Abstract
In healthcare, integration of artificial intelligence (AI) holds strong promise for facilitating clinicians’ work, especially in clinical imaging. We aimed to assess the impact of AI implementation for medical imaging on efficiency in real-world clinical workflows and conducted a systematic review searching six medical databases. Two reviewers double-screened all records. Eligible records were evaluated for methodological quality. The outcomes of interest were workflow adaptation due to AI implementation, changes in time for tasks, and clinician workload. After screening 13,756 records, we identified 48 original studies to be incuded in the review. Thirty-three studies measured time for tasks, with 67% reporting reductions. Yet, three separate meta-analyses of 12 studies did not show significant effects after AI implementation. We identified five different workflows adapting to AI use. Most commonly, AI served as a secondary reader for detection tasks. Alternatively, AI was used as the primary reader for identifying positive cases, resulting in reorganizing worklists or issuing alerts. Only three studies scrutinized workload calculations based on the time saved through AI use. This systematic review and meta-analysis represents an assessment of the efficiency improvements offered by AI applications in real-world clinical imaging, predominantly revealing enhancements across the studies. However, considerable heterogeneity in available studies renders robust inferences regarding overall effectiveness in imaging tasks. Further work is needed on standardized reporting, evaluation of system integration, and real-world data collection to better understand the technological advances of AI in real-world healthcare workflows. Systematic review registration: Prospero ID CRD42022303439, International Registered Report Identifier (IRRID): RR2-10.2196/40485.
Subject terms: Medical imaging, Health services
Introduction
With a rising number of patients and limited staff available, the need for changes in healthcare is a pressing issue1. Artificial intelligence (AI) technologies promise to alleviate the current burden by taking over routine tasks, such as monitoring patients, documenting care tasks, providing decision support, and prioritizing patients by analyzing clinical data2,3. AI-facilitated innovations are claimed to significantly reduce the workload of healthcare professionals4,5.
Several medical specialties have already introduced AI into their routine work, particularly in data-intensive domains, such as genomics, pathology, and radiology4. In particular, image-based disciplines have seen substantial benefits from the pattern recognition abilities of AI, positioning them at the forefront of AI integration in clinical care3,6. AI technologies expedite the processing of an increasing number of medical images, being used for detecting artifacts, malignant cells or other suspicious structures, and optionally for the succeeding prioritization of patients7–9.
To successfully adopt AI in everyday clinical practice, different ways for effective workflow integration can be conceived, largely depending on the specific aim, that is, enhancing the quality of diagnosis, providing reinsurance, or reducing human workload10,11. Efficiency outcomes related to AI implementation include shorter reading times or a reduced workload of clinicians to meet the growing demand for interpreting an increasing number of images12–14. Thus, whether AI fulfills these aims and enables higher efficiency in everyday clinical work remains largely unknown.
Healthcare systems are complex, combining various components and stakeholders that interact with each other15. While the success of AI technology implementation highly depends on the setting, processes, and users, current studies largely focus on the technical features and capabilities of AI, not on its actual implementation and consequences in the clinical landscape2,3,6,16,17. Therefore, this systematic review aimed to examine the influence of AI technologies on workflow efficiency in medical imaging tasks within real-world clinical care settings to account for effects that stem from the complex and everyday demands in real-world clinical care, all not being existent in experimental and laboratory settings18.
Results
Study selection
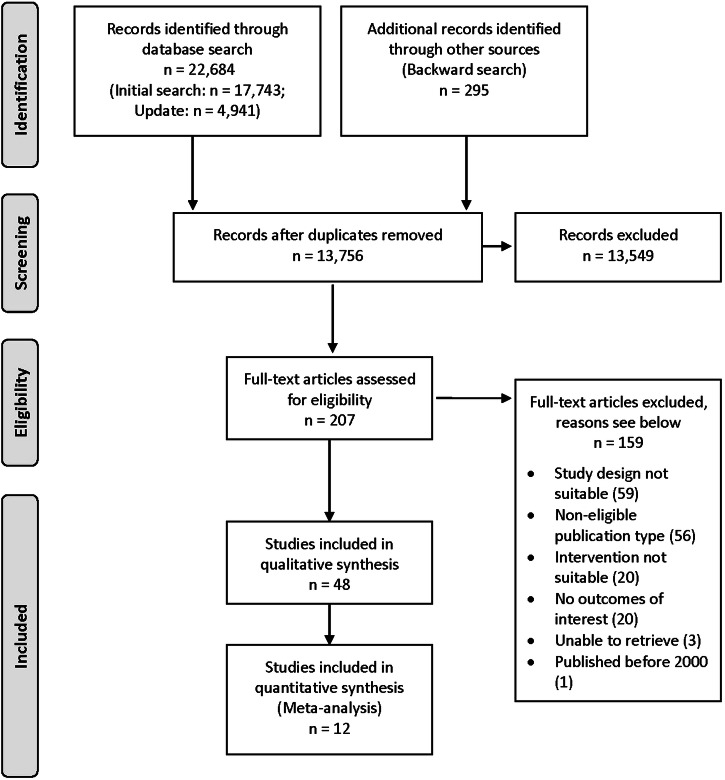
We identified 22,684 records in databases and an additional 295 articles through backward search. After the removal of duplicates, the 13,756 remaining records were included in the title/abstract screening. Then, 207 full texts were screened, of which 159 were excluded primarily because of inadequate study designs or not focusing on AI for interpreting imaging data (Supplementary Table 1). Finally, 48 studies were included in the review and data extraction. Twelve studies underwent additional meta-analyses. A PRISMA flow chart is presented in Fig. 1.
Fig. 1. PRISMA flowchart.
Visual representation of the search strategy, data screening and selection process of this systematic review.
Study characteristics
Of the 48 extracted studies, 30 (62.5%) were performed in a single institution, whereas the 18 (37.5%) remaining studies were multicenter studies. One study was published in 2010, another in 2012, and all other included studies were published from 2018 onward. Research was mainly conducted in North America (n = 21), Europe (n = 12), Asia (n = 11), and Australia (n = 3). Furthermore, one study was conducted across continents. The included studies were stemming from the medical departments of radiology (n = 26), gastroenterology (n = 6), oncology (n = 4), emergency medicine (n = 4), ophthalmology (n = 4), human genetics (n = 1), nephrology (n = 1), neurology (n = 1), and pathology (n = 1). Most studies used computed tomography (CT) for imaging, followed by X-ray and colonoscopes. The most prominent indications were intracranial hemorrhage, followed by pulmonary embolism, and cancer screening. Table 1 presents the key characteristics of all included studies.
Table 1.
Key characteristics of included studies
| Study | Year | Country | Setting | Medical specialty | Number of professionals | Imaging modality | Number of cases/ patients/ scans |
|---|---|---|---|---|---|---|---|
| Arbabshirani et al.7 | 2018 | USA | Single-Center | Radiology | 1 | CT | 347 patients |
| Batra et al.34 | 2023 | USA | Single-Center | Radiology | 32 | CT | 2501 examinations of 2197 patients |
| Carlile et al.80 | 2020 | USA | Multi-Center | ED | 112 | X-Ray | 1855 scans, survey on 202 scans |
| Cha et al.38 | 2021 | USA | Single-Center | Oncology | 18 | CT | 173 patients |
| Cheikh et al.81 | 2022 | France | Multi-Center | ED | 79 | CT | 7323 examinations |
| Chen et al.53 | 2022 | China | Multi-Center | Radiology | 4 | CT | 85 patients |
| Conant et al.28 | 2019 | USA | Single-Center | Radiology | 24 | DBT | 260 cases |
| Davis et al.39 | 2022 | USA | Multi-Center | Radiology / ED | NI | CT | 50,658 cases |
| Diao et al.20 | 2022 | China | Multi-Center | Radiology | 7 | CT | 251 patients |
| Duron et al.21 | 2021 | France | Multi-Center | Radiology / ED | 12 | X-Ray | 600 cases |
| Elijovich et al.82 | 2022 | USA | Multi-Center | Neurology | NI | CT | 680 patients |
| Ginat83 | 2021 | USA | Single-Center | Radiology | 5 | CT | 8723 scans |
| Hassan et al.40 | 2022 | USA | Single-Center | Radiology / Neurology | NI | CT | 63 patients |
| Hong et al.84 | 2022 | South Korea | Single-Center | Radiology | 60 | X-Ray | 1352 chest radiographs of 1319 patients |
| Jones et al.85 | 2021 | Australia | Multi-Center | Radiology | 11 | X-Ray | 2972 scans of 2665 patients |
| Kanagasingam et al.22 | 2018 | Australia | Single-Center | Ophthalmology | 4 | Photographs | 386 images of 216 patients |
| Kiljunen et al.86 | 2020 | Finland/ Estonia/ Singapore | Multi-Center | Oncology | 13 | CT | 45 scans of 30 patients |
| Ladabaum et al.41 | 2023 | USA | Multi-Center | Gastroenterology | 52 | Colonoscopy | 2329 patients |
