Abstract
The adhesion of bacteria to surfaces is associated with physicochemical and biological interactions. The present investigations provide new results about the differential adhesion levels of skin bacteria using a representative 3D skin model which mainly relies on the different physicochemical properties of the respective surfaces. Modulation of the adhesion of bacteria and thus their colonization, may occur by adjusting the physicochemical properties of the epidermal and bacterial surfaces. Lewis acid and hydrophobicity were the most strongly correlated parameters with the antiadhesion properties of the tested compounds. Modulation of physicochemical properties appears to be the primary driver of reduced Staphylococcus aureus adhesion in this study, with no significant changes observed in the expression of genes associated with classical adhesion pathways.
Subject terms: Biofilms, Applied microbiology, Microbiome, Antimicrobials
Introduction
The skin microbiota is a delicate balance of bacteria, fungi and viruses that colonize the human skin and its appendages. Depending on the skin site physiology, colonization by microorganisms differs in terms of diversity and relative abundance. Dry sites present the highest bacterial diversity with the four main skin bacterial phyla: Actinobacteria, Firmicutes, Proteobacteria and Bacteroidetes. Sebaceous sites, which show the lowest bacterial diversity yet the highest abundance, are primarily colonized by lipophilic Actinobacteria such as Cutibacterium and Corynebacterium species. Moist skin sites present with an intermediate diversity and abundance, largely dominated by Staphylococcus and Corynebacterium species1,2. Overall, Staphylococcus spp. and Cutibacterium acnes are the most abundant and ubiquitous genera within the human skin microbiome3.
The skin is an organ with a complex surface, especially its most superficial layer, the stratum corneum, a structure undergoing constant renewal. Microbial adhesion to the skin is strongly influenced by the host skin immune system, the skin substrate and the physicochemical conditions of the skin site (i.e. pH, temperature, humidity, topography, exposomes,…)4–6. Adhesion depends on the different physicochemical properties of the surface and adhering microorganisms. In this context, microbial colonization of the skin surface is an extremely complex and very difficult to predict mechanism. To successfully colonize the skin, microorganisms have developed different mechanisms that are mainly involved in the first stage of colonization: adhesion to the host cells is performed in 2 steps. First, a contact and a reversible adhesion, modulated by attractive and repulsive forces generated between the two surfaces, combined or not with mechanical trapping (due to the roughness of the skin). Many studies highlight the fundamental role of hydrophobic/hydrophilic potential, the Lewis acid-base balance and van der Waals electrostatic interactions in the initiation of the adhesion process of microorganisms to any surface (biotic or abiotic). The second step is an irreversible adhesion, which promotes the adaptation and persistence of the microorganism within the skin. This step generally requires the presence of specific molecules or structures on the surface of the microorganisms (i.e. pili, flagella, surface proteins, carbohydrates)7–12.
It is well established that the adhesion of exogenous microorganisms to the skin surface and their colonization may cause skin infections or dysbiosis associated to skin disorders, such as atopic dermatitis2,13–15 or acne2,15–17. It has also been confirmed that modifications in the physicochemical properties of microorganisms or surfaces negatively or positively affect the adhesive properties of a microorganism to the skin11,12.
In dermatology, the use and the formulation of ingredients that may help to modulate the physicochemical interactions between the skin and the exogenous micro-organisms constitute a new approach to prevent skin infections and dysbiosis. Several studies have characterized the adhesion modes of microorganisms to skin cells using human corneocytes or keratinocytes in suspension18–20. This system mimics in situ phenomena but remains insufficiently representative of the in vivo situation. Three-dimensional (3D) reconstructed skin models have emerged as a valuable tool in dermatological research. These models contain a fully differentiated epidermal barrier, mimicking the morphological and molecular characteristics of normal human epidermis21,22. 3D skin models are standardized, easily accessible and, unlike skin explants, available in a sterile environment. Their accuracy and relevance have been confirmed in toxicology and efficacy testing of cosmetic products23. These models can be colonized by microorganisms in order to study human skin-microbiota interactions24,25. Thus, the 3D reconstructed skin models, in addition to being able to bypass animal experimentation, are an ideal model to understand the mechanisms driving the first step of colonization and how to modulate the adhesion of microorganisms to the skin surface using specific compounds.
Therefore, in the present study, a 3D reconstructed human skin model was used to investigate the physicochemical properties of adhering microorganisms and the skin surface driving the adhesion profile at the mechanistic level11,12. Moreover, the physicochemical modification induced by the tested compounds was assessed to demonstrate its capacity to modulate the adhesion of skin bacteria. A transcriptomic analysis was conducted to determine the molecular and physicochemical mechanisms involved in the modulation of bacterial adhesion to the skin.
Results
Topical formulation-intended compounds modulate bacterial adhesion differently
Firstly, the native bacterial adhesion to the 3D skin model was assessed. Results revealed significant differences in the native adhesion of skin major bacterial strains (Fig. 1). The strongest adhesion was achieved by S. aureus with an average of 5.7 Log CFU/cm2, followed by S. epidermidis with 5.3 Log CFU/cm2 and C. acnes with 5.0 Log CFU/cm2. This native adhesion observed on ATCC strains is representative of the adhesion behavior of wild-type strains (Supplementary Fig. 1, Supplementary Table 1).
Fig. 1. Native adhesion of representative ATCC strains skin bacteria after 2 h of incubation on the 3D skin model.
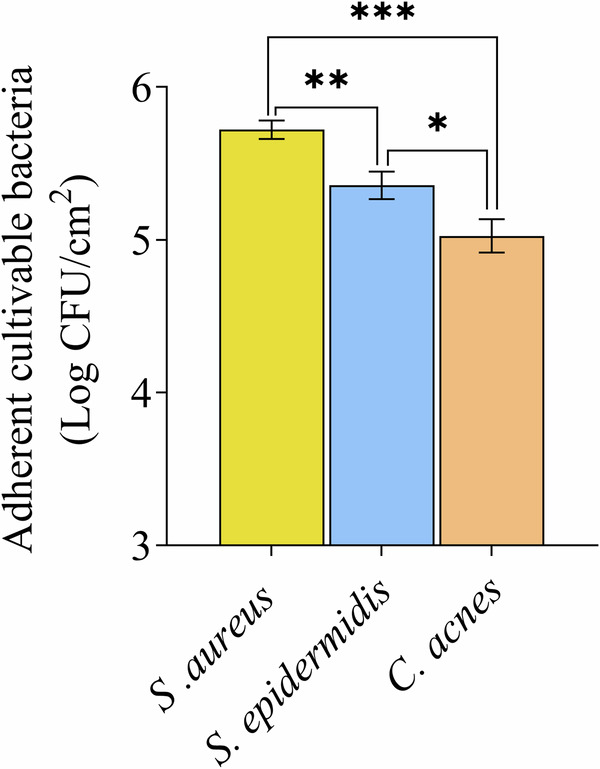
Results are expressed in Log10 of the number of bacteria adhered per cm2 of reconstructed skin of the 3D skin model. Error bars show standard error of the mean (SEM) of independent treatments (n > 3). Statistical significance was calculated by a multiple comparison of strains using the Tukey’s test following one-way ANOVA. (*) for p < 0.05, (**) for p < 0.01 and (***) for p < 0.001.
