Abstract
Parvoviral rolling hairpin replication generates palindromic genomic concatemers whose junctions are resolved to give unit-length genomes by a process involving DNA replication initiated at origins derived from each viral telomere. The left-end origin of minute virus of mice (MVM), oriL, contains binding sites for the viral initiator nickase, NS1, and parvovirus initiation factor (PIF), a member of the emerging KDWK family of transcription factors. oriL is generated as an active form, oriLTC, and as an inactive form, oriLGAA, which contains a single additional nucleotide inserted between the NS1 and PIF sites. Here we examined the interactions on oriLTC which lead to activation of NS1 by PIF. The two subunits of PIF, p79 and p96, cooperatively bind two ACGT half-sites, which can be flexibly spaced. When coexpressed from recombinant baculoviruses, the PIF subunits preferentially form heterodimers which, in the presence of ATP, show cooperative binding with NS1 on oriL, but this interaction is preferentially enhanced on oriLTC compared to oriLGAA. Without ATP, NS1 is unable to bind stably to its cognate site, but PIF facilitates this interaction, rendering the NS1 binding site, but not the nick site, resistant to DNase I. Varying the spacing of the PIF half-sites shows that the distance between the NS1 binding site and the NS1-proximal half-site is critical for nickase activation, whereas the position of the distal half-site is unimportant. When expressed separately, both PIF subunits form homodimers that bind site specifically to oriL, but only complexes containing p79 activate the NS1 nickase function.
Parvoviruses infect a broad range of invertebrate and vertebrate species, including humans (14). The viral genome is a linear single-stranded DNA molecule of around 5 kb that has short unique terminal palindromes, which fold back on themselves to form characteristic hairpin duplexes and which are central to the viral replication strategy (1, 2, 11–13, 16). Their limited coding capacity means that these viruses must rely predominantly upon host cell replication factors, augmented and orchestrated by the virus-encoded pleiotropic initiator protein NS1 (3, 10, 26, 29, 30).
The viral replication strategy, dubbed rolling hairpin replication, is an evolutionary modification of the more widely employed rolling circle replication (RCR) mechanism (24), adapted for the amplification of linear molecules. A single, unidirectional, leading-strand-specific replication fork, assembled on the 3′ hairpin (by convention called the left-end hairpin), first copies the incoming linear genome, creating a monomer duplex intermediate in which the original right-end hairpin has been unfolded and copied to form a duplex terminal palindrome. Sequential unfolding and refolding of the two palindromic viral termini then allow the direction of the advancing fork to be switched back and forwards along the molecule, copying the strand it has just finished synthesizing while displacing its previous complement. This process generates a series of dimeric and tetrameric replication intermediates in which duplex copies of the unit-length linear genome are linked in head-to-head and tail-to-tail configurations by duplex copies of the appropriate terminal hairpin. As in other RCR systems, minute virus of mice (MVM) encodes, in NS1, a site-specific nickase, which recognizes disparate origin sequences present in the two viral termini and is able to nick and resolve these concatemers into unit-length molecules (12, 13, 15). To do this, NS1 first binds to a specific recognition site in the duplex viral origin and then introduces a single-strand nick at an adjacent consensus site and/or structure. This reaction leaves NS1 covalently attached to the 5′ end of the DNA at the nick site via a phosphotyrosine bond and generates a base-paired 3′ nucleotide with a free hydroxyl group, which, in turn, primes an additional round of unidirectional DNA synthesis (17). For imperfectly palindromic termini, this mechanism would be predicted to generate two forms, which would be inverted complements of one another. While the hairpin at the 5′ end of the MVM genome is found in both conformations, called “flip” and “flop,” the hairpin at the 3′ end is conserved in one sequence orientation, called “flip.”
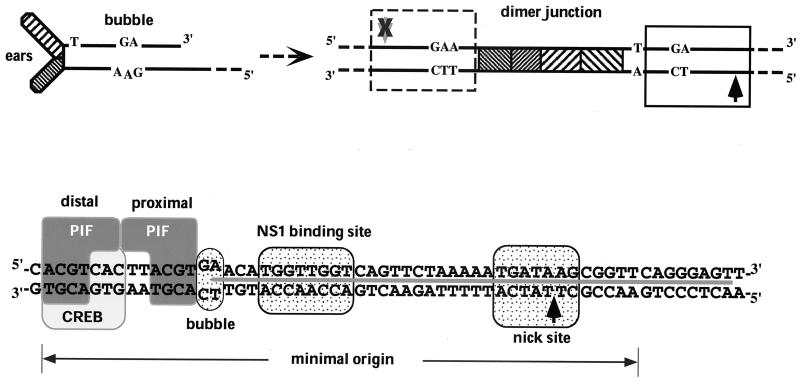
In single-stranded viral DNA, the terminal palindrome at the “left” end can be folded into the Y-shaped structure shown in Fig. 1 (top left). This Y-shaped turnaround structure contains a sequence mismatch in the middle of its stem, called the “bubble,” where a GA dinucleotide on one strand is positioned opposite a GAA trinucleotide on the other. In this folded configuration, the left-end palindrome is not an active origin. However, after being unfolded and copied to form the dimer junction sequence (Fig. 1, top right), the two strands of the original hairpin, containing their particular bubble nucleotides, are separated on each side of the axis of symmetry. One of these, the TC arm, contains an active NS1-dependent origin, termed oriLTC. The minimal sequence from this arm required to support initiation is approximately 50 bp long and contains the TC bubble dinucleotide, as detailed at the bottom of Fig. 1. The equivalent sequence from the other arm, termed oriLGAA, is identical to oriLTC, except that the GAA bubble trinucleotide replaces the TC dinucleotide, rendering it inactive as a DNA replication origin. It is not the sequence of the bubble per se but rather the insertion of the single extra nucleotide which stops NS1 from nicking this arm (16). By an unknown mechanism, but apparently as a consequence of this arrangement of active origin opposing inactive origin, the resolution of the dimer junction occurs asymmetrically, such that the products are invariably one newly synthesized, extended terminus and one hairpin in the “flip” orientation. While this neatly explains the conservation of “flip” at the left end of the genome, it is not yet known why this is of importance in the viral life cycle.
FIG. 1.
Formation and organization of MVM oriL. (Top left) Structure of the left-end hairpin showing the 3′ OH used for priming replication and the mismatched bubble sequence as present in the parental single-stranded viral genome. (Top right) Organization of the left-end hairpin sequences within the duplex dimer junction generated by replication through the hairpin. Hatched boxes represent the palindromic sequences which were originally folded to give the ears of the hairpin form. The boxed sequence (expanded below) represents the minimum active replication origin on the outboard arm, oriLTC, while the sequence in the dashed box represents the corresponding origin on the inboard arm, oriLGAA, which is inactive. The potential nick sites on each side of the junction are indicated by arrows (black arrow, active site; grey arrow with X, inactive site). (Bottom panel) Sequence of oriLTC, showing the different elements involved in replication. The PIF binding site (see the text) overlaps a consensus binding site for the CREB/ATF family of host transcription factors. The two tetranucleotide motifs bound by PIF are designated “distal” and “proximal” to indicate their positions relative to the NS1 binding site. Other boxes indicate sequences involved in the bubble dinucleotide (or trinucleotide) spacer element described in the text, the (ACCA)2 NS1 binding site, and the nick site, a specific sequence required for nicking and covalent attachment of NS1. The heavy line between the DNA strands indicates sequences protected by NS1 from DNase I digestion.