| Levy et al.87 | 2022 | Israel | Single-Center | Gastroenterology | 30 | Colonoscopy | 4414 patients |
| Liu et al.35 | 2022 | China | Multi-Center | Ophthalmology | 2 | OCT | 1257 patients |
| Marwaha et al.88 | 2021 | Canada | Single-Center | Human Genetics | 15 | Photographs | 72 patients |
| Mueller et al.8 | 2022 | Denmark | Single-Center | Radiology | 2 | CT | 90 scans |
| Nehme et al.29 | 2023 | USA | Single-Center | Gastroenterology | 39 | Colonoscopy | 1041 patients |
| O’Neill et al.89 | 2021 | USA | Single-Center | Radiology | NI | CT | 6696 cases |
| Oppenheimer et al.90 | 2023 | Germany | Single-Center | Radiology | 2 | X-Ray | 1163 exams of 735 patients |
| Pierce et al.19 | 2021 | USA | Single-Center | Radiology | NI | X-Ray | 30,847 examinations |
| Potrezke et al.54 | 2023 | USA | Single-Center | Nephrology | 12 | MRI | 170 cases of 161 patients |
| Quan et al.91 | 2022 | USA | Multi-Center | Gastroenterology | 6 | Colonoscopy | 600 patients |
| Raya-Povedano et al.36 | 2021 | Spain | Single-Center | Radiology | 5 | DM/DBT | 15,986 patients |
| Repici et al.24 | 2020 | Italy | Multi-Center | Gastroenterology | 6 | Colonoscopy | 685 patients |
| Ruamviboonsuk et al.92 | 2022 | Thailand | Multi-Center | Ophthalmology | 12 | Photographs | 7651 patients |
| Sandbank et al.93 | 2022 | Israel | Single-Center | Pathology | NI | Microscope | 5954 cases |
| Schmuelling et al.94 | 2021 | Switzerland | Single-Center | Radiology | 3 | CT | 1808 scans of 1770 patients |
| Seyam et al.95 | 2022 | Switzerland | Single-Center | Radiology | NI | CT | 4450 patients |
| Sim et al.96 | 2022 | Singapore | Single-Center | Radiology | NI | X-Ray | 9431 datasets |
| Strolin et al.97 | 2023 | Italy | Single-Center | Oncology | NI | CT | 111 patients |
| Sun et al.55 | 2022 | USA | Multi-Center | Radiology | NI | X-Ray | 5335 images |
| Tchou et al.31 | 2010 | USA | Single-Center | Radiology | 5 | DM | 267 cases |
| Tricarico et al.56 | 2022 | Italy | Single-Center | Radiology | NI | X-Ray | 2942 scans |
| Vassallo et al.32 | 2019 | Italy | Single-Center | Radiology | 3 | CT | 225 patients |
| Wang et al.26 | 2019 | China | Single-Center | Gastroenterology | 8 | Colonoscopy | 1058 patients |
| Wang et al.98 | 2020 | China | Multi-Center | Radiology | 2 | CT | 2120 patients |
| Wittenberg et al.33 | 2012 | Netherlands | Single-Center | Radiology | 6 | CT | 209 patients |
| Wong et al.99 | 2021 | Canada | Multi-Center | Oncology | 39 | CT | 606 radiotherapy plans |
| Wong et al.100 | 2023 | USA | Single-Center | Radiology | 17 | X-Ray | 214 scans |
| Yacoub et al.37 | 2022 | USA | Single-Center | Radiology | 3 | CT | 390 scans |
| Yang et al.101 | 2022 | China | Multi-Center | Ophthalmology | NI | Photographs | 1001 patients |
| Zia et al.30 | 2022 | Australia | Single-Center | Radiology | 49 | CT | 1446 scans |
ED Emergency Department, CT Computed Tomography, DBT Digital Breast Tomosynthesis, DM Digital Mammography, MRI Magnetic Resonance Imaging, OCT Optical Coherence Tomography.
Concerning the purpose of using AI tools in clinical work, we classified the studies into three main categories. First, five studies (10.4%) described an AI tool used for segmentation tasks (e.g., determining the boundaries or volume of an organ). Second, 25 studies (52.1%) used AI tools to examine detection tasks to identify suspicious cancer nodules or fractures. Third, 18 studies (37.5%) investigated the prioritization of patients according to AI-detected critical features (e.g., reprioritizing the worklist or notifying the treating clinician via an alert).
Regarding the AI tools described in the studies, 34 studies (70.8%) focused on commercially available solutions (Table 2). Only Pierce et al. did not specify which commercially available algorithm was used19. Thirteen studies (27.1%) used non-commercially available algorithms, detailed information on these algorithms is provided in Table 3. Different measures were used to evaluate the accuracy of these AI tools, including sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV), and area under the curve (AUC). Sensitivity and specificity were the most commonly reported measures (see Tables 2 and 3).
Table 2.
Overview of the commercial AI tools used in the included studies
| Source | Clearance | Body part | Purpose | Technology | Study | Sensitivity | Specificity | Processing time |
|---|---|---|---|---|---|---|---|---|
| Aidoc Medical, Tel Aviv, Israel/New York, NY, USA | FDA | Head | Prioritization | Convolutional neural network | Davis et al.39 | 95.0% | 99.0% | near real-time |
| Ginat 83 | 88.4% | 96.1% | 3 min | |||||
| O’Neill et al.89 | 95.0% | 99.0% | 30–45 sec | |||||
| Seyam et al.95 | 87.2% | 93.9% | NI | |||||
| Zia et al.30 | 85.7% | 96.8% | NI | |||||
| Aidoc Medical, Tel Aviv, Israel | CE, FDA | Chest | Prioritization | Convolutional neural network | Batra et al.34 | 83.3% | 97.1% | NI |
| Cheikh et al.81 | 92.6% | 95.8% | NI | |||||
| Schmuelling et al.94 | 79.6% | 95.0% | 12.6 min | |||||
| AITEM Solutions, Turin, Italy | NI | Chest | Prioritization | Convolutional neural network | Tricarico et al.56 | 78.2% | 64.2% | NI |
| Annalise AI, Sydney, Australia | Pre-existing regulatory approval | Chest | Detection | Convolutional neural network | Jones et al.85 | NI | NI | NI |
| Digital Diagnostics, Coralville, IA, USA | FDA | Eye | Prioritization | Deep learning and rule-based models | Kanagasingam et al.22 | NI | 92.0% | <3 min |
| EndoVigilant Inc., MD, USA | NI | Colon | Detection | NI | Quan et al.91 | 90.0% | 97.0% | 30 frames per sec |
| FDNA Inc., Sunrise, FL, USA | NI | Face | Detection | NI | Marwaha et al.88 | NI | NI | NI |
| Gleamer, Paris, France | NI | Whole body | Detection | Convolutional neural network | Duron et al.21 |
79.4% (reader + AI, patient-wise) |
93.6% (reader + AI, patient-wise) |
NI |
| Oppenheimer et al.90 | 86.9% | 84.7% | 3 min | |||||
| Hologic, Marlborough, MA, USA | NI | Breast | Detection | NI | Tchou et al.31 | NI | NI | NI |
| iCAD, Nashua, NH, USA | NI | Breast | Detection | Convolutional neural network | Conant et al.28 | 85.0% (reader + AI) | 69.6% (reader + AI) | NI |
| Infervision Technology Co., Ltd., Beijing, China | CE, FDA | Chest | Detection | Deep learning | Diao et al.20 | NI | NI | NI |
| Limbus AI, Regina, Saskatchewan, Canada | NI | Whole body | Segmentation | Deep learning | Wong et al.99 | NI | NI | NI |
| Lunit, Seoul, South Korea | NI | Chest | Detection | Deep learning | Hong et al.84 | 74.8% | 99.8% | NI |
| Medtronic, Minneapolis, MN, USA | FDA | Colon | Detection | NI | Ladabaum et al.41 | NI | NI | NI |
| Levy et al.87 | NI | NI | NI | |||||
| Nehme et al.29 | NI | NI | NI | |||||
| Repici et al.24 | 99.7% | NI | real-time | |||||
| MVision AI Oy, Helsinki, Finland | CE, FDA | Whole body | Segmentation | Convolutional neural network | Kiljunen et al.86 | NI | NI | NI |
| Strolin et al.97 | NI | NI | 2.3 min | |||||
| Philipps Healthcare, Best, The Netherlands | NI | Chest | Detection | NI | Wittenberg et al.33 | 96.0% | 22.0% | NI |
| ScreenPoint Medical, Nijmegen, The Netherlands | CE, FDA | Breast | Prioritization | Deep learning | Raya-Povedano et al.36 | 84.1% (reader + AI) | NI | NI |
| Shanghai Wision AI Co., Ltd., Shanghai, China | NI | Colon | Detection | Deep learning | Wang et al.26 | 94.4% per image | 95.9% per image | real-time |
| Shenzhen SiBright CO. Ltd., Shenzen, China | NIFDC | Eye | Detection | Ensemble of 3 convolutional neural networks | Yang et al.101 | 86.7% | 96.1% | 24 sec per eye |
| Siemens Healthcare, Erlangen, Germany | FDA | Chest | Detection | NI | Mueller et al.8 | NI | NI | NI |
| Yacoub et al.37 | NI | NI | NI | |||||
| Viz.ai, San Francisco, CA, USA | FDA | Head | Prioritization | NI | Elijovic et al.82 | 81.0% | NI | NI |
| Hassan et al.40 | 87.6% | 88.5% | NI |
NI No information, CE Conformité Européenne, FDA Food and Drug Administration, NIFDC National Institutes for Food and Drug Control.
Table 3.