Then, to assess the modulation of bacterial adhesion compounds to be formulated topically, the absence of any anti-bacterial activity was confirmed. This was done by quantifying viable bacteria after a 2-h treatment of the inoculum with the tested compounds (Fig. 2). With the exception of Sodium Laureth Sulfate (SLS) and Rhamnolipid (RHM), the compounds used in this study did not show any antibacterial activity at the tested concentrations. SLS caused a full loss of viability on C. acnes (7.4 Log reduction) and a moderate loss of viability on S. aureus (2.0 Log reduction), while RHM induced a slight loss of viability on C. acnes (0.98 Log reduction). To ensure the integrity of the adhesion process, the absence of cytotoxic effects induced by tested compounds on the 3D skin cells was also confirmed (Supplementary Fig. 2), as cell viability is crucial for this process. All tested compounds exhibit a viability percentage ≥89%.
Fig. 2. Antibacterial activity of tested compounds after 2 h of contact time.
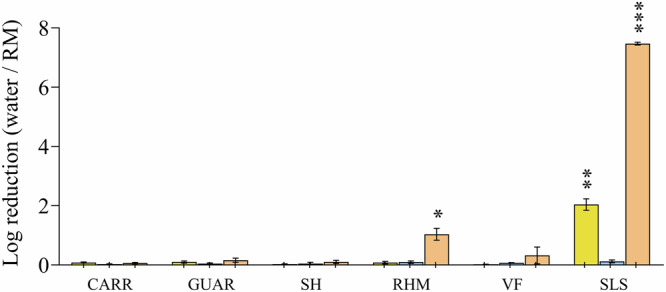
Results are expressed as the logarithm of reduction, i.e., the Log10 of the ratio of viable bacteria counted between the compounds-treated conditions and the sterile distilled water-treated condition (negative control). SLS: Sodium Laureth Sulfate ; CARR: Carrageenan ; GUAR: Guar Hydroxypropyltrimonium Chloride ; SH: Sodium Hyaluronate ; RHM: Rhamnolipids ; VF: Vitreoscilla filiformis extract. Error bars show standard error of the mean (SEM) of independent treatments (n ≥ 3). Statistical significance was calculated by a paired t-test. (*) for p < 0.05, (**) for p < 0.01 and (***) for p < 0.001.
Bacterial adhesion on the 3D skin model after topical application of compounds was only assessed on those compounds without antibacterial activity (i.e. Log reduction lower than 0.5). The comparison with the water control condition showed that certain compounds induced a significant inhibition of S. aureus, S. epidermidis and C. acnes adhesion to the 3D skin model’s epidermal surface (Fig. 3). However, a clear dynamic in the antiadhesion behavior was observed, which varied depending on the bacterial species and tested compounds. RHM strongly inhibited S. aureus adhesion, with an adhesion logarithmic reduction of 3.3 Log, at least 2 Log higher than any other tested compounds (P < 0.001). Carrageenan (CARR), Sodium Hyaluronate (SH), Guar Hydroxypropyltrimonium Chloride (GUAR) and Vitreoscilla filiformis extract (VF) induced low to medium S. aureus adhesion inhibition ranging from 1.1 Log (CARR) to 0.6 Log (GUAR) with no significant difference between those 4 compounds. All compounds induced a low to medium inhibition of S. epidermidis, with no significant differences between the tested compounds. Maximum S. epidermidis adhesion inhibition was reached with CARR (1.3 Log) and minimum with VF extract (0.7 Log). The maximum adhesion inhibition of C. acnes was obtained with CARR (0.6 Log) and the minimum with GUAR (0.4 Log) with no significant difference between tested compounds. Despite the moderate microbiological Log reduction (lower than 0.5 Log) these differences were statistically significant in comparison to negative control.
Fig. 3. Influence of tested compounds on the adhesion of skin bacteria to the 3D skin model.
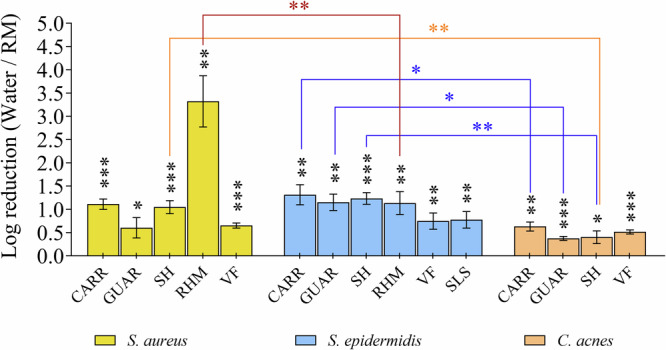
The differential effect of compounds on bacterial adhesion was assessed using a 3D skin model. Bacteria were introduced after 2 h of compound exposure on the 3D skin model and incubated for an additional 2 h. Results are expressed as the logarithm of reduction, i.e. the Log10 of the ratio of bacteria numbers adhered per cm2 of reconstructed skin of the 3D model between the RM-treated conditions and the sterile distilled water-treated condition (negative control). SLS: Sodium Laureth Sulfate ; CARR: Carrageenan ; GUAR: Guar Hydroxypropyltrimonium Chloride ; SH: Sodium Hyaluronate ; RHM: Rhamnolipids ; VF: Vitreoscilla filiformis extract. Error bars show standard error of the mean (SEM) of independent treatments (n ≥ 3). Statistical significance was calculated by a paired t-test and by a multiple comparison of strains using the Tukey’s test following one-way ANOVA. (*) for p < 0.05, (**) for p < 0.01 and (***) for p < 0.001.
