Abstract
In cattle, bovine respiratory syncytial virus (BRSV) is associated with secondary bacterial infections; however, the mechanisms of the interaction between BRSV and bacteria are unclear. Trueperella pyogenes (T. pyogenes) causes pneumonia in cattle and is involved in secondary infections following viral infections. In this study, we evaluated the effect of BRSV infection on the adhesion of T. pyogenes to BRSV-infected cells. BRSV infection significantly enhanced the adhesion of T. pyogenes to cells in a multiplicity of infection- and time-dependent manner. The BRSV-mediated change in the adhesion of T. pyogenes was widely observed in various cell types and bacterial strains. The results from the gentamicin protection assay showed that BRSV infection did not affect the intracellular invasion ability of T. pyogenes. Furthermore, adhesion assays conducted using BRSV G protein-expressing cells and anti-BRSV G antibodies revealed that the increased adhesion of T. pyogenes to cells was mediated by the G protein of BRSV. In addition, immunofluorescence assay revealed the colocalization of BRSV G protein and T. pyogenes. Thus, BRSV infection can potentially lead to bovine respiratory disease complex by promoting the adhesion of T. pyogenes to the infected cells.
Keywords: bacterial adhesion, bovine respiratory disease complex, bovine respiratory syncytial virus, co-infection, Trueperella pyogenes
Bovine respiratory disease complex (BRDC) is a respiratory disease caused by complex interactions between multiple pathogens, in addition to environmental factors such as temperature fluctuations and weaning [4]. Compared to infections caused by a single pathogen, BRDC was reported to have a worse prognosis because of the mutual influence of multiple pathogens, such as viruses and bacteria [10]. Bovine alphaherpesvirus 1, bovine respiratory syncytial virus (BRSV), and bovine parainfluenza virus 3 are the most common viral causes of respiratory diseases in cattle [4]. Bacteria such as Pasteurella multocida (P. multocida), Mannheimia haemolytica (M. haemolytica), Mycoplasma bovis, Histophilus somni, and Trueperella pyogenes (T. pyogenes) can cause diseases independently and cause secondary infections [14].
BRSV causes respiratory diseases in cattle that are lethal to some calves [25]. Many calves develop fever, nasal discharge, and cough due to BRSV infection, whereas others exhibit no or few clinical signs [28]. BRSV infections causes significant economic losses to the cattle industry in many countries [8, 13, 21]. BRSV can independently cause clinical diseases and promote the development of secondary bacterial infections in the lower respiratory tract [23, 26]. Respiratory systems harbor a plethora of resident bacteria. Because of the frequent occurrence of severe illnesses through viral and bacterial co-infection, recent studies have focused on the interactions between viruses and bacteria in human infectious diseases [1, 5, 6]. Studies reported that the relevant mechanisms included pathogen stability, enhanced adhesion, immune tolerance, and immune response suppression [15, 18]. Some mechanisms of the interaction between viruses and bacteria have been elucidated in the development of BRDC. It has been reported that the adhesion of P. multocida to respiratory epithelial cells infected with BRSV or bovine coronavirus is greater than that to noninfected cells [9, 23].
T. pyogenes is a gram-positive, pleomorphic, non-spore-forming, non-motile, non-capsulated, facultatively anaerobic rod [19]. T. pyogenes is commonly found in the mucous membranes of the upper respiratory, gastrointestinal, and urogenital tracts of cattle [11]. In cattle, T. pyogenes can cause infections of the reproductive tract and mammary gland, pneumonia, and liver abscessation [19]. T. pyogenes has been identified as a secondary pathogen in BRDC [17]. In cattle with BRDC, a significant increase in T. pyogenes infection was reported, indicating a substantial association between BRDC and T. pyogenes [7]. In a survey conducted in Northeast China, Zhou et al. reported that 28% of cattle infected with T. pyogenes infected with BRSV. This value was significantly higher than that of cattle not infected with T. pyogenes (12%) [29]. Thus, BRSV infection appears to be associated with an increase of T. pyogenes infection. However, the molecular mechanisms of why BRSV infection promotes T. pyogenes infection are unclear. In this study, the effect of BRSV infection on T. pyogenes adhesion to cells was investigated.
MATERIALS AND METHODS
Cell lines
All the cell lines were cultured at 37°C in a humidified atmosphere containing 5% CO2. Madin-Darby bovine kidney (MDBK) cells and human embryonic kidney-derived 293T cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) (Nissui, Tokyo, Japan) supplemented with 10% fetal bovine serum (FBS). Human lung cancer-derived A549 cells were maintained in Eagle’s minimal essential medium (Nissui) supplemented with 10% FBS. Primary cultures of bovine epithelial culture (bLEC) cells were kindly provided by Dr. T. Okabayashi (Department of Veterinary Science, Faculty of Agriculture, University of Miyazaki, Miyazaki, Japan) [22]. bLEC cells were seeded in a plate coated with collagen type I (Sumitomo Bakelite Co., Ltd., Tokyo, Japan) and maintained in DMEM/F12 (Thermo Fisher Scientific, Waltham, MA, USA), supplemented with 2% FBS, 100 U/mL penicillin, 100 mg/mL streptomycin, 10 ng/mL epidermal growth factor, 1% insulin-transferrin-selenium, 1% non-essential amino acid, and 2 mM L-glutamine.