Non-commercially available AI algorithms
| Study | Developers | Body part | Purpose | Technology | Sensitivity | Specificity | Processing time | Notes |
|---|---|---|---|---|---|---|---|---|
| Arbabshirani et al.7 | SD | Head | Prioritization | Convolutional neural network | 70.0% | 87.0% | 2.3 sec | |
| Carlile et al.80 | SD | Lung | Detection | Convolutional neural network | 82.8% | 72.6% | real-time | |
| Cha et al.38 | Elguindi et al.102 | Multiple | Segmentation | Deep learning | NI | NI | NI | |
| Chen et al.53 | SD | Head | Detection | Deep learning | 33.3% (reader + AI) | 91.5% (reader + AI) | NI | |
| Liu et al.35 | Wang et al.103 | Eye | Detection | Deep learning and rule decision models | 98.5% | 96.2% | 21.4 hours | |
| Potretzke et al.54 | SD | Kidney | Segmentation | NI | NI | NI | NI | |
| Ruamviboonsuk et al.92 | Gulshan et al.104 | Eye | Detection | Deep learning | 91.4% | 95.4% | real-time | Validated in Krause et al.105, Ruamviboonsuk et al.106 |
| Sandbank et al.93 | SD | Breast | Detection | Multilayered convolutional neural networks | 98.1% | 96.2% | real-time | |
| Sim et al.96 | SD | Lung | Detection | Deep learning | 78.8% | 97.1% | NI | |
| Sun et al.55 | SD | Lung | Detection | NI | 67.0% | 77.0% | real-time | |
| Vassallo et al.32 | Retico et al.107 | Lung | Detection | NI | 85.0% for lesions >3 mm | NI | 19 min | Validated in Torres et al.108 |
| Wang et al.98 | SD | Lung | Prioritization | U-Net-based deep learning model | 92.3% | 85.1% | 0.55 min | |
| Wong et al.100 | Brown et al.109 | Chest | Detection | Open-Source framework (SimpleMind) | 88.0% | NI | 3–4 min |
NI No information, SD Self-developed by authors, as described in respective publication.
In total only four studies followed a reporting guideline, three studies20–22 used Standards for Reporting of Diagnostic Accuracy (STARD) reporting guideline23 and Repici et al.24 followed the CONSORT guidelines for randomized controlled trials25. Only two studies24,26 pre-registered their protocol and none of the included studies provided or used an open-source available algorithm.
Appraisal of methodological quality
When assessing the methodological quality of the 45 non-randomized studies only one (2.2%) was rated with an overall “low” risk of bias. Four studies (8.9%) were rated “moderate”, 28 studies (62.2%) were rated “serious”, and 12 studies (26.7%) were rated “critical”. All three randomized studies were appraised with an overall high risk of bias. Summary plots of the risk of bias assessments are shown in Fig. 2, full assessments can be found in Supplementary Figs. 1 and 2. The assessment of the quality of reporting using the Methodological Index for Non-randomized Studies (MINORS) is included in Supplementary Figs. 3 and 4. Higher scores indicate higher quality of reporting, with the maximum score being 24 for comparative studies and 16 for non-comparative studies27. Comparative studies reported a Median of 9 of 12 criteria with a median overall score of 15 (range: 9–23) and noncomparative studies reported a Median of 7 of 8 checklist items, with a median overall score of 7 (range: 6–14).
Fig. 2. Quality assessment of included articles.
Summary plots of the risk of bias assessments via Risk of Bias in Non-randomized Studies of Interventions tool (ROBINS-I) for non-randomized studies and the Cochrane Risk of Bias tool (Rob 2) for randomized studies.
Outcomes
Of all included studies, 33 (68.8%) surveyed the effects of AI implementation on clinicians’ time for task execution. The most frequently reported outcomes included (1) reading time (i.e., time the clinicians required to interpret an image); (2) report turnaround time (i.e., the time from completing the scan until the report is finalized); and (3) total procedure time (i.e., the time needed for colonoscopy)28–30. Times were assessed via surveys, recorded by researchers or staff, retrieved via time stamps, or self-recorded. Seventeen studies did not describe how they obtained the reported times.
Regarding our research question, whether AI use improves efficiency, 22 studies (66.6%) reported a reduction in time for task completion due to AI use, with 13 of these studies proving the difference to be statistically significant (see Table 4). Eight studies (24.2%) reported that AI did not reduce the time required for tasks. The remaining three studies (9.1%) chose a design or implementation protocol in which the AI was used after the normal reading, increasing the task time measured by study design31–33.
Table 4.
Outcomes organized by time type measured
| Study | Time Type | Assessment Method | Statistical measures | Pre/Without AI | Post/With AI | Absolute Difference (%) | Significance | Workflow Adaptation |
|---|---|---|---|---|---|---|---|---|
| Batra et al.34 | Reading timea | Timestamps extracted from EMR and radiologist dictation system | Mean | 00:26:30 | 00:26:18 | −00:00:12 (−0.75%) | n.s. | Triage |
| Cheikh et al.81 | Reading timea | Survey | Mean (SD) | 00:14:33 (00:09:05) | 00:15:36 (00:09:46) | +00:01:03 ( + 7.22%) | *** | Triage |
| Chen et al.53 | Reading time | NI | Mean (SD) | 00:03:39 (00:00:24) | 00:02:56 (00:00:01) | −00:00:43 (−19.77%) | * | NI |
| Conant et al.28 | Reading time | NI | Mean (CI) | 00:01:04 (00:00:25) | 00:00:30 (00:00:12) | −00:00:34 (−52.57%) | ** | Second reader, concurrent |
| Diao et al.20 | Reading time | Automatically recorded | Mean (SD) | 00:04:30 (00:02:24) | 00:03:43 (00:02:26) | −00:00:47 (−17.41%) | *** | Second reader, sequential |
| Duron et al.21 | Reading time | Automatically recorded | Mean | 00:01:07 | 00:00:57 | −00:00:10 (−14.93%) | n.s. | Second reader, concurrent |
| Mueller et al.8 | Reading time − resident | NI | Mean (SD) | 00:06:10 (00:02:49) | 00:07:17 (00:02:29) | +00:01:07 ( + 18.11%) | n.s. | Depending on radiologist’s choice |
| Mueller et al.8 | Reading time − consultant | NI | Mean (SD) | 00:06:06 (00:01:50) | 00:06:20 (00:02:01) | +00:00:14 ( + 3.83%) | n.s. | Depending on radiologist’s choice |
| O’Neill et al.89 | Reading timeb | NI | Median (CI) | 00:04:50 (00:00:27) | 00:06:14 (00:05:28) | +00:01:23 ( + 28.73%) | n.s. | Triage |
| Schmuelling et al.94 | Reading timea | Timestamps in the clinical information system | Mean (SD) | 01:25:30 (04:42:00) | 01:18:30 (04:33:00) | −00:07:00 (-8.19%) | n.s. | Triage |
| Vassallo et al.32 | Reading time | Recorded by investigator | Mean (SD) | 00:04:56 (00:01:20) | 00:05:29 (00:01:23) | +00:00:33 ( + 11.15%) | * | Sequential due to study design |
| Yacoub et al.37 | Reading time | Self-measured with digital stopwatch | Mean (SD) | 00:07:01 (00:02:55) | 00:05:28 (00:02:02) | −00:01:33 (−22.09%) | *** | Second reader, concurrent |
| Cha et al.38 | Contouring time | Self-report | Median (IQR) | 00:40:00 (00:43:00) | 00:28:00 (00:10:00) | −00:12:00 (−30.00%) | ** | First reader |
| Kiljunen et al.86 | Contouring time | NI | Mean | 00:27:00 | 00:15:00 | −00:12:00 (−44.44%) | NI | First reader |
| Strolin et al.97 | Contouring time | NI | Median (Range) | 00:25:00 (01:47:00) | 00:12:18 (00:46:54) | −00:12:42 (−50.80%) | *** | First reader |
| Potretzke et al.54 | Segmentation timec | .. | .. | .. | .. | .. | NI | First reader |
| Tchou et al.31 | Time to review AI results | Timestamp macro in Excel/ recording by investigator | Mean (SE) | 00:01:58 (00:00:04) | .. | 00:00:23d (00:00:02) | NI | Sequential due to study design |
| Wittenberg et al.33 | Time to review AI results | NI | Mean (Range) | 00:01:15 (00:01:02) | .. | 00:00:22d (00:00:18) | NI | Sequential due to study design |
| Arbabshirani et al.7 | Time to interpretationb | NI | Median (IQR) | 08:32:00 (01:51:00) | 00:19:00 (00:22:00) | −08:13:00 (−96.29%) | *** | Triage |
| Ginat83 | Wait time (ED cases)b | Automatically recorded | Mean (SD) | 01:25:00 (03:14:00) | 01:12:00 (02:57:00) | −00:13:00 (−15.29%) | n.s. | Triage |
| Ginat83 | Wait time (inpatient cases)b | Automatically recorded | Mean (SD) |
06:30:00 (06:08:00) |
05:52:00 (05:15:00) |
−00:38:00 (−9.74%) |
** | Triage |
| Ginat83 | Wait time (outpatient cases)b | Automatically recorded | Mean (SD) |
11:14:00 (13:45:00) |
01:10:00 (02:21:00) |
−10:04:00 (−89.61%) |
*** | Triage |
| O’Neill et al.89 | Wait timeb | NI | Median (CI) | 00:15:45 (00:00:46) | 00:12:01 (00:01:55) | −00:03:44 (−23.75%) | *** | Triage |
| Elijovich et al.82 | Time to notification | Retrospective documentation | Median (IQR) | 00:26:00 (00:14:00) | 00:07:00 (00:04:00) | −00:19:00 (−73.08%) | *** | Triage |
| Hong et al.84 | Time to treatment | Retrospectively through analysis of electronic medical records | Mean (SD) | 02:30:00 (03:24:00) | 01:12:00 (19:30:00) | −01:18:00 (−4.91%) | n.s. | Second reader, concurrent |
| Batra et al.34 | Report turnaround time | Timestamps | Mean | 00:59:54 | 00:47:36 | −00:12:18 (−20.53%) | *** | Triage |
| Davis et al.39 | Report turnaround timea | NI | Mean (SD) | 01:03:30 (01:02:36) | 00:52:30 (00:53:55) | −00:11:00 (−17.32%) | ** | Triage |
| Seyam et al.95 | Report turnaround timeb | Timestamps extracted from the electronic medical record and PACS | Mean (CI) | 01:00:00 (00:17:00) | 01:03:00 (00:11:00) | +00:03:00 ( + 5.00%) | NI | Triage |
| Sim et al.96 | Report turnaround time | Extracted timestamps from the hospital’s RIS | Mean | 00:09:00 | 00:07:00 | −00:02:00 (−22.22%) | NI | Triage |