The inhibition of bacterial adhesion by topical formulation-intended compounds is driven by modulation of the physicochemical properties of epidermal and bacterial surfaces
Tested compounds increase the skin’s wettability
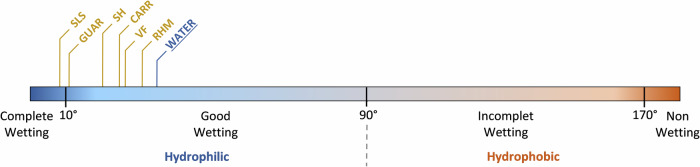
Contact angle measurements on the 3D skin model’s epidermal surface defined the surface wettability after the topical compound application (Fig. 4, Supplementary Table 2). A good wettability was observed for the water-treated control condition, and a trend towards increased wettability for all the compounds was observed. This increase in wettability was more marked for GUAR and SLS, which made the epidermal surface of 3D skin completely wettable. To exclude any bias introduced to static contact angle measurements by the skin surface structure/roughness, we measured the roughness of the 3D skin model surface in the presence of water, RHM and GUAR to have both extremes in terms of wettability shift represented. As shown on Supplementary Figure 3, no significant differences in the roughness of the 3D skin model were observed in the presence of these 3 compounds.
Fig. 4. Overview of the influence of the tested compounds on the wetting of the 3D skin model epidermal surface after 2 h of contact time.
Wettability was evaluated by measuring contact angle expressed in degrees using the sessile drop method. Values are defined from at least three independent experiments (n ≥ 3).
Since the adhesion process of bacteria also relies on the physicochemical properties of their external layer, the physicochemical properties of the bacterial surface in the native condition and after contact with tested compounds was characterized.
Tested compounds modulate the physicochemical properties at the surface of bacteria
Table 1 shows the affinity of the bacterial skin strains used in this study to MATS solvents in the condition of native adhesion. The tested bacteria showed a different solvent affinity profile, reflecting differences in their surface physicochemistry: S. aureus has a highly hydrophobic envelope, with no significant Lewis polarity. S. epidermidis has a hydrophobic, Lewis strongly acid (electron pair-accepting/γ+) and slightly base (electron pair-donating/γ−) envelope. The C. acnes envelope exhibits a slightly hydrophilic, Lewis slightly acid and slightly basic attributes.
Table 1.
Surface polarity and Lewis acid-base characteristics of non-compound-treated skin bacteria
| Bacteria | Physicochemical parametersa | Water | ||
|---|---|---|---|---|
| S. aureus | Hydrophobic (%) | 93.9 | ± | 1.3 |
| Lewis acid γ+ | 5.2 | ± | 0.9 | |
| Lewis base γ− | 8.1 | ± | 1.7 | |
| S. epidermidis | Hydrophobic (%) | 87.0 | ± | 3.0 |
| Lewis acid γ+ | 54.9 | ± | 1.5 | |
| Lewis base γ− | 18.9 | ± | 5.7 | |
| C. acnes | Hydrophobic (%) | 68.3 | ± | 0.5 |
| Lewis acid γ+ | 21.2 | ± | 3.3 | |
| Lewis base γ− | 19.6 | ± | 1.2 | |
aParameters are expressed in percent for hydrophobicity and in arbitrary units for Lewis acid/base. Values are defined from at least three independent MATS experiments ± standard error of the mean (SEM).
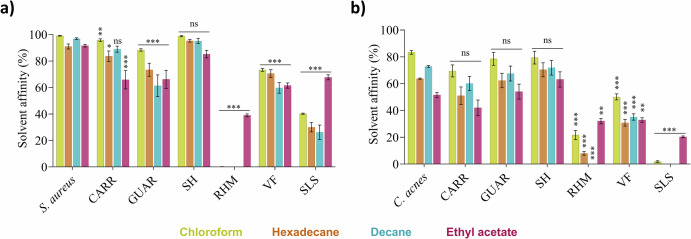
The influence of compounds on this native affinity was assessed on S. aureus (Fig. 5a) and C. acnes (Fig. 5b), given their opposite native surface physicochemistry (Table 1) and their involvement in skin disorders2. All compounds, except SH, significantly (p ≤ 0.05) influenced the solvent affinity profile of S. aureus, while for C. acnes, only RHM, VF and SLS have a significant influence on its affinity profile.
Fig. 5. Influence of tested compounds after 2 h of contact time on native affinity profile of bacterial surface with MATS solvents.
Affinity profile of (a) S. aureus and (b) C. acnes surface to MATS solvents in the compounds-treated conditions. SLS: Sodium Laureth Sulfate ; CARR: Carrageenan ; GUAR: Guar Hydroxypropyltrimonium Chloride ; SH: Sodium Hyaluronate ; RHM: Rhamnolipids ; VF: Vitreoscilla filiformis extract. Error bars show standard error of the mean (SEM) of independent treatments (n ≥ 3). Statistical significance was calculated by a multiple comparison of the conditions using the Tukey’s test following one-way ANOVA. (*) for p < 0.05, (**) for p < 0.01 and (***) for p < 0.001.
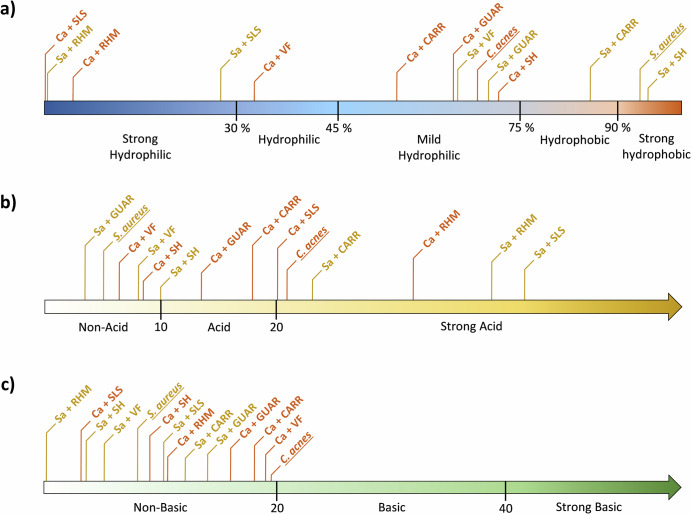
These changes in affinity reflect, for S. aureus (Fig. 6a, Supplementary Table 3), a decrease of hydrophobicity with CARR, a radical change to mild hydrophily for GUAR and VF, or to strong hydrophily for SLS and RHM. The hydrophily of C. acnes increased with VF and increased very strongly with RHM and SLS. In Fig. 6b, S. aureus significantly increased its capacity to accept electron-pair (Lewis acid) with CARR, and very strongly with RHM and SLS, while C. acnes increases it moderately with RHM or loses it with GUAR, SH and CARR (Supplementary Table 3). The gamma- component (Lewis base) was not significantly impacted for S. aureus and disappeared for C. acnes with RHM, SH and SLS (Fig. 6c, Supplementary Table 3).