The minimal active origin contains three distinct recognition elements: the NS1 binding site, the nick site, and the binding site for an essential cellular cofactor. In the presence of ATP, a multimeric NS1 complex binds site specifically to the (ACCA)2 motif, extending in an asymmetric fashion over the nick site and protecting it from DNase I digestion (10). However, by itself NS1 cannot nick the origin. For this to occur a cellular factor, which we have dubbed parvovirus initiation factor (PIF), is required (4). PIF binds coordinately to two copies of its tetranucleotide half-site, ACGT, at one end of the origin, separated from the NS1 binding site by the bubble dinucleotide. Although the equivalent region from oriLGAA is not an active origin, both it and oriLTC appear to bind NS1 and purified PIF equally well (5, 6). PIF is a widely expressed heterodimer consisting of two related polypeptides, p79 and p96, encoded by two separate genes, located at human chromosomes 1p34 and 20qter, respectively (34; J. Christensen, E. Burnett, S. F. Cotmore, C. M. Ticknor, and P. Tattersall, unpublished data). Database searches indicate that PIF is the human homologue of a rat factor dubbed the glucocorticoid-modulating element binding protein (GMEB), which was identified because it binds in vitro to a region in the promoter of the tyrosine amino transferase gene known to enhance its sensitivity to glucocorticoids (23, 27, 28, 33). The optimal PIF binding site has recently been defined, and high-affinity binding sites for this complex have been identified in the promoters of a wide range of cellular genes (E. Burnett, J. Christensen, and P. Tattersall, submitted for publication). Both PIF subunits are transcriptionally active (6) and are members of an emerging family of site-specific DNA-binding proteins which have a characteristic conserved domain containing the amino acid motif KDWK, first described for the Drosophila homeotic activator DEAF-1 (20).
We have cloned both subunits of the PIF heterodimer from HeLa cells and expressed them in insect cells from recombinant baculoviruses (6). In this study we compared the abilities of recombinant and HeLa-derived PIF subunits to heterodimerize and to interact with NS1 and viral origin DNA. We explore the nature of PIF-NS1 interactions which are required to activate the MVM origin and examined differences between the complexes assembled on each form of oriL that ultimately lead to both asymmetric resolution of the dimer junction and the sequence conservation of the left MVM hairpin.
MATERIALS AND METHODS
Purification of HeLa PIF complexes.
Nuclear extracts from HeLa S3 cells were prepared and fractionated by Q-Sepharose, Zn2+-metal chelate Sepharose, and DNA affinity chromatography as described previously (6).
Recombinant baculoviruses.
To optimize expression of wild-type p96 and p79, PCR fragments, beginning three nucleotides upstream of the open reading frame, were cloned into the baculovirus transfer vector pVL1393 (Invitrogen) to generate pVL1393-p96 and pVL1393-p79 as previously described (6). For glutathione S-transferase (GST)-NS1, the NS1 open reading frame was isolated from pKO NS1/NS2, a gift from D. Pintel, Department of Molecular Microbiology and Immunology, School of Medicine, University of Missouri, Columbia, and cloned into pAcGHLT-A. Recombinant baculoviruses were produced as previously described (8).
Purification of rPIF.
Recombinant PIF (rPIF) p79, rPIF p96, and rPIF p96-p79 complexes were generated by infecting or coinfecting High Five insect cells at a high multiplicity (∼10 PFU per cell) with recombinant baculoviruses. Nuclear extracts were prepared essentially as previously described (7), and after absorption on Q-Sepharose to remove contaminating DNA, the extracts were diluted to 75 mM NaCl and purified using MonoQ (HR5/5) or a 1-ml HiTrap Q-Sepharose column equilibrated in buffer A (25 mM Tris-HCl, 1 mM EDTA, 0.1 mM phenylmethylsulfonyl fluoride, 0.01% NP-40, 1.0 mM dithiothreitol [DTT], and 10% glycerol) containing 50 mM NaCl. Bound proteins were eluted with a 25-ml linear NaCl gradient (0.05 to 0.5 M) in buffer A. Recombinant MVM GST-NS1 and His-tagged MVM NS1 were expressed in Sf9 insect cells using baculovirus vectors and purified essentially as previously described (3).
Electrophoretic mobility shift assay (EMSA).
DNA-binding reactions were carried out in 25 μl of 20 mM HEPES-KOH (pH 7.8)–100 mM NaCl–0.5 mM EDTA–1 mM DTT–10% glycerol supplemented with 0.5 μl of rabbit reticulocyte lysate and 50 μg of bovine serum albumin/ml as nonspecific protein carriers. Proteins were incubated for 10 min at room temperature in the presence of nonspecific carrier DNA before addition of the radiolabeled probe (5 × 104 cpm) and then incubated at room temperature for a further 20 min. For supershift analysis, incubations were continued for 20 min in the presence of 1 μl of antibody, as indicated in the figure legends, before being separated by electrophoresis through 6% native polyacrylamide gels at 4°C for 3 h at 200 V. Gels were fixed in 10% acetic acid, dried, and exposed for autoradiography. Polyclonal antibodies used for supershift analyses were raised in rabbits by immunization with specific peptides, G P G V P V P Q L T S V P L G K (p79 residues 385 to 400) and D T E G K A V I L E T E L R T (p96 residues 534 to 548), which will be reported in detail elsewhere (Christensen et al., unpublished).
Plasmids.
The minimal origin templates used for nicking and DNase I protection assays were derived from pL1-2TC and pL1-2GAA, which contain MVM oriLTC and oriLGAA, respectively (16). Origins with mutations in the PIF binding site were generated by PCR using primers with the desired mutations and cloned into pCRII (Invitrogen, San Diego, Calif.). All sequences were verified by standard dideoxy DNA sequencing. To make substrates for nicking assays, origins were excised by digestion with EcoRI, purified from agarose gels, and 3′-end labeled with [32P]dATP using Sequenase (4).
Nicking assays.
Nicking assays were performed essentially as previously described (4). Briefly, purified HeLa or rPIF was incubated for 20 min at 37°C in the presence of ATP, variable amounts of GST-NS1 (specified in the figure legends), and a 32P-labeled DNA fragment corresponding to the minimal 3′ origin region of MVM. Reactions were terminated by adding an equal volume of 25 mM Tris-HCl (pH 8.0)–5 mM EDTA–100 mM NaCl–1% sodium dodecyl sulfate (SDS)–10% glycerol and incubating at 60°C for 15 min, and the products were analyzed by electrophoresis through 6% acrylamide gels equilibrated in 0.2% SDS. Nicked DNA products were also deproteinized and analyzed on sequencing gels to confirm that the shifted bands detected in these assays were nicked at the correct site (data not shown).