| Zia et al.30 | Report turnaround timeb | NI | Mean (SD) | 01:06:42 (00:41:30) | 01:20:00 (01:04:24) | +00:13:18 ( + 19.94%) | * | Second reader, sequential |
| Schmuelling et al.94 | ED turnaround timea | Timestamps in the clinical information system | Mean (SD) | 02:06:00 (01:04:12) | 01:59:00 (01:41:00) | −00:07:00 (−5.56%) | n.s. | Triage |
| Hassan et al.40 | DIDO time at PSC | NI | Mean (SD) | 03:46:42 (04:02:54) | 02:04:24 (00:57:36) | −01:42:18 (−45.13%) | * | Triage |
| Yang et al.101 | Time for diagnosis | NI | Mean (SD) | 00:00:38 (00:00:32) | 00:00:24 (00:00:08) | −00:00:14 (−36.84%) | NI | NI |
| Ladabaum et al.41 | Withdrawal time | NI | Mean (CI) | 00:17:30 (00:01:30) | 00:18:00 (00:01:36) | +00:00:30 ( + 2.86%) | n.s. | NI |
| Nehme et al.29 | Withdrawal time | NI | Median (IQR) | 00:17:00 (00:15:00) | 00:18:00 (00:16:00) | +00:01:00 ( + 5.88%) | n.s. | NI |
| Repici et al.24 | Withdrawal time | Stopwatch | Mean (SD) | 00:07:15 (00:02:29) | 00:06:57 (00:01:41) | −00:00:18 (−4.14%) | n.s. | NI |
| Wang et al.26 | Withdrawal time | NI | Mean (SD) | 00:06:23 (00:01:13) | 00:06:53 (00:01:47) | +00:00:30 ( + 7.82%) | *** | Second reader, concurrent |
| Ladabaum et al.41 | Total procedure time | NI | Mean (CI) | 00:26:06 (00:01:36) | 00:26:42 (00:01:48) | +00:00:36 ( + 2.30%) | n.s. | NI |
| Levy et al.87 | Total procedure time | Recorded by endoscopy nurse | Median (IQR) | 00:24:00 (00:17:00) | 00:22:00 (00:12:00) | −00:02:00 (−8.33%) | *** | NI |
| Nehme et al.29 | Total procedure time | NI | Median (IQR) | 00:23:00 (00:16:00) | 00:24:00 (00:19:00) | +00:01:00 ( + 4.35%) | n.s. | NI |
| Quan et al.91 | Total procedure time | NI | Mean (SD) | 00:19:30 (00:07:12) | 00:21:24 (00:09:06) | +00:01:54 ( + 9.74%) | ** | NI |
| Wang et al.26 | Total procedure time | NI | Mean (SD) | 00:12:06 (00:04:05) | 00:12:31 (00:04:23) | +00:00:25 ( + 3.47%) | n.s. | Second reader, concurrent |
| Carlile et al.80 | .. | .. | .. | .. | .. | .. | .. | Second reader, concurrent |
| Jones et al.85 | .. | .. | .. | .. | .. | .. | .. | Second reader, concurrent |
| Kanagasingam et al.22 | .. | .. | .. | .. | .. | .. | .. | Triage + notification |
| Liu et al.35 | .. | .. | .. | .. | .. | .. | .. | Triage + notification |
| Marwaha et al.88 | .. | .. | .. | .. | .. | .. | .. | Sequential |
| Oppenheimer et al.90 | .. | .. | .. | .. | .. | .. | .. | Second reader, sequential |
| Pierce et al.19 | .. | .. | .. | .. | .. | .. | .. | Triage |
| Raya-Povedano et al.36 | .. | .. | .. | .. | .. | .. | .. | Gatekeeper |
| Ruamviboonsuk et al.92 | .. | .. | .. | .. | .. | .. | .. | Gatekeeper |
| Sandbank et al.93 | .. | .. | .. | .. | .. | .. | .. | Second reader, sequential |
| Sun et al.55 | .. | .. | .. | .. | .. | .. | .. | Second reader, sequential |
| Tricarico et al.56 | .. | .. | .. | .. | .. | .. | .. | Triage |
| Wang et al.98 | .. | .. | .. | .. | .. | .. | .. | Triage + notification |
| Wong et al.99 | .. | .. | .. | .. | .. | .. | .. | First reader |
| Wong et al.100 | .. | .. | .. | .. | .. | .. | .. | Second reader, concurrent |
n.s. Not significant, AI Artificial intelligence, CI 95% confidence interval, DIDO Door-in-door out time, ED Emergency department, EMR Electronic medical record, IQR Interquartile range, NI No information, PACS Picture archiving and communication system, PSC Primary stroke center, RIS Radiology information system, SD Standard deviation, SE Standard error.
Time formats are hh:mm:ss. *p < 0.05, **p < 0.01, ***p < 0.001.
a Time measurements for scans that have been classified positive for pulmonary embolism.
b Time measurements for scans that have been classified positive for intracranial hemorrhage.
c Potretzke et al. reported a reduction in segmentation time but no concrete numbers.
d Additional reading time for AI use.
For our meta-analyses, we established clusters with studies deploying similar methods, outcomes, and specific purposes. Concerning studies on detection tasks, we identified two main subgroups: studies using AI for interpreting CT scans (n = 7) and those using AI for colonoscopy (n = 6). Among studies using AI for interpreting CT images, a meta-analysis was performed for four studies reporting clinicians’ reading times. As shown in Fig. 3a, the reading times for interpreting CT images did not differ between the groups: standardized mean error (SMD): −0.60 (95% confidence interval, −2.02 to 0.82; p = 0.30). Furthermore, the studies showed significantly high heterogeneity: Q = 109.72, p < 0.01, I2 = 96.35%. This heterogeneity may be associated with the different study designs included or the risk of bias ratings, with only one study being rated having a low risk of bias. Furthermore, Mueller et al.8 reported no overall reading time but separated it for resident and attending physician, which we included separately in our meta-analysis. Concerning the use of AI for colonoscopy, five studies reported comparable measures. Our random effects meta-analysis showed no significant difference between the groups: SMD: −0.04 (95% CI, −0.76 to 0.67; p = 0.87), with significant heterogeneity: Q = 733.51, p < 0.01, I2 = 99.45% (Fig. 3b). Four of the included studies had a serious risk of bias, whereas one randomized study included was rated with a high risk of bias. Among 11 studies that reported AI use for the prioritization of patients’ scans, four measured the turnaround time. The study by Batra et al.34 did not report variance measures and was therefore excluded from the meta-analysis. The remaining three studies used the AI tool Aidoc (Tables 2 and 4) to detect intracranial hemorrhage and reported the turnaround time for cases flagged positive. The meta-analysis showed no significant difference in turnaround time between cases with and without AI use: SMD: 0.03 (95% CI, −0.50 to 0.56; p = 0.84), with a significant heterogeneity across studies: Q = 12.31, p < 0.01, I2 = 83.75% (Fig. 3c). All included studies were non-randomized studies, with two studies being rated with a serious risk of bias and one with a moderate risk of bias.
Fig. 3. Results of meta-analyses.
Graphical display and statistical results of the three meta-analyses: a Studies using AI for detection tasks in CT images and reported clinicians’ reading time. b Studies using AI to detect polyps during colonoscopy and measured the total procedure time. c Studies that used AI for reprioritization and measured the turnaround times for cases flagged positive. All included studies used AIDOC for intracranial hemorrhage detection.
In total, 37 studies reported details on the actual workflow adaptations due to AI implementation, which we classified into four main variants (depicted exemplarily in Fig. 4). 16 studies (43.2%) used an AI tool as a triage system, i.e., the AI tool reprioritized the worklist or the AI tool sent an alert to the clinician or referred the patient to a specialist for further examination (Fig. 4a: AI triage). In two studies (5.4%), the AI tool acted as a gatekeeper, only referring cases labeled as suspicious to the clinician for further review, while excluding the remaining cases (Fig. 4a: AI gatekeeper). In 13 studies (35.1%), AI tools were used as a second reader for detection tasks in two variants (Fig. 4b: AI second reader). Eight studies reported that the AI tool functioned as a second reader in a concurrent mode, presenting additional information during the task to clinicians (e.g., in colonoscopy studies, where the workflow remained the same as before displaying additional information during the procedure). Five studies described a workflow in which the AI tool was used additionally after the normal detection task, resulting in a sequential second reader workflow. In five segmentation studies (13.5%), the AI tool served as a first reader with the clinician reviewing and then correcting the AI-provided contours (Fig. 4c: AI first reader).
Fig. 4. Prototypical workflows after AI implementation.
Visual representation of the different workflows when using AI as reported in the included studies: a Workflows when using AI for prioritization tasks. b Workflow when using AI for detection. c Workflow when using AI for segmentation tasks. Figure created with Canva (Canva Pty Ltd, Sydney, Australia).
In a single study (2.7%), the type of actual workflow implementation was at the radiologist’s choice. Three studies used a study design with the AI tool as a second reader in a pre-specified reading sequence; therefore, we did not classify them as workflow adaptations. The remaining studies did not provide sufficient information on workflow implementation.
In our initial review protocol, we also aimed to include investigations on clinician workload14. Apart from three studies, Liu et al.35, Raya-Povedano et al.36, and Yacoub et al.37, which calculated the saved workload in scans or patients because of AI use, no other study reported AI implementation effects on clinicians’ workload (besides the time for tasks effects, see above). Other reported outcomes included evaluations of the AI performing the task (i.e., satisfaction)8,38; frequency of AI use29,30; patient outcomes, such as length of stay or in-hospital complications39,40; and sensitivity or specificity changes8,21,24,28,41.
