Abstract
Retroviruses containing inserts of exogenous sequences frequently eliminate the inserted sequences upon spread in susceptible cells. We have constructed replication-competent murine leukemia virus (MLV) vectors containing internal ribosome entry site (IRES)-transgene cassettes at the env-3′ untranslated region boundary in order to examine the effects of insert sequence and size on the loss of inserts during viral replication. A virus containing an insertion of 1.6 kb replicated with greatly attenuated kinetics relative to wild-type virus and lost the inserted sequences in a single infection cycle. In contrast, MLVs containing inserts of 1.15 to 1.30 kb replicated with kinetics only slightly attenuated compared to wild-type MLV and exhibited much greater stability, maintaining their genomic integrity over multiple serial infection cycles. Eventually, multiple species of deletion mutants were detected simultaneously in later infection cycles; once detected, these variants rapidly dominated the population and thereafter appeared to be maintained at a relative equilibrium. Sequence analysis of these variants identified preferred sites of recombination in the parental viruses, including both short direct repeats and inverted repeats. One instance of insert deletion through recombination with an endogenous retrovirus was also observed. When specific sequences involved in these recombination events were eliminated, deletion variants still arose with the same kinetics upon virus passage and by apparently similar mechanisms, although at different locations in the vectors. Our results suggest that while lengthened, insert-containing genomes can be maintained over multiple replication cycles, preferential deletions resulting in loss of the inserted sequences confer a strong selective advantage.
Replicating retrovirus populations are characterized by a high degree of genetic change (6). This genetic diversity is the product of high frequencies of base misincorporations (4, 13, 40), rearrangements (7, 57), and both homologous (5, 18, 48) and nonhomologous (33, 58, 59) recombination events in the viral genome. Such genetic variability gives retroviruses the ability to adapt quickly to changes in selective pressures.
A variety of replication-competent retroviral vectors have been created by the insertion of heterologous sequences into full-length viral genomes. Such vectors have been constructed from several retrovirus species, including Rous sarcoma virus (RSV) (3, 29, 36), murine leukemia virus (MLV) (10, 28, 39, 49), spleen necrosis virus (12), human immunodeficiency virus (21, 27, 32, 55), and human foamy virus (42). In studies in which the structure of these lengthened viruses was examined subsequent to replication, the inserted sequences were usually found to have been partially or completely lost from the population within three or fewer passages through cultures of susceptible cells (12, 24, 28, 39, 42, 49). The tendency of retroviruses to rapidly delete insertions has been observed with various different insert sequences, indicating that the sequence requirements for efficient deletion are fairly permissive.
Previous studies that have analyzed in detail deletion mutants of nondefective retroviruses utilized RSV, which loses most its src coding sequence upon replication in culture (2, 31, 33, 58). However, differences in the stability of the RSV src gene in transformed versus nontransformed cells have been reported, suggesting that the presence of this oncogene may result in selective pressures on cultured viruses based not only on the virus's replicative fitness, but also on the cytoproliferative function of the protein encoded by this gene (2, 30). In this system, examination of virus stability in the absence of such selective pressures is therefore difficult.
We have recently shown that an internal ribosome entry site (IRES)-transgene expression cassette inserted at the env-3′ untranslated region (UTR) boundary in the MLV genome results in a fully replication-competent vector that can be used as a tool to efficiently and reliably transmit transgenes in single-passage mammalian cell culture (28a, 46). In the present studies, we used this system to examine the genetic stability of insert-containing MLVs and analyzed the effects of particular sequences in these viruses on the emergence of deletion variants during virus passage.
MATERIALS AND METHODS
Retroviral vector plasmid construction.
An infectious Moloney MLV proviral clone was excised with NheI, which cuts once within each long terminal repeat (LTR), from plasmid pZAP (45) (kindly provided by John A. Young, University of Wisconsin) in order to eliminate flanking rat genomic sequences and recloned in the plasmid backbone of MLV vector g1ZIN to produce plasmid pZAP2. The region of the env gene from the unique NsiI site to the termination codon was amplified by PCR and fused to the encephalomyocarditis virus IRES (22) amplified from plasmid pEMCF by overlap extension PCR (16), introducing the restriction sites BstBI and NotI at the 3′ end. Plasmids g1ZIN and pEMCF were kindly provided by W. French Anderson, University of Southern California. The region from the env termination codon to the 3′ end of the 3′ LTR was also amplified by PCR, introducing NotI and AflIII sites at the 5′ and 3′ ends of the amplification product, respectively. A three-way ligation was used to insert this PCR product and the overlap extension PCR product into pZAP2 at its NsiI site and an AflIII site in the plasmid backbone, producing plasmid pZAPd. The puromycin acetyltransferase gene (pac) from plasmid pPUR (Clontech), the hygromycin phosphotransferase gene (hph) from plasmid pTK-hygro (Clontech), and the green fluorescent protein (GFP) cDNA (9) of plasmid pEGFP (Clontech) were each amplified by PCR and inserted into the BstBI and NotI sites of pZAPd, in frame with the authentic start codon of the IRES, producing pZAPd-puro, pZAPd-hygro, and pZAPd-GFP, respectively. All regions generated by PCR were verified by sequencing. A pZAPd-GFP-based construct in which an 11-bp repeat sequence flanking the IRES-GFP insert was eliminated and replaced by an MluI site was also generated by site-directed mutagenesis and designated pZAPm-GFP. An additional construct in which the Moloney MLV ecotropic envelope was replaced with the amphotropic envelope from 4070A was generated by overlap extension PCR and designated pAZE-GFP.
Cell culture and virus production.
293T (11), NIH 3T3 (20), and XC (51) cells were cultivated in Dulbecco's modified Eagle's medium with 10% fetal bovine serum. Virus stock was produced by transfection of 293T cells using calcium phosphate precipitation with pZAP2, pZAPd-GFP, pZAPd-puro, or pZAPd-hygro (47). Virus-containing supernatant was collected 48 h following transfection and passed through 0.45-μm syringe filters before use.
Viral assays.
Reverse transcriptase (RT) activity in supernatants of infected cell cultures was assayed as described previously (53). Quantitation of the reaction products was carried out using a Storm Phosphorlmager (Molecular Dynamics). Virus titers were determined by the UV-XC syncytium assay (41).
Single-cycle infection with replicating viruses in culture.
NIH 3T3 cells at 20% confluence in 6-cm dishes were infected with stock virus at a multiplicity of infection (MOI) of 0.0005. At 3, 5, and 8 days postinfection, the cells were examined by microscopy and split 1:5, and an aliquot of the cells was analyzed for GFP expression by flow cytometry as described below.
Multiple-cycle infections with replicating viruses in culture.
NIH 3T3 cells at 20% confluence in 6-cm dishes were infected with stock virus at an MOI of 0.001. At 2 days postinfection, the cells were split 1:5, and in the case of ZAPd-GFP, aliquots were analyzed for GFP expression by flow cytometry as described below. At 4 days postinfection, 100-fold dilutions of cell culture supernatant were used to infect fresh NIH 3T3 cultures. At the 4-day time point, we subcultivated each culture for preparation of unintegrated proviral DNA and analyzed ZAPd-GFP-infected cells by flow cytometry. This cycle was repeated several additional times. Proviral DNA was extracted from each culture by a modified Hirt procedure (26).