DNase I protection assays.
DNase I protection assays were performed essentially as previously described (4). Briefly, 50 ng of rPIF p79-p96 complex and 25, 50, 100, and 225 ng of His-tagged NS1 were incubated for 20 min at room temperature followed by 20 min on ice in 25 μl of a mixture containing 25 mM HEPES-KOH, 100 mM NaCl, 2 mM MgCl2, 2 mM γ-S-ATP, 1 mM DTT, 50 μg of bovine serum albumin/ml, 250 ng of Scram (a nonspecific double-stranded oligonucleotide), 10% glycerol, and DNA fragments 3′ end labeled on one strand with the appropriate 32P-labeled deoxynucleoside triphosphate (approximately 105 cpm). Reaction mixtures were then incubated for 1 min at room temperature before the addition of DNase I (0.4 U). Incubations were continued for a further minute at room temperature and terminated by the addition of 0.15 ml of 10 mM Tris-HCl (pH 8)–10 mM EDTA–0.5% SDS–proteinase K (300 μg) and incubation at 50°C for 45 min. Samples were extracted with phenol-chloroform and ethanol precipitated before electrophoresis through 6% denaturing acrylamide gels.
RESULTS
PIF p79 and p96 subunits preferentially associate into heterodimers.
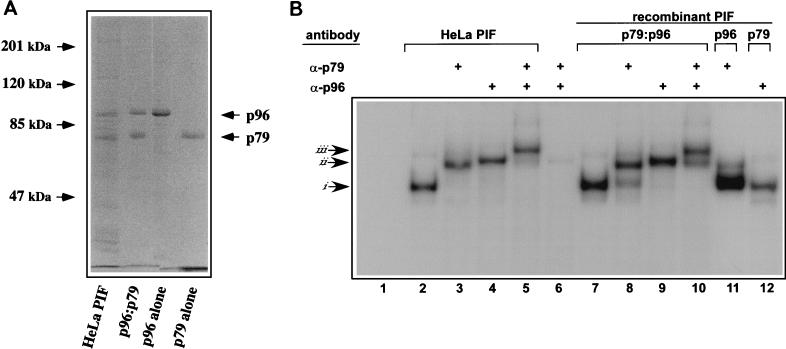
We have previously described the purification of an active PIF complex from HeLa S3 nuclear extracts using a combination of standard resins and site-specific DNA affinity chromatography. The purified complex contains two major polypeptides, p96 and p79, with apparent molecular masses of around 96 and 79 kDa, in equimolar concentrations (Fig. 2A), suggesting that the factor is heteromeric (6). When recombinant baculoviruses were used to express p96 and p79 in insect cells, either separately or in combination, the purified subunits bound DNA with similar specificity to one another and to both recombinant and HeLa p79-p96 heterocomplexes (Burnett et al., submitted). Recombinant complexes shifted the probe to approximately the same position in native gels as did the complex purified from HeLa cells, suggesting that the complexes from both sources were dimeric (Fig. 2B, lanes 2 and 7). To investigate the extent to which purified recombinant and HeLa p79-p96 preparations were a mixture of hetero- and homodimers, purified samples were analyzed in electrophoretic mobility supershift assays using rabbit polyclonal antisera raised against peptide sequences unique for p79 or p96. Figure 2B shows that addition of p79- and p96-specific antisera supershifted each complex to similar positions in the gel (Fig. 2B, lanes 3 and 8 or 4 and 9, respectively), suggesting that both preparations were predominantly heteromeric. In the example shown in Fig. 2B, some of the rPIF complex was not shifted by the p79 antibody (lane 8), indicating a slight excess of p96, presumably in the form of the homodimer, in this preparation,. In the negative control, neither p79 nor p96 complexes were shifted by p96 or p79 antibodies, respectively (Fig. 2B, lanes 11 and 12). When both antibodies were added to heterocomplex binding reactions at the same time, a “super-duper” shift was observed, with most of the supershifted complexes being retarded still further in the gel, thus confirming their heteromeric nature. These data suggest that when p79 and p96 are expressed at similar concentrations in HeLa and insect cells, they preferentially associate as heterodimers.
FIG. 2.
Characterization of rPIF subunits expressed from separate baculovirus vectors. (A) SDS-polyacrylamide gel electrophoresis analysis of purified rPIF. DNA-affinity purified PIF from HeLa S3 cells was compared to purified rPIF subunits, either coexpressed (p96-p79) or expressed alone in insect cells using baculovirus vectors. The gel was stained with Coomassie brilliant blue. (B) Autoradiograph of an EMSA gel of HeLa cell derived- and rPIF complexes showing complexes generated when 32P-labeled duplex oligonucleotides covering the PIF binding region in the MVM 3′ origin (5′-T C A T C A C G T C A C T T A C G T G A A-3′) were incubated with PIF purified from HeLa S3 cells by DNA-affinity chromatography (lanes 2 to 5) or with human PIF complexes purified from High Five insect cells infected with recombinant baculoviruses (lanes 7 to 12). PIF p79-p96 heterocomplexes purified from coinfected cells are shown in lanes 7 to 10, while lanes 11 and 12 contain p79 and p96 complexes purified from cells singly infected with the appropriate baculovirus. Preformed complexes were incubated with rabbit antisera specific for one of the PIF subunits, as indicated at the top of each lane. Lanes 6, 11, and 12 are negative controls (lane 6 contains both antibodies and the probe, but no PIF). Arrows at the side mark the positions of shifted (i), supershifted (ii), and super-duper-shifted (iii) PIF-oriL probe complexes.
rPIF heterocomplex activates NS1 to nick oriLTC but not oriLGAA
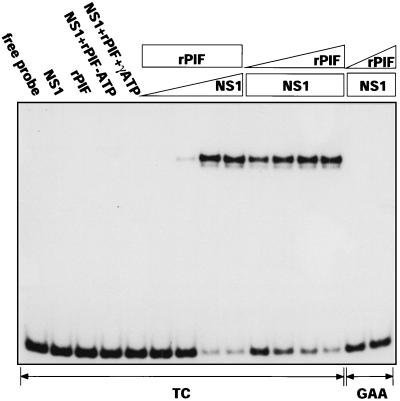
We next tested recombinant p96-p79 heterocomplexes to see if they were able to activate the nickase function of NS1 under standard reaction conditions and with the precise spacing constraints exhibited by the cellular activator isolated from HeLa extracts. For this, 32P-, 3′-end-labeled DNA fragments containing either the minimal active (TC) or inactive (GAA) origin sequences were incubated in the presence or absence of purified recombinant NS1, PIF, and ATP. Reaction products from these assays were heated to 60°C in 0.5% SDS and analyzed on SDS-polyacrylamide gels to abolish noncovalent protein-DNA interactions. As a result, while the nicking assay resembles the EMSA, it exclusively monitors the covalent conversion of substrate DNA to a new retarded species. This product carries a single-strand nick at the initiation site and has NS1 covalently attached to the new 5′ end generated at the nick, as previously described (4, 5). The assay results presented in Fig. 3 show that, like HeLa PIF, recombinant p96-p79 complexes can activate the viral nickase, that nicking requires ATP hydrolysis, and that the efficiency of the reaction varies with the concentration of NS1 and PIF. It also indicates that the spacing between the NS1 and PIF binding sites remains critical, since when separated by the correct number of nucleotides, as in the TC substrate, NS1 and the PIF heterodimer interact in a way that activates the nickase function of NS1, while the insertion of a single extra base between the two binding sites, as in the GAA sequence, prevents this interaction.
