated precursors. Mol Cell Proteomics. 2020:19(7):1236–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S, Sanchez-Bernabeu A, Demmers LC, Wu W, Heck AJR. The HLA Ligandome comprises a limited repertoire of O-GlcNAcylated antigens preferentially associated with HLA-B*07:02. Front Immunol. 2021:12:796584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagliuca S, Gurnari C, Rubio MT, Visconte V, Lenz TL. Individual HLA heterogeneity and its implications for cellular immune evasion in cancer and beyond. Front Immunol. 2022:13:944872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker R, Partridge T, Wormald C, Kawahara R, Stalls V, Aggelakopoulou M, Parker J, Powell Doherty R, Ariosa Morejon Y, Lee E, et al. Mapping the SARS-CoV-2 spike glycoprotein-derived peptidome presented by HLA class II on dendritic cells. Cell Rep. 2021:35(8):109179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters K, Peters M. The role of lectin receptors and their ligands in controlling allergic inflammation. Front Immunol. 2021:12:635411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinho SS, Alves I, Gaifem J, Rabinovich GA. Immune regulatory networks coordinated by glycans and glycan-binding proteins in autoimmunity and infection. Cell Mol Immunol. 2023:20(10):1101–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polasky DA, Nesvizhskii AI. Recent advances in computational algorithms and software for large-scale glycoproteomics. Curr Opin Chem Biol. 2023:72:102238. [DOI] [PubMed] [Google Scholar]
- Polasky DA, Yu F, Teo GC, Nesvizhskii AI. Fast and comprehensive N- and O-glycoproteomics analysis with MSFragger-Glyco. Nat Methods. 2020:17(11):1125–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell AW, Ramarathinam SH, Ternette N. Mass spectrometry-based identification of MHC-bound peptides for immunopeptidomics. Nat Protoc. 2019:14(6):1687–1707. [DOI] [PubMed] [Google Scholar]
- Racle J, Michaux J, Rockinger GA, Arnaud M, Bobisse S, Chong C, Guillaume P, Coukos G, Harari A, Jandus C, et al. Robust prediction of HLA class II epitopes by deep motif deconvolution of immunopeptidomes. Nat Biotechnol. 2019:37(11):1283–1286. [DOI] [PubMed] [Google Scholar]
- Reynisson B, Barra C, Kaabinejadian S, Hildebrand WH, Peters B, Nielsen M. Improved prediction of MHC II antigen presentation through integration and motif deconvolution of mass spectrometry MHC eluted ligand data. J Proteome Res. 2020:19(6):2304–2315. [DOI] [PubMed] [Google Scholar]
- Saleh H, Liloglou T, Rigden DJ, Parsons JL, Grundy GJ. KH-like domains in PARP9/DTX3L and PARP14 coordinate protein-protein interactions to promote cancer cell survival. J Mol Biol. 2024:436(4):168434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santambrogio L, Franco A. The yin/yang balance of the MHC-self-immunopeptidome. Front Immunol. 2022:13:1035363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma P, Zhang X, Ly K, Kim JH, Wan Q, Kim J, Lou M, Kain L, Teyton L, Winau F. Hyperglycosylation of prosaposin in tumor dendritic cells drives immune escape. Science. 2024:383(6679):190–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley P, Moremen KW, Lewis NE, Taniguchi N, Aebi M. N-Glycans. In: Varki A, Cummings RD, Esko JD, Stanley P, Hart GW, Aebi M, Mohnen D, Kinoshita T, Packer NH, Prestegard JH, et al., editors. Essentials of Glycobiology. 4th ed. Cold Spring Harbor (NY); 2022. pp. 103–116. [Google Scholar]
- Stavenhagen K, Hinneburg H, Thaysen-Andersen M, Hartmann L, Varon Silva D, Fuchser J, Kaspar S, Rapp E, Seeberger PH, Kolarich D. Quantitative mapping of glycoprotein micro-heterogeneity and macro-heterogeneity: an evaluation of mass spectrometry signal strengths using synthetic peptides and glycopeptides. J Mass Spectrom. 2013:48(6):i. [DOI] [PubMed] [Google Scholar]
- Tjondro HC, Loke I, Chatterjee S, Thaysen-Andersen M. Human protein paucimannosylation: cues from the eukaryotic kingdoms. Biol Rev Camb Philos Soc. 2019:94(6):2068–2100. [DOI] [PubMed] [Google Scholar]
- Ugonotti J, Chatterjee S, Thaysen-Andersen M. Structural and functional diversity of neutrophil glycosylation in innate immunity and related disorders. Mol Asp Med. 2021:79:100882. [DOI] [PubMed] [Google Scholar]
- Ugonotti J, Kawahara R, Loke I, Zhu Y, Chatterjee S, Tjondro HC, Sumer-Bayraktar Z, Neelamegham S, Thaysen-Andersen M. N-acetyl-beta-D-hexosaminidases mediate the generation of paucimannosidic proteins via a putative noncanonical truncation pathway in human neutrophils. Glycobiology. 2022:32(3):218–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente MM, Alves I, Fernandes A, Dias AM, Santos-Pereira B, Perez-Anton E, Santos S, Yang T, Correia A, Munster-Kuhnel A, et al. Mannosylated glycans impair normal T-cell development by reprogramming commitment and repertoire diversity. Cell Mol Immunol. 2023:20(8):955–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West CM, Malzl D, Hykollari A, Wilson IBH. Glycomics, Glycoproteomics, and Glycogenomics: an inter-taxa evolutionary perspective. Mol Cell Proteomics. 2021:20:100024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Li S, Xiao W, Zhang C, Li T, Liao Z, Liu H, Xing R, Yao W, Yang J. Tumoral C2 regulates the tumor microenvironment by increasing the ratio of M1/M2 macrophages and tertiary lymphoid structures to improve prognosis in melanoma. Cancers (Basel). 2024:16(5):908. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Immunopeptidomics LC–MS/MS datasets available through the ProteomeXchange Consortium via the PRIDE repository (PXD012308) in were analysed in this study.