Virus and bacteria
The BRSV rs-52 strain, which was isolated from a BRSV vaccine (Bovine RS live Vaccine, Kyoto Biken, Uji, Japan), was propagated for 3 days in MDBK cells cultured in DMEM, supplemented with 2% FBS. BRSV in the supernatant was collected after serial passages. Infectious titers were determined by performing plaque-forming assay using MDBK cells overlaid with DMEM supplemented with 0.8% carboxymethyl-cellulose and 2% FBS, and expressed as plaque-forming units. The RNA of the BRSV 28-1356 strain, which was kindly provided by Dr. G. Yaegashi (Iwate Prefecture Central Livestock Hygiene Service Center, Iwate, Japan), was derived from the nasal swabs of cattle infected with BRSV.
The T. pyogenes B-1 strain was isolated from the synovial fluid of cattle with arthritis in 2001 in Japan. The B-2 strain was isolated from the purulent discharge of cattle with uterine pyometra in 2002 in Japan. The B-3 strain was isolated from the purulent discharge of cattle with omphalitis in 2017 in Japan. All samples were isolated on 5% sheep blood agar at 37°C for 24–48 hr in an aerobic culture.
Plasmids
Total RNA was isolated from the two strains of BRSV using the RNeasy Mini Kit (Qiagen, Hilden, Germany) following the manufacturer’s protocol. Total RNA was reverse transcribed to cDNA using Random 6 mers with the PrimeScript II 1st strand cDNA Synthesis Kit (TaKaRa Bio Inc., Kusatsu, Japan), following the manufacturer’s protocol. The BRSV G genes of the two strains and Aequorea coerulescens green fluorescent (AcGFP) gene were amplified by PCR using primers (BRSV G forward primer 5′-cgctaccggtctcgagaccATGTCCAACCATACCCATCAT-3′, rs-52-G reverse primer 5′-cgatgttaactct agaTTAGATCTGTGTAGTTGATTGAT-3′, 28–1356-G reverse primer 5′-cgatgttaactctagaTTAGATCTGTGTAGTTGAGTT-3′, cGFP forward primer 5′-cgctaccggtctcgagaccATGGTGAGCAAGGGCGCCGA-3′, AcGFP reverse primer 5′-cgatgttaactctagaTTACTTGTACAG CTCATCCATGC-3′). The lowercase letters are the sequences for the In-Fusion reaction, and the uppercase letters are the sequences for elongating the insert. The pAcGFP1 (TaKaRa Bio Inc.) was used as the template for AcGFP. The CSII-EF-RfA was provided by Dr. Hiroyuki Miyoshi (the RIKEN BRC through the National BioResource Project of the MEXT, Japan) (cat. RDB04387) for cDNA expression using the EF-1 alpha promoter. After amplification of the target gene, PCR products were inserted between the XhoI and XbaI sites of CSII-EF-RfA using the In-Fusion HD Cloning Kit (TaKaRa Bio Inc.). The resulting plasmid was designated pCSII-EF-rs-52-G, pCSII-EF-28-1356-G, and pCSII-EF-AcGFP.
To create a recombinant BRSV G protein, the BRSV G gene was amplified by PCR using primers (forward primer 5′-attaat tcggatccgaatATGGACCACAACAACTCCCAAAC-3′ and reverse primer 5′-ggtggtggtgctcgagGATCTGTGTAGTTGATTG-3′) and BRSV cDNA as a template. The lowercase letters are sequences for In-Fusion reaction, and the uppercase letters are sequences for elongating the insert. After amplification of the target gene, the PCR products were inserted between the BamHI and EcoRI sites of pET22b (+) (TaKaRa Bio Inc.), which has a His-tag at the C-terminus, using the In-Fusion HD Cloning Kit (TaKaRa Bio Inc.). The resulting plasmid was designated pET22b (+)-BRSV-G.
Purification of BRSV G protein
The plasmid pET22b (+)-BRSV-G was transformed to Escherichia coli (E. coli) strain Rosetta-gami B competent cells (Novagen, Madison, WI, USA). Transformed E. coli was cultured into LB broth containing 50 µg/mL ampicillin. Recombinant BRSV G protein was induced by the addition of 1 mM IPTG. Induced bacteria were harvested and resuspended in sonication buffer (300 mM NaCl, 20 mM phosphate buffer; pH 7.5). After sonication, the supernatant was recovered and applied to a HisTALON Superflow Cartridge (TaKaRa Bio, Inc.). The column was washed extensively with 10 mM imidazole in the sonication buffer and eluted with 10–150 mM imidazole in the sonication buffer. The protein fraction was collected and dialyzed using Regenerated Cellulose Dialysis Tubing Nominal MWCO:3500d (Life Technologies, Carlsbad, CA, USA). After dialysis, the samples were subjected to Macro-Prep High-S chromatography (Bio-Rad Laboratories, Inc., Hercules, CA, USA). The column was washed with 20 mM Tris-HCl (pH 8.0) and eluted using 0–500 mM NaCl. The protein fraction was collected and concentrated using an ULTRA FILTER: Molecular Weight cut-off of 10,000 Da (Advantech, Tokyo, Japan). The buffer was then replaced with phosphate-buffered saline (PBS) by using Sephadex G-25 (Sigma-Aldrich, St. Louis, MO, USA). The purity of BRSV G protein was determined by silver staining analysis using SDS-PAGE.