FIG. 3.
rPIF activates NS1 to nick the viral origin. A 32P-labeled DNA fragment containing the oriLTC sequence (TC) was incubated in the absence or presence of NS1 (100 ng), rPIF (25 ng), and ATP (3 mM) or γ-S-ATP (3 mM). In the titrations, a constant amount of rPIF (25 ng) or NS1 (100 ng) was incubated with increasing amounts of NS1 (10, 25, 50, and 100 ng) or rPIF (0.5 2.5, 12.5, and 50 ng). Only covalent NS1-DNA complexes are retarded in the SDS-polyacrylamide gel system used for this assay (see Materials and Methods). GAA, reactions performed with oriLGAA with NS1 and rPIF at inputs of 100 ng and 2.5 to 50 ng.
PIF enhances NS1 binding to oriLTC but not to oriLGAA
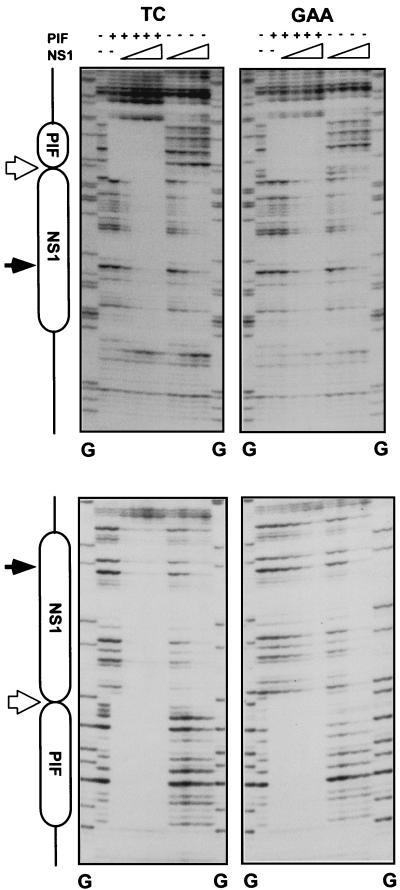
We have previously shown that when analyzed individually, rPIF and NS1 both bind to the active (TC) and inactive (GAA) origin sequences equally well and protect equivalent regions of both sequences from DNase I digestion (5, 10). However, it remained possible that these proteins might show cooperative binding to origin DNA. To test this possibility, TC and GAA sequences were digested with DNase I in the presence of rPIF p79-p96 and increasing amounts of NS1 and in the presence of γ-S-ATP, which is required for stable NS1-DNA binding. By itself, rPIF protected the sequences containing its ACGT half-sites equally well on both the active and inactive origins (Fig. 4), while NS1 complexes similarly protected equivalent regions of approximately 43 bp, encompassing both its (ACCA)2 binding motif and the nick site, on both substrates. The addition of increasing amounts of NS1 (25 to 225 ng) in the presence of PIF eventually protected the entire minimal origin regions of both the TC and GAA sequences from DNase I digestion in a concentration-dependent manner. The TC origin was totally protected at the lowest input of NS1 tested, a concentration that was insufficient to protect the NS1 site in the absence of PIF. In contrast, the higher NS1 concentrations required to protect the inactive GAA sequence from digestion were the same, irrespective of whether PIF was present. NS1 concentrations required to protect the NS1 binding site and nick site of the active origin were two- to fourfold lower in the presence of PIF than in its absence. This suggests that these two complexes interact to form a stable ternary complex when positioned the correct number of nucleotides apart on the origin, but that insertion of a single extra nucleotide between their two binding sites prevents the interaction between the two proteins.
FIG. 4.
PIF enhances NS1 binding to oriLTC but not to oriLGAA. Diagrams of the major elements in oriL are shown on the left. Sequences protected from DNase I digestion by PIF and NS1 are indicated by boxes. Positions of the nick site and bubble sequence are indicated by filled and open arrows, respectively. TC and GAA denote which form of oriL was used as the substrate. PIF (top), 50 ng of purified rPIF p79-p96 complex; NS1 (top), increasing amounts of purified His-tagged NS1 (25, 50, 100, and 225 ng). All assays were performed in the presence of 2 mM γ-S-ATP. Upper and lower pairs of autoradiographs show the protection patterns obtained when the DNA strands were 32P-, 3′-end-labeled on the upper and lower strands, respectively, of the oriL diagrammed in Fig. 1. Lanes G, products of a G-specific chemical sequencing reaction of each labeled substrate (25), used for aligning each autoradiograph with its relevant protection diagram.
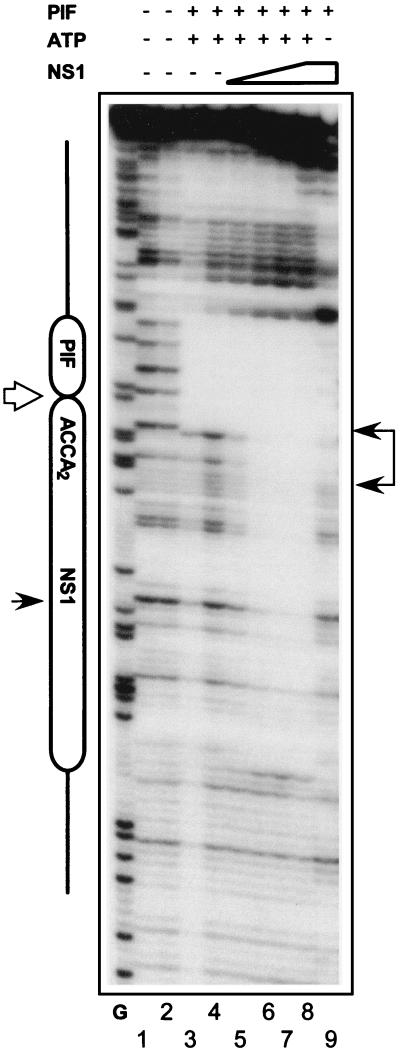
PIF promotes NS1 binding to the minimal active TC origin in the absence of ATP.
