Abstract
A characterization of the A45R gene from vaccinia virus (VV) strain Western Reserve is presented. The open reading frame is predicted to encode a 125-amino-acid protein (Mr, of 13,600) with 39% amino acid identity to copper-zinc superoxide dismutase (Cu-Zn SOD). Sequencing of the A45R gene from other orthopoxviruses, here and by others, showed that the protein is highly conserved in all viruses sequenced, including 16 strains of VV, 2 strains of cowpox virus, camelpox virus, and 4 strains of variola virus. In all cases the protein lacks key residues involved in metal ion binding that are important for the catalytic activity. The A45R protein was expressed in Escherichia coli, purified, and tested for SOD activity, but neither enzymatic nor inhibitory SOD activity was detected. Additionally, no virus-encoded SOD activity was detected in infected cells or purified virions. A monoclonal antibody raised against the A45R protein expressed in E. coli identified the A45R gene product as a 13.5-kDa protein that is expressed late during VV infection. Confocal microscopy of VV-infected cells indicated that the A45R protein accumulated predominantly in cytoplasmic viral factories. Electron microscopy and biochemical analyses showed that the A45R protein is incorporated into the virion core. A deletion mutant lacking the majority of the A45R gene and a revertant virus in which the deleted gene was restored were constructed and characterized. The growth properties of the deletion mutant virus were indistinguishable from those of wild-type and revertant viruses in all cell lines tested, including macrophages. Additionally, the virulence and pathogenicity of the three viruses were also comparable in murine and rabbit models of infection. A45R is unusual in being the first VV core protein described that affects neither virus replication nor virulence.
Vaccinia virus (VV) is the most extensively studied member of the poxvirus family, a group of large DNA viruses that replicate in the cell cytoplasm (37). It contains a double-stranded DNA genome encoding about 200 proteins (19, 27). Many of these proteins are essential for virus growth in tissue culture; these include enzymes required for virus replication in the cytoplasm and structural proteins needed for the formation of the two infectious forms of the virus, the intracellular mature virus (IMV) and the extracellular enveloped virus (EEV) (6, 24). Other proteins, mostly encoded by genes located near the ends of the virus genome, facilitate virus replication in vivo or interfere with host immune functions. The latter group deals with nonspecific immune mechanisms, such as complement, interferon, and the inflammatory response, which are induced rapidly and constitute the first host response against the invading organism. Additionally, VV expresses proteins that block apoptosis and an enzyme (3β-hydroxysteroid dehydrogenase [3β-HSD]) that synthesizes steroid hormones and contributes to VV virulence (reviewed in reference 52).
The A45R open reading frame (ORF), previously called SalF8R in VV strain Western Reserve (WR), is predicted to encode a 13.6-kDa protein with 39% amino acid identity with copper-zinc superoxide dismutase (Cu-Zn SOD) (51), an enzyme that catalyzes the conversion of superoxide to oxygen and hydrogen peroxide (35). There are three known forms of SOD that contain either Mn, Fe, or both Cu and Zn. The MnSODs are found in prokaryotes and in the matrix of mitochondria, the FeSODs are present in prokaryotes and in a few families of plants, and the Cu-Zn SODs occur primarily in the cytosol of eukaryotic cells and in chloroplasts but have also been found in a few species of bacteria. All SODs catalyze the same reaction with high efficiency, and all operate by a similar mechanism in which the metal is the catalytic factor in the active site (reviewed in reference 18). The cytosolic Cu-Zn SOD is a dimeric metalloprotein composed of identical, noncovalently linked subunits, each of 16 kDa. However, mammalian extracellular fluids contain a tetrameric glycosylated Cu-Zn SOD. The three-dimensional structure of Cu-Zn SOD shows the protein to contain eight antiparallel β-strands with a Greek key topology and three protruding loops of nonrepetitive structure, one of which binds the Zn atom (54).
Superoxide radicals arise during numerous oxidations in both living and nonliving systems and can act directly as oxidants or generate other reactive products that are toxic to cells, causing damage to lipid membranes, nucleic acid, carbohydrates, and proteins. To overcome this problem, life forms have developed an effective defensive system, including SOD, which scavenges active oxygen species generated during aerobic metabolism. Consequently, aerobic existence is accompanied by a persistent state of oxidative siege, and the survival of a given cell is determined by its balance of reactive oxygen intermediates and antioxidants. Disturbance of this balance can lead to disease (20). Superoxide is also generated deliberately by professional phagocytes (neutrophils, eosinophils, and macrophages) during the respiratory burst to kill microorganisms (7, 58). In the case of activated neutrophils, the superoxide released also produces a chemotaxin by reacting with a component of blood plasma (42), allowing one activated neutrophil to recruit others and thus to produce local inflammation. Therefore, a VV SOD activity might be advantageous by enhancing virus survival in, and in the presence of, phagocytes.
The nucleotide sequence of the A45R ORF has been determined in VV strains WR (51), Copenhagen (19), modified virus Ankara (MVA) (5), and Tian Tan (GenBank accession number AAF34056); in variola major virus strains Harvey (1), Bangladesh 1975 (33), and India 1967 (49); and in variola minor virus strain Garcia 1966 (50). It is highly conserved, showing more than 96% amino acid identity. Alignment of the amino acid sequences of Cu-Zn SODs with those from VV and variola virus indicates that although the viral protein is predicted to contain the eight strands of antiparallel β-structure and a similar overall globular structure, it lacks the Zn-binding region located near the C terminus and the majority of histidine residues necessary for ion binding. These observations make it unlikely that this protein has SOD activity, but it is quite possible that the comparable protein in other VV strains or other orthopoxviruses might be active.