Antibodies
Mouse monoclonal antibody against β-actin (Sc-69879) was purchased from Sigma-Aldrich. Rabbit anti-BRSV antibody (PA1-23354) was purchased from Thermo Fisher Scientific. Rat anti-HA antibody (15645900) was purchased from Roche (Basel, Switzerland). Rabbit anti-GFP antibody (598) was purchased from Medical and Biological Laboratories Co., LTD (Tokyo, Japan). Peroxidase affinipure goat anti-mouse antibody (115-035-044) and peroxidase affinipure goat anti-rabbit antibody (111-035-003) were purchased from Jackson ImmunoResearch Laboratories, Inc. (West Grove, PA, USA). Goat anti-rat IgG antibody (HRP) (ab97057) was purchased from Abcam (Cambridge, UK). Alexa Flour 594-conjugated anti-rabbit IgG antibody (R37119) was purchased from Thermo Fisher Scientific.
To produce the anti-BRSV G protein polyclonal antibody, Balb/c mice (7-week-old females) were immunized twice at 1-month intervals with purified BRSV G (25 µg) and adjuvant TiterMax Gold (Sigma-Aldrich), following the manufacturer’s instructions. Three days after the final immunization, the antiserum was collected and used as a polyclonal antibody. All animal experiments were conducted in accordance with the Regulations for the Care and Use of Laboratory Animals of Kitasato University. The animal experimentation protocol was approved by the President of Kitasato University based on the judgment made by the Institutional Animal Care and Use Committee of Kitasato University (Approval ID: 22-071). The polyclonal antibody could detect the correct size of G protein form the lysate of BRSV-infected cells (data not shown).
Adhesion assay
The adhesion assay was performed as previously described [23], with slight modifications. MDBK, A549, and bLEC cells were inoculated with BRSV (multiplicity of infection [MOI] 0.01, 0.1, or 1) or transfected with plasmids using Trans IT LT-1 (Mirus Bio LCC, Madison, WI, USA). After 2 hr, the inoculation medium was replaced with a culture medium containing 2% FBS. After 24, 48, and 72 hr, the cells were exposed to 108 colony forming units (CFU) T. pyogenes for 2 hr at 37°C. The cells were washed three times with PBS and collected by trypsinization. The cells and attached bacteria were centrifuged at 8,000 × g for 5 min. T. pyogenes was diluted with PBS and plated on 5% sheep blood agar. The CFU/mL was evaluated after 48 hr at 37°C. The number of adherent bacteria per cell was calculated by dividing the total number of CFU by the total number of monolayer cells.
Gentamicin protection assay
MDBK cells were inoculated with BRSV (MOI 1). After 2 hr, the inoculation medium was replaced with culture medium containing 2% FBS. At 48 hr after infection, the cells were exposed 108 CFU of T. pyogenes for 2 hr at 37°C. To kill the extracellular bacteria, the cell surfaces were washed three times DMEM containing 2% FBS and 40 µg/mL gentamicin and incubated with DMEM containing 2% FBS and 40 µg/mL gentamicin for 1 hr at 37°C. Cells were washed three times with PBS and collected by trypsinization. Cells and internalized bacteria were centrifuged at 8,000 × g for 5 min. T. pyogenes was diluted with PBS and plated on 5% sheep blood agar. The number of CFU/mL was counted after 48 hr at 37°C. The number of bacteria with or without gentamicin was calculated.
Immunoblotting
Cells were lysed in buffer (100 mM Tris-HCl [pH 6.8], 4% SDS, 1.2% 2-Mercaptoethanol, and 20% glycerol), boiled, and subjected to 12% SDS-PAGE. The proteins were transferred to polyvinylidene difluoride membranes (GE Healthcare, Chicago, IL, USA) and incubated with the appropriate antibodies. The immune complexes were visualized using ECL prime Western Blotting Detection Reagents (Cytiva, Tokyo, Japan) and detected using an i-Bright 1500 system (Thermo Fisher Scientific).
Immunofluorescence assay
For the immunofluorescence assay, T. pyogenes was incubated with 2 µg/mL fluorescein isothiocyanate (FITC; Dojindo, Tokyo, Japan) for 30 min at 25°C. FITC-labeled bacteria were washed with PBS and adjusted with DMEM. MDBK cells were cultured on glass slides were fixed with 4% paraformaldehyde in PBS at 25°C for 30 min. After three washes with PBS, the cells were permeabilized for 20 min at 25°C in PBS containing 0.2% Triton-X100 and blocked with PBS containing 2% bovine serum albumin for 1 hr at 25°C. The cells were incubated with blocking buffer containing mouse anti-BRSV G protein antibodies at 25°C for 1 hr, washed three times with PBS, and incubated with blocking buffer containing Alexa Flour 594-conjugated secondary antibodies at 25°C for 1 hr. Finally, the cells were washed three times with PBS, mounted using Mounting Medium with 4′,6-diamidono-2-phenylindole (DAPI) (H-1200) (Vector Laboratories, Inc., Newark, CA, USA) and observed using an LSM710 laser-scanning confocal microscope (ZEISS, Oberkochen, Germany).