Although PIF only appeared to increase NS1 binding to the TC origin approximately two- to fourfold, TC sequences are at least 100-fold more active than their GAA counterparts when assayed as substrates for initiation in vitro (16). This suggests that PIF may have additional roles in the replication initiation process, over and above its ability to promote formation of the active ternary complex. The nicking reaction is a dynamic process. In the presence of either ATP or its nonhydrolyzable analogue γ-S-ATP, NS1 forms a multimeric complex which can be shown to bind site specifically to the (ACCA)2 repeat, but this interaction is unstable under conditions which promote ATP hydrolysis (3, 10). PIF could thus potentially stabilize NS1 molecules at the origin in the absence of ATP and/or after its hydrolysis. To test this hypothesis, the TC and GAA sequences were subjected to DNase I digestion in the presence and absence of γ-S-ATP. In both situations PIF, by itself, protected sequences containing its ACGT half-sites from digestion (Fig. 5, lanes 3 and 4). In the presence of γ-S-ATP and PIF, increasing amounts of NS1 rendered the origin totally protected from DNase I (Fig. 5, lanes 5 to 8), but in the absence of γ-S-ATP the protection pattern changed so that NS1 protected the (ACCA)2 repeat but not the region around the nick site (lane 9). The (ACCA)2 region was not protected at all by NS1 in the absence of PIF or on the GAA sequence even in the presence of PIF (data not shown).
FIG. 5.
PIF stabilizes NS1 binding the to oriLTC in the absence of ATP. An autoradiograph of DNase I protection analysis of rPIF and NS1 binding to an oriLTC probe in the presence or absence of γ-S-ATP is shown. The major elements of oriL are indicated on the left, as described in the legend to Fig. 4. PIF (top), 50 ng of purified rPIF p79-p96 complex; NS1 (top), increasing amounts of purified His-tagged NS1 (25, 50, 100, and 225 ng). Samples were digested with 0.8 (lanes 2 and 4 through 9) or 0.4 (lanes 1 and 3) U of DNase I. Lane G, G-specific digest of the probe, as described in the legend to Fig. 4.
Spacing between the NS1 binding site and the proximal, but not distal, PIF half-site is critical for origin activation.
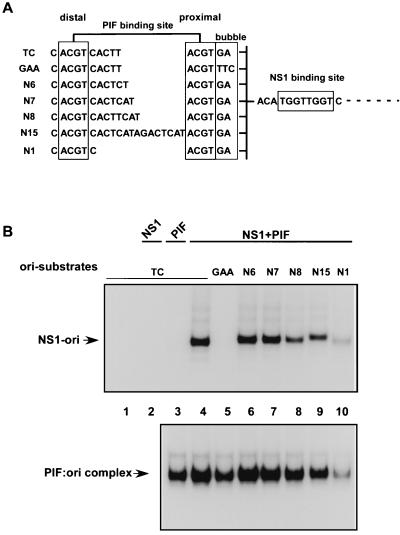
Insertion of a single nucleotide between the NS1 and PIF binding sites, as found in the GAA sequence, abolishes cooperative binding between PIF and NS1 and prevents activation of the NS1 nickase function. However, PIF binding sites are very unusual because they are made up from two palindromic ACGT half-sites which, although they are spaced five nucleotides apart in the origin, can still bind PIF when they are spaced anywhere from 1 to 15 nucleotides apart (6). Indeed, recent binding studies indicate that PIF would bind the origin with higher affinity if the two ACGT sites were 6 rather than 5 bp apart (Burnett et al., submitted). This led us to ask whether the disposition of the PIF dimers around the DNA helix was critical for activation or whether the ACGT half-site distal to the NS1 binding site could be moved provided that the proximal site was maintained in its original position. Since changing the spacing between the ACGT motifs influences the PIF binding affinity to some extent, we correlated NS1-dependent nicking with PIF DNA binding, as measured by EMSA, for oriLGAA and a series of mutant oriLTC sequences in which the position of the distal ACGT motif in the PIF binding site varied, as shown in Fig. 6A. Surprisingly, PIF was able to activate NS1-mediated nicking on all of these origin sequences with the exception of oriLGAA (Fig. 6B), and in each case, except that of oriLGAA, the nicking efficiency correlated directly with the binding efficiency of PIF for that particular sequence (Fig. 6). Thus, the differences in nicking efficiency seen in Fig. 6B for the active origins appear to reflect differences in PIF binding activity, rather than changes in the ability of PIF to cooperate with NS1 in the nicking reaction. Significantly, the mutant of oriLTC with a single extra nucleotide inserted between the PIF ACGT motifs (N6 in Fig. 6) has its distal ACGT half-site in the same position, relative to the NS1 binding and nicking sites, as it is in oriLGAA, but, unlike oriLGAA, the mutant arrangement is fully competent for nicking. Thus, the spacing between NS1 and the proximal PIF ACGT half-site is critical, but the position of the distal half-site can vary through a complete turn of the helix without affecting the ability of the active ternary complex to form.
FIG. 6.
Only the position of the NS1 proximal PIF half-site is important for oriL activation. (A) Sequences of the PIF site in the top strand of 32P-labeled, double-stranded, minimal origin probes used as substrates in nicking and gel mobility shift assays. The proximal and distal PIF half-sites are boxed, and the nucleotides between the half-sites are indicated. (B, top) Autoradiograph of nicking assays performed in the presence of purified GST-tagged NS1 (100 ng) and purified recombinant p79-p96 complex (10 ng). 32P-labeled double-stranded DNA substrates used in each lane are indicated, and the nicking assays were analyzed as described in the legend to Fig. 3. The reaction products (NS1-ori) are covalent NS1-DNA complexes. Lanes 1 and 2, samples in which the probe was incubated alone or in the presence of only NS1, respectively. (B, bottom) Autoradiograph of an EMSA gel demonstrating the binding of purified recombinant p79-p96 complex (2 ng) to the 32P-labeled mutant origin substrates. Under these assay conditions, the shifted reaction products (PIF:ori complex) are not covalently modified.
The PIF p79 subunit, but not the p96 subunit, activates the NS1 nickase.