In this report, a characterization of the A45R gene of VV strain WR is provided. We show that the gene is highly conserved in all orthopoxviruses tested, is expressed late during infection, and encodes a protein that is present in the viral core. We also show that the protein expressed in Escherichia coli does not have SOD activity. The role of the A45R gene in VV biology was investigated by deleting the gene from the virus genome and analyzing the biological effect in culture and in infected mice and rabbits.
MATERIALS AND METHODS
Cells and viruses.
Monkey kidney BSC-1 and CV-1 cells, rabbit kidney (RK)13 cells, HeLa cells, and D980R cells (a hypoxanthine phosphoribosyltransferase [HPRT]-negative HeLa derivative) were obtained from the Cell Bank of the Sir William Dunn School of Pathology (University of Oxford, Oxford, United Kingdom). The monocyte-macrophage cell lines P388D1 (mouse) and U937 (human) were kindly provided by S. Gordon (Sir William Dunn School of Pathology). Cells were grown in minimal essential medium (MEM) (GIBCO) supplemented with 10% fetal bovine serum (FBS), except for D980R cells and the monocyte-macrophage cell lines (P388D1 and U937), for which Dulbecco's modified Eagle's medium (DMEM) (GIBCO) and RPMI 1640 (GIBCO) with 10% FBS were used, respectively. The growth conditions and the sources of VV strains and other orthopoxviruses have been described previously (2, 3). Working stocks of virus were prepared by Dounce homogenization of infected BSC-1 cells, removal of nuclei by centrifugation, and sedimentation of virus in the supernatant through a sucrose cushion as described elsewhere (32). Virus infectivity was titrated by plaque assay on BSC-1 cells (32). IMV and EEV were prepared from infected BSC-1 or RK13 cells by sedimentation through sucrose gradients as described elsewhere (41).
Sequencing of the A45R ORF in other orthopoxviruses.
A fragment containing the entire A45R ORF plus 81 nucleotides upstream and 126 nucleotides downstream was amplified by PCR using DNA extracted from cells infected with different orthopoxviruses as the template and the oligonucleotides 5′-GCGGGGATCCGGCACCACCAGTAACCGCGTAC-3′ and 5′-GCGCGTCGACTTTGTCCCTATCAAATTCGACAG-3′ (the BamHI and SalI restriction sites, respectively, are underlined), based on the VV WR strain sequence. The PCR products (generated from multiple PCRs to avoid potential PCR errors) were sequenced using the same primers and the ABI PRISM dye terminator cycle sequencing ready reaction kit. Samples were run on an automatic ABI 373 DNA sequencer, and DNA sequences were analyzed by using the University of Wisconsin Genetics Computer Group (GCG) program package, version 9.0.
Plasmid constructions. (i) Plasmids used for constructing recombinant viruses.
A 2.4-kb EcoRI fragment containing the A45R gene and flanking sequences was excised from the SalI F fragment of VV WR (pSalIF) (51) and was cloned into EcoRI-cut pUC119 to form plasmid pFAT1. The same fragment was also cloned into EcoRI-cut pSJH7 (23), containing the E. coli guanine phosphoribosyltransferase (Ecogpt) gene under the control of the VV 7.5 promoter. The resulting plasmid, termed pFAT2, was used to generate the A45R revertant virus by transient dominant selection (TDS) (16).
Plasmid pFAT6 was used to generate the A45R deletion virus by TDS (16) and was constructed in three steps. First, a 935-bp EcoRI-NcoI fragment (the NcoI site was made blunt ended by treatment with Klenow enzyme), containing the first 64 nucleotides of the A45R ORF and 871 nucleotides upstream, was excised from pFAT1 and cloned into pSJH7 (23), which had been digested with EcoRI and SmaI, to form plasmid pFAT3. This regenerated the NcoI site. Second, to eliminate the first 64 nucleotides of the A45R ORF, a 304-bp fragment was removed by digestion of pFAT3 with Eco47III and NcoI and was replaced by a PCR product lacking the first 64 nucleotides of A45R to generate plasmid pFAT4. This PCR product was formed using a forward oligonucleotide spanning the Eco47III restriction site (5′-GCGCAGCGCTTCTCTCACCTTATCAAAG-3′) and a reverse oligonucleotide complementary to the 24 nucleotides upstream of the A45R ORF, which contained an additional 5′ NcoI restriction site (5′-GCGCCATGGATTATTTAGTAAAATAGAATAAGTAG-3′) (restriction sites are underlined). The nucleotide sequence of the cloned PCR fragment was verified by DNA sequencing. Finally, a 3′ flanking region of 1,345 nucleotides, containing the 3′-end 152 nucleotides of A45R ORF and 1,193 nucleotides downstream, was obtained from pFAT1 by NdeI digestion, Klenow treatment, and HindIII digestion (which cut downstream of A45R within the pUC119 polylinker). This fragment was then cloned into pFAT4 that had been digested with SalI, end filled with Klenow enzyme, and digested with HindIII. The resulting plasmid, pFAT6, had lost the first 223 nucleotides (60% of the A45R coding sequence) of the A45R ORF. The 152 nucleotides at the 3′ end of the A45R ORF were retained to avoid interference with the expression of the A46R ORF, which overlaps the 3′ end of the A45R ORF. The remaining 50-amino-acid peptide would not be expressed because it lacked a translation initiation codon.
(ii) Plasmids for expression of A45R in bacteria.