Fig. 6. Overview of the influence of tested compounds after 2 h of contact time on physicochemical parameters of the bacterial surface.
Figure shows (a) surface polarity, (b) Lewis acid, and (c) Lewis base of S. aureus (yellow) and C. acnes (orange) before and after compounds treatment. SLS: Sodium Laureth Sulfate ; CARR: Carrageenan ; GUAR: Guar Hydroxypropyltrimonium Chloride ; SH: Sodium Hyaluronate ; RHM: Rhamnolipids ; VF: Vitreoscilla filiformis extract. Lewis acids/bases are expressed in arbitrary units related to the difference of adhesion percent to solvent couples, respectively, ethyl acetate/decane and chloroform/hexadecane. Hydrophobicity is expressed in mean percent of adhesion to decane and hexadecane.
To explain this behavior, a multidimensional correlation analysis between the different physicochemical properties and the anti-adhesion profile of tested compounds was performed.
Correlation between the inhibition of bacterial adhesion and the modulation of the bacterial physicochemistry
The Pearson correlation matrix (Fig. 7) revealed a very strong linear correlation between the Lewis acid interaction and the adhesion inhibition of S. aureus (0.926). Furthermore, this matrix also demonstrated a strong anti-correlation between bacterial surface hydrophobicity and adhesion inhibition of S. aureus (−0.861). Lewis acid interaction also strongly anti-correlated with the hydrophobic properties of S. aureus (−0.722). The Lewis acid trait reflects the high presence of electron-hole hydrophilic molecules in the bacterial envelope. Conversely, the Lewis base characteristic significantly correlated with other matrix parameters.
Fig. 7. Pearson correlation matrix between the physicochemical parameters of the bacterial surface and its adhesion to the 3D skin model.
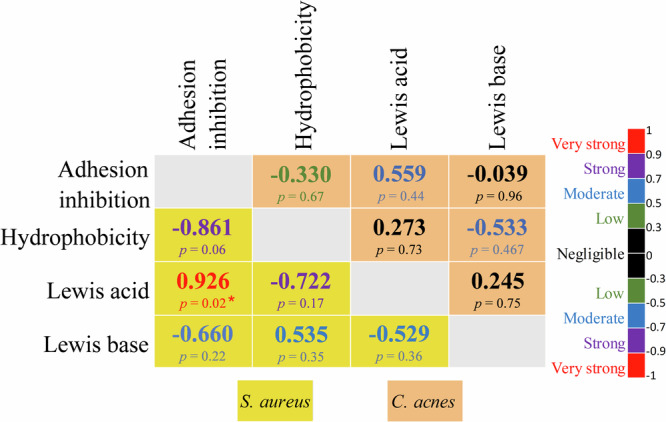
The scale of the colors is denoted as follows: Very strong (red), Strong (purple), Moderate (blue), Low (green) and Negligible (dark) positive or negative correlation. Correlations for S. aureus and C. acnes are indicated in the yellow and salmon boxes respectively. Statistical significance was calculated by a multiple comparison of the conditions using the Tukey’s test following one-way ANOVA. (*) for p < 0.05, (**) for p < 0.01 and (***) for p < 0.001.
Apart from a tendency in the correlation between the Lewis acid interaction and the adhesion inhibition (0.559), no other parameter correlated with C. acnes. This indicates that the modulation of physicochemical properties of the bacterial surface may induce a strain-specific antiadhesion effect.
Inhibition of the adhesion of S. aureus on the skin by topical formulation-intended compounds induce minor, if any, changes to the S. aureus transcriptome
As the capacity of the tested compounds to inhibit adhesion was more important with the S. aureus strain (Fig. 3) and strongly correlated with the physicochemical mechanisms (Fig. 7), an RNAseq study was carried out on S. aureus to investigate whether this strong anti-adhesion effect was also driven by specific biological mechanisms.
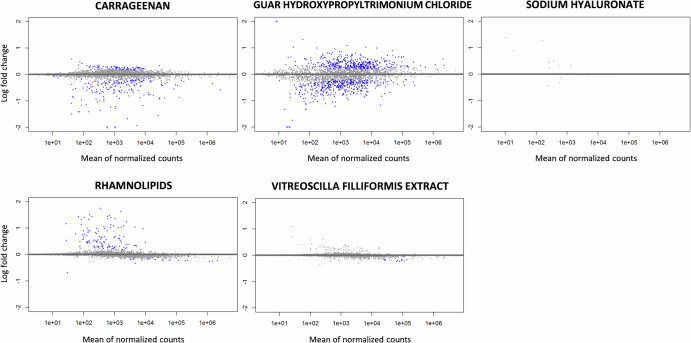
A differential gene expression analysis highlighted that the tested compounds did not significantly modify the S. aureus transcriptome after 2 h of contact in planktonic growth conditions, which was considered enough to induce a significant anti-adhesion effect (Fig. 8 and Table 2).
Fig. 8. MA plot of genes differentially expressed in S. aureus between tested compounds conditions and water control condition.
Each point represents a gene transcript, the x-axis shows the normalized count of these transcripts and the y-axis shows the logarithm of the fold change. Genes with an adjusted p-value lower than 0.1 are shown in blue.
Table 2.
Differential RNA expression analysis (≥ 2-fold ; padj < 0.1) of S. aureus between compounds-treated conditions and water-treated condition
| Locus tag | Encoded protein | log2FoldChange | ||||
|---|---|---|---|---|---|---|
| CARR | GUAR | SH | RHM | VF | ||
| SAOUHSC_00465 | Biofilm formation stimulator VEG | −2.25 | ||||
| SAOUHSC_00975 | DoxX family transmembrane protein | −2.21 | ||||
| SAOUHSC_02320 | Hypothetical unknow protein | 3.15 | ||||
| SAOUHSC_02853 | Hypothetical protein belongs to the UPF0346 family | −2.09 | ||||
| SAOUHSC_03017 | N-acethyltransferase | −2.24 | ||||
| SAOUHSC_T00012 | tRNAasp | −2.98 | ||||
SH, RHM and VF did not induce significant differential expression in any of the ≈3050 genes encoded in the S. aureus genome, CARR significantly repressed the expression of 3 genes. One of them is the biofilm stimulator VEG. GUAR significantly induced the expression of 3 genes. This very small alteration in the gene expression after treatment was confirmed after a less stringent criteria for significance of differential gene expression. After the significance threshold for differential expression was set at 1.5 Log, SH and VF did not induce any differential expression in any of the genes. The number of genes differentially expressed for CARR, GUAR and RHM was 11, 9 and 6 respectively (Supplementary Table 4). Neither the gene coding for the microbial surface components recognizing adhesive matrix molecules (MSCRAMMs) nor those in the icaADBC operon coding for the biofilm components or agr quorum sensing system were differentially expressed.