Flow cytometry.
NIH 3T3 cells were washed with phosphate-buffered saline, trypsinized, and collected by low-speed centrifugation. Cells were resuspended and analyzed with a Becton Dickinson FACScan fluorescence-activated cell sorter (FACS) using the FL1 emission channel to monitor green fluorescence.
Southern blot analysis of viral genomes.
Hirt DNA was digested with NheI, separated by electrophoresis, and blotted onto nylon membranes. Probes were generated by [32P]dCTP-labeled random priming of restriction fragments from each specific transgene or a common 2-kb NheI-XhoI MLV LTR-gag fragment. The blots were hybridized and washed under standard conditions and analyzed by PhosphorImager.
PCR analysis of viral deletion mutants.
PCR amplification of Hirt DNA from infected NIH 3T3 cells was performed using upstream primers hybridizing to the ecotropic (in the case of ZAPd-hygro, ZAPd-GFP, and ZAPm-GFP) or the amphotropic (in the case of AZE-GFP) env gene and a common downstream primer hybridizing at the 3′ UTR-3′ LTR border. Upon electrophoresis, PCR products that were smaller than the expected size for full-length virus genomes were gel purified and sequenced.
RESULTS
Generation of lengthened MLV-based retroviruses.
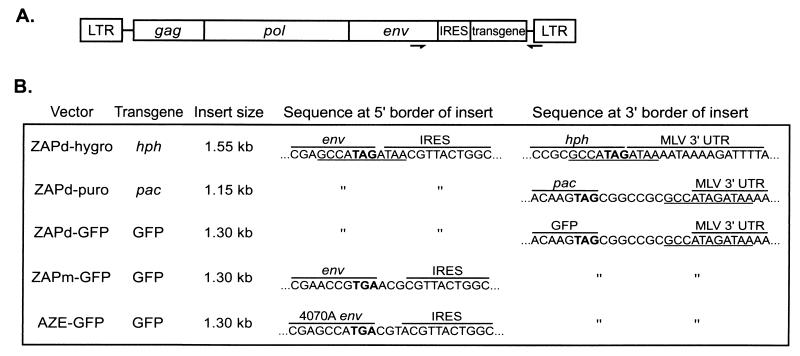
The encephalomyocarditis virus IRES linked to sequences encoding puromycin acetyltransferase, GFP, or hygromycin phosphotransferase was inserted into the ecotropic MLV genome, positioning the insertion immediately after the env termination codon. These plasmids were designated pZAPd-puro, pZAPd-GFP, and pZAPd-hygro and contained IRES transgene insertions of 1.15, 1.3, and 1.55 kb, respectively, positioned at the env-UTR boundary (Fig. 1). Infection of NIH 3T3 cells with ZAPd-puro and ZAPd-hygro stock virus conferred resistance to puromycin and hygromycin, respectively, and cells infected with pZAPd-GFP exhibited bright green fluorescence when observed by UV light microscopy, demonstrating that the vectors mediated functional expression of each transgene (data not shown).
FIG. 1.
Insert-containing viruses used in this study. (A) General structure of viruses, showing location of IRES-transgene cassettes within the MLV genome. Arrows indicate location of primers used in PCR amplification of proviral DNA of passaged virus. (B) Structural details of each virus, including sizes of IRES-transgene inserts and nucleotide sequences at 5′ and 3′ borders of insert. Nucleotides in bold show the position of the env stop codon. An 11-bp repeat sequence that flanks the inserts of ZAPd-hygro, ZAPd-puro, and ZAPd-GFP is underlined.
Replication kinetics of insert-containing viruses through a single infection cycle.
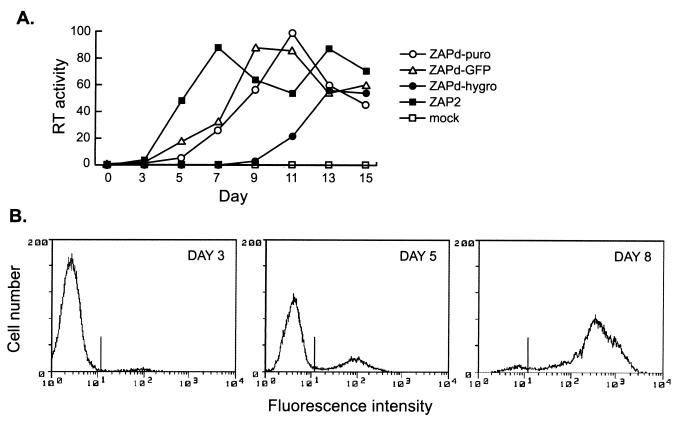
To examine the ability of ZAPd-puro, ZAPd-GFP, and ZAPd-hygro to replicate in cultured cells, we monitored RT activity in transfected cultures over a 15-day period. The parental wild-type virus, ZAP2, was used in parallel as a control. Both ZAPd-puro and ZAPd-GFP showed approximately the same lag period (3 days) as wild-type MLV prior to the appearance of detectable levels of RT activity, and thereafter exhibited a time course slightly attenuated compared to that of wild-type MLV (Fig. 2), suggesting that the insert-containing viruses replicated with moderately slower kinetics than wild-type virus. In contrast, ZAPd-hygro was greatly attenuated compared to wild-type MLV or the other insert-containing viruses, exhibiting a lag period of 9 days prior to the appearance of detectable RT. Thereafter, the rise in RT activity in the ZAPd-hygro-infected cultures was robust, suggesting that it may have derived from the exponential growth of an initially small revertant population. When propagated on NIH 3T3 cells, the titer of ZAPd-GFP reached 1.2 × 105 to 3.8 × 105 PFU/ml, while that of wild-type MLV on NIH 3T3 cells was 2.1 × 106 to 5.0 × 106 PFU/ml, indicating that the presence of the 1.3-kb IRES-GFP insert reduced production of infectious particles approximately 10-fold.
FIG. 2.
In vitro replication kinetics of viruses. (A) NIH 3T3 cells were transfected with pZAPd-puro, pZAPd-GFP, pZAPd-hygro, or pZAP2 and passaged for 15 days. Culture medium was harvested from confluent cells every 2 days starting on day 3 and assayed for RT activity as described in Materials and Methods. RT activities are expressed in arbitrary units. Values are the means obtained from two independent experiments. (B) Spread of ZAPd-GFP through a single culture as detected by flow cytometry. NIH 3T3 cells were infected at an MOI of 0.0005 and examined at 3, 5, and 8 days postinoculation. Histograms show fluorescence intensity versus cell number for each day indicated.
We also assessed the replication kinetics of ZAPd-GFP by following the spread of GFP through cells inoculated at low MOI. GFP fluorescence was detected in only a small percentage (∼3%) of the cells 3 days postinoculation, while at 5 days, approximately one-quarter of the population exhibited fluorescence. By day 8, approximately 95% of the cells fluoresced (Fig. 2B), demonstrating that the virus transmitted the GFP marker gene with high efficiency.