Statistical analysis
The data for statistical analyses were the averages of three independent experiments. Results were expressed as the mean ± standard deviation. Data from only two groups were determined by Student’s t-test (Welch’s t-test) and those of multiple groups were analyzed by one-way analysis of variance followed by Tukey’s test.
RESULTS
BRSV infection enhances the adhesion of T. pyogenes in an MOI- and time-dependent manner
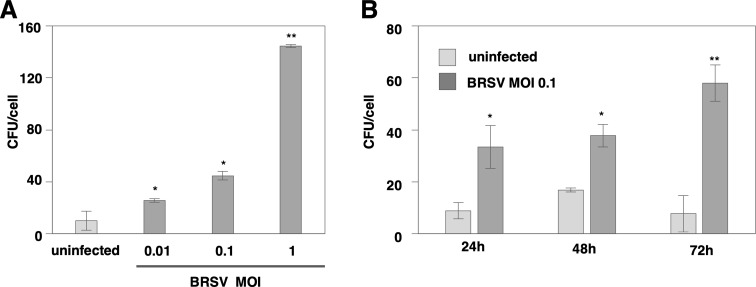
MDBK cells were infected with BRSV, and the number of T. pyogenes adhering per cell was measured after 48 hr (Fig. 1A). The adhesion of T. pyogenes was significantly increased by BRSV infection in an MOI-dependent manner. Next, the adhesion of T. pyogenes was measured at 24, 48, and 72 hr after BRSV infection (Fig. 1B). A significant increase in adhesion was recorded at all time points, with a further increase observed at 72 hr post-infection. These results indicated that BRSV infection enhanced the adhesion of T. pyogenes in an MOI- and time-dependent manner.
Fig. 1.
Bovine respiratory syncytial virus (BRSV) infection enhances the adhesion of Truperella pyogenes (T. pyogenes) in an multiplicity of infection (MOI-) and time-dependent manner. (A) The number of adherent T. pyogenes to Madin-Darby bovine kidney (MDBK) cells infected with BRSV at an MOI of 0, 0.01, 0.1, or 1 for 48 hr was measured by an adhesion assay. (B) The number of adherent T. pyogenes on MDBK cells infected with BRSV at an MOI of 0.1 for 24, 48, or 72 hr was measured by an adhesion assay. In all cases, asterisks indicate significant differences (*P<0.05, **P<0.01) versus the results for control cells.
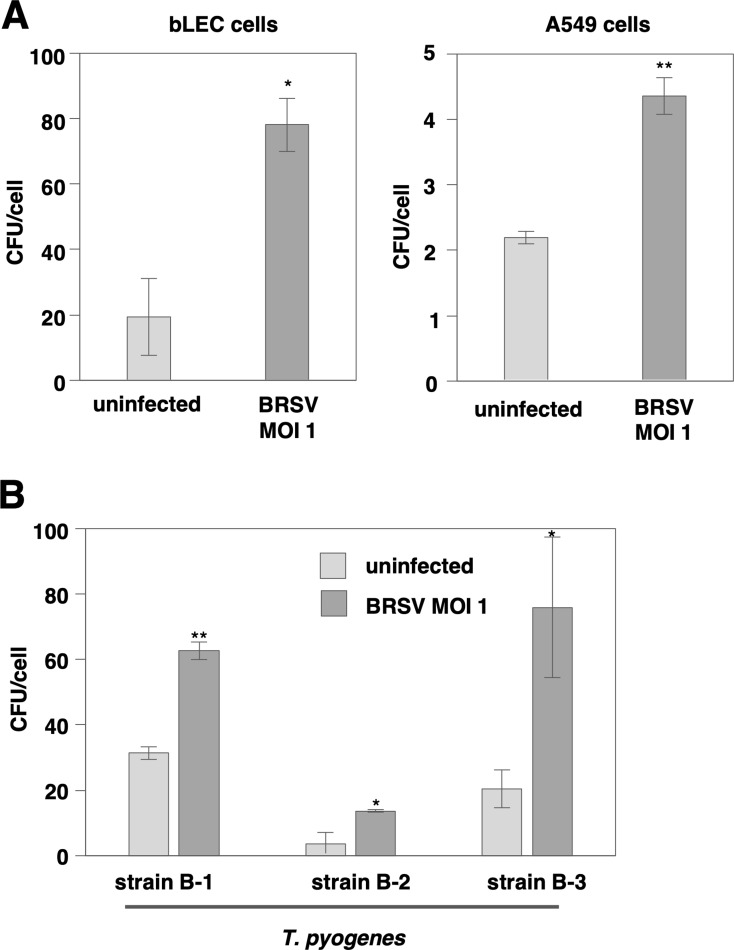
The enhanced adhesion due to BRSV infection was also observed in bLEC and A549 cells (Fig. 2A). Furthermore, a similar enhancement was observed when the three different strains of T. pyogenes were used (Fig. 2B). The enhanced adhesion of T. pyogenes due to BRSV infection occur widely regardless of the cell type including primary culture and of the bacterial strain.