Modulating only the physicochemical properties of the bacterial or the skin surface is sufficient to delay or decrease initial bacterial adhesion and thus colonization. RHM shows a very strong anti-adhesion effect on S. aureus by modulating only the physicochemical properties of its surface, without any significant differential expression of the transcriptome.
Discussion
This study shows a significant difference in the native adhesion to the 3D skin model for the 3 major bacteria strains, with a maximum adhesion for S. aureus, an intermediate adhesion for S. epidermidis and a minimum adhesion and for C. acnes (Fig. 1, Supplementary Fig. 1). Similar trends were previously observed for S. aureus and S. epidermidis on the 3D skin model24.
Initial bacterial colonization of the skin or of any other surface is always initiated by an initial stage of reversible bacterial adhesion to the substrate, followed by a second stage of irreversible adhesion. Reversible adhesion is conditioned by the distance separating the partners and by the sum of the repulsive and attractive forces generated between them, as initially described for abiotic systems by the Derjaguin-Landau-Verwey-Overbeek (DLVO) theory26. This was extended to XDLVO, also considering Lewis acid-base interactions to the model bacterial adhesion more accurately26,27. Lewis acid-base interactions are considered to be the predominant forces governing the observed behavior, with the resulting hydrophobicity effectively evaluated using the apolar solvents of MATS solvent pairs28.
Under native conditions, the 3 bacterial strains also showed notable different surface physicochemical profiles. S. aureus exhibits a strongly hydrophobic surface without significant Lewis interactions, S. epidermidis is slightly less hydrophobic, but has an important Lewis acid and slightly basic characteristics. C. acnes is slightly less hydrophobic, but presents a weaker electron pair-accepting characteristic, and thus a weaker Lewis acid (Table 1). These physicochemical differences can be explained by structural differences of the bacterial envelopes. Indeed, the composition in membrane lipids and proteins, teichoic and lipoteichoic acids, oligosaccharides and fimbriae are all components that influence the hydrophobicity and Lewis acid-base balance of bacterial envelopes11,24.
The 3D skin model exhibits similar surface properties to normal and hyperseborrheic human skin, with a relatively hydrophilic, electron-donating surface21. Its hydrophilicity is mainly due to the adsorption of water molecules on the surface (Fig. 4, Supplementary Table 2), while the electron-donating component is supported by the COOH groups of free fatty acids24,29,30.
Lewis acid–base interactions typically represent about 90% of all non-covalent interactions in water, as a very polar medium, regardless of their attractive or repulsive properties31,32. In water, a net repulsion between hydrophilic entities (i.e. bacteria and skin model surface) can occur if their attraction to water molecules is stronger than between water molecules themselves, as supported by increased gamma+ and gamma- for C. acnes and slightly less for S. epidermidis31. This hydration layer makes other interactions difficult28, compromising the approach of any other molecule or (a)biotic surfaces. This phenomenon results in hydrophilic repulsion33. Furthermore, no significant Lewis interactions of S. aureus (γ+≈5 and γ-≈8) are responsible for the maximum adhesion observed on 3D skin model, due to the absence of hydrophilic repulsion.
We have herewith demonstrated that all tested compounds without antibacterial activity induce a significant inhibition of bacterial adhesion to the skin, and consequently a perturbation of bacteria-skin interactions. They also increased the wettability of 3D skin model. The adhesion of the genus Staphylococcus to the 3D skin model appears to be easier to prevent than that of C. acnes. These results can be explained by the MATS experiment on S. aureus and C. acnes. As the hydrophobicity and the Lewis acid-base balance are natively very different between bacteria of the genus Staphylococcus and C. acnes, the observed difference in the ability of compounds to inhibit their adhesion seems to be consistent. Moreover, as C. acnes is slightly hydrophilic, the ability of compounds to increase its hydrophilicity is limited compared to S. aureus, which is natively strongly hydrophobic and becomes more hydrophilic under the influence of certain tested compounds. Similarly, the compound effect on the Lewis acid-base balance can only be greater for S. aureus, which is naturally deprived of Lewis charges compared to C. acnes, which has a slight Lewis acid-base, underlining the complexity of the interactions that lead to adhesion or its inhibition.
In order to better understand the process, the Pearson’s statistical test highlighted several insights: a very strong linear correlation between adhesion inhibition by tested compounds and the Lewis acid component, as well as a strong anti-correlation of adhesion inhibition with hydrophobicity for S. aureus and not for C. acnes. This shows the possibility of specific modulation of targeted bacterial adhesion implicated in skin disorders by specific ingredients.
However, despite three runs performing the RNA-Seq experiments under the same conditions as for the anti-adhesion assay and after detaching bacteria from the 3D skin model cells, no appropriate bacterial RNA quantity/quality to perform the analysis (RIN < 7) could be obtained. As an alternative, a bacterial transcriptome analysis in a static 2D culture was performed. Current literature confirms that the expression of the same biological pathways involved in the adhesion process to biotic surfaces as those obtained under our RNAseq experimental conditions where the adhesion matrix was an abiotic surface34,35. While not directly tested in our system, literature further supports the involvement of MSCRAMMs and other virulence loci in S. aureus adhesion to biological surfaces. For example, a study by Cruz et al. found upregulation of clfA, ebps, and fnbA within 2–3 h of infection in a skin explant model. This suggests their potential role in the early stages of adhesion, even though their expression wasn’t significantly altered by the compounds within our tested timeframe36.
To the best of our knowledge, this is the first time that the possibility has been shown to modulate the initial adhesion of skin bacteria by only adjusting the physicochemical properties of the bacterial surface without any interference with biological pathways involved in adhesion/colonization of bacteria. These findings reveal a fascinating prospect for the prevention of skin colonization by pathogenic bacteria, without recourse to antimicrobials.
Methods
Bacterial strains and culture conditions
In this study, Staphylococcus aureus ATCC 6538, Staphylococcus epidermidis ATCC 12228 and Cutibacterium acnes ATCC 6919 from the American Type Culture Collection (ATCC) were used as representative members of Firmicutes and Actinobacteria, the two main phyla of human skin, regardless of the skin site. Staphylococcal strains, received freeze-dried, were subcultured three times for 24 h in Trypto-casein-soy broth (TSB) at 37 °C to make “−80 °C stock cultures”. Strains were renewed every 18 months. In order to provide a “−20 °C working stock”, the contents of a cryotube from the −80 °C stock cultures were subcultured twice for 24 h in TSB and frozen in TSB supplemented with glycerol (15 g/l). Staphylococcal working stocks were renewed every 6 months. For C. acnes, lyophilizates was subcultured twice on Trypto-casein-soy Agar (TSA) slant for 4 days à 35 °C and stock at 4 °C to make a “working stock” which was renewed every 2 weeks.