Genetic stability of ZAPd-puro, ZAPd-GFP, and ZAPd-hygro upon replication through serial infection cycles.
We serially reinoculated fresh plates of NIH 3T3 cells with cell-free ZAPd-puro, ZAPd-GFP, or ZAPd-hygro virus supernatants, using 100-fold dilutions of conditioned medium from the previous cycle for each subsequent infection, to examine stability over multiple replication cycles. No antibiotic selection pressure was applied during these infections.
Hirt DNA from each serially infected cell population, digested with NheI, which cleaves once within each LTR and thus yields a full-length linearized genome, was analyzed by Southern blot using probes specific for the corresponding transgene sequence, (Fig. 3A, C, and E, respectively) or a common probe for the 5′ LTR-gag region of MLV (Fig. 3B, D, and F).
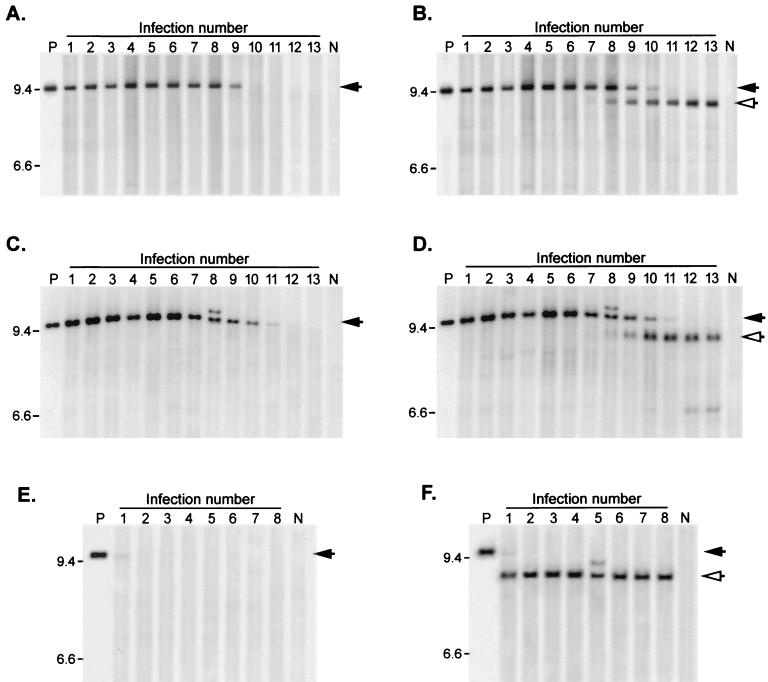
FIG. 3.
Stability of insert-containing genomes over multiple serial infections. Viruses were subjected to repeated serial passage through NIH 3T3 cultures as described in Materials and Methods. Unintegrated proviral DNA was isolated from each virus passage, digested with NheI, and subjected to Southern blotting. (A and B) DNA from ZAPd-puro serial infections, using pac-specific probe (A) or MLV LTR-gag probe (B). (C and D) DNA from ZAPd-GFP serial infections, using GFP-specific probe (C) or LTR-gag probe (D). (E and F) DNA from ZAPd-hygro serial infections, using hph-specific probe (E) or LTR-gag probe (F). Lanes P, NheI-digested plasmid DNA encoding the corresponding virus. Lanes N, DNA isolated from mock-infected cells. Intact full-length provirus signals are indicated by solid arrows, and deletion mutants are indicated by open arrows. Expected full-length fragment sizes: ZAPd-puro, 9,437 bp; ZAPd-GFP, 9559 bp; ZAPd-hygro, 9851 bp. Positions of size standards are shown on the left (in kilobases).
The ZAPd-puro virus, which contains a 1.15-kb insert, showed no sign of deletion for the first six infection cycles, with only the full-length genome containing the insert sequence being detectable during this interval (Fig. 3A and B). At the seventh infection cycle, a variant population was detected at very low levels. This variant population hybridized to the LTR-gag probe (Fig. 3B) but not the pac-specific probe (Fig. 3A), and the genome size of this deletion mutant population appeared to be roughly similar to that of wild-type MLV. Over the subsequent five infections, the levels of this deleted population grew, while that of full-length ZAPd-puro decreased, indicating that loss of the insert imparted a replicative advantage.
Similarly, the full-length ZAPd-GFP signal was detected using either a GFP-specific probe (Fig. 3C) or the common LTR-gag probe (Fig. 3D) throughout more than eight serial infection cycles. This high level of stability was reproducibly and quite consistently observed through repeated experiments, each conducted with more than 10 serial passages. However, as observed with the ZAPd-puro virus, a progressive diminution in the full-length ZAPd-GFP signal was observed after the eighth infection cycle, corresponding with the progressive emergence of a variant virus population similar in size to wild-type MLV (Fig. 3D), and which was not detected by the GFP probe (Fig. 3C).
In contrast, proviral DNA from serial infections with ZAPd-hygro exhibited deletions from the first passage (Fig. 3E and F). Among the unintegrated proviral species produced upon the first infection, only a very small fraction proved to be full-length ZAPd-hygro (Fig. 3E), and by the second infection a deletion variant similar in size to wild-type MLV had completely outgrown the vector (Fig. 3F).
GFP fluorescence serves as a reliable surrogate marker for genomic stability of the ZAPd-GFP virus over multiple infection cycles.
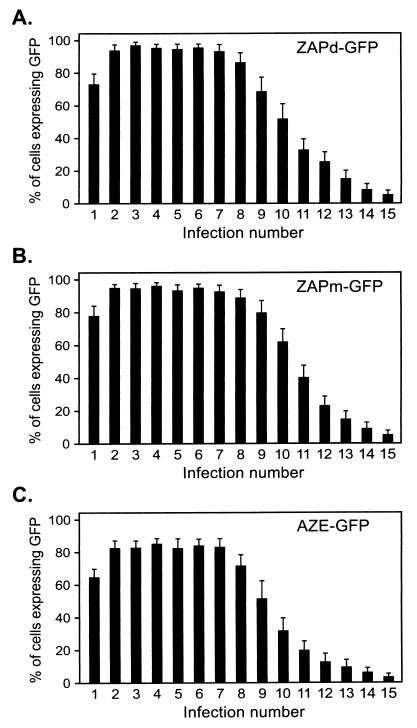
Based on the results of single-passage FACS analysis data and the extent of the deletions observed in ZAPd-GFP over serial passage, we hypothesized that the GFP reporter would greatly facilitate monitoring of virus replication and stability over multiple replication cycles. Therefore, we again performed serial infections with ZAPd-GFP, examining the cells by FACS at 2 and 4 days of each infection cycle. Only a small percentage of cells in each cycle expressed GFP by day 2 after inoculation (data not shown). However, the percentage of GFP-positive cells increased by day 4 of each cycle, indicating that GFP transduction was the result of progressive viral transmission rather than high initial levels of infection. The percentage of cells transduced by the virus at each day 4 time point, as determined by flow cytometry, is shown in Fig. 4A. Each serial infection up to the seventh or eighth cycle consistently generated transduction levels approaching 100% by day 4 postinoculation, after which the efficiency of GFP marker gene transmission was observed to decrease progressively, and almost no spread of GFP fluorescence was observed by the 15th cycle (Fig. 4A).