Risk of bias across studies
Funnel plots for the studies included in the meta-analyses were created (Supplementary Figs. 5–7). 19 studies declared a relevant conflict of interest and six other studies had potential conflicts of interest, which sum up to more than 50% of the included studies.
Additionally, we ran several sensitivity analyses to evaluate for potential selection bias. We first searched the dblp computer science bibliography, yielding 1159 studies for title and abstract screening. Therein, we achieved perfect interrater reliability (100%). Subsequently, only thirteen studies proceeded to full-text screening, with just one meeting our review criteria. This study by Wismueller & Stockmaster42 was also part of our original search. Notably, this study was the only conference publication providing a full paper (refer to Supplementary Table 2).
Moreover, to ensure comprehensive coverage and to detect potentially missed publications due to excluding conference proceedings, we screened 2614 records from IEEE Xplore, MICCAI, and HICSS. Once again, our title and abstract screening demonstrated perfect interrater reliability (100%). However, despite including 31 publications in full-text screening, none met our inclusion criteria upon thorough assessment. Altogether, this additionally searches showed no significant indication for a potential selection bias and potentially missing out key work in other major scientific publication outlets.
Using AMSTAR-2 (A MeaSurement Tool to Assess Systematic Reviews)43, we rated the overall confidence in the results as low, mainly due to our decision to combine non-randomized and randomized studies within our meta-analysis (Supplementary Fig. 8).
Discussion
Given the widespread adoption of AI technologies in clinical work, our systematic review and meta-analysis assesses efficiency effects on routine clinical work in medical imaging. Although most studies reported positive effects, our three meta-analyses with subsets of comparable studies showed no evidence of AI tools reducing the time on imaging tasks. Studies varied substantially in design and measures. This high heterogeneity renders robust inferences. Although nearly 67% of time-related outcome studies have shown a decrease in time with AI use, a noteworthy portion of these studies revealed conflicts of interest, potentially influencing study design or outcome estimation44. Our findings emphasize the need for comparable and independent high-quality studies on AI implementation to determine its actual effect on clinical workflows.
Focusing on how AI tools were integrated into the clinical workflow, we discovered diverse adoptions of AI applications in clinical imaging. Some studies have provided brief descriptions that lack adequate details to comprehend the process. Despite predictions of AI potentially supplanting human readers or serving as gatekeepers, with humans primarily reviewing flagged cases to enhance efficiency10,11, we noted a limited adoption of AI in this manner across studies. In contrast, most studies reported AI tools as supplementary readers, potentially extending the time taken for interpretation when radiologists must additionally incorporate AI-generated results18,45. Another practice involved concurrent reading, which seems beneficial because it guides clinicians’ attention to crucial areas, which potentially improves reading quality and safety without lengthening reading times45,46. Regardless of how AI was used, a crucial factor is its alignment with the intended purpose and task15.
Although efficiency stands out in the current literature, we were also interested in whether AI affects clinicians’ workload, besides the time measurements, such as number of tasks or cognitive load. We only found three studies on AI’s impact on clinicians’ workload, but no study assessed workload separately (e.g., in terms of cognitive workload changes)18,35–37. This gap in research is remarkable since human–technology interaction and human factors assessment will be a success factor for the adoption of AI in healthcare47,48.
Our study included a vast variety of AI solutions reported in the publications. The majority was a large number of commercially available AI solutions which mostly had acquired FDA or CE clearance, ensuring safety of use in a medical context49. Nevertheless, it is desirable that future studies provide more detailed information about the accuracy of the AI solutions in their use case or processing times, which both can be crucial to AI adoption50. Regarding included studies which used non-commercially available algorithms, some of the studies did not specify the origin or source of the algorithm (i.e., developer). Especially with the specific characteristics and potential bias being introduced through the specific algorithm (e.g., for example stemming from a training bias or gaps in the underlying data), it is essential to provide information about the origins and prior validation steps of the algorithm in clinical use51,52. Interestingly, only four included studies discussed the possibility of bias in the AI algorithm53–56. Open science principles, such as data or code sharing, aid to mitigate the impact of bias. Yet, none of the studies in our review used open-source solutions or provided their algorithm52. Additionally, guidelines such as CONSORT-AI or SPIRIT-AI provide recommendations for the reporting of clinical studies using AI solutions57, as previous systematic reviews have also identified serious gaps in the reporting on clinical AI solutions58,59. Our results corroborate this shortcoming, as none of the studies reporting non-commercial algorithms and only four studies overall followed a reporting guideline. Notwithstanding, for some included studies, AI-specific reporting guidelines were published after their initial publication. Nevertheless, comprehensive and transparent reporting remains insufficient.
With our review, we were able to replicate some of the findings by Yin et al., who provided a first overview on AI solutions in clinical practice, e.g., insufficient reporting in included studies60. By providing time for tasks and meta-analyses as well as workflow descriptions our review substantially extends the scope of their review, providing a robust and detailed overview on the efficiency effects of AI solutions. In 2020, Nagendran et al. provided a review comparing AI algorithms for medical imaging and clinicians, concluding that only few prospective studies in clinical settings exist59. Our systematic review demonstrated an increase in real-world studies in previous years and provides an up-to-date and comprehensive overview on AI solutions currently used in medical imaging practice. Our study thereby addresses one of the previously mentioned shortcomings, that benefits of the AI algorithm in silico or in retrospective studies might not transfer into clinical benefit59. This is also recognized by Han et al.61 who evaluated randomized controlled trials evaluating AI in clinical practice and who argued that efficiency outcomes will strongly depend on implementation processes in actual clinical practice.
The complexities of transferring AI solutions from research into practice were explored in a review by Hua et al.62 who evaluated the acceptability AI for medical imaging by healthcare professionals. We believe that for AI to unfold its full potential, it is essential to pay thorough attention to the adoption challenges and work system integration in clinical workplaces. Notwithstanding the increasing number of studies on AI use in real-world settings during the last years, many questions on AI implementation and workflow integration remain unanswered. On the one hand, limited consideration prevails on acceptance of AI solutions by professionals62. Although studies even discuss the possibility of AI as a teammate in the future63,64, most available studies rarely include perceptions of affected clinicians60. On the other hand, operational and technical challenges as well as system integration into clinical IT infrastructures are major challenges, as many of the described algorithms are cloud-based. Smooth interoperability between new AI technologies and local clinical information systems as well as existing IT infrastructure is key to efficient clinical workflows50. For example, the combination of multimodal data, such as imaging and EHR data, could be beneficial for future decision processes in healthcare65.
Our review has several limitations. First, publication bias may have contributed to the high number of positive findings in our study. Second, despite searching multiple databases, selection bias may have occurred, particularly as some clinics implementing AI do not systematically assess or publish their processes in scientific formats60. Moreover, we excluded conference publications which could be the source for potential biases. Nevertheless, we ran different sensitivity analyses for publication and selection bias, and did not find evidence for major bias introduced due to our search and identification strategy. Yet, aside from one conference paper, all other conference publications merely provided abstracts or posters, lacking a comprehensive base for the extraction of required details. Third, we focused exclusively on medical imaging tasks to enhance the internal validity of clinical tasks across diverse designs, AI solutions, and workflows. Fourth, the low quality rating of our review on the AMSTAR-2 checklist, which is due to the diverse study designs we included, calling for more comparable high quality studies in this field. Nevertheless, we believe that our review provides a thorough summary of the available studies matching our research question. Finally, our review concentrated solely on efficiency outcomes stemming from the integration of AI into clinical workflows. Yet, the actual impact of AI algorithms on efficiency gains in routine clinical work can be influenced by further, not here specified local factors, e.g., existent IT infrastructure, computational resources, processing times. Next to the testing of the AI solutions under standardized conditions or in randomized controlled trials, which can indicate whether AI solution are suitable for the transfer into routine medical care, careful evaluations of how AI solutions fit into everyday clinical workflow should be expanded, i.e., ideally before implementation. Exploring adoption procedures along with identifying key implementation facilitators and barriers provides valuable insights into successful AI technology use in clinical routines. However, it is important to note that AI implementation can address a spectrum of outcomes, including but not limited to enhancing patient quality and safety, augmenting diagnostic confidence, and improving healthcare staff satisfaction8.
In conclusion, our review showed a positive trend toward research on actual AI implementation in medical imaging, with most studies describing efficiency improvements in course of AI technology implementation. We derive important recommendations for future studies on the implementation of AI in clinical settings. The rigorous use of reporting guidelines should be encouraged, as many studies reporting time outcomes did not provide sufficient details on their methods. Providing a protocol or clear depiction of how AI tools modify clinical workflows allows comprehension and comparison between pre- and post-adoption processes while facilitating learning and future implementation practice. Considering the complexity of healthcare systems, understanding the factors contributing to successful AI implementation is invaluable. Our review corroborates the need for comparable evaluations to monitor and quantify efficiency effects of AI in clinical real-world settings. Finally, future research should therefore explore success and potential differences between different AI algorithms in controlled trials as well as in real-world clinical practice settings to inform and guide future implementation processes.
Methods
Registration and protocol
Before its initiation, our systematic literature review was registered in a database (PROSPERO, ID: CRD42022303439), and the review protocol was peer-reviewed (International Registered Report Identifier RR2-10.2196/40485)14. Our reporting adheres to the Preferred Reporting Items for Systematic Review and Meta-Analysis (PRISMA) statement reporting guidelines (Supplementary Table 3). During the preparation of this work, we used ChatGPT (version GPT-3.5, OpenAI) to optimize the readability and wording of the manuscript. After using this tool, the authors reviewed and edited the content as required and take full responsibility for the content of the publication.