Inoculums calibrated at 107 CFU/ml of bacteria in post-exponential phase growth phase were prepared in physiological water from subcultures of working stock (Table 3) performed in TSB at 37°C for Staphylococcal strains or 35 °C for C. acnes. Post-exponential phase was chosen to avoid variability when measuring adhesion and to mimic the bacterial physiological state on the skin surface24.
Table 3.
Duration of subcultures performed from working stock
| S. aureus ATCC 6538 | S. epidermidis ATCC 12228 | C. acnes ATCC 6919 | |
|---|---|---|---|
| 1st subculture | 24 h | 24 h | 48 h |
| 2nd subculture | 8 h | 8 h | NA |
| 3rd subculture | 16 h | 24 h | NA |
NA Not applicable.
Preparation of the tridimensional (3D) skin model
Tridimensional skin model samples (EPISKIN, Lyon, France) were received on nutritive transport agar on the 14th day of culture22. Upon receipt, the EPISKIN D-13 samples were placed in new 12-well plates containing 2 ml of fresh maintenance medium that was non-supplemented with antibiotics (EPISKIN, Lyon, France) and was incubated for 24 h at 37 °C and 5% CO2. The maintenance medium was renewed every 24 h and the EPISKIN D-13 samples were used from day 15 to day 17 of culture for the physicochemical characterization and bacterial adhesion assessment.
Preparation of tested compounds
All compounds used in this study are listed in Table 4. They were diluted in water to obtain a final concentration similar to that used in formula. Solutions were prepared the day before the experiments.
Table 4.
Tested compounds
| Abbreviation | INCI name | Concentration (%) | Comments | Origin |
|---|---|---|---|---|
| SLS | Sodium Laureth Sulfate | 1% | Anionic surfactant | Vegetal |
| CARR | Carrageenan | 2% | Anionic sulfated polysaccharides | Vegetal |
| GUAR | Guar Hydroxypropyltrimonium Chloride | 1% | Quaternary ammonium derived from Guar gum, cationic | Vegetal |
| SH | Sodium Hyaluronate | 1% | Sodium salt of hyaluronic acid, anionic | Bioprocess |
| RHM | Rhamnolipids | 0,5% | Anionic biosurfactant | Bioprocess |
| VF | Vitreoscilla filiformis extract | 1% | Biomass | Bacterial |
Definition of the surface energy characteristic of the 3D skin model
According to previous work, the surface energy characteristics of 3D skin samples with/without 2 h treatment by compounds were obtained by contact angle measurement using the sessile drop method with a G40 goniometer (Krüss, Hamburg, Germany) and three pure solvents with known surface tension properties: distilled water MilliQ (Millipore Corporation, Bedford, MA, USA), formamide and diiodomethane (Sigma, St. Louis, MO, USA)21.
3D Skin surface roughness analysis
Surface roughness of treated 3D skin models was characterized using optical profilometry. Following a 2-h incubation with tested compounds at 37 °C and 5% CO2 3D skin models were punched and fixed to glass slides using Vetbond™ Tissue Adhesive (3 M™, Cergy, France). Surface roughness was assessed using a ContourGT-K optical profilometer (Bruker, Billerica, MA, USA) in Vertical Scanning Interferometry (VSI) mode.
Assessment of 3D skin cell viability
The viability of 3D skin models following a 2-h topical exposure to tested compounds was assessed using an MTT cell viability assay. Briefly, 25 µL of each tested compound was applied to the surface of the 3D skin models and incubated for 2 h at 37 °C and 5% CO2. Following incubation, the surface of each model was washed with 25 mL of PBS+ and gently wiped with a sterile swab. Each model was then transferred to a 12-well plate containing 1 mg/mL tetrazolium (Sigma, St. Louis, MO, USA) and incubated for a further 3 h (37 °C, 5% CO2). Finally, 3D skin models were punched, transferred to acidified isopropanol, and incubated at 4 °C for 48 h. Absorbance at 570 nm was measured and cell viability was calculated as a percentage relative to the water-treated control using the following formula: Cell viability (%) = (OD of the RM-treated condition / OD of the water-treated condition) × 100.
Antibacterial activity assessment of tested compounds
Before starting the analysis of the effect of compounds on the adhesion of skin strains used in this study, antibacterial activity was characterized. To this end, 25 µl of each compound tested solution was added to 1 ml of each bacterial inoculum calibrated to 107 CFU/ml. The mixtures were incubated for 2 h and enumerated on TSA. Antibacterial activity was evaluated by calculating the logarithmic reduction of viable bacteria incubated with the compound against bacteria incubated with sterile distilled water (negative control).
Characterization of the bacterial surface polarity and Lewis acid-base traits
The surface polarity and Lewis acid-base balance of the bacteria with/without 2 h treatment by compounds were assessed using the Microbial Adhesion To Solvents (MATS) method37 and pure solvent couples with known surface tension properties: chloroform/hexadecane and ethyl acetate/decane (Sigma, St. Louis, MO, USA). For each strain, 2,4 mL of calibrated suspensions at OD400nm = 0.8 (OD400nm(init)) in physiological water were prepared. Suspensions were supplemented with 60 µL of compounds or distilled water and incubated for 2 h at 37 °C. Then, 2.4 ml of each bacterial suspension was mixed with 400 μl of each solvent. The tubes were vigorously shaken for 90 seconds to emulsify and incubated without shaking at room temperature for 15 min to ensure a complete separation of the two phases and OD400nm (15 min) was measured in the aqueous phase. The adhesion percentages of bacteria to each solvent were calculated using the following equation: % of adhesion = (1−OD400nm(15 min)/OD400nm(init)) × 100. Lewis acidity and basicity values, expressed in arbitrary units, are determined based on the contrasting affinities observed between these two specific solvent pairs. Lewis acidity is established from the affinity difference between chloroform and hexadecane, while Lewis basicity is determined from the affinity difference between decane and ethyl acetate. Hydrophobicity, expressed in percentage (%), is calculated based on the average affinity to decane and hexadecane.