Fig. 2.
The enhanced adhesion of Truperella pyogenes (T. pyogenes) attributable to bovine respiratory syncytial virus (BRSV) infection is observed in various cell lines and bacterial strains. (A) The number of adherent T. pyogenes specimens on bLEC, or A549 cells infected with BRSV at an multiplicity of infection (MOI) of 1 for 48 hr was measured by an adhesion assay. (B) The number of specimens of three different strains of T. pyogenes adhering to Madin-Darby bovine kidney (MDBK) cells infected with BRSV at an MOI of 1 for 48 hr was measured by an adhesion assay. In all cases, asterisks indicate significant differences (*P<0.05, **P<0.01) versus the results for control cells.
BRSV infection is not related to intracellular invasion by T. pyogenes
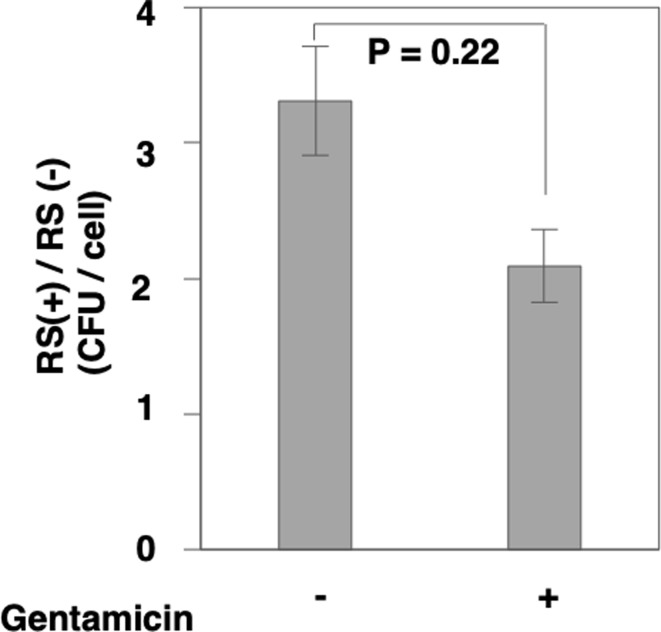
It was reported that T. pyogenes could invade cells [11]. Therefore, to determine whether BRSV infection is involved in intracellular invasion of T. pyogenes, we conducted a gentamicin protection assay. The results showed that the extent of adhesion enhancement caused by BRSV infection was not altered in the presence of gentamicin (P=0.22) (Fig. 3). This result suggests that BRSV infection does not contribute to the intracellular invasion of T. pyogenes. In fact, viruses enhance the adhesion of the bacterium to the cell surface.
Fig. 3.
Bovine respiratory syncytial virus (BRSV) infection is not related to intracellular invasion by Truperella pyogenes (T. pyogenes). The number of T. pyogenes specimens adhering to Madin-Darby bovine kidney (MDBK) cells infected with BRSV at an multiplicity of infection (MOI) of 1 for 48 hr was measured by the gentamicin protection assay. The number of adhering bacteria in the presence or absence of gentamicin was calculated.
Glycoproteins of BRSV correlate with bacterial attachment
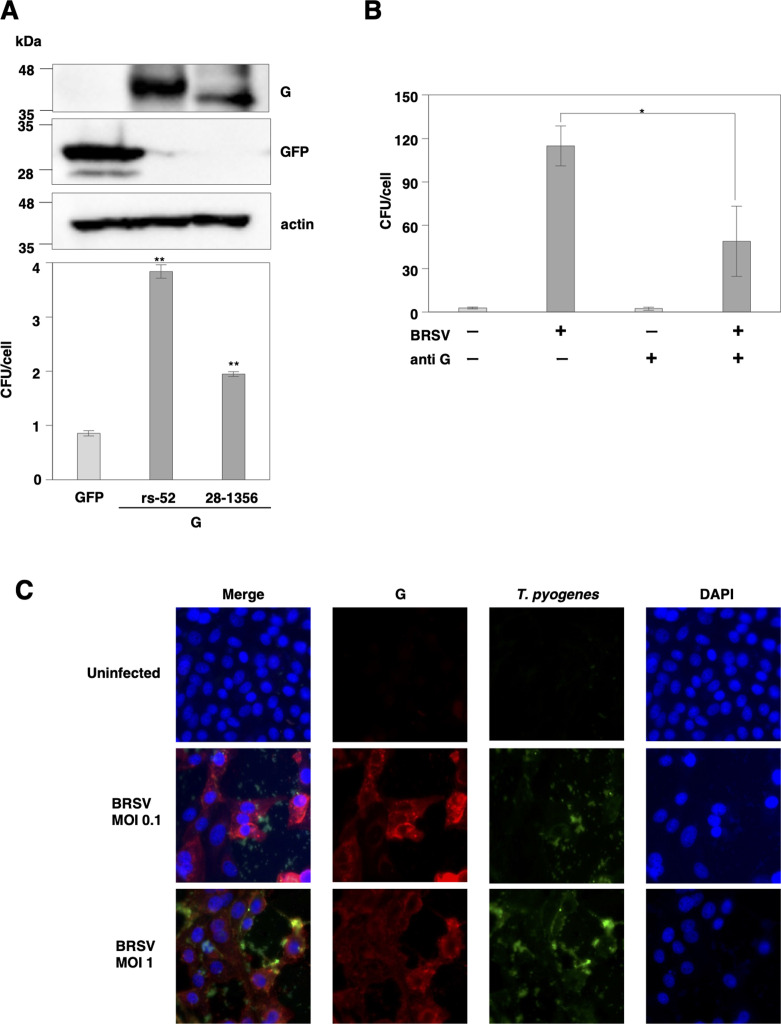
G protein, a glycoprotein present on the surface of BRSV, is primarily involved in cell adhesion [3, 27]. Several bacteria can bind to respiratory syncytial viruses (RSV) through G protein, contributing to secondary bacterial infections [20]. Therefore, to confirm whether the G protein of BRSV is involved in the enhancement of T. pyogenes adhesion, we conducted an adhesion assay using cells expressing the BRSV G protein. The results revealed a considerable increase in the adhesion of T. pyogenes to cells expressing BRSV G protein derived from two strains belonging to other subgenotypes (Fig. 4A). Furthermore, when polyclonal antibodies against BRSV G protein were used, the adhesion enhancement caused by BRSV infection was inhibited (Fig. 4B). The immunofluorescence assay demonstrated an increase in the number of T. pyogenes adhering to cells following BRSV infection, indicated the colocalization of bacteria with BRSV G protein (Fig. 4C). These results suggest that G protein of BRSV is involved in the cell adhesion enhancement of T. pyogenes.
Fig. 4.
Glycoproteins of bovine respiratory syncytial virus (BRSV) are related to the enhancement of bacterial attachment. (A) BRSV G protein derived from two strains was exogenously expressed in 293T cells through plasmid transfection. The expression of BRSV G protein in cells was determined by immunoblotting (upper panel). The number of Truperella pyogenes (T. pyogenes) specimens adhering to cells infected with BRSV at an multiplicity of infection (MOI) of 1 for 48 hr was measured by the adhesion assay (lower panel). (B) The number of T. pyogenes specimens adhering to cells infected with BRSV at an MOI of 1 for 48 hr with or without anti-G antibody was measured by the adhesion assay. (C) Fluorescence localization in cells infected with BRSV and bound by FITC- conjugated T. pyogenes (green) was observed with a confocal microscope. BRSV G (red) was stained with an antibody, and nuclei were stained with 4′,6-diamidono-2-phenylindole (DAPI). In all cases, asterisks indicate significant differences (*P<0.05, **P<0.01) versus the results for control cells.