Search strategy and eligibility criteria
Articles were retrieved through a structured literature search in the following electronic databases: MEDLINE (PubMed), Embase, PsycINFO, Web of Science, IEEE Xplore, and Cochrane Central Register of Controlled Trials. We included original studies on clinical imaging, written in German or English, retrieved in full-text, and published in peer-reviewed journals from the 1st of January 2000 onward, which marked a new area of AI in healthcare with the development of deep learning14,66. The first search was performed on July 21st, 2022, and was updated on May 19th, 2023. Furthermore, a snowball search screening of the references of the identified studies was performed to retrieve relevant studies. Dissertations, conference proceedings, and gray literature were excluded. This review encompassed observational and interventional studies, such as randomized controlled trials and nonrandomized studies on interventions (e.g., before–after studies). Only studies that introduced AI to actual real-life clinical workflows were eligible, that is, those not conducted in an experimental setting or in a laboratory. The search strategy followed the PICO framework:
Population: This review included studies conducted in real-world healthcare facilities, such as hospitals and clinics, using medical imaging and surveying healthcare professionals of varying expertise and qualifications.
Exposure/interventions: This review encompassed studies that focused on various AI tools for diagnostics and their impact on healthcare professionals’ interaction with the technology across various clinical imaging tasks67. We exclusively focused on AI tools that interpret image data for disease diagnosis and screening5. For data extraction, we used the following working definition of AI used for clinical diagnostics: “any computer system used to interpret imaging data to make a diagnosis or screen for a disease, a task previously reserved for specialists”14.
Comparators: This review emphasized studies comparing the workflow before AI use with that after AI use or the workflow with AI use with that without AI use, although this was not a mandatory criterion to be included in the review.
Outcomes: The primary aim of this study was to evaluate how AI solutions impact workflow efficiency in clinical care contexts. Thus, we focused on three outcomes of interest: (1) changes in time required for task completion, (2) workflow adaptation, and (3) workload.
Changes in time for completion of imaging tasks were considered, focusing on reported quantitative changes attributed to AI usage (e.g., throughput times and review duration).
Workflow adaptation encompasses changes in the workflow that result from the introduction of new technologies, particularly in the context of AI implementation (i.e., specifying the time and purpose of AI use).
Workload refers to the demands of tasks on human operators and changes associated with AI implementation (e.g., cognitive demands or task load).
The detailed search strategy following the PICO framework can be found in Supplementary Table 4 and Supplementary Note 1.
Screening and selection procedure
All retrieved articles were imported into the Rayyan tool68,69 for title and abstract screening. In the first step, after undergoing a training, two study team members (KW and JK/MW/NG) independently screened the titles and abstracts to establish interrater agreement. In the second step, the full texts of all eligible publications were screened by KW and JK. Any potential conflicts regarding the inclusion of articles were resolved through discussions with a third team member (MW). Reasons for exclusion were documented, as depicted in the flow diagram in Fig. 170.
Data extraction procedure
Two authors (JK and KW/FZ) extracted the study data and imported them into MS Excel which then went through random checks by a study team member (MW). To establish agreement all reviewers extracted data from the first five studies based on internal data extraction guidelines.
Study quality appraisal and risk of bias assessment
To evaluate the methodological quality of the included studies, two reviewers (KW and JK) used three established tools. The Risk of Bias in Non-randomized Studies of Interventions tool (ROBINS-I) for non-randomized studies and the Cochrane Risk of Bias tool (Rob 2) for randomized studies were used71,72. To assess the reporting quality of the included studies, the MINORS was used27. The MINORS was used instead of the Quality of Reporting of Observational Longitudinal Research checklist73, as pre-specified in the review protocol, because this tool was more adaptable to all included studies. Appraisals were finally established through discussion until consensus was achieved.
Strategy for data synthesis
First, we describe the overall sample and the key information from each included study. Risk of bias assessment evaluations are presented in narrative and tabular formats. Next, where comparable studies were sufficient, a meta-analysis was performed to examine the effects of AI introduction. We used the method of Wan et al.74 to estimate the sample mean and standard deviation from the sample size, median, and interquartile range because the reported measures varied across the included studies. Furthermore, we followed the Cochrane Handbook for calculating the standard deviation from the confidence interval (CI)75. The metafor package in R76 was used to quantitatively synthesize data from the retrieved studies. Considering the anticipated heterogeneity of effects, a random-effects model was used to estimate the average effect across studies. Moreover, we used the DerSimonian and Laird method to determine cross-study variance and the Hartung–Knapp method to estimate the variance of the random effect77,78. Heterogeneity was assessed using Cochran’s Q test79 and the I2 statistic75. In cases where a meta-analysis was not feasible, the results were summarized in narrative form and presented in tabular format.
Meta-biases
Potential sources of meta-bias, such as publication bias and selective reporting across studies, were considered. Funnel plots were created for the studies included in the meta-analyses.
To assess whether our review is subject to selection bias due to the choice of databases and publication types, we conducted an additional search in the dblp computer science bibliography (with our original search timeframe). As this database did not allow our original search string, the adapted version is found in Supplementary Note 2. Additionally, we performed searches on conference proceedings of the last three years, spanning publications from the January 1st 2020 until May 15th 2023. We surveyed IEEE Xplore and two major conferences not included in the database: International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI) and Hawaii International Conference on System Sciences (HICSS). We conducted an initial screening of titles and abstracts, with one reviewer (KW) screening all records and JK screening 10% to assess interrater reliability. Full-text assessments for eligibility were then performed by one of the reviewers, respectively (KW or JK). Furthermore, the AMSTAR-2 critical appraisal tool for systematic reviews of randomized and/or non-randomized healthcare intervention studies was used43.
Supplementary information
Acknowledgements
We sincerely thank Dr. Nikoloz Gambashidze (Institute for Patient Safety, University Hospital Bonn) for helping with the title and abstract screening. We thank Annika Strömer (Institute for Medical Biometry, Informatics and Epidemiology, University of Bonn) for her statistical support. This research was financed through institutional budget, i.e., no external funding.
Author contributions
K.W.: conceptualization, data curation, formal analysis, investigation, methodology, project administration, software, visualization, writing – original draft, writing – preparation, review, and editing; J.K.: data curation, investigation, visualization, writing – review and editing; F.Z.: investigation, writing – review and editing; M.W.: conceptualization, funding acquisition, supervision, validation. All authors have read and approved the manuscript.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Data availability
All data generated or analyzed during this study is available from the corresponding author upon reasonable request.
Code availability
Code for meta-analyses available via https://github.com/katwend/metaanalyses.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41746-024-01248-9.
References
- 1.Yeganeh, H. An analysis of emerging trends and transformations in global healthcare. IJHG24, 169–180 (2019). [Google Scholar]
- 2.Asan, O., Bayrak, A. E. & Choudhury, A. Artificial intelligence and human trust in healthcare: focus on clinicians. J. Med. Internet Res.22, e15154 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Park, C.-W. et al. Artificial intelligence in health care: current applications and issues. J. Korean Med. Sci.35, e379 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ahmad, Z., Rahim, S., Zubair, M. & Abdul-Ghafar, J. Artificial Intelligence (ai) in medicine, current applications and future role with special emphasis on its potential and promise in pathology: present and future impact, obstacles including costs and acceptance among pathologists, practical and philosophical considerations. a comprehensive review. Diagn. Pathol.16, 24 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.He, J. et al. The practical implementation of artificial intelligence technologies in medicine. Nat. Med.25, 30–36 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wong, S. H., Al-Hasani, H., Alam, Z. & Alam, A. Artificial intelligence in radiology: how will we be affected? Eur. Radiol.29, 141–143 (2019). [DOI] [PubMed] [Google Scholar]
- 7.Arbabshirani, M. R. et al. Advanced machine learning in action: identification of intracranial hemorrhage on computed tomography scans of the head with clinical workflow integration. npj Digit. Med.1, 9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mueller, F. C. et al. Impact of concurrent use of artificial intelligence tools on radiologists reading time: a prospective feasibility study. Acad. Radiol.29, 1085–1090 (2022). [DOI] [PubMed] [Google Scholar]
- 9.Pumplun, L., Fecho, M., Wahl, N., Peters, F. & Buxmann, P. Adoption of machine learning systems for medical diagnostics in clinics: qualitative interview study. J. Med. Internet Res.23, e29301 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dahlblom, V., Dustler, M., Tingberg, A. & Zackrisson, S. Breast cancer screening with digital breast tomosynthesis: comparison of different reading strategies implementing artificial intelligence. Eur. Radiol.33, 3754–3765 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miyake, M. et al. Comparative performance of a primary-reader and second-reader paradigm of computer-aided detection for CT colonography in a low-prevalence screening population. Jpn J. Radio.31, 310–319 (2013). [DOI] [PubMed] [Google Scholar]