Bacterial anti-adhesion effect assessment of topical formulation-intended compounds on 3D skin model
To evaluate the bacterial anti-adhesion effect, 25 µl of tested compound solutions or sterile distilled water (negative control) were gently spread on the epidermal layer of 3D skin samples and incubated for 2 h at 37 °C and 5% CO2. After this first incubation, without rinsing, 1 mL of bacterial inoculum was added on the epidermal layer and incubated a second time for 2 h at 37 °C and 5% CO2. Then, non-adherent bacteria were eliminated by five consecutive rinses with 1 mL of sterile distilled water. To retrieve adhering bacteria, 3D skin samples were cut from their nacelle, placed in a glass tube containing 9 ml of neutralizing media EUGON LTSup (BioMérieux, Marcy-l’Étoile, France) and sonicated for 5 min at 35 kHz. The resulting solution containing the bacteria removed from the epidermal surface of the 3D skin samples was incubated for 20 min at room temperature, and bacteria were enumerated on TSA. Finally, the bacterial anti-adhesion effect of compounds was evaluated by calculating the logarithmic reduction between bacteria adhering to the 3D skin sample with tested compound compared to bacteria adhering to the 3D skin sample with sterile distilled water (negative control).
RNA sequencing analysis
Sample preparation for RNAseq
Total RNA extraction was performed using the RNeasy Plus Micro (Qiagen®, Germany) on 1 ml of S. aureus suspension at 107 CFU/mL, previously incubated with tested compounds for 2 h at 37 °C without shaking. Ribosomal RNA was depleted with a Ribo-Zero rRNA Depletion Kit (Illumina, USA). The quantity and integrity of the total RNA was assessed with an HS RNA Analysis Kit on fragment analyzer (Agilent Technologies, UK).
Libraries were prepared using Illumina’s TruSeq Stranded mRNA kit (Illumina, USA). Briefly, mRNAs were selected using poly-T beads. Following this, RNA was fragmented to generate double-stranded cDNA and Illumina-specific adapters were ligated (Dual Indexing with IDTs for Illumina – TruSeq RNA UD Indexes). Eleven PCR cycles were applied to amplify the libraries. The quality of the libraries was checked on a Fragment Analyzer (Agilent). The libraries and pool were quantified by qPCR using the Library Quantification Kit - Illumina (KAPA). Finally, sequencing was performed on an Illumina NovaSeq 6000 using a read length of 150 bp paired-ends with Illumina NovaSeq 6000 sequencing kits (Illumina, USA).
Transcriptomic analysis
For gene and transcript quantification, a pseudo-alignment strategy based on a reference genome was chosen. The ASM202514v1 version of the Staphylococcus aureus genome (ATCC 6538 strain) was used (https://www.ncbi.nlm.nih.gov/datasets/genome/GCF_002025145.1/). The RNA-Seq raw data were analyzed using the nf-core/rnaseq v3.12.0 analysis pipeline (10.5281/zenodo.1400710) which is part of the NF-core pipeline collection38. This is an up-to-date and classically selected set of tools for performing RNA-Seq analyses. The main stages of the pipeline follow FastQC of raw read QC, TrimGalore for adapter and quality trimming and Salmon for Transcriptome quantification without alignment.
RNAseq differential expression analysis
A differential analysis was performed with the DESeq2 v1.38.3 tool, running on R-Studio v2022.07.0 with R v4.2.2 with default settings after the quality control. In this analysis, transcripts with fewer than 10 reads in total were removed, as well as those present in fewer than 3 samples with 10 reads. A value of 0.1 was used as the adjusted P-value threshold to determine whether a gene’s results were significant. Functional analyses were carried out using the Eggnog mapper tool v2.1.10 (http://eggnog5.embl.de/#/app/home), which classifies genes into ortholog classes.
Statistical analysis
For the analysis and graphical presentation, GraphPad Prism® Software (V9.5, San Diego, CA, United States) was used. The Shapiro-Wilk normality test was used to verify the normality of the data and statistical significance was obtained using a paired t-test to compare control conditions with treated conditions. Multiple comparisons were performed using Tukey’s test following one-way ANOVA. The threshold of significance was fixed at (*) for p < 0.05, (**) for p < 0.01 and (***) for p < 0.001.
Supplementary information
Acknowledgements
Metys INRAE transfer consulting platform for conducting and analyzing RNA-seq experiments. Marie-Noëlle Bellon-Fontaine for her advice in the initial phases of the project, Alban Ott for his critical review of the statistical analysis.
Author contributions
X.J. conducted the experiments, participated in the study design, performed data analysis, generated figures and participated in the manuscript editing and review. S.J., C.P., and N.K. conducted the experiments and analyzed data. F.Z. conducted the experiments and generated/analyzed complementary data. C.D. participated in the manuscript editing and review, data analysis, and provided physicochemical expertise. S.C. participated in the study design and reviewed the manuscript, A.K. participated in the study design, supervised and administred the project and reviewed and edited the manuscript. All authors read and approved the final manuscript.
Data availability
All data used and analyzed during the study are available from the corresponding author on request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Severine Jansen, Charleyne Prenom.
These authors jointly supervised this work: Cécile Duclairoir-Poc, Sylvie Cupferman.
Supplementary information
The online version contains supplementary material available at 10.1038/s41522-024-00568-8.