Plasmid pFAT8 was used to express a thioredoxin-A45R fusion protein in E. coli. A DNA fragment containing the A45R ORF and terminal BamHI sites was amplified by PCR from pFAT1 DNA using oligonucleotides 5′-GCGCGGATCCATGGCTGTTTGTATAATAGACC-3′ and 5′-GCGCGGATCCTCAAACGCCATTCTCGTTAATTG-3′ as primers (the BamHI sites are underlined, and the initiation and termination codons are boldfaced). The PCR fragment was digested with BamHI and cloned into BglII-cut pThioHis C (His-Patch ThioFusion Expression System; Invitrogen, BV) to form pFAT8. The nucleotide sequence of the cloned PCR fragment was verified by DNA sequencing.
Construction of A45R deletion mutant and revertant viruses.
A VV deletion mutant lacking 60% of the A45R ORF was isolated by TDS using the Ecogpt gene as the transient selectable marker (16) as described elsewhere (29). CV-1 cells were infected with VV strain WR at 0.05 PFU/cell and transfected with pFAT6. Progeny virus was plated onto monolayers of BSC-1 cells in the presence of mycophenolic acid (MPA). After a further round of plaque purification under this selection, MPA-resistant viruses were resolved to wild-type virus (vWTA45R) and A45R deletion virus (vΔA45R) by plating onto D980R cells in the presence of 6-thioguanine (25, 28). vWTA45R and vΔA45R viruses were plaque purified twice more. The revertant virus, in which the A45R locus was restored to that of wild-type virus, was constructed using TDS as described above. Plasmid pFAT2 was transfected into CV-1 cells infected with vΔA45R, and after isolation of MPA-resistant intermediate viruses, a revertant virus (vRA45R) was isolated on D980R cells in the presence of 6-thioguanine.
The genomic structures of vWTA45R, vΔA45R, and vRA45R were analyzed by Southern blotting and PCR and were found to be as predicted.
Expression of the A45R ORF in E. coli.
The A45R ORF was expressed as a thioredoxin fusion protein using the His-Patch Thiofusion Expression System (Invitrogen, BV) according to the manufacturer's specifications with some modifications. Plasmid pFAT8 was transformed into E. coli TOP 10 cells, and expression of the thioredoxin-A45R fusion protein was induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 6 h at 30°C. Cells from a 50-ml culture were pelleted, resuspended in 10 ml of 20 mM sodium phosphate (pH 7.8), and lysed by three sonication-freeze-thaw cycles. Insoluble cellular material was removed by centrifugation (at 3,000 × g for 15 min at 4°C), and the soluble fusion protein was purified by application to a 2-ml Ni2+ column (ProBond) as instructed by the manufacturer. An imidazole gradient was unable to elute the protein, so 50 mM EDTA was used instead. The purified fusion protein was dialyzed against phosphate-buffered saline (PBS) prior to quantification by the bicinchoninic acid (BCA) (Pierce) protein assay. Finally, to overcome the possible loss of metal ions during the purification process, the purified fusion protein was reconstituted by simultaneous addition of Cu2+ and Zn2+ in solution as described previously (26).
Activity assay for SOD.
The SOD activities of the purified A45R fusion protein, VV-infected cells, and IMV particles were measured using the SOD assay kit (Calbiochem) according to the manufacturer's instructions. BSC-1, RK13, HeLa, P388D1, and U937 cells (5 × 106) were mock infected or infected with vWTA45R, vΔA45R, or vRA45R at 10 PFU/cell. Cells were harvested at 14 h postinfection (p.i.), pelleted, resuspended in 250 μl of water, and lysed by three cycles of freeze-thawing. After centrifugation (at 15,000 × g for 10 min at 4°C) the supernatant was collected, treated with 400 μl of ice-cold ethanol-chloroform at a 62.5/37.5 (vol/vol) ratio, and centrifuged at 3,000 × g for 5 min at 4°C, and different amounts of the resulting supernatant were used in the SOD assay. The same treatment was performed for 150 μg of highly purified IMV particles. In all cases, the SOD assay was carried out in triplicate and bovine Cu-Zn SOD was used as a standard.
Generation of MAbs against A45R.
Murine monoclonal antibodies (MAbs) specific for A45R were obtained by standard procedures (21). BALB/c mice, 8 to 12 weeks old, were immunized subcutaneously with 50 μg of purified thioredoxin-A45R fusion protein in complete Freund's adjuvant, followed by similar injections in incomplete Freund's adjuvant at 4-week intervals. After a final boost, the spleen cells were fused with mouse hypoxanthine-aminopterin-thymidine (HAT)-sensitive NS1/1 myeloma cells and the resultant hybridomas were screened for A45R-specific antibody by indirect immunofluorescence and immunoblotting. After three cycles of cloning by limiting dilution, a clone that secreted an A45R-reactive antibody was selected and called MAb 2.B.11. This MAb was purified over a protein A-Sepharose column (Sigma) according to the manufacturer's specifications. The subclass of MAb 2.B.11 was identified as immunoglobulin G2a (IgG2a) using an immunoglobulin typing kit (Sigma).
Virus growth curves.
Confluent monolayers of different cell lines were infected at either 0.01 or 10 PFU/cell. After 1 h, the inoculum was removed, and the monolayers were first washed three times with PBS and then overlaid with either MEM containing 2.5% FBS or RPMI supplemented with 10% FBS. At different times p.i., the cells were scraped into the growth medium, freeze-thawed three times, and sonicated, and the infectivity was titrated on BSC-1 cells.
Production of IMV and EEV was determined as follows. Cell monolayers were infected with 10 PFU/cell. After 24 h, the culture supernatant was removed and centrifuged to pellet detached cells, and the supernatant was retained as the EEV fraction. To obtain the IMV fraction, cells were scraped into MEM or RPMI containing 2.5 or 10% FBS, respectively, added to the pelleted cells from above, lysed by three freeze-thaw cycles, and sonicated. Virus titers were determined by plaque assay on BSC-1 cells in duplicate.