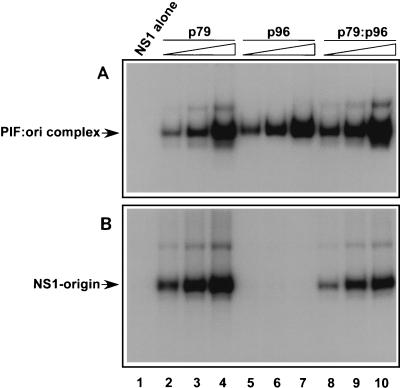
The PIF gene products isolated from HeLa cells, or from recombinant baculoviruses, preferentially form heterodimers which bind the minimal origin, activating the nickase function of NS1, and hence are presumably essential for MVM replication. However, p79 and p96 are expressed from unlinked genes, and their transcripts accumulate at different ratios in different cell types (34; Christensen et al., unpublished). Thus, if only heterodimers could support initiation, differential expression of the two subunits might be a determinant of cell-specific susceptibility to MVM. Using recombinant proteins, we have shown here that p79 and p96 can homodimerize when expressed alone and that each homodimer can bind the minimal origin (Fig. 2B, lanes 11 and 12). This finding has allowed us to dissect the individual roles of each polypeptide in activating NS1. The ability of increasing concentrations of p79 and p96 homodimers and p79-p96 heterodimers to bind the TC origin was monitored by EMSA, as shown in Fig. 7A. Homodimers comprising either p79 or p96 bound the minimal origin with efficiencies that were comparable to that of the heterodimer (Fig. 7A). However, as shown in Fig. 7B, only complexes that contained p79 (heterodimers or homodimers) were able to activate nicking. No nicking was observed, at any input tested, when p96 homodimer was complexed with NS1 and oriL DNA, even though efficient p96 DNA binding was observed throughout this concentration range. Neither p79 nor p96 homodimers supported NS1-mediated nicking of the inactive GAA origin (data not shown).
FIG. 7.
The PIF p79 subunit activates the NS1 nickase. (A) Autoradiograph of EMSA gels demonstrating the binding of purified GST-tagged NS1 (100 ng) alone and of increasing amounts of purified recombinant p79 (0.9, 2.7, and 8.1 ng [lanes 2 to 4, respectively]), p96 (0.9, 2.7, and 8.1 ng [lanes 5 to 7, respectively]), or p79-p96 heterocomplexes (0.9, 2.7, and 8.1 ng [lanes 8 to 10, respectively]) to a 32P-labeled duplex oligonucleotide probe containing the PIF recognition site in oriL. Shifted reaction products (PIF:ori complex) are not covalently modified. (B) Autoradiograph of gels from nicking assays performed in the presence of purified GST-tagged NS1 (100 ng) alone and in the presence of increasing amounts of purified recombinant homodimers and heterodimers, as in the top panel. The nicking assay was performed as described in the legend to Fig. 3, using a 32P-labeled DNA fragment containing oriLTC. Reaction products (NS1-origin) represent covalent NS1-DNA complexes.
DISCUSSION
Many eukaryotic origins function poorly, if at all, unless supplemented by one or more auxiliary transcription factor binding site, but the molecular mechanisms by which these factors potentiate replication are diverse. In some cases they have been shown to change the activity of an initiator protein, recruit replication proteins for the initiation complex, or change the nucleosomal architecture of the origin by allowing it to bend or loop (reviewed in reference 19). In the MVM left origin, the viral initiator nickase, NS1, is activated by a cellular heteromeric protein complex, PIF, both subunits of which belong to the emerging KDWK transcription factor family. As part of the present study, we have shown that purified rPIF heterodimers efficiently reconstitute origin binding and nicking reactions, with the same sequence specificity and constraints as the purified HeLa complex, and thus confirm that we have identified and cloned all components of this essential cofactor.
NS1 resembles other members of the RCR initiator protein superfamily in that it binds site specifically to sequences present in the viral origins, nicks single-stranded DNA at an adjacent site, and shares certain protein consensus catalytic site motifs. It is unusual, however, because it has also become modified to fulfill multiple roles in the viral life cycle, and to this end, it binds site specifically at many other positions in replicative-form viral DNA (3; S. F. Cotmore and P. Tattersall, unpublished data). Presumably as a consequence of this promiscuity, its nickase function is kept strictly suppressed when bound at most sites in the genome, and it becomes activated at the viral origins only in the presence of specific cellular cofactors. Surprisingly, although both MVM hairpins contain sequences that are capable of serving as NS1-dependent origins, the structure of these two origins and the types of interaction used to activate NS1 at the two ends of the genome appear to be very different. Thus, NS1 can nick oriR in its imperfectly base-paired hairpin configuration with the help of a sequence nonspecific DNA-bending protein from the high-mobility group 1 (HMG1) family (18). However, NS1 can nick oriL only after the left hairpin has been opened and copied into a fully base-paired, linear duplex, where it requires activation by the very unusual PIF heterocomplex.
In contrast to the results reported here for PIF, there is no evidence that HMG1 potentiates NS1 binding to a single site in the DNA at the right hairpin. Instead, it changes the structure of the NS1-DNA complex by interacting with, and coordinating, two separate NS1 complexes bound to specific binding sites spaced at opposite ends of the structure, some 100 nucleotides apart. This interaction creates a double-helical loop at a specific site in the origin stem that appears to be necessary for initiation. This distortion of the DNA suggests that a specific three-dimensional nucleoprotein complex has been established in which all constituents are stabilized by their interactions with each other and the active site of the nickase has become accessible (11). The use of HMG1 as a cofactor for replication initiation has also been described in the hairpin origins of another parvovirus, adeno-associated virus 2, although in this case HMG potentiates but is not absolutely required for initiation, at least under particular reaction conditions (9).
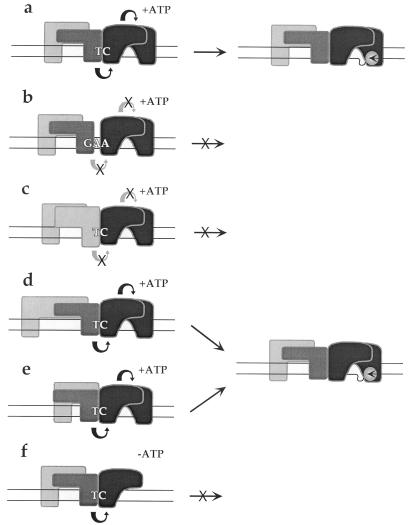
As discussed above, the active MVM left origin, oriLTC, is embedded in one arm of a palindromic dimer bridge sequence which spans adjacent genomes in a duplex MVM replication intermediate. The other arm of the bridge contains a single additional nucleotide in the bubble sequence, which we have shown previously (16) renders oriLGAA inactive. While individually PIF and NS1 bind equally well to both forms of oriL, we show here that together they bind only oriLTC in a cooperative fashion, interfacing across the TC bubble sequence, as diagrammed in Fig. 8a. In this context they form a higher-affinity ternary complex that effectively protects all of the minimal origin from DNase I digestion. Cooperative binding of replication initiator proteins and sequence-specific cofactors at replication origins has been observed with many other DNA viruses and appears to be a common mechanism for enhancing the stability or activity of initiation complexes. Thus, for example, binding of E1, the virus-encoded initiator protein of bovine papillomavirus, is enhanced approximately 40-fold in the presence of the E2 viral gene product (31). Likewise the polyomavirus initiator, large T antigen, shows enhanced binding in the presence of the cellular transcription factor AP1 (21, 22), and the terminal protein-polymerase complex of adenovirus binds cooperatively at the origin with the cellular transcription factor Oct-1 (32). The cooperative molecular interplay between PIF, NS1, and oriLTC is unusual because the spatial constraints are so stringent that insertion of even a single extra nucleotide between the two binding sites is sufficient to totally prevent complex formation (Fig. 8b). This observation therefore explains earlier mutagenesis studies showing that the number, rather than the sequence, of nucleotides in the bubble spacer region was critical for initiation (16).