References
- 1.Oh, J. et al. Biogeography and individuality shape function in the human skin metagenome. Nature514, 59–64 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Byrd, A. L., Belkaid, Y. & Segre, J. A. The human skin microbiome. Nat. Rev. Microbiol.16, 143–155 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Ahle, C. M. et al. Interference and co-existence of staphylococci and Cutibacterium acnes within the healthy human skin microbiome. Commun. Biol.5, 923 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bojar, R. A. & Holland, K. T. Review: the human cutaneous microflora and factors controlling colonisation. World J. Microbiol. Biotechnol.18, 889–903 (2002). [Google Scholar]
- 5.Grice, E. A. et al. Topographical and temporal diversity of the human skin microbiome. Science324, 1190–1192 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Costello, E. K. et al. Bacterial community variation in human body habitats across space and time. Science326, 1694–1697 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Katsikogianni, M. & Missirlis, Y. F. Concise review of mechanisms of bacterial adhesion to biomaterials and of techniques used in estimating bacteria-material interactions. Eur. Cell Mater.8, 37–57 (2004). [DOI] [PubMed] [Google Scholar]
- 8.Habimana, O., Semião, A. J. C. & Casey, E. The role of cell-surface interactions in bacterial initial adhesion and consequent biofilm formation on nanofiltration/reverse osmosis membranes. J. Membr. Sci.454, 82–96 (2014). [Google Scholar]
- 9.Hori, K. & Matsumoto, S. Bacterial adhesion: from mechanism to control. Biochem. Eng. J.48, 424–434 (2010). [Google Scholar]
- 10.Van Loosdrecht, M. C. M., Norde, W. & Zehnder, A. J. B. Physical chemical description of bacterial adhesion. J. Biomater. Appl.5, 91–106 (1990). [DOI] [PubMed] [Google Scholar]
- 11.Krasowska, A. & Sigler, K. How microorganisms use hydrophobicity and what does this mean for human needs? Front. Cell Infect. Microbiol.4, 112 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Muhammad, M. H. et al. Beyond risk: bacterial biofilms and their regulating approaches. Front Microbiol11, 928 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pothmann, A., Illing, T., Wiegand, C., Hartmann, A. A. & Elsner, P. The microbiome and atopic dermatitis: a review. Am. J. Clin. Dermatol.20, 749–761 (2019). [DOI] [PubMed] [Google Scholar]
- 14.Weidinger, S., Beck, L. A., Bieber, T., Kabashima, K. & Irvine, A. D. Atopic dermatitis. Nat. Rev. Dis. Prim.4, 1 (2018). [DOI] [PubMed] [Google Scholar]
- 15.Schommer, N. N. & Gallo, R. L. Structure and function of the human skin microbiome. Trends Microbiol.21, 660–668 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bojar, R. A. & Holland, K. T. Acne and Propionibacterium acnes. Clin. Dermatol.22, 375–379 (2004). [DOI] [PubMed] [Google Scholar]
- 17.Fitz-Gibbon, S. et al. Propionibacterium acnes strain populations in the human skin microbiome associated with acne. J. Invest. Dermatol.133, 2152–2160 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cole, G. W. & Silverberg, N. L. The adherence of Staphylococcus aureus to human corneocytes. Arch. Dermatol.122, 166–169 (1986). [PubMed] [Google Scholar]
- 19.Feuillie, C. et al. Adhesion of Staphylococcus aureus to corneocytes from atopic dermatitis patients is controlled by natural moisturizing factor levels. mBio9, e01184-18 (2018) [DOI] [PMC free article] [PubMed]
- 20.Lizardo, M., Magalhães, R. M. & Tavaria, F. K. Probiotic adhesion to skin keratinocytes and underlying mechanisms. Biology (Basel)11, 1372 (2022). [DOI] [PMC free article] [PubMed]
- 21.Lerebour, G., Cupferman, S., Cohen, C. & Bellon-Fontaine, M. N. Comparison of surface free energy between reconstructed human epidermis and in situ human skin. Ski. Res Technol.6, 245–249 (2000). [DOI] [PubMed] [Google Scholar]
- 22.Tinois, E. et al. In vitro and post-transplantation differentiation of human keratinocytes grown on the human type IV collagen film of a bilayered dermal substitute. Exp. Cell Res.193, 310–319 (1991). [DOI] [PubMed] [Google Scholar]
- 23.Burden, N., Sewell, F. & Chapman, K. Testing chemical safety: what is needed to ensure the widespread application of non-animal approaches? PLOS Biol.13, e1002156 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lerebour, G., Cupferman, S. & Bellon-Fontaine, M. N. Adhesion of Staphylococcus aureus and Staphylococcus epidermidis to the Episkin reconstructed epidermis model and to an inert 304 stainless steel substrate. J. Appl. Microbiol.97, 7–16 (2004). [DOI] [PubMed] [Google Scholar]
- 25.Rademacher, F., Simanski, M., Gläser, R. & Harder, J. Skin microbiota and human 3D skin models. Exp. Dermatol.27, 489–494 (2018). [DOI] [PubMed] [Google Scholar]
- 26.Derjaguin, B. & Landau, L. Theory of the stability of strongly charged lyophobic sols and of the adhesion of strongly charged particles in solutions of electrolytes. Prog. Surf. Sci.43, 30–59 (1993). [Google Scholar]
- 27.Van Oss, C. J., Good, R. J. & Chaudhury, M. K. The role of van der Waals forces and hydrogen bonds in “hydrophobic interactions” between biopolymers and low energy surfaces. J. Colloid Interface Sci.111, 378–390 (1986). [Google Scholar]
- 28.Moriarty, T. F., Poulsson, A. H. C., Rochford, E. T. J. & Richards, R. G. in Comprehensive Biomaterials (ed P. Ducheyne) 75–100 (Elsevier, 2011).
- 29.Mavon, A. et al. Sebum and stratum corneum lipids increase human skin surface free energy as determined from contact angle measurements: A study on two anatomical sites. Colloids Surf. B: Biointerfaces8, 147–155 (1997). [Google Scholar]
- 30.Mavon, A., Redoules, D., Humbert, P., Agache, P. & Gall, Y. Changes in sebum levels and skin surface free energy components following skin surface washing. Colloids Surf. B: Biointerfaces10, 243–250 (1998). [Google Scholar]
- 31.van Oss, C. J. Long-range and short-range mechanisms of hydrophobic attraction and hydrophilic repulsion in specific and aspecific interactions. J. Mol. Recognit.16, 177–190 (2003). [DOI] [PubMed] [Google Scholar]
- 32.Grasso, D., Subramaniam, K., Butkus, M., Strevett, K. & Bergendahl, J. A review of non-DLVO interactions in environmental colloidal systems. Rev. Environ. Sci. Biotechnol.1, 17–38 (2002). [Google Scholar]
- 33.Sun, Q., Zhang, M. & Cui, S. The structural origin of hydration repulsive force. Chem. Phys. Lett.714, 30–36 (2019). [Google Scholar]
- 34.Dufrêne, Y. F. & Persat, A. Mechanomicrobiology: how bacteria sense and respond to forces. Nat. Rev. Microbiol.18, 227–240 (2020). [DOI] [PubMed] [Google Scholar]
- 35.Kreve, S. & Reis, A. C. D. Bacterial adhesion to biomaterials: what regulates this attachment? A review. Jpn. Dent. Sci. Rev.57, 85–96 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cruz, A. R., van Strijp, J. A. G., Bagnoli, F. & Manetti, A. G. O. Virulence gene expression of Staphylococcus aureus in human skin. Front. Microbiol.12, 692023 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bellon-Fontaine, M. N., Rault, J. & van Oss, C. J. Microbial adhesion to solvents: a novel method to determine the electron-donor/electron-acceptor or Lewis acid-base properties of microbial cells. Colloids Surf. B: Biointerfaces7, 47–53 (1996). [Google Scholar]
- 38.Ewels, P. A. et al. The nf-core framework for community-curated bioinformatics pipelines. Nat. Biotechnol.38, 276–278 (2020). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data used and analyzed during the study are available from the corresponding author on request.