Western blot analysis.
BSC-1 cells (106) were mock infected or infected with the appropriate viruses at 10 PFU/cell and maintained for 18 h in the presence or absence of 40 μg of cytosine arabinoside (araC)/ml. Cells were harvested, collected by centrifugation, resuspended in 200 μl of Laemmli buffer with (reducing conditions) or without (nonreducing conditions) 2-β-mercaptoethanol (2-ME), and sonicated to shear DNA. Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred electrophoretically to nitrocellulose membranes (56). After being blocked for 1 h at room temperature (RT) with TBS (20 mM Tris-HCl–500 mM NaCl [pH 7.5]) containing 3% (wt/vol) nonfat dry milk, blots were probed with the purified anti-A45R (α-A45R) MAb 2.B.11 (diluted 1/10,000) in TTBS (TBS–0.2% Tween 20), 1% (w/v) nonfat dry milk. Rabbit polyclonal antisera against the VV protein BIR (diluted 1/2,500) (8) or F13L (diluted 1/100,000) (22) or the murine MAb AB1.1 against the D8L gene product (diluted 1/2,500) (41) was used as a control in some experiments. The blots were then incubated with horseradish peroxidase-conjugated goat α-mouse or α-rabbit Ig (Sigma) diluted 1/2,000 in TTBS–1% nonfat dry milk for 1 h at RT, and the immune complexes were detected using the enhanced chemiluminescence (ECL) detection system (Amersham) according to the manufacturer's instructions. For analysis of virion proteins, 20 μg of purified IMV and EEV particles was resolved by SDS-PAGE, transferred to nitrocellulose membranes, and immunoblotted as above.
Indirect immunofluorescence staining for conventional and confocal microscopy.
Subconfluent BSC-1 cells grown on glass coverslips were infected at 1 PFU/cell. At different times p.i., cells were washed in ice-cold PBS and fixed by incubation with 4% paraformaldehyde (PFA) in 250 mM HEPES (pH 7.4) for 10 min on ice, followed by 40 min at RT in 8% PFA in the same buffer. Cells were then washed with PBS, permeabilized with 0.2% Triton X-100 for 15 min at RT, and treated for 20 min at RT with PBS containing 20 mM glycine. After being washed in PBS, cells were incubated in blocking solution (PBS, 1% bovine serum albumin [BSA], 0.2% fish skin gelatin [pH 8]) for 1 h at RT, incubated with the purified α-A45R MAb 2.B.11 (diluted 1/250) in blocking solution for 1 h at RT, washed extensively with blocking solution, and then incubated with fluorescein isothiocyanate (FITC)-conjugated goat α-mouse Ig (Sigma) at a 1/320 dilution in blocking solution for 30 min at RT. The cells were rinsed three times in PBS and once in water, and then the coverslips were mounted in Mowiol containing 1 μg of 4′,6′-diamidino-2-phenylindole (DAPI)/ml. Samples were analyzed with an Axiophot microscope (Zeiss). Confocal microscopy was performed using a Bio-Rad MCR 1024 laser scanning microscope, and images were collected and processed with COMOS-7 and Adobe Photoshop software.
Fractionation of IMV by detergent treatment.
Viral envelopes were separated from the cores as described previously (14) with some modifications. Purified IMV (1010 particles) was disaggregated by sonication and incubated in lysis buffer (1% NP-40, 50 mM Tris-HCl, 5 mM MgCl2 [pH 7.9]) for 30 min at RT. After centrifugation at 13,000 × g for 15 min at 4°C, the supernatant containing the membrane protein fraction was collected. The pellet was resuspended in lysis buffer containing 50 mM dithiothreitol (DTT) and incubated for 30 min at RT, and then the soluble fraction (membrane protein–NP-40–DTT fraction) was released from the cores by centrifugation under the same conditions. The cores were treated with 1% NP-40–0.5% sodium deoxycholate–0.1% SDS at 4°C for 30 min, and the solubilized core fraction was separated from the insoluble fraction by centrifugation at 13,000 × g for 15 min at 4°C. Samples were resolved by SDS-PAGE, transferred to nitrocellulose membranes, and immunoblotted as above.
Electron microscopy. (i) Immunogold labeling and negative staining of purified IMV particles.
Carbon-coated nickel 400-mesh grids were glow discharged and floated on 108 PFU of purified IMV particles for 5 min at RT. The grids were washed with PBS and fixed by incubation with 4% PFA in 250 mM HEPES (pH 7.4) for 45 min at RT. After being washed in PBS, the samples were either permeabilized with 0.2% Triton X-100 in PBS for 15 min at RT or not permeabilized and treated for 20 min at RT with PBS containing 20 mM glycine. Virus-coated grids were blocked with 3% BSA–0.45% fish skin gelatin in PBS (pH 8) (blocking solution) for 1 h at RT, incubated with the purified α-A45R MAb 2.B.11 (diluted 1/25) in blocking solution for 1 h at RT, and washed extensively with PBS. Bound Ig was detected by incubation for 1 h at RT with colloidal-gold-conjugated rabbit α-mouse Ig (BioCell) diluted 1/10 in blocking solution. The murine MAb C3 against A27L (diluted 1/25) (44) was used as a control in some experiments. After a final wash with PBS, samples were fixed in PBS containing 2.5% glutaraldehyde and washed in double-distilled water, and virus particles were negatively stained with 2.5% uranyl acetate. Samples were examined in a Zeiss Omega 912 electron microscope (LEO Electron Microscope Ltd., Oberkochen, Germany). Digital images were captured with the integrated Soft Imaging Software (SIS) image package (SIS Imaging Software, GmbH, Munster, Germany) and were processed using Adobe Photoshop software.