FIG. 8.
Proposed interactions of PIF and NS1 at different forms of oriL. PIF subunits p79 and p96 are shown as dark and light gray boxes, respectively, while the subunits of the putative NS1 dimer are depicted in black. Functional intersubunit interactions are depicted as curved arrows (black arrows, interactions which occur; grey arrows with X, interactions which fail to occur). Potential substrate complexes are shown on the left, and the nicked strand with its covalent NS1-oriL product is shown on the right.
During the initiation process a tight interface and/or steric alignment between PIF and NS1 over the dinucleotide bubble sequence appears to be necessary, suggesting that direct protein-protein interactions may be involved. As we have shown in this paper, the p79 subunit, acting as a homodimer, is sufficient to activate nicking (Fig. 8c). Preliminary functional dissection of the PIF subunits indicates that a specific string of nonconserved amino acids in the otherwise conserved KDWK DNA-binding domain is responsible for the ability of p79 to cooperate with NS1 (Christensen et al., unpublished). This suggests that the critical interaction, or signal, between p79 and NS1 required to activate nicking (Fig. 8) takes place on, or very close to, the DNA. Moreover, our analysis of mutant origins in which the NS1-proximal PIF half-site was kept constant while insertions and deletions were introduced between the distal and proximal half-sites (Fig. 8d and e) indicates that only the position of the proximal ACGT site is important. Since PIF is still able to activate NS1 to nick origins in which the spacing between its two half-sites varies from 1 to 15 nucleotides, we conclude (i) that the distal PIF half-site is simply required to stabilize binding of the complex to the origin and (ii) that the disposition of most of the PIF heterodimer around the DNA helix is of little, if any, importance. Taken together, these observations strongly suggest that the PIF-NS1 interaction is a highly localized event, which may well be restricted to a p79 DNA-binding domain occupying the proximal site.
Although the level of cooperativity observed in the protection assays was modest, such assays are carried out under relatively fixed conditions and may inadequately reflect the changing environment of the host cell during viral infection. For example, in vitro NS1 can be shown to bind site specifically to DNA only in the presence of ATP or one of its nonhydrolyzable analogues, and the available evidence suggests that this is because ATP promotes oligomerization and only oligomeric forms of NS1 complex with the DNA in a way which is stable enough to survive purification. In the presence of ATP, the DNA sequence protected by NS1 is approximately 43 nucleotides long, extending from its specific (ACCA)2 recognition element to a region downstream of the nick site (10). However, here we show that PIF can mediate the binding of NS1 to the TC origin in the absence of ATP and that under these conditions the NS1 footprints are much smaller and are confined to its (ACCA)2 recognition motif. This suggests that PIF may allow NS1 to bind DNA stably as monomers or lower-order oligomers, as suggested in Fig. 8f, and may facilitate the retention of NS1 in the complex following ATP hydrolysis. While origin binding does not require ATP hydrolysis, covalent attachment of NS1 during the nicking reaction does, presumably to fuel the unwinding activity needed to present the nick site to the nickase as single-stranded DNA. Thus, in the absence of PIF, the necessary ATP hydrolysis might be expected to cause NS1 to dissociate from the DNA, terminating the initiation process. Accordingly, it has been observed that artificial MVM origin substrates which present the nick site in a single-stranded form, and thus eliminate the need for NS1 helicase activity, can be nicked in the absence of both ATP hydrolysis and PIF, albeit with limited efficiency (J. P. F. Nuesch, J. Christensen, and J. Rommelaere, submitted for publication).
The data presented here therefore suggest a model for replication initiation on oriLTC that involves coordinate binding of NS1 to a p79 PIF subunit across the TC-containing intersite duplex and possible retention of at least one NS1 molecule following completion of the nicking reaction. Whether these ternary complexes dissociate after nicking occurs or are necessary for subsequent events such as recruitment of the DNA polymerase and its auxiliary factors to the new 3′ end or unwinding of the nontemplate strand to allow fork progression will be the focus of future studies.
ACKNOWLEDGMENTS
We thank Ulla Toftegaard for excellent technical assistance.
This work was supported by U.S. Public Health Service grant AI26109 (to P.T.) from the National Institutes of Health. J.C. was supported by grants from the Danish Center for Biotechnology and the Royal Veterinary and Agricultural University Research Council.
REFERENCES
- 1.Astell C R, Chow M B, Ward D C. Sequence analysis of the termini of virion and replicative forms of minute virus of mice DNA suggests a modified rolling hairpin model for autonomous parvovirus DNA replication. J Virol. 1985;54:171–177. doi: 10.1128/jvi.54.1.171-177.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baldauf A Q, Willwand K, Mumtsidu E, Nüesch J P, Rommelaere J. Specific initiation of replication at the right-end telomere of the closed species of minute virus of mice replicative-form DNA. J Virol. 1997;71:971–980. doi: 10.1128/jvi.71.2.971-980.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Christensen J, Cotmore S F, Tattersall P. Minute virus of mice transcriptional activator protein NS1 binds directly to the transactivation region of the viral P38 promoter in a strictly ATP-dependent manner. J Virol. 1995;69:5422–5430. doi: 10.1128/jvi.69.9.5422-5430.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Christensen J, Cotmore S F, Tattersall P. A novel cellular site-specific DNA-binding protein cooperates with the viral NS1 polypeptide to initiate parvovirus DNA replication. J Virol. 1997;71:1405–1416. doi: 10.1128/jvi.71.2.1405-1416.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Christensen J, Cotmore S F, Tattersall P. Parvovirus initiation factor PIF: a novel human DNA-binding factor which coordinately recognizes two ACGT motifs. J Virol. 1997;71:5733–5741. doi: 10.1128/jvi.71.8.5733-5741.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Christensen J, Cotmore S F, Tattersall P. Two new members of the emerging KDWK family of combinatorial transcription modulators bind as a heterodimer to flexibly spaced PuCGPy half-sites. Mol Cell Biol. 1999;19:7741–7750. doi: 10.1128/mcb.19.11.7741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Christensen J, Pedersen M, Aasted B, Alexandersen S. Purification and characterization of the major nonstructural protein (NS-1) of Aleutian mink disease virus. J Virol. 