FIG. 4.
Transmission of GFP by GFP-encoding viruses over multiple serial infection cycles. Each virus was serially passaged through multiple NIH 3T3 cultures as in Fig. 3. Four days after exposure to virus, each culture was examined for GFP expression by flow cytometry. Shown are the percentages of cells expressing GFP at each passage. Values were obtained from three independent experiments, and error bars represent standard deviations.
This progressive decrease in GFP transmission observed by FACS correlated closely with the emergence of the deletion mutant observed by Southern blot above, suggesting that the wild-type deletion mutant competes successfully with the insert-containing genome. This competition presumably occurred via superinfection resistance, as in the later infection cycles the maximum percentage of GFP-positive cells was not further increased beyond day 4 levels by additional cultivation of the cells (data not shown). The precise correlation between the loss of GFP expression and the loss of full-length forms also suggests that the level of GFP fluorescence can serve as a reliable surrogate marker for genomic stability and persistence of the ZAPd-GFP virus over multiple infection cycles.
Effect of repeat sequence deletion and envelope sequence replacement on genetic stability of lengthened MLV through multiple replication cycles.
The IRES-transgene insert of ZAPd-GFP is flanked by an 11-bp repeat that might predispose the virus to a recombination event that would reconstitute the wild-type MLV sequence. We therefore constructed a variant of ZAPd-GFP, designated ZAPm-GFP, in which mutations were introduced into the upstream 11-bp repeat to eliminate homology with the downstream repeat sequence; these changes consisted of seven point mutations, including three silent mutations in the last three codons of env (Fig. 1). An additional variant of ZAPd-GFP in which the Moloney MLV ecotropic envelope was replaced with the 4070A amphotropic envelope was generated and designated AZE-GFP (Fig. 1). This vector, like ZAPm-GFP, lacks the upstream copy of the 11-bp repeat found in ZAPd-GFP.
ZAPm-GFP replicated with kinetics indistinguishable from those of ZAPd-GFP, as determined by transmission of the GFP marker through NIH 3T3 cultures after inoculation at low MOI (data not shown). Surprisingly, elimination of the upstream 11-bp repeat homology did not appear to prolong the stability of the virus over multiple infection cycles. Upon serial infection using the same protocol as above, ZAPm-GFP still showed progressively decreasing levels of day 4 postinoculation GFP fluorescence starting from infection cycle 8 (Fig. 4B), indicating that the same progressive loss of the full-length genome occurred from this cycle onward.
The AZE-GFP virus also showed efficient replication and GFP transmission, demonstrating that the 1.3-kb IRES-GFP insertion at the env-UTR boundary is relatively stable even in the context of different upstream envelope sequences. However, the replication kinetics of AZE-GFP were found to be somewhat attenuated compared to ZAPd-GFP and ZAPm-GFP, and during serial infection experiments the percentage of GFP-positive cells after infection by the amphotropic virus only reached 80% by each day 4 time point up to cycle 7 (Fig. 4C), whereas its ecotropic counterparts had consistently reached around 95% by the same time point in previous experiments (Fig. 4A and B). In each of the first seven serial infection cycles, the percentage of GFP-positive cells did subsequently reach 100% within the next 2 days, confirming that the lower percentage of fluorescent cells was due to delayed replication of full-length AZE-GFP and not to early emergence of deletion mutants (data not shown); however, the same time point (day 4) was used throughout for the serial infection experiments in order to preserve consistency in the assay. From cycle 8 onward, a progressive decrease in the percentage of fluorescent cells at each day 4 time point was observed, similar to that observed with the ecotropic viruses; this progressive decline in GFP fluorescence in later cycles could not be rescued by prolonged culture of each serially infected cell population, again indicating the overgrowth of deletion mutants.
Analysis of deletion mutants.
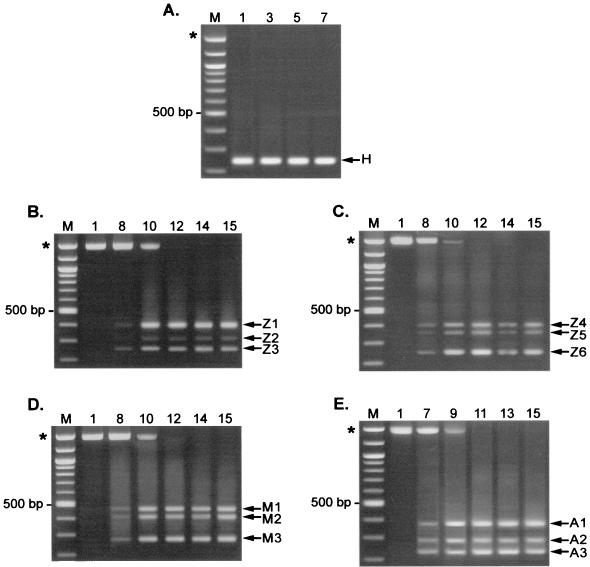
We further characterized the deletion mutant populations that arose during the first infection cycle of ZAPd-hygro and after the seventh or eighth infection cycles of ZAPd-GFP, ZAPm-GFP, and AZE-GFP by PCR amplification and sequencing of Hirt DNA from each cycle. In the case of ZAPd-hygro, the PCR results indicate that the variant population observed on Southern blot consisted of a single major species of deletion mutant (Fig. 5A). In contrast, DNA from passaged ZAPd-GFP, ZAPm-GFP, and AZE-GFP revealed three major deletion species. However, none of these could be detected prior to the seventh or eighth infection cycles (Fig. 5B to E and data not shown), and all deleted forms appeared to emerge roughly simultaneously.
FIG. 5.
PCR analysis of transgene insert region of serially passaged viruses. Hirt DNA from each indicated infection cycle was used as the template in PCR using an upstream primer specific for the appropriate env gene and a common downstream primer specific for the 3′ UTR-3′ LTR border region. (A) Amplification of passaged ZAPd-hygro. Overexposure of this gel (not shown) revealed the presence of a faint band of approximately the size expected for full-length ZAPd-hygro only in cycle 1. (B and C) Amplification of proviral DNA from two independent infection series using ZAPd-GFP. (D) Passaged ZAPm-GFP. (E) Passaged AZE-GFP. Lanes M, 100-bp molecular size markers. Asterisks indicate size of product expected for undeleted, full-length virus.