(ii) Immunogold labeling of thin sections of purified IMV.
Purified IMV particles were pelleted by centrifugation for 40 min at 14,000 × g in a Eppendorf centrifuge and then were fixed with 8% PFA in 250 mM HEPES (pH 7.4) for 1 h at RT. After being washed in PBS, the pellet was incubated for 20 min with 20 mM glycine in PBS, dehydrated in a graded ice-cold ethanol series, and embedded in LR White (polymerization for 12 h at 56°C; London Resin Company), and ultrathin sections on nickel grids were immunolabeled and analyzed as described above.
Virulence assays. (i) Murine intranasal model.
Female BALB/c mice (5 to 6 weeks old) weighing between 15 and 20 g were anesthetized and infected intranasally with either 104, 105, or 106 PFU per animal in 25 μl of PBS (61). Mice were weighed daily and monitored for signs of illness. Animals that had lost more than 30% of their original body weight were sacrificed by cervical dislocation. Virus infectivity in different organs was determined by plaque assay on BSC-1 cells.
(ii)Murine intradermal model.
Female BALB/c mice (8 weeks old) were anesthetized and injected intradermally in the left ear pinnae with either 104, 105, or 106 PFU per animal in 10 μl of PBS. The diameters of lesions produced on infected ears were determined daily using a micrometer. Virus titers in ears at different times after inoculation were determined as described previously (57).
(iii) Rabbit skin model.
A female New Zealand White rabbit (weighing 1.5 kg) was inoculated subcutaneously along the left and right flanks with either 104, 105, or 106 PFU/site in 100 μl of PBS. The diameters of lesions produced were determined daily. For histological examination of VV-induced lesions, a similar experiment was carried out, in which the rabbit was sacrificed 3 days after inoculation.
Statistical analysis.
Student's t test was used to test for the significance of the results (P < 0.05).
Nucleotide sequence accession numbers.
The nucleotide sequences presented in this paper have been assigned GenBank accession numbers AF349002, AF349003, AF349004, AF349005, AF349006, AF349006, AF349007, AF349008, AF349009, AF349010, AF349011, AF349012, AF349013, AF3749014, AF349015, and AF349016.
RESULTS
The A45R gene is conserved in orthopoxviruses.
The A45R gene in the WR strain of VV is located within the HindIII A fragment at the right end of the VV genome. The sequence of the A45R gene predicts a primary translation product of 125 amino acids with a molecular size of 13.6 kDa and an estimated isoelectric point of 6.5. The protein does not contain any potential signal peptide or transmembrane anchor sequence. As noted previously, A45R is predicted to encode a polypeptide with 39% amino acid identity with Cu-Zn SODs from eukaryotic sources (51).
Nucleotide sequence data are available for the A45R gene of several VV and variola virus strains (see the introduction). The predicted protein sequence is highly conserved among these viruses, but comparison with eukaryotic Cu-Zn SODs indicated that A45R might not function as a SOD (51). Therefore, we investigated if the gene is conserved in other orthopoxviruses, and in particular, whether the missing regions at the C terminus are present. The nucleotide sequences of the A45R genes from 12 strains of VV, cowpox virus (Brighton Red strain), elephantpox virus (considered a cowpox virus strain), and camelpox virus were determined. Alignment of the deduced amino acid sequences showed that A45R is highly conserved (94 to 100% amino acid identity) and has the same length in each virus with the exception of VV strains MVA (containing a deletion of amino acids 27 to 30) and Tashkent (containing a base insertion near the 3′ end of the gene that produces a frameshift so that the last 12 amino acids are absent and are replaced by an unrelated sequence of 7 amino acids, NNTFSYN). In all cases, the protein lacked the Zn-binding region of loop 6, 5 and the loop 7, 8 near the C terminus. However, the highly conserved nature of the A45R gene suggests that it might have an important function in the virus life cycle.
The A45R protein expressed in E. coli does not have SOD activity.
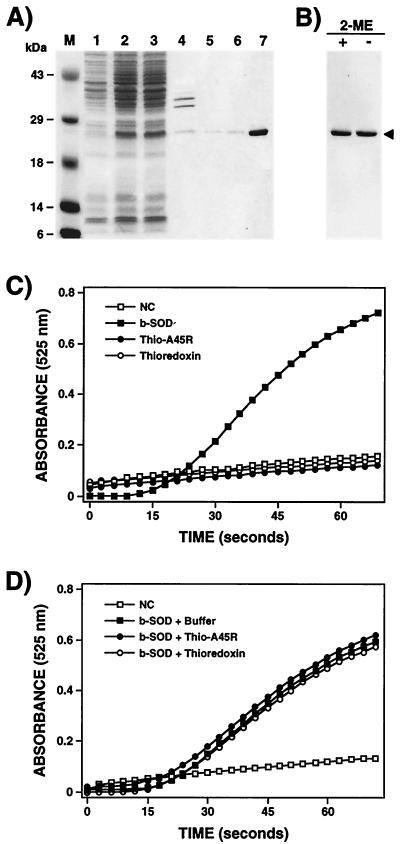
To characterize biochemically the A45R protein and to produce greater amounts of protein for functional analysis and antibody production, the protein was expressed as a fusion protein in an IPTG-inducible bacterial vector and purified to near-homogeneity by chromatography on a nickel agarose resin (see Materials and Methods). Expression of large amounts of soluble fusion protein was possible only when the induction step was carried out at 30°C, the affinity purification was performed in the absence of NaCl, and elution from the nickel column was done with 50 mM EDTA (see Materials and Methods). As shown in Fig. 1A, bacteria carrying the pFAT8 plasmid expressed an abundant polypeptide of 25 kDa after, but not before, induction with IPTG. Because two cysteine residues are present in the amino acid sequence of A45R, we investigated whether the affinity-purified fusion protein formed disulfide-bonded dimers. The purified protein was boiled in Laemmli buffer with (reducing conditions) or without (nonreducing conditions) 2-ME and was analyzed by SDS-PAGE, but the migration of the protein was unaltered (Fig. 1B).