DISCUSSION
In this study, we demonstrated that BRSV infection enhanced the adhesion of T. pyogenes to cells via expression of BRSV G protein. BRSV causes respiratory diseases in cattle, and co-infection with bacteria exacerbates disease severity. T. pyogenes is an important secondary pathogen in bovine respiratory infections [29], leading to the development of aggravated respiratory symptoms following BRSV infection. We found that the enhancement of T. pyogenes adhesion to cells due to BRSV infection was dependent on the MOI and time after infection in vitro. Additionally, BRSV infection did not affect intracellular invasion by T. pyogenes, indicating increased adhesion to the cell surface. Consequently, it can be suggested that BRSV infection promoted T. pyogenes infection in cattle.
In this study, we observed an enhancement in the adhesion of T. pyogenes to MDBK, bLEC and A549 cells after BRSV infection. We hypothesized that the viral proteins expressed on the cell membrane following BRSV infection are involved in the enhancement of bacterial adhesion. We found that BRSV G proteins expressed on the surface of BRSV-infected cells enhance cell adhesion of T. pyogenes. BRSV G protein is involved in the attachment to host cell receptors [27]. In BRSV-infected cells, BRSV G protein co-localized with T. pyogenes. These findings suggested that BRSV G protein acted as a receptor for T. pyogenes or that its expression influenced the expression of host factors.
A previous study reported that BRSV infection induced the overexpression of platelet-activating factor receptors, thereby enhancing the adhesion of P. multocida to cells [23]. Furthermore, BRSV infection downregulate intercellular adhesion molecule-1, thereby suppressing the attachment of P. multocida to the upper respiratory tract, and promoting bacterial invasion into the lower respiratory tract [24]. The BRSV-mediated change in the adhesion of P. multocida to cells are attributed to alterations in the expression of host factors associated with BRSV, however, the involvement of viral proteins, such as G protein, in this change has not been investigated. It is possible that changes in the expression levels of host factors altered the adhesion properties of T. pyogenes.
On the other hand, as an example of viral proteins functioning as bacterial receptors, influenza virus infection induce an increased expression of hemagglutinin membrane glycoprotein on the host cell surface, enhancing the adhesion of Streptococcus pyogenes [16]. Furthermore, the RSV G protein enhances cell adhesion by binding to penicillin-binding protein 1a of Streptococcus pneumoniae [2, 20]. Further detailed investigation is required to understand the mechanism by which the BRSV G protein influences adhesion of T. pyogenes. Additionally, the potential for changes in adhesion properties owing to the expression of BRSV G protein in other bacteria cannot be ruled out, and further investigation is necessary.