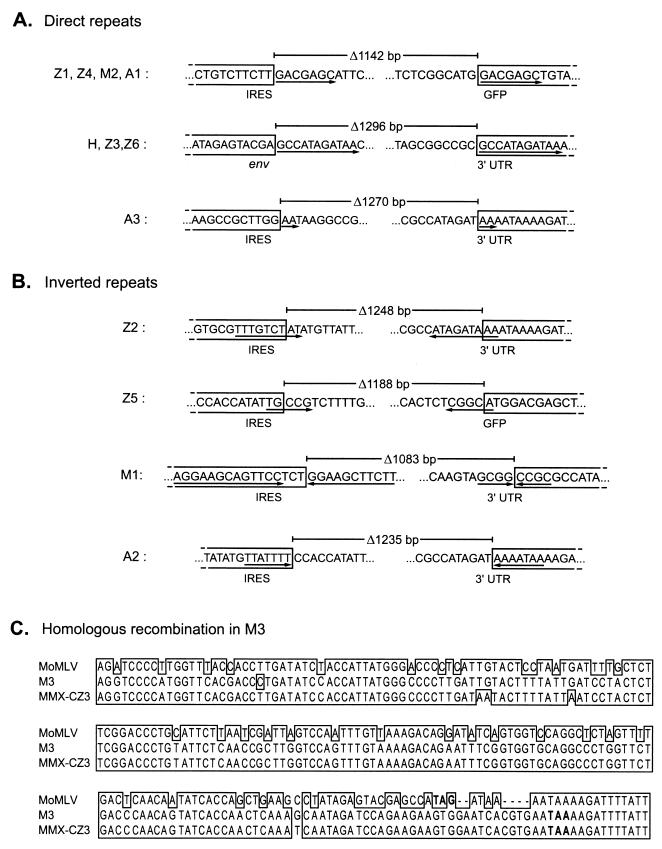
Sequencing of the amplified PCR products indicated that deletions occurred through recombination events between both direct and inverted repeats (Fig. 6A and B). Particular deletion patterns occurred repeatedly in independent experiments using different virus constructs (Fig. 6A, H/Z3/Z6 and Z1/Z4/M2/A1) and hence might represent preferred recombinational forms, while the other deletion species were unique to a single experiment or vector (Fig. 6A, A3, and Fig. 6B and C). One of the recurring deletion species, occurring in both ZAPd-GFP and ZAPd-hygro, exactly matched that of the wild-type env-UTR junction sequence (Fig. 6A, H/Z3/Z6). It is likely that this revertant derived from recombination between the 11-bp direct repeat sequences flanking the IRES-GFP insert (Fig. 1 and 6A), resulting in deletion of the entire 1,296-bp insert and reconstitution of the wild-type sequence. This species emerged rapidly and was the only one present after a single passage of ZAPd-hygro but, in the case of ZAPd-GFP, emerged only after multiple replication cycles and was not the only species present. As the upstream copy of the 11-bp direct repeat had been eliminated in ZAPm-GFP and AZE-GFP, we did not expect to find precise reconstitution of the wild-type sequence, and indeed this did not occur.
FIG. 6.
Sequence analysis of deletion junctions of variants identified by PCR. Each of the PCR products shown in Fig. 5 was purified and sequenced. The sequences remaining in each deletion variant are boxed, and the intervening sequences deleted from the parental virus are unboxed. Lengths of deletions are indicated above each variant. (A) Deletion junctions of variants whose deletions were flanked by direct repeats in the parental genome. Direct repeats are underlined with arrows. (B) Deletion junctions characterized by inverted repeats in parental virus. Inverted repeats are underlined with arrows, and the orientation of each repeat is indicated by arrow direction. (C) Alignment of ZAPm-GFP variant M1 with Moloney MLV and endogenous virus MMX-CZ3 by the Clustal X program (23). Envelope stop codons are indicated in bold.
Another deletion species emerged repeatedly in all three GFP-encoding viruses (Fig. 6A, Z1/Z4/M2/A1). In this variant, recombination occurred between 7-bp direct repeats, one of which was located in the IRES and the other within the GFP sequence, leading to the loss of 1,142 bp of the insert.
The remaining deletion species differed in sequence and were not consistently observed to arise from experiment to experiment. Sequence analysis of one such variant species, A3, implicated recombination occurring between two dinucleotide repeats (Fig. 6A). Four other deletion species were associated with inverted repeats in the parental virus sequences (Fig. 6B). In three of these species, one copy of an inverted repeat was present on each side of the deletion junction: variant Z2 had a 1,248-bp deletion associated with 9-bp inverted repeat sequences, Z5 had a 1,188-bp deletion associated with 5-bp inverted repeat sequences, and A2 had a 1,235-bp deletion associated with 7-bp inverted repeat sequences, situated at the recombination breakpoints (Fig. 6B). The fourth inverted repeat deletion (M1) was associated with two complete palindromes with the potential to form hairpins aligned at each recombination breakpoint (Fig. 6B). The remaining variant species, M3, generated a PCR product approximating the wild-type MLV env-UTR sequence in size (Fig. 5D). However, sequence alignment analysis showed significant disparities between the M3 sequence and the parental Moloney MLV sequence, and a BLAST search of GenBank revealed extremely high homology with an endogenous mouse retrovirus sequence (56) (GenBank accession no. AF017530) (Fig. 6C), suggesting that it was derived by recombination of ZAPm-GFP with endogenous retrovirus present in the NIH 3T3 cells.
DISCUSSION
We have constructed a series of replication-competent MLVs containing insertions at the env-3′ UTR boundary to examine virus stability over multiple serial infection cycles. Our results suggest that an insertion at this position can be well tolerated as long as the insert size does not exceed a certain threshold or contain recombinogenic sequences. This construct design thus allowed us to examine the population dynamics of lengthened MLV genomes over repeated passage. Use of the IRES-GFP insert allowed us to track virus spread by flow cytometry, with loss of fluorescence serving as a reliable surrogate marker for genetic instability and the concomitant emergence of deletion mutants. In collaborative studies, we have also successfully employed a similar IRES-GFP insertion in feline leukemia virus (FeLV) to follow FeLV-A–to–FeLV-B conversion, suggesting that this strategy may prove to be generally useful in studies of retrovirus replication (Z. Chang, J. Pan, C. Logg, N. Kasahara, and P. Roy-Burman, submitted for publication).
On comparing replication-competent MLV vectors containing insertions between 1.15 and 1.55 kb long, we found striking differences in stability which correlated with differences in replication kinetics observed in single-cycle infections. ZAPd-hygro, which contains an insert of 1.55 kb, displayed the most attenuated replication kinetics. When this virus was propagated, it was almost completely overgrown after a single passage by a fully deleted revertant possessing wild-type MLV sequence. In contrast, ZAPd-puro and ZAPd-GFP, with 1.15- and 1.3-kb inserts, respectively, exhibited replication kinetics much closer to those of wild-type virus and retained their inserts for much longer periods. Presumably, the replicative fitness of ZAPd-puro and ZAPd-GFP relative to ZAPd-hygro account, at least in large part, for their greater stability, as they were likely better able to compete with deletion variants that arose during replication.