FIG. 1.
Expression, purification, and biochemical characterization of A45R protein. (A) Expression of A45R in E. coli. The A45R protein was expressed in E. coli as a thioredoxin fusion protein with the His-Patch ThioFusion Expression System (Invitrogen, BV) and purified by affinity chromatography on a nickel column as described in Materials and Methods. Samples were analyzed by SDS-PAGE (12% gel), and proteins were visualized by staining with Coomassie brilliant blue. Lanes: 1, uninduced whole-cell extract; 2, induced whole-cell extract; 3, induced soluble lysate; 4, induced insoluble lysate; 5, 6, and 7, proteins eluted from the Ni2+ column with 350 or 500 mM imidazole or 50 mM EDTA, respectively. Molecular size markers (M) are shown in kilodaltons. (B) Analysis of the purified fusion protein under reducing and nonreducing conditions. Before electrophoresis on a 12% polyacrylamide gel, samples were boiled for 3 min in Laemmli buffer with (+) or without (−) 2% 2-ME. Proteins were detected by staining with Coomassie brilliant blue. The arrowhead indicates the position of the fusion protein. (C and D) Investigation of possible enzymatic and inhibitory activity of the A45R protein expressed in E. coli. The SOD activity assay (C) was carried out with 5 μg of the purified fusion protein (Thio-A45R) as described in Materials and Methods. The same assay was done without protein (NC) or with 5 μg of thioredoxin as a negative control and with 5 μg of bovine Cu-Zn SOD (b-SOD) as a positive control. The inhibitory effect of the purified protein on the enzymatic activity of the bovine Cu-Zn SOD was also analyzed (D). In this case, before the SOD assay, 5 μg of bovine Cu-Zn SOD was incubated for 30 min at 37°C with either buffer, 5 μg of the purified fusion protein, or 5 μg of thioredoxin. Mean values from three experiments are shown.
The purified protein was tested for enzymatic or inhibitory SOD activity using a commercial SOD assay kit (see Materials and Methods), based on a molecule (5,6,6a,11b-tetrahydro-3,9,10-trihydroxybenzo[c]fluorene) that undergoes alkaline auto-oxidation, which is accelerated by SOD. This auto-oxidation yields a chromophore that absorbs maximally at 525 nm. Figures 1C and D show that the purified protein did not have either SOD activity or an inhibitory effect on the enzymatic activity of bovine Cu-Zn SOD. Identical results were obtained when the experiments were carried out with purified A45R protein separated from the thioredoxin by cleavage with enterokinase (data not shown).
Disruption of the A45R gene in the virus genome.
To help identify and to study the function of the A45R gene product in the virus life cycle, a deletion mutant virus lacking 60% of the ORF (vΔA45R), a plaque-purified wild-type virus (vWTA45R), and a revertant virus (vRA45R) were constructed (see Materials and Methods). The genome structures of the recombinant viruses were analyzed by Southern blotting and PCR and were shown to be as predicted. In addition, expression of the A44L and A46R genes was analyzed by Western and Northern blotting respectively, and no differences were detected among the three viruses (data not shown). Finally, the absence of A45R expression by vΔA45R was demonstrated by immunoblotting extracts from cells infected with this virus (see below). Collectively, these data showed that the recombinant viruses have the predicted genome structures and gene expression patterns.
A45R is not essential for virus growth in culture.
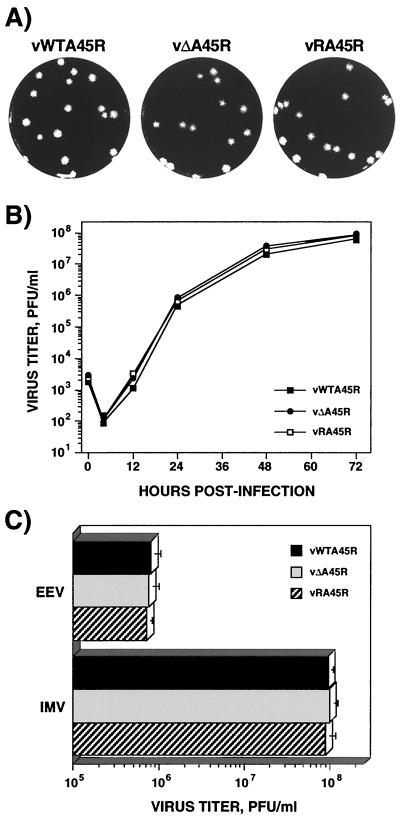
Isolation of the deletion mutant virus vΔA45R showed that the A45R gene is dispensable for virus replication in vitro. In fact, the size and morphology of plaques formed by vΔA45R on BSC-1 cells were indistinguishable from those of vWTA45R and vRA45R (Fig. 2A). Nevertheless, it was possible that there were differences in the virus yield that would not be reflected in plaque size. To test this possibility, the growth kinetics of these viruses were compared. After infection of BSC-1 cells at 0.01 PFU/cell (Fig. 2B) or 10 PFU/cell (data not shown), no differences in total-virus yields were detected for the three viruses. To compare the amounts of IMV and EEV produced by these viruses, BSC-1 cells were infected at 10 PFU/cell and the virus in the cells or supernatant was determined at 24 h p.i. as described in Materials and Methods. Similar titers were produced by vΔA45R, vWTA45R, and vRA45R (Fig. 2C), confirming that loss of the A45R gene did not affect the production of normal amounts of either IMV or EEV. The three viruses also share similar growth characteristics after infection of CV-1, RK13, HeLa, and D980R cells (data not shown).