- 12.Hosny, A., Parmar, C., Quackenbush, J., Schwartz, L. H. & Aerts, H. J. W. L. Artificial intelligence in radiology. Nat. Rev. Cancer18, 500–510 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Leeuwen, K. G., de Rooij, M., Schalekamp, S., van Ginneken, B. & Rutten, M. J. C. M. How does artificial intelligence in radiology improve efficiency and health outcomes? Pediatric Radiol.52, 2087–2093 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wenderott, K., Gambashidze, N. & Weigl, M. Integration of artificial intelligence into sociotechnical work systems—effects of artificial intelligence solutions in medical imaging on clinical efficiency: protocol for a systematic literature review. JMIR Res. Protoc.11, e40485 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Salwei, M. E. & Carayon, P. A Sociotechnical systems framework for the application of artificial intelligence in health care delivery. J. Cogn. Eng. Decis. Making16, 194–206 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wolff, J., Pauling, J., Keck, A. & Baumbach, J. Success factors of artificial intelligence Implementation in Healthcare. Front. Digit. Health3, 594971 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Felmingham, C. M. et al. The importance of incorporating human factors in the design and implementation of artificial intelligence for skin cancer diagnosis in the real world. Am. J. Clin. Dermatol.22, 233–242 (2021). [DOI] [PubMed] [Google Scholar]
- 18.Wenderott, K., Krups, J., Luetkens, J. A., Gambashidze, N. & Weigl, M. Prospective effects of an artificial intelligence-based computer-aided detection system for prostate imaging on routine workflow and radiologists’ outcomes. Eur. J. Radiol.170, 111252 (2024). [DOI] [PubMed] [Google Scholar]
- 19.Pierce, J. et al. Seamless integration of artificial intelligence into the clinical environment: our experience with a novel pneumothorax detection artificial intelligence algorithm. J. Am. Coll. Radiol.18, 1497–1505 (2021). [DOI] [PubMed] [Google Scholar]
- 20.Diao, K. et al. Diagnostic study on clinical feasibility of an AI-based diagnostic system as a second reader on mobile CT images: a preliminary result. Ann. Transl. Med.10, 668 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Duron, L. et al. Assessment of an AI aid in detection of adult appendicular skeletal fractures by emergency physicians and radiologists: a multicenter cross-sectional diagnostic study. Radiology300, 120–129 (2021). [DOI] [PubMed] [Google Scholar]
- 22.Kanagasingam, Y. et al. Evaluation of artificial intelligence–based grading of diabetic retinopathy in primary care. JAMA Netw. Open1, e182665 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bossuyt, P. M. et al. STARD 2015: an updated list of essential items for reporting diagnostic accuracy studies. Radiology277, 826–832 (2015). [DOI] [PubMed] [Google Scholar]
- 24.Repici, A. et al. Efficacy of real-time computer-aided detection of colorectal neoplasia in a randomized trial. Gastroenterology159, 512–520.e7 (2020). [DOI] [PubMed] [Google Scholar]
- 25.Schulz, K. F., Altman, D. G. & Moher, D. CONSORT Group CONSORT 2010 Statement: updated guidelines for reporting parallel group randomised trials. BMJ340, c332–c332 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang, P. et al. Real-time automatic detection system increases colonoscopic polyp and adenoma detection rates: a prospective randomised controlled study. Gut68, 1813–1819 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Slim, K. et al. Methodological index for non-randomized studies (MINORS): development and validation of a new instrument: methodological index for non-randomized studies. ANZ J. Surg.73, 712–716 (2003). [DOI] [PubMed] [Google Scholar]
- 28.Conant, E. F. et al. Improving accuracy and efficiency with concurrent use of artificial intelligence for digital breast tomosynthesis. Radiol. Artif. Intell.1, e180096 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nehme, F. et al. Performance and attitudes toward real-time computer-aided polyp detection during colonoscopy in a large tertiary referral center in the United States. Gastrointest. Endosc.98, 100–109.e6 (2023). [DOI] [PubMed]
- 30.Zia, A. et al. Retrospective analysis and prospective validation of an Ai-based software for intracranial haemorrhage detection at a high-volume trauma centre. Sci. Rep.12, 19885 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tchou, P. M. et al. Interpretation time of computer-aided detection at screening mammography. Radiology257, 40–46 (2010). [DOI] [PubMed] [Google Scholar]
- 32.Vassallo, L. et al. A cloud-based computer-aided detection system improves identification of lung nodules on computed tomography scans of patients with extra-thoracic malignancies. Eur. Radiol.29, 144–152 (2019). [DOI] [PubMed] [Google Scholar]
- 33.Wittenberg, R. et al. Acute pulmonary embolism: effect of a computer-assisted detection prototype on diagnosis—an observer study. Radiology262, 305–313 (2012). [DOI] [PubMed] [Google Scholar]
- 34.Batra, K., Xi, Y., Bhagwat, S., Espino, A. & Peshock, R. Radiologist worklist reprioritization using artificial intelligence: impact on report turnaround times for CTPA examinations positive for acute pulmonary embolism. Am. J. Roentgenol221, 324–333 (2023). [DOI] [PubMed] [Google Scholar]
- 35.Liu, X. et al. Evaluation of an OCT-AI-based telemedicine platform for retinal disease screening and referral in a primary care setting. Transl. Vis. Sci. Technol.11, 4 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Raya-Povedano, J. L. et al. AI-based strategies to reduce workload in breast cancer screening with mammography and tomosynthesis: a retrospective evaluation. Radiology300, 57–65 (2021). [DOI] [PubMed] [Google Scholar]
- 37.Yacoub, B. et al. Impact of artificial intelligence assistance on chest CT interpretation times: a prospective randomized study. Am. J. Roentgenol.219, 743–751 (2022). [DOI] [PubMed] [Google Scholar]
- 38.Cha, E. et al. Clinical implementation of deep learning contour auto segmentation for prostate radiotherapy. Radiother. Oncol.159, 1–7 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Davis, M. A., Rao, B., Cedeno, P. A., Saha, A. & Zohrabian, V. M. Machine learning and improved quality metrics in acute intracranial hemorrhage by noncontrast computed tomography. Curr. Probl. Diagn. Radiol.51, 556–561 (2022). [DOI] [PubMed] [Google Scholar]
- 40.Hassan, A., Ringheanu, V. & Tekle, W. The implementation of artificial intelligence significantly reduces door-in-door-out times in a primary care center prior to transfer. Interv. Neuroradiol.29, 631–636 (2022). [DOI] [PMC free article] [PubMed]
- 41.Ladabaum, U. et al. Computer-aided detection of polyps does not improve colonoscopist performance in a pragmatic implementation trial. Gastroenterol. 164, 481–483 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wismüller, A. & Stockmaster, L. A Prospective randomized clinical trial for measuring radiology study reporting time on artificial intelligence-based detection of intracranial hemorrhage in emergent Care Head CT (2020).
- 43.Shea, B. J. et al. Amstar 2: a critical appraisal tool for systematic reviews that include randomised or non-randomised studies of healthcare interventions, or both. BMJ358, j4008 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boutron, I. et al. Considering bias and conflicts of interest among the included studies. In Cochrane Handbook for Systematic Reviews of Interventions (eds Higgins, J. P. T. et al.) 177–204 (Wiley, 2019).
- 45.Beyer, F. et al. Comparison of sensitivity and reading time for the use of computer-aided detection (CAD) of pulmonary nodules at MDCT as concurrent or second reader. Eur. Radio.17, 2941–2947 (2007). [DOI] [PubMed] [Google Scholar]
- 46.Fujita, H. AI-based computer-aided diagnosis (AI-CAD): the latest review to read first. Radio. Phys. Technol.13, 6–19 (2020). [DOI] [PubMed] [Google Scholar]
- 47.Asan, O. & Choudhury, A. Research trends in artificial intelligence applications in human factors health care: mapping review. JMIR Hum. Factors8, e28236 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Herrmann, T. & Pfeiffer, S. Keeping the organization in the loop: a socio-technical extension of human-centered artificial intelligence. AI Soc.38, 1523–1542 (2023). [Google Scholar]
- 49.Allen, B. The role of the FDA in ensuring the safety and efficacy of artificial intelligence software and devices. J. Am. Coll. Radiol.16, 208–210 (2019). [DOI] [PubMed] [Google Scholar]
- 50.Wenderott, K., Krups, J., Luetkens, J. A. & Weigl, M. Radiologists’ perspectives on the workflow integration of an artificial intelligence-based computer-aided detection system: a qualitative study. Appl. Ergon.117, 104243 (2024). [DOI] [PubMed] [Google Scholar]
- 51.Nazer, L. H. et al. Bias in artificial intelligence algorithms and recommendations for mitigation. PLOS Digit Health2, e0000278 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Norori, N., Hu, Q., Aellen, F. M., Faraci, F. D. & Tzovara, A. Addressing bias in big data and AI for health care: a call for open science. Patterns2, 100347 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen, W. et al. Improving the diagnosis of acute ischemic stroke on non-contrast Ct using deep learning: a multicenter study. Insights Imaging13, 184 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Potretzke, T. et al. Clinical implementation of an artificial intelligence algorithm for magnetic resonance-derived measurement of total kidney volume. Mayo Clin. Proc.98, 689–700 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sun, J. et al. Performance of a chest radiograph AI diagnostic tool for COVID-19: a prospective observational study. Radiol. Artif. Intell.4, e210217 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tricarico, D. et al. Convolutional neural network-based automatic analysis of chest radiographs for the detection of COVID-19 pneumonia: a prioritizing tool in the emergency department, phase i study and preliminary ‘real life’ results. Diagnostics12, 570 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ibrahim, H. et al. Reporting guidelines for clinical trials of artificial intelligence interventions: the SPIRIT-AI and CONSORT-AI guidelines. Trials22, 11 (2021). [DOI] [PMC free article] [PubMed]
- 58.Liu, X. et al. A comparison of deep learning performance against health-care professionals in detecting diseases from medical imaging: a systematic review and meta-analysis. Lancet Digit. Health1, e271–e297 (2019). [DOI] [PubMed] [Google Scholar]
- 59.Nagendran, M. et al. Artificial intelligence versus clinicians: systematic review of design, reporting standards, and claims of deep learning studies. BMJ m689 (2020). [DOI] [PMC free article] [PubMed]
- 60.Yin, J., Ngiam, K. Y. & Teo, H. H. Role of artificial intelligence applications in real-life clinical practice: systematic review. J. Med. Internet Res.23, e25759 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Han, R. et al. Randomised controlled trials evaluating artificial intelligence in clinical practice: a scoping review. Lancet Digit. Health6, e367–e373 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hua, D., Petrina, N., Young, N., Cho, J.-G. & Poon, S. K. Understanding the factors influencing acceptability of AI in medical imaging domains among healthcare professionals: a scoping review. Artif. Intell. Med.147, 102698 (2024). [DOI] [PubMed] [Google Scholar]
- 63.Bruni, S., Freiman, M. & Riddle, K. Beyond the tool vs. teammate debate: exploring the sidekick metaphor in human-AI Dyads. In: Julia Wright and Daniel Barber (eds) Human Factors and Simulation. AHFE (2023) International Conference. AHFE Open Access, 83 (2023).