Sequence analysis of the deleted variants indicated that some of the deletions may have occurred through inter- or intramolecular template switching (25, 35, 54) between short direct repeat sequences within and flanking the transgene cassette. Recombination between an 11-bp direct repeat sequence that flanks the insertions of ZAPd-hygro and ZAPd-GFP resulted in exact reconstitution of wild-type MLV sequence. Unexpectedly, when variants of ZAPd-GFP lacking the upstream copy of the 11-bp repeat were serially passaged, no improvement in stability was observed.
One deletion species arising by recombination between 7-bp direct repeats within the insert occurred in all IRES-GFP-containing vectors. Notably, while sequence analysis revealed that the IRES-GFP cassette contains 77 pairs of perfect direct repeats of 7 bp or more, only this particular repeat pair was involved in formation of a deletion mutant. One possibility is that the sequence context of this specific 7-bp repeat might be particularly recombinogenic. Alternatively, as this repeat is spaced further apart than any of the other 77 pairs, the frequent occurrence of this deletion species may simply reflect a higher rate of recombination between homologous sequences that are spaced further apart (19), although this idea has recently been challenged (1). A third possibility is that, while other deletion species might have arisen through recombination at the other repeats, this particular species exhibits the largest deletion and its genome size is closest to that of wild-type MLV, allowing it to compete more effectively during virus passage. This particular deletion was also associated with recognition sequences for murine topoisomerases Ib and II in the parental virus, in which the cleavage sites were precisely aligned with the deletion endpoints. The significance of this association, however, is unknown.
We also observed frequent deletion between inverted repeats. This type of deletion has been proposed to occur when RT jumps over hairpins formed by such repeats in viral RNA during reverse transcription (33). Interestingly, of the many deletion junctions that have been characterized in previous studies of defective retrovirus vectors, the majority occurred between sites with directly repeated sequences, but remarkably few occurred between sites having inverted repeat homology (14, 25, 34, 35, 38, 52, 57, 59). In contrast, about half of the deletion species that we observed occurred by recombination at inverted repeats.
We observed one instance of deletion via homologous recombination with an endogenous polytropic retroviral sequence, which resulted in loss of the entire IRES-GFP insert. Similar “patch repair” of an exogenous virus by an endogenous sequence has been observed previously (8, 43), although in the earlier cases the repair was of a lethal deletion rather than a nonlethal insertion.
It is notable that these multiple species of deletion mutants detected in the ZAPd-GFP, ZAPm-GFP, and AZE-GFP infections were in each case undetectable over the first six to seven infection cycles and subsequently appeared to emerge simultaneously. Neither Southern blot nor PCR analysis revealed any major deletion intermediates during the serial infections, suggesting that these short deletion mutants were the only major species to arise and that these were generated without a stepwise series of progressive deletions. While it remains possible that longer deletion intermediates were present but not amplified efficiently in this assay, this is unlikely, as the full-length IRES-GFP sequence could be amplified efficiently and showed a progressive loss of amplification signal beginning at cycle 7 or 8, consistent with the Southern blot results.
The lack of intermediate forms could be a consequence of recombination occurring at an early stage. If reversion was an early event, it occurred at low frequency, and it is likely that very few such deletion mutants were present from the first infection cycle. In the case of ZAPd-hygro, it is clear that revertants were present from the initial infection. However, for the other vectors, the advantage in replicative fitness of the various deletion mutants may not have been strong enough for these to have become apparent until many cycles had passed. An alternative, though not mutually exclusive, explanation is that at least some of the revertant species arose simultaneously in later cycles due to the requirement for some prior initiating event which resulted in increased rates of subsequent recombination. Previous studies have suggested that retrovirus recombination occurs within a distinct subpopulation (17) and that one predisposing factor might be the emergence of variant forms of RT that exhibit an increased frequency of template switching (37, 50).
As no external selection pressure was applied on any of the insert-containing virus populations, all deletion mutants presumably gained predominance through natural selection processes favoring those species that replicated most efficiently. Once detected, the major deletion mutant species all appeared to persist and gain dominance together. Presumably these multiple species, which are all similar in size, were collectively represented by the deletion mutant signal on Southern blot, which also showed no apparent intermediate forms. While the PCR analysis employed is not absolutely quantitative, the relative amounts of the amplification products from these deletion species would be predicted to change over multiple infection cycles if any particular species gained dominance over the others; however, this was not observed, suggesting that all three major deletion species could replicate with similar efficiency, and thus remained at an apparent equilibrium.
The fact that each of the deletion variants observed, using various pathways of recombination, had lost at least 84% of the IRES-GFP sequence indicates a strong selective advantage for either the natural genome length or the loss of insert sequences not contributing to efficient replication. While it has become clear that there is no absolute limitation on genome length within a certain range (15, 44), cumulative inefficiencies at multiple stages in the retroviral life cycle may result in a significant overall replicative disadvantage for viruses harboring exogenous sequences, and thereby subject such genomes to stringent limitations by the process of natural selection.
ACKNOWLEDGMENTS
We thank Pradip Roy-Burman and W. French Anderson for critical review of the manuscript.
This work was supported by a pilot project grant through the USC/Norris Breast Cancer Research Program, Department of Defense Breast Cancer Research Program grant BC980554, and NIH grant R01 CA85908. C.R.L. is the recipient of a Breast Cancer Research Project Fellowship through the Norris Comprehensive Cancer Center, and C.-K.T. is the recipient of a predoctoral fellowship from the Susan G. Komen Breast Cancer Foundation.