Because the G protein is the most variable protein among all BRSV proteins, BRSV is classified into subgenotypes I-X based on the sequence of the G protein [12]. The amino acid homology of the BRSV G protein between the rs-52 (subgenotype II) and 28-1356 strains (subgenotype X) used in this study was approximately 84%. Consequently, adhesion assays conducted with BRSV G protein-expressing cells from each strain revealed that BRSV G proteins from both strains enhanced the adhesion of T. pyogenes; however, the extent of adhesion varied. The reason for the difference in the number of T. pyogenes adhesion between the two strains is unknown, however it might be due to the difference in the gene transfer efficiency of the BRSV G gene or the expression levels of BRSV G protein. Further investigation is needed to evaluate the gene transfer efficiency and the expression levels. Because BRSV G sequences continue to accumulate mutations, leading to the emergence of new subgenotypes, it is possible that viral strains that promote enhanced bacterial adhesion will arise. Consequently, it is essential to further elucidate the detailed mechanisms and continuously monitor the trends in mutations.
In conclusion, we elucidated that BRSV infection enhanced the adhesion of T. pyogenes to cells and identified the involvement of the BRSV G protein in this mechanism. The strengthened adhesion of T. pyogenes potentially contributed to the exacerbation of BRDC.
Conflicts of Interest
The authors declare no conflicts of interest.
Acknowledgments
We thank Dr. H. Miyoshi, Dr. T. Okabayashi and Dr. G. Yaegashi for providing experimental materials.
REFERENCES
- 1.Almand EA, Moore MD, Jaykus LA. 2017. Virus-bacteria interactions: an emerging topic in human infection. Viruses 9: 58. doi: 10.3390/v9030058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Avadhanula V, Wang Y, Portner A, Adderson E. 2007. Nontypeable Haemophilus influenzae and Streptococcus pneumoniae bind respiratory syncytial virus glycoprotein. J Med Microbiol 56: 1133–1137. doi: 10.1099/jmm.0.47086-0 [DOI] [PubMed] [Google Scholar]
- 3.Battles MB, McLellan JS. 2019. Respiratory syncytial virus entry and how to block it. Nat Rev Microbiol 17: 233–245. doi: 10.1038/s41579-019-0149-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bell RL, Turkington HL, Cosby SL. 2021. The bacterial and viral agents of BRDC: immune evasion and vaccine developments. Vaccines (Basel) 9: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bosch AATM, Biesbroek G, Trzcinski K, Sanders EAM, Bogaert D. 2013. Viral and bacterial interactions in the upper respiratory tract. PLoS Pathog 9: e1003057. doi: 10.1371/journal.ppat.1003057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brealey JC, Sly PD, Young PR, Chappell KJ. 2015. Viral bacterial co-infection of the respiratory tract during early childhood. FEMS Microbiol Lett 362: 362. doi: 10.1093/femsle/fnv062 [DOI] [PubMed] [Google Scholar]
- 7.Centeno-Martinez RE, Glidden N, Mohan S, Davidson JL, Fernández-Juricic E, Boerman JP, Schoonmaker J, Pillai D, Koziol J, Ault A, Verma MS, Johnson TA. 2022. Identification of bovine respiratory disease through the nasal microbiome. Anim Microbiome 4: 15. doi: 10.1186/s42523-022-00167-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dubrovsky SA, Van Eenennaam AL, Aly SS, Karle BM, Rossitto PV, Overton MW, Lehenbauer TW, Fadel JG. 2020. Preweaning cost of bovine respiratory disease (BRD) and cost-benefit of implementation of preventative measures in calves on California dairies: The BRD 10K study. J Dairy Sci 103: 1583–1597. doi: 10.3168/jds.2018-15501 [DOI] [PubMed] [Google Scholar]
- 9.Fahkrajang W, Sudaryatma PE, Mekata H, Hamabe S, Saito A, Okabayashi T. 2021. Bovine respiratory coronavirus enhances bacterial adherence by upregulating expression of cellular receptors on bovine respiratory epithelial cells. Vet Microbiol 255: 109017. doi: 10.1016/j.vetmic.2021.109017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gershwin LJ, Berghaus LJ, Arnold K, Anderson ML, Corbeil LB. 2005. Immune mechanisms of pathogenetic synergy in concurrent bovine pulmonary infection with Haemophilus somnus and bovine respiratory syncytial virus. Vet Immunol Immunopathol 107: 119–130. doi: 10.1016/j.vetimm.2005.04.004 [DOI] [PubMed] [Google Scholar]
- 11.Jost BH, Billington SJ. 2005. Arcanobacterium pyogenes: molecular pathogenesis of an animal opportunist. Antonie van Leeuwenhoek 88: 87–102. doi: 10.1007/s10482-005-2316-5 [DOI] [PubMed] [Google Scholar]