- 64.Flathmann, C. et al. Examining the impact of varying levels of AI teammate influence on human-AI teams. Int. J. Hum.-Comput. Stud.177, 103061 (2023). [Google Scholar]
- 65.Huang, S.-C., Pareek, A., Seyyedi, S., Banerjee, I. & Lungren, M. P. Fusion of medical imaging and electronic health records using deep learning: a systematic review and implementation guidelines. npj Digit. Med.3, 136 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kaul, V., Enslin, S. & Gross, S. A. History of artificial intelligence in medicine. Gastrointest. Endosc.92, 807–812 (2020). [DOI] [PubMed] [Google Scholar]
- 67.Dias, R. & Torkamani, A. Artificial intelligence in clinical and genomic diagnostics. Genome Med.11, 70 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ouzzani, M., Hammady, H., Fedorowicz, Z. & Elmagarmid, A. Rayyan-a web and mobile app for systematic reviews. Syst. Rev.5, 210 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ouzzani, M., Hammady, H., Fedorowicz, Z. & Elmagarmid, A. Rayyan-a web and mobile app for systematic reviews. Syst Rev.5, 210 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Page, M. J. et al. The Prisma 2020 statement: an updated guideline for reporting systematic reviews. BMJ372, n71 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sterne, J. A. et al. ROBINS-I: a tool for assessing risk of bias in non-randomised studies of interventions. BMJ355, i4919 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sterne, J. A. C. et al. RoB 2: a revised tool for assessing risk of bias in randomised trials. BMJ366, l4898 (2019). [DOI] [PubMed]
- 73.Tooth, L., Ware, R., Bain, C., Purdie, D. M. & Dobson, A. Quality of reporting of observational longitudinal research. Am. J. Epidemiol.161, 280–288 (2005). [DOI] [PubMed] [Google Scholar]
- 74.Wan, X., Wang, W., Liu, J. & Tong, T. Estimating the sample mean and standard deviation from the sample size, median, range and/or interquartile range. BMC Med Res Methodol.14, 135 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Higgins, J. P. T., Thompson, S. G., Deeks, J. J. & Altman, D. G. Measuring inconsistency in meta-analyses. BMJ327, 557–560 (2003). [DOI] [PMC free article] [PubMed]
- 76.Viechtbauer, W. Conducting meta-analyses in R with the metafor Package. J Stat Softw.36, 1–48 (2010). [Google Scholar]
- 77.DerSimonian, R. & Laird, N. Meta-analysis in clinical trials. Control. Clin. Trials7, 177–188 (1986). [DOI] [PubMed] [Google Scholar]
- 78.Hartung, J. An alternative method for meta-analysis. Biom. J. J. Math. Methods Biosci.41, 901–916 (1999). [Google Scholar]
- 79.Cochran, W. G. The combination of estimates from different experiments. Biometrics10, 101 (1954). [Google Scholar]
- 80.Carlile, M. et al. Deployment of artificial intelligence for radiographic diagnosis of COVID-19 pneumonia in the emergency department. J. Am. Coll. Emerg. Phys. Open1, 1459–1464 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cheikh, A. B. et al. How artificial intelligence improves radiological interpretation in suspected pulmonary embolism. Eur. Radiol.32, 5831–5842 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Elijovich, L. et al. Automated emergent large vessel occlusion detection by artificial intelligence improves stroke workflow in a hub and spoke stroke system of care. J. NeuroIntervent Surg.14, 704–708 (2022). [DOI] [PubMed] [Google Scholar]
- 83.Ginat, D. Implementation of machine learning software on the radiology worklist decreases scan view delay for the detection of intracranial hemorrhage on CT. Brain Sci.11, 832 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hong, W. et al. Deep learning for detecting pneumothorax on chest radiographs after needle biopsy: clinical implementation. Radiology303, 433–441 (2022). [DOI] [PubMed] [Google Scholar]
- 85.Jones, C. M. et al. Assessment of the effect of a comprehensive chest radiograph deep learning model on radiologist reports and patient outcomes: a real-world observational study. BMJ Open11, e052902 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kiljunen, T. et al. A deep learning-based automated CT segmentation of prostate cancer anatomy for radiation therapy planning-A retrospective multicenter study. Diagnostics10, 959 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Levy, I., Bruckmayer, L., Klang, E., Ben-Horin, S. & Kopylov, U. Artificial intelligence-aided colonoscopy does not increase adenoma detection rate in routine clinical practice. Am. J. Gastroenterol.117, 1871–1873 (2022). [DOI] [PubMed] [Google Scholar]
- 88.Marwaha, A., Chitayat, D., Meyn, M., Mendoza-Londono, R. & Chad, L. The point-of-care use of a facial phenotyping tool in the genetics clinic: enhancing diagnosis and education with machine learning. Am. J. Med. Genet. A185, 1151–1158 (2021). [DOI] [PubMed] [Google Scholar]
- 89.O’Neill, T. J. et al. Active reprioritization of the reading worklist using artificial intelligence has a beneficial effect on the turnaround time for interpretation of head CT with intracranial hemorrhage. Radiol. Artif. Intell.3, e200024 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Oppenheimer, J., Lüken, S., Hamm, B. & Niehues, S. A prospective approach to integration of AI fracture detection software in radiographs into clinical workflow. Life (Basel, Switzerland)13, 223 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Quan, S. Y. et al. Clinical evaluation of a real-time artificial intelligence-based polyp detection system: a US multi-center pilot study. Sci. Rep.12, 6598 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ruamviboonsuk, P. et al. Real-time diabetic retinopathy screening by deep learning in a multisite national screening programme: a prospective interventional cohort study. Lancet Digit. Health4, e235–44 (2022). [DOI] [PubMed] [Google Scholar]
- 93.Sandbank, J. et al. Validation and real-world clinical application of an artificial intelligence algorithm for breast cancer detection in biopsies. npj Breast Cancer8, 129 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Schmuelling, L. et al. Deep learning-based automated detection of pulmonary embolism on CT pulmonary angiograms: no significant effects on report communication times and patient turnaround in the emergency department nine months after technical implementation. Eur. J. Radiol.141, 109816 (2021). [DOI] [PubMed] [Google Scholar]
- 95.Seyam, M. et al. Utilization of artificial intelligence-based intracranial hemorrhage detection on emergent noncontrast CT images in clinical workflow. Radiol. Artif. Intell.4, e210168 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sim, J. Z. T. et al. Diagnostic performance of a deep learning model deployed at a National COVID-19 screening facility for detection of pneumonia on frontal chest radiographs. Healthcare10, 175 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Strolin, S. et al. How smart is artificial intelligence in organs delineation? Testing a CE and FDA-approved deep-learning tool using multiple expert contours delineated on planning CT images. Front. Oncol.13, 1089807 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wang, M. et al. Deep learning-based triage and analysis of lesion burden for COVID-19: a retrospective study with external validation. Lancet Digit. Health2, e506–e515 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wong, J. et al. Implementation of deep learning-based auto-segmentation for radiotherapy planning structures: a workflow study at two cancer centers. Radiat. Oncol.16, 101 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wong, K. et al. Integration and evaluation of chest X-ray artificial intelligence in clinical practice. J. Med. Imaging10, 051805 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yang, Y. et al. Performance of the AIDRScreening system in detecting diabetic retinopathy in the fundus photographs of Chinese patients: a prospective, multicenter, clinical study. Ann. Transl. Med.10, 1088 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Elguindi, S. et al. Deep learning-based auto-segmentation of targets and organs-at-risk for magnetic resonance imaging only planning of prostate radiotherapy. Phys. Imaging Radiat. Oncol.12, 80–86 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wang, L. et al. An intelligent optical coherence tomography-based system for pathological retinal cases identification and urgent referrals. Trans. Vis. Sci. Tech.9, 46 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Gulshan, V. et al. Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. JAMA316, 2402 (2016). [DOI] [PubMed] [Google Scholar]
- 105.Krause, J. et al. Grader variability and the importance of reference standards for evaluating machine learning models for diabetic retinopathy. Ophthalmology125, 1264–1272 (2018). [DOI] [PubMed] [Google Scholar]
- 106.Ruamviboonsuk, P. et al. Deep learning versus human graders for classifying diabetic retinopathy severity in a nationwide screening program. npj Digit. Med.2, 25 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Retico, A., Delogu, P., Fantacci, M. E., Gori, I. & Preite Martinez, A. Lung nodule detection in low-dose and thin-slice computed tomography. Comput. Biol. Med.38, 525–534 (2008). [DOI] [PubMed] [Google Scholar]
- 108.Lopez Torres, E. et al. Large scale validation of the M5L lung CAD on heterogeneous CT datasets. Med. Phys.42, 1477–1489 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Brown, M. S. et al. Automated endotracheal tube placement check using semantically embedded deep neural networks. Acad. Radiol.30, 412–420 (2023). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study is available from the corresponding author upon reasonable request.
Code for meta-analyses available via https://github.com/katwend/metaanalyses.