REFERENCES
- 1.Anderson J A, Bowman E H, Hu W S. Retroviral recombination rates do not increase linearly with marker distance and are limited by the size of the recombining subpopulation. J Virol. 1998;72:1195–1202. doi: 10.1128/jvi.72.2.1195-1202.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barnier J V, Marx M, Dezelee P, Laugier D, Poirier F, Calothy G, Hillova J, Hill M. Transformation-defective mutants with 5′ deletions of the src gene are frequently generated during replication of Rous sarcoma virus in established quail fibroblasts. Virology. 1990;177:505–514. doi: 10.1016/0042-6822(90)90515-s. [DOI] [PubMed] [Google Scholar]
- 3.Barsov E V, Hughes S H. Gene transfer into mammalian cells by a Rous sarcoma virus-based retroviral vector with the host range of the amphotropic murine leukemia virus. J Virol. 1996;70:3922–3929. doi: 10.1128/jvi.70.6.3922-3929.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Battula N, Loeb L A. The infidelity of avian myeloblastosis virus deoxyribonucleic acid polymerase in polynucleotide replication. J Biol Chem. 1974;249:4086–4093. [PubMed] [Google Scholar]
- 5.Clavel F, Hoggan M D, Willey R L, Strebel K, Martin M A, Repaske R. Genetic recombination of human immunodeficiency virus. J Virol. 1989;63:1455–1459. doi: 10.1128/jvi.63.3.1455-1459.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coffin J M. Genetic diversity and evolution of retroviruses. Curr Top Microbiol Immunol. 1992;176:143–164. doi: 10.1007/978-3-642-77011-1_10. [DOI] [PubMed] [Google Scholar]
- 7.Coffin J M. Genetic variation in AIDS viruses. Cell. 1986;46:1–4. doi: 10.1016/0092-8674(86)90851-2. [DOI] [PubMed] [Google Scholar]
- 8.Colicelli J, Goff S P. Isolation of a recombinant murine leukemia virus utilizing a new primer tRNA. J Virol. 1986;57:37–45. doi: 10.1128/jvi.57.1.37-45.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cormack B P, Valdivia R H, Falkow S. FACS-optimized mutants of the green fluorescent protein (GFP) Gene. 1996;173:33–38. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- 10.Dillon P J, Lenz J, Rosen C A. Construction of a replication-competent murine retrovirus vector expressing the human immunodeficiency virus type 1 Tat transactivator protein. J Virol. 1991;65:4490–4493. doi: 10.1128/jvi.65.8.4490-4493.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DuBridge R B, Tang P, Hsia H C, Leong P M, Miller J H, Calos M P. Analysis of mutation in human cells by using an Epstein-Barr virus shuttle system. Mol Cell Biol. 1987;7:379–387. doi: 10.1128/mcb.7.1.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gelinas C, Temin H M. Nondefective spleen necrosis virus-derived vectors define the upper size limit for packaging reticuloendotheliosis viruses. Proc Natl Acad Sci USA. 1986;83:9211–9215. doi: 10.1073/pnas.83.23.9211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gojobori T, Yokoyama S. Rates of evolution of the retroviral oncogene of Moloney murine sarcoma virus and of its cellular homologues. Proc Natl Acad Sci USA. 1985;82:4198–4201. doi: 10.1073/pnas.82.12.4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hajjar A M, Linial M L. A model system for nonhomologous recombination between retroviral and cellular RNA. J Virol. 1993;67:3845–3853. doi: 10.1128/jvi.67.7.3845-3853.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Herman S A, Coffin J M. Efficient packaging of readthrough RNA in ALV: implications for oncogene transduction. Science. 1987;236:845–848. doi: 10.1126/science.3033828. [DOI] [PubMed] [Google Scholar]
- 16.Horton R M, Hunt H D, Ho S N, Pullen J K, Pease L R. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene. 1989;77:61–68. doi: 10.1016/0378-1119(89)90359-4. [DOI] [PubMed] [Google Scholar]
- 17.Hu W S, Bowman E H, Delviks K A, Pathak V K. Homologous recombination occurs in a distinct retroviral subpopulation and exhibits high negative interference. J Virol. 1997;71:6028–6036. doi: 10.1128/jvi.71.8.6028-6036.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu W S, Temin H M. Genetic consequences of packaging two RNA genomes in one retroviral particle: pseudodiploidy and high rate of genetic recombination. Proc Natl Acad Sci USA. 1990;87:1556–1560. doi: 10.1073/pnas.87.4.1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu W S, Temin H M. Retroviral recombination and reverse transcription. Science. 1990;250:1227–1233. doi: 10.1126/science.1700865. [DOI] [PubMed] [Google Scholar]
- 20.Jainchill J L, Aaronson S A, Todaro G J. Murine sarcoma and leukemia viruses: assay using clonal lines of contact-inhibited mouse cells. J Virol. 1969;4:549–553. doi: 10.1128/jvi.4.5.549-553.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jamieson B D, Zack J A. In vivo pathogenesis of a human immunodeficiency virus type I reporter virus. J Virol. 1998;72:6520–6526. doi: 10.1128/jvi.72.8.6520-6526.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jang S K, Krausslich H G, Nicklin M J, Duke G M, Palmenberg A C, Wimmer E. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J Virol. 1988;62:2636–2643. doi: 10.1128/jvi.62.8.2636-2643.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jeanmougin F, Thompson J D, Gouy M, Higgins D G, Gibson T J. Multiple sequence alignment with Clustal X. Trends Biochem Sci. 1998;23:403–405. doi: 10.1016/s0968-0004(98)01285-7. [DOI] [PubMed] [Google Scholar]
- 24.Jespersen T, Duch M, Carrasco M L, Warming S, Pedersen F S. Expression of heterologous genes from an IRES translational cassette in replication competent murine leukemia virus vectors. Gene. 1999;239:227–235. doi: 10.1016/s0378-1119(99)00402-3. [DOI] [PubMed] [Google Scholar]
- 25.Jones J S, Allan R W, Temin H M. One retroviral RNA is sufficient for synthesis of viral DNA. J Virol. 1994;68:207–216. doi: 10.1128/jvi.68.1.207-216.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Joseph D R. Efficient production of xenotropic murine leukemia virus unintegrated proviral DNA by cocultivation. J Virol. 1981;38:1095–1098. doi: 10.1128/jvi.38.3.1095-1098.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee A H, Han J M, Sung Y C. Generation of the replication-competent human immunodeficiency virus type 1 which expresses a jellyfish green fluorescent protein. Biochem Biophys Res Commun. 1997;233:288–292. doi: 10.1006/bbrc.1997.6304. [DOI] [PubMed] [Google Scholar]
- 28.Lobel L I, Patel M, King W, Nguyen-Huu M C, Goff S P. Construction and recovery of viable retroviral genomes carrying a bacterial suppressor transfer RNA gene. Science. 1985;228:329–332. doi: 10.1126/science.2984770. [DOI] [PubMed] [Google Scholar]
- 28a.Logg C R, Tai C K, Logg A, Anderson W F, Kasahara N. A uniquely stable replication-competent retrovirus vector achieves efficient gene delivery in vitro and in solid tumors. Hum Gene Ther. 2001;12:921–932. doi: 10.1089/104303401750195881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murakami M, Watanabe H, Niikura Y, Kameda T, Saitoh K, Yamamoto M, Yokouchi Y, Kuroiwa A, Mizumoto K, Iba H. High-level expression of exogenous genes by replication-competent retrovirus vectors with an internal ribosomal entry site. Gene. 1997;202:23–29. doi: 10.1016/s0378-1119(97)00468-x. [DOI] [PubMed] [Google Scholar]
- 30.Neiman P E, Stone J, Gilbert L, Fincham V J, McMillin-Helsel C, Wyke J A. Restriction of replication of Rous sarcoma virus mediated by the src region of the genome: analysis of the formation and integration of viral DNA and the effects of conditional and nonconditional mutations in src. Virology. 1981;114:516–525. doi: 10.1016/0042-6822(81)90231-2. [DOI] [PubMed] [Google Scholar]