- 12.Kumagai A, Kawauchi K, Andoh K, Hatama S. 2021. Sequence and unique phylogeny of G genes of bovine respiratory syncytial viruses circulating in Japan. J Vet Diagn Invest 33: 162–166. doi: 10.1177/1040638720975364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Larson RL. 2005. Effect of cattle disease on carcass traits. J Anim Sci 83: E37–E43. doi: 10.2527/2005.8313_supplE37x [DOI] [Google Scholar]
- 14.Mosier D. 2014. Review of BRD pathogenesis: the old and the new. Anim Health Res Rev 15: 166–168. doi: 10.1017/S1466252314000176 [DOI] [PubMed] [Google Scholar]
- 15.Neu U, Mainou BA. 2020. Virus interactions with bacteria: Partners in the infectious dance. PLoS Pathog 16: e1008234. doi: 10.1371/journal.ppat.1008234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Okamoto S, Kawabata S, Nakagawa I, Okuno Y, Goto T, Sano K, Hamada S. 2003. Influenza A virus-infected hosts boost an invasive type of Streptococcus pyogenes infection in mice. J Virol 77: 4104–4112. doi: 10.1128/JVI.77.7.4104-4112.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pardon B, Buczinski S. 2020. Bovine respiratory disease diagnosis: what progress has been made in infectious diagnosis? Vet Clin North Am Food Anim Pract 36: 425–444. doi: 10.1016/j.cvfa.2020.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pfeiffer JK, Virgin HW. 2016. Viral immunity. Transkingdom control of viral infection and immunity in the mammalian intestine. Science 351: aad5872–aad5872. doi: 10.1126/science.aad5872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rzewuska M, Kwiecień E, Chrobak-Chmiel D, Kizerwetter-Świda M, Stefańska I, Gieryńska M. 2019. Pathogenicity and Virulence of Trueperella pyogenes: A Review. Int J Mol Sci 20: 2737. doi: 10.3390/ijms20112737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith CM, Sandrini S, Datta S, Freestone P, Shafeeq S, Radhakrishnan P, Williams G, Glenn SM, Kuipers OP, Hirst RA, Easton AJ, Andrew PW, O’Callaghan C. 2014. Respiratory syncytial virus increases the virulence of Streptococcus pneumoniae by binding to penicillin binding protein 1a. A new paradigm in respiratory infection. Am J Respir Crit Care Med 190: 196–207. doi: 10.1164/rccm.201311-2110OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Snowder GD, Van Vleck LD, Cundiff LV, Bennett GL, Koohmaraie M, Dikeman ME. 2007. Bovine respiratory disease in feedlot cattle: phenotypic, environmental, and genetic correlations with growth, carcass, and longissimus muscle palatability traits. J Anim Sci 85: 1885–1892. doi: 10.2527/jas.2007-0008 [DOI] [PubMed] [Google Scholar]
- 22.Sudaryatma PE, Mekata H, Kubo M, Subangkit M, Goto Y, Okabayashi T. 2019. Co-infection of epithelial cells established from the upper and lower bovine respiratory tract with bovine respiratory syncytial virus and bacteria. Vet Microbiol 235: 80–85. doi: 10.1016/j.vetmic.2019.06.010 [DOI] [PubMed] [Google Scholar]
- 23.Sudaryatma PE, Nakamura K, Mekata H, Sekiguchi S, Kubo M, Kobayashi I, Subangkit M, Goto Y, Okabayashi T. 2018. Bovine respiratory syncytial virus infection enhances Pasteurella multocida adherence on respiratory epithelial cells. Vet Microbiol 220: 33–38. doi: 10.1016/j.vetmic.2018.04.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sudaryatma PE, Saito A, Mekata H, Kubo M, Fahkrajang W, Okabayashi T. 2020. Bovine respiratory syncytial virus decreased Pasteurella multocida adherence by downregulating the expression of intercellular adhesion molecule-1 on the surface of upper respiratory epithelial cells. Vet Microbiol 246: 108748. doi: 10.1016/j.vetmic.2020.108748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taylor JD, Fulton RW, Lehenbauer TW, Step DL, Confer AW. 2010. The epidemiology of bovine respiratory disease: What is the evidence for predisposing factors? Can Vet J 51: 1095–1102. [PMC free article] [PubMed] [Google Scholar]
- 26.Timsit E, Christensen H, Bareille N, Seegers H, Bisgaard M, Assié S. 2013. Transmission dynamics of Mannheimia haemolytica in newly-received beef bulls at fattening operations. Vet Microbiol 161: 295–304. doi: 10.1016/j.vetmic.2012.07.044 [DOI] [PubMed] [Google Scholar]
- 27.Valarcher JF, Taylor G. 2007. Bovine respiratory syncytial virus infection. Vet Res 38: 153–180. doi: 10.1051/vetres:2006053 [DOI] [PubMed] [Google Scholar]
- 28.Zewde D, Gemeda G, Ashagrie T. 2022. Review on bovine respiratory syncytial virus characteristic, pathogenesis and control methods applied for the disease. Austin J Vet Sci Anim Husb 9: 1103. [Google Scholar]
- 29.Zhou Y, Shao Z, Dai G, Li X, Xiang Y, Jiang S, Zhang Z, Ren Y, Zhu Z, Fan C, Zhang G. 2023. Pathogenic infection characteristics and risk factors for bovine respiratory disease complex based on the detection of lung pathogens in dead cattle in Northeast China. J Dairy Sci 106: 589–606. doi: 10.3168/jds.2022-21929 [DOI] [PubMed] [Google Scholar]