FIG. 2.
Growth properties of vΔA45R in cell culture. (A) Plaque phenotype. vWTA45R, vΔA45R, and vRA45R were plated onto monolayers of BSC-1 cells and were overlaid with MEM supplemented with 2.5% FBS and 1.5% carboxymethylcellulose for 2 days before being stained with 0.1% crystal violet in 15% ethanol. (B) Growth curves. BSC-1 cells were infected at 0.01 PFU/cell with vWTA45R, vΔA45R, or vRA45R. At different times p.i. cells were scraped into the growth medium, and the production of infectious virus was determined by plaque assay on BSC-1 cells. Data shown are mean values from triplicate samples. The standard deviation was lower than 20% in all cases (not shown). (C) IMV and EEV production. Monolayers of BSC-1 cells were infected with 10 PFU of either vWTA45R, vΔA45R, or vRA45R/cell. After 24 h, the yields of IMV and EEV were determined by plaque assay on BSC-1 cells. Data are means from three experiments. Error bars, standard deviations.
Finally, the growth properties of vΔA45R, vWTA45R and vRA45R were analyzed in murine resident peritoneal macrophages or bone marrow-derived macrophages, and in the mouse or human monocyte-macrophage cell lines P388D1 and U937, respectively. After infection with 0.01 or 10 PFU/cell, no virus production was detected in either peritoneal or bone marrow-derived murine macrophages. In contrast, virus replication was observed in P388D1 and U937 cells, but the yields of infectious virus were indistinguishable for the three viruses (data not shown).
Overall, these results indicate that the A45R gene is dispensable for replication of VV in tissue culture and does not lead to a host range restriction on any of the monkey, rabbit, or human cell lines tested.
The A45R gene encodes a 13.5-kDa polypeptide expressed late during VV infection.
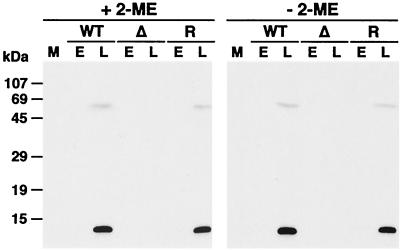
The nucleotide sequence of the WR A45R gene revealed a potential late transcriptional start site (TAAAT) (15) close to the initiation codon, suggesting that the gene might be expressed late during infection. To investigate this experimentally, the expression of A45R protein in VV-infected cells was analyzed by Western blotting using the A45R-specific MAb 2.B.11. Parallel blots were probed with a rabbit polyclonal antiserum against the B1R protein (8) to show that virus-specific protein was present in all samples (data not shown). Extracts from BSC-1 cells 18 h after infection with vWTA45R and vRA45R contained a protein of 13.5 kDa that was present only in the absence of araC and was absent from uninfected cells and cells infected with vΔA45R (Fig. 3). In addition, there was a minor higher-molecular-size polypeptide (∼65 kDa) also recognized by the MAb 2.B.11 that might represent A45R oligomers. In the absence of reducing agents, no differences in the electrophoretic mobility or in the protein pattern were detected (Fig. 3), suggesting that the putative A45R oligomers are not due to the formation of disulfide bonds. Finally, analysis of the proteins present in the tissue culture supernatant of VV-infected cells indicated that A45R protein was not secreted from the cell (data not shown).
FIG. 3.
Identification of the A45R protein in VV-infected cells. BSC-1 cells were mock infected (M) or infected with vWTA45R (WT), vΔA45R (Δ), or vRA45R (R) at 10 PFU/cell in the presence (E) or absence (L) of 40 μg of araC/ml. At 18 h p.i. cell lysates were resolved by SDS-PAGE (15% gel) under reducing (+ 2-ME) or nonreducing (− 2-ME) conditions, transferred to nitrocellulose, and incubated with MAb 2.B.11 (diluted 1/10,000). Bound antibodies were detected by using horseradish peroxidase-conjugated goat α-mouse Ig (Sigma) and ECL reagents as described in Materials and Methods. Positions of molecular size markers are shown.
Subcellular localization of A45R protein in VV-infected cells.
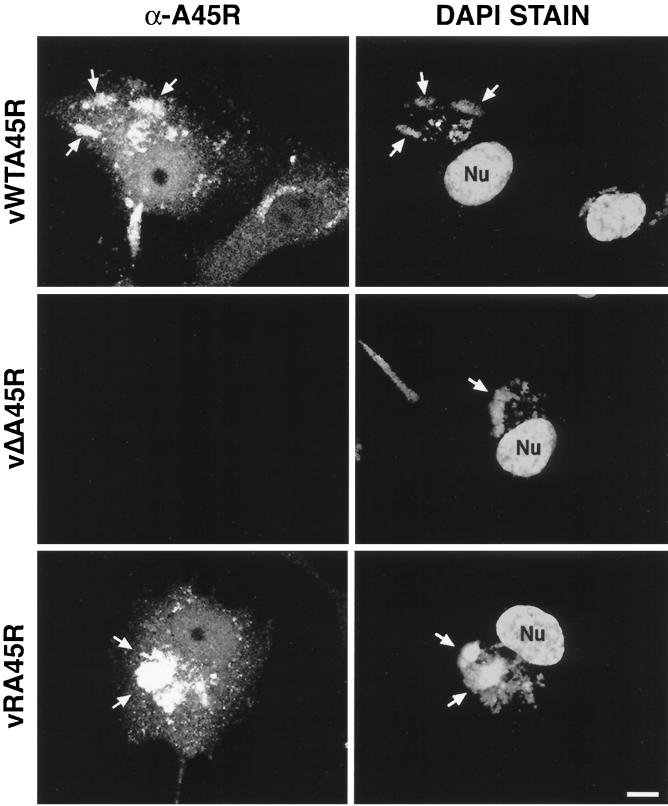
The intracellular localization of A45R during VV infection was determined by confocal microscopy (Fig. 4). In cells infected with either vWTA45R or vRA45R, the A45R protein was distributed diffusely throughout the nucleus and cytoplasm of infected cells but showed a more intense staining pattern in perinuclear cytoplasmic regions that colocalized with the viral DNA factories, which were visualized by DAPI staining (Fig. 4). In addition, MAb 2.B.11 decorated punctate structures throughout the cell, which probably represent individual virus particles. In cells infected with vΔA45R no staining was detected, demonstrating the specificity of MAb 2.B.11. As a control, immunofluorescence analysis was carried out with MAb AB1.1, which recognizes the D8L protein, and identical staining patterns were observed for the three viruses (data not shown).