- 31.Omer C A, Pogue-Geile K, Guntaka R, Staskus K A, Faras A J. Involvement of directly repeated sequences in the generation of deletions of the avian sarcoma virus src gene. J Virol. 1983;47:380–382. doi: 10.1128/jvi.47.2.380-382.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Page K A, Liegler T, Feinberg M B. Use of a green fluorescent protein as a marker for human immunodeficiency virus type 1 infection. AIDS Res Hum Retroviruses. 1997;13:1077–1081. doi: 10.1089/aid.1997.13.1077. [DOI] [PubMed] [Google Scholar]
- 33.Parvin J D, Wang L H. Mechanisms for the generation of src-deletion mutants and recovered sarcoma viruses: identification of viral sequences involved in src deletions and in recombination with c-src sequences. Virology. 1984;138:236–245. doi: 10.1016/0042-6822(84)90348-9. [DOI] [PubMed] [Google Scholar]
- 34.Pathak V K, Temin H M. 5-Azacytidine and RNA secondary structure increase the retrovirus mutation rate. J Virol. 1992;66:3093–3100. doi: 10.1128/jvi.66.5.3093-3100.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pathak V K, Temin H M. Broad spectrum of in vivo forward mutations, hypermutations, and mutational hotspots in a retroviral shuttle vector after a single replication cycle: deletions and deletions with insertions. Proc Natl Acad Sci USA. 1990;87:6024–6028. doi: 10.1073/pnas.87.16.6024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Petropoulos C J, Hughes S H. Replication-competent retrovirus vectors for the transfer and expression of gene cassettes in avian cells. J Virol. 1991;65:3728–3737. doi: 10.1128/jvi.65.7.3728-3737.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pfeiffer J K, Georgiadis M M, Telesnitsky A. Structure-based Moloney murine leukemia virus reverse transcriptase mutants with altered intracellular direct-repeat deletion frequencies. J Virol. 2000;74:9629–9636. doi: 10.1128/jvi.74.20.9629-9636.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pulsinelli G A, Temin H M. Characterization of large deletions occurring during a single round of retrovirus vector replication: novel deletion mechanism involving errors in strand transfer. J Virol. 1991;65:4786–4797. doi: 10.1128/jvi.65.9.4786-4797.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reik W, Weiher H, Jaenisch R. Replication-competent Moloney murine leukemia virus carrying a bacterial suppressor tRNA gene: selective cloning of proviral and flanking host sequences. Proc Natl Acad Sci USA. 1985;82:1141–1145. doi: 10.1073/pnas.82.4.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roberts J D, Bebenek K, Kunkel T A. The accuracy of reverse transcriptase from HIV-1. Science. 1988;242:1171–1173. doi: 10.1126/science.2460925. [DOI] [PubMed] [Google Scholar]
- 41.Rowe W P, Pugh W E, Hartley J W. Plaque assay techniques for murine leukemia viruses. Virology. 1970;42:1136–1139. doi: 10.1016/0042-6822(70)90362-4. [DOI] [PubMed] [Google Scholar]
- 42.Schmidt M, Rethwilm A. Replicating foamy virus-based vectors directing high level expression of foreign genes. Virology. 1995;210:167–178. doi: 10.1006/viro.1995.1328. [DOI] [PubMed] [Google Scholar]
- 43.Schwartzberg P, Colicelli J, Goff S P. Recombination between a defective retrovirus and homologous sequences in host DNA: reversion by patch repair. J Virol. 1985;53:719–726. doi: 10.1128/jvi.53.3.719-726.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shin N H, Hartigan-O'Connor D, Pfeiffer J K, Telesnitsky A. Replication of lengthened Moloney murine leukemia virus genomes is impaired at multiple stages. J Virol. 2000;74:2694–2702. doi: 10.1128/jvi.74.6.2694-2702.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shoemaker C, Hoffman J, Goff S P, Baltimore D. Intramolecular integration within Moloney murine leukemia virus DNA. J Virol. 1981;40:164–172. doi: 10.1128/jvi.40.1.164-172.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smith E, Redman R A, Logg C R, Coetzee G A, Kasahara N, Frenkel B. Glucocorticoids inhibit developmental stage-specific osteoblast cell cycle: dissociation of cyclin A-cyclin-dependent kinase 2 from E2F4–p130 complexes. J Biol Chem. 2000;275:19992–20001. doi: 10.1074/jbc.M001758200. [DOI] [PubMed] [Google Scholar]
- 47.Soneoka Y, Cannon P M, Ramsdale E E, Griffiths J C, Romano G, Kingsman S M, Kingsman A J. A transient three-plasmid expression system for the production of high titer retroviral vectors. Nucleic Acids Res. 1995;23:628–633. doi: 10.1093/nar/23.4.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stuhlmann H, Berg P. Homologous recombination of copackaged retrovirus RNAs during reverse transcription. J Virol. 1992;66:2378–2388. doi: 10.1128/jvi.66.4.2378-2388.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stuhlmann H, Jaenisch R, Mulligan R C. Construction and properties of replication-competent murine retroviral vectors encoding methotrexate resistance. Mol Cell Biol. 1989;9:100–108. doi: 10.1128/mcb.9.1.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Svarovskaia E S, Delviks K A, Hwang C K, Pathak V K. Structural determinants of murine leukemia virus reverse transcriptase that affect the frequency of template switching. J Virol. 2000;74:7171–7178. doi: 10.1128/jvi.74.15.7171-7178.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Svoboda J. Presence of chicken tumour virus in the sarcoma of the adult rat inoculated after birth with Rous sarcoma tissue. Nature. 1960;186:980–981. doi: 10.1038/186980b0. [DOI] [PubMed] [Google Scholar]
- 52.Swain A, Coffin J M. Mechanism of transduction by retroviruses. Science. 1992;255:841–845. doi: 10.1126/science.1371365. [DOI] [PubMed] [Google Scholar]
- 53.Telesnitsky A, Blain S, Goff S P. Assays for retroviral reverse transcriptase. Methods Enzymol. 1995;262:347–362. doi: 10.1016/0076-6879(95)62029-x. [DOI] [PubMed] [Google Scholar]
- 54.Temin H M. Retrovirus variation and reverse transcription: abnormal strand transfers result in retrovirus genetic variation. Proc Natl Acad Sci USA. 1993;90:6900–6903. doi: 10.1073/pnas.90.15.6900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Terwilliger E F, Godin B, Sodroski J G, Haseltine W A. Construction and use of a replication-competent human immunodeficiency virus (HIV-1) that expresses the chloramphenicol acetyltransferase enzyme. Proc Natl Acad Sci USA. 1989;86:3857–3861. doi: 10.1073/pnas.86.10.3857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tomonaga K, Coffin J M. Structure and distribution of endogenous nonecotropic murine leukemia viruses in wild mice. J Virol. 1998;72:8289–8300. doi: 10.1128/jvi.72.10.8289-8300.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Varela-Echavarria A, Prorock C M, Ron Y, Dougherty J P. High rate of genetic rearrangement during replication of a Moloney murine leukemia virus-based vector. J Virol. 1993;67:6357–6364. doi: 10.1128/jvi.67.11.6357-6364.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Voynow S L, Coffin J M. Evolutionary variants of Rous sarcoma virus: large deletion mutants do not result from homologous recombination. J Virol. 1985;55:67–78. doi: 10.1128/jvi.55.1.67-78.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang J, Temin H M. Rate and mechanism of nonhomologous recombination during a single cycle of retroviral replication. Science. 1993;259:234–238. doi: 10.1126/science.8421784. [DOI] [PubMed] [Google Scholar]