1995;69:1802–1809. doi: 10.1128/jvi.69.3.1802-1809.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Christensen J, Storgaard T, Bloch B, Alexandersen S, Aasted B. Expression of Aleutian mink disease parvovirus proteins in a baculovirus vector system. J Virol. 1993;67:229–238. doi: 10.1128/jvi.67.1.229-238.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Costello E, Saudan P, Winocour E, Pizer L, Beard P. High mobility group chromosomal protein 1 binds to the adeno-associated virus replication protein (Rep) and promotes Rep-mediated site-specific cleavage of DNA, ATPase activity and transcriptional repression. EMBO J. 1997;16:5943–5954. doi: 10.1093/emboj/16.19.5943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cotmore S F, Christensen J, Nüesch J P, Tattersall P. The NS1 polypeptide of the murine parvovirus minute virus of mice binds to DNA sequences containing the motif [ACCA]2–3. J Virol. 1995;69:1652–1660. doi: 10.1128/jvi.69.3.1652-1660.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cotmore S F, Christensen J, Tattersall P. Two widely spaced initiator binding sites create an HMG1-dependent parvovirus rolling-hairpin replication origin. J Virol. 2000;74:1332–1341. doi: 10.1128/jvi.74.3.1332-1341.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cotmore S F, Nüesch J P, Tattersall P. In vitro excision and replication of 5′ telomeres of minute virus of mice DNA from cloned palindromic concatemer junctions. Virology. 1992;190:365–377. doi: 10.1016/0042-6822(92)91223-h. [DOI] [PubMed] [Google Scholar]
- 13.Cotmore S F, Nüesch J P, Tattersall P. Asymmetric resolution of a parvovirus palindrome in vitro. J Virol. 1993;67:1579–1589. doi: 10.1128/jvi.67.3.1579-1589.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cotmore S F, Tattersall P. The autonomously replicating parvoviruses of vertebrates. Adv Virus Res. 1987;33:91–174. doi: 10.1016/s0065-3527(08)60317-6. [DOI] [PubMed] [Google Scholar]
- 15.Cotmore S F, Tattersall P. In vivo resolution of circular plasmids containing concatemer junction fragments from minute virus of mice DNA and their subsequent replication as linear molecules. J Virol. 1992;66:420–431. doi: 10.1128/jvi.66.1.420-431.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cotmore S F, Tattersall P. An asymmetric nucleotide in the parvoviral 3′ hairpin directs segregation of a single active origin of DNA replication. EMBO J. 1994;13:4145–4152. doi: 10.1002/j.1460-2075.1994.tb06732.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cotmore S F, Tattersall P. DNA replication in the autonomous parvoviruses. Semin Virol. 1995;6:271–281. [Google Scholar]
- 18.Cotmore S F, Tattersall P. High-mobility group 1/2 proteins are essential for initiating rolling-circle-type DNA replication at a parvovirus hairpin origin. J Virol. 1998;72:8477–8484. doi: 10.1128/jvi.72.11.8477-8484.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DePamphilis M L. Origins of DNA replication. In: DePamphilis M L, editor. DNA replication in eukaryotic cells. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 45–86. [Google Scholar]
- 20.Gross C T, McGinnis W. DEAF-1, a novel protein that binds an essential region in a Deformed response element. EMBO J. 1996;15:1961–1970. [PMC free article] [PubMed] [Google Scholar]
- 21.Guo W, Tang W J, Bu X, Bermudez V, Martin M, Folk W R. AP1 enhances polyomavirus DNA replication by promoting T-antigen-mediated unwinding of DNA. J Virol. 1996;70:4914–4918. doi: 10.1128/jvi.70.8.4914-4918.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ito K, Asano M, Hughes P, Kohzaki H, Masutani C, Hanaoka F, Kerppola T, Curran T, Murakami Y, Ito Y. c-Jun stimulates origin-dependent DNA unwinding by polyomavirus large T antigen. EMBO J. 1996;15:5636–5646. [PMC free article] [PubMed] [Google Scholar]
- 23.Kaul S, Blackford J A, Chen J, Ogryzko V V, Simons S S. Properties of the glucocorticoid modulatory element binding proteins GMEB-1 and -2: potential new modifiers of glucocorticoid receptor transactivation and members of the family of KDWK proteins. Mol Endocrinol. 2000;14:1010–1027. doi: 10.1210/mend.14.7.0494. [DOI] [PubMed] [Google Scholar]
- 24.Kornberg A, Baker T A. DNA replication. 2nd ed. New York, N.Y: W. H. Freeman; 1991. [Google Scholar]
- 25.Maxam A M, Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65:499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- 26.Naeger L K, Cater J, Pintel D J. The small nonstructural protein (NS2) of the parvovirus minute virus of mice is required for efficient DNA replication and infectious virus production in a cell-type-specific manner. J Virol. 1990;64:6166–6175. doi: 10.1128/jvi.64.12.6166-6175.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oshima H, Simons S S., Jr Modulation of transcription factor activity by a distant steroid modulatory element. Mol Endocrinol. 1992;6:416–428. doi: 10.1210/mend.6.3.1584217. [DOI] [PubMed] [Google Scholar]
- 28.Oshima H, Szapary D, Simons S S., Jr The factor binding to the glucocorticoid modulatory element of the tyrosine aminotransferase gene is a novel and ubiquitous heteromeric complex. J Biol Chem. 1995;270:21893–21901. doi: 10.1074/jbc.270.37.21893. [DOI] [PubMed] [Google Scholar]
- 29.Rhode S L., III trans-activation of parvovirus P38 promoter by the 76K noncapsid protein. J Virol. 1985;55:886–889. doi: 10.1128/jvi.55.3.886-889.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rhode S L., III Both excision and replication of cloned autonomous parvovirus DNA require the NS1 (rep) protein. J Virol. 1989;63:4249–4256. doi: 10.1128/jvi.63.10.4249-4256.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seo Y S F, Muller, Lusky M, Gibbs E, Kim H Y, Phillips B, Hurwitz J. Bovine papilloma virus (BPV)-encoded E2 protein enhances binding of E1 protein to the BPV replication origin. Proc Natl Acad Sci USA. 1993;90:2865–2869. doi: 10.1073/pnas.90.7.2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van Leeuwen H C, Rensen M, van der Vliet P C. The Oct-1 POU homeodomain stabilizes the adenovirus preinitiation complex via a direct interaction with the priming protein and is displaced when the replication fork passes. J Biol Chem. 1997;272:3398–3405. doi: 10.1074/jbc.272.6.3398. [DOI] [PubMed] [Google Scholar]
- 33.Zeng H, Jackson D A, Oshima H, Simons S S., Jr Cloning and characterization of a novel binding factor (GMEB-2) of the glucocorticoid modulatory element. J Biol Chem. 1998;273:17756–17762. doi: 10.1074/jbc.273.28.17756. [DOI] [PubMed] [Google Scholar]
- 34.Zeng H, Kaul S, Simons S S., Jr Genomic organization of human GMEB-1 and rat GMEB-2: structural conservation of two multifunctional proteins. Nucleic Acids Res. 2000;28:1819–1829. doi: 10.1093/nar/28.8.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]