FIG. 4.
Localization of VV A45R protein in infected cells by immunofluorescence. Subconfluent BSC-1 cells were infected with the indicated viruses at 1 PFU/cell. At 14 h p.i., cells were fixed, permeabilized, and incubated with MAb 2.B.11. Bound antibody was detected with FITC-conjugated secondary antibody as described in Materials and Methods. Representative cells are shown. Viral and cellular DNA were stained with DAPI. Arrows indicate viral factories. Nu, nucleus. Bar, 10 μm.
A45R protein is encapsidated and localizes in the viral core.
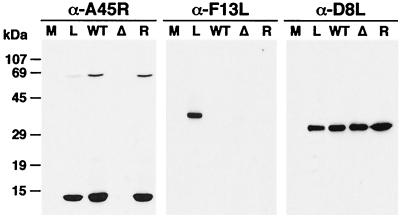
To investigate whether the A45R protein was present in virus particles, vWTA45R, vΔA45R, and vRA45R were grown in RK13 cells and IMV was purified by sucrose density gradient centrifugation. Virus proteins were then analyzed in parallel with extracts of vWTA45R-infected cells by immunoblotting with MAb 2.B.11 (Fig. 5). Proteins of 13.5 and 65 kDa were detected in infected cells and in IMV particles from vWTA45R and vRA45R but not in IMV particles from vΔA45R. The absence of the 65-kDa polypeptide in mock-infected cells and in IMV particles from vΔA45R indicates that it is A45R specific. As in the case of infected-cell extracts, the presence or absence of reducing agents did not alter the electrophoretic mobility or pattern of virion proteins. Whether the high-molecular-weight product is a homo- or a heteromultimeric complex is not known. The blot was stripped and reprobed with MAb AB1.1 against the IMV protein D8L to confirm equivalent protein loading. The purity of IMV preparations was confirmed by probing the blot with anti-F13L antibody, which recognizes an EEV-specific protein (22). These data demonstrate that A45R is incorporated into virus particles.
FIG. 5.
The A45R protein is incorporated into virions. IMVs from vWTA45R (WT), vΔA45R (Δ), or vRA45R (R) were analyzed by SDS-PAGE together with extracts from BSC-1 cells mock infected (M) or infected with vWTA45R at 10 PFU/cell for 18 h (L). After transfer to nitrocellulose filters, proteins were detected by immunoblotting with MAb 2.B.11. As controls, the blot was reprobed with a rabbit α-F13L antibody or with the mouse α-D8L MAb AB1.1. Positions of molecular size markers are shown.
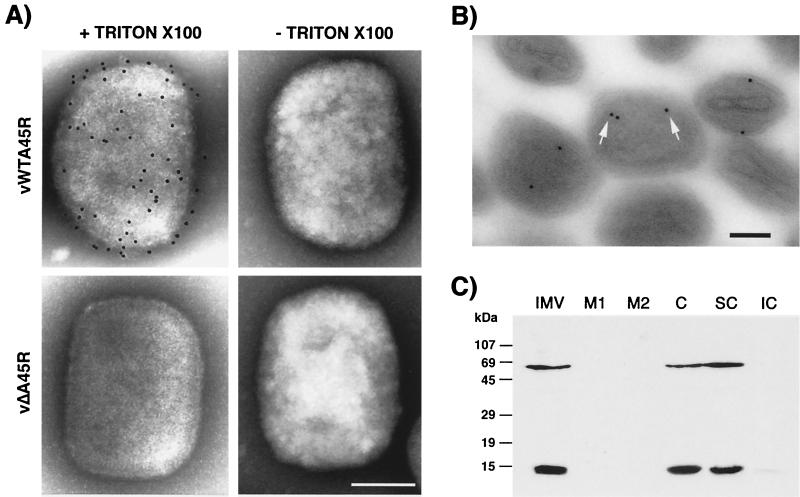
To study the subviral localization of A45R, we used three different approaches. First, purified IMV preparations were analyzed by immunoelectron microscopy before and after permeabilization (Fig. 6A). Intact vWTA45R and vΔA45R IMV particles were not labeled with MAb 2.B.11, suggesting that the A45R protein is not exposed on the virion surface. In contrast, vWTA45R and vΔA45R particles were labeled by MAb C3 against the VV A27L protein, which is exposed on the surface of the virion (data not shown) (53). When IMV particles were permeabilized by treatment with Triton X-100 to allow the antibody access to epitopes protected by the virus envelope, immunogold labeling of vWTA45R IMV but not of vΔA45R IMV was observed, suggesting that the A45R protein is associated with the viral core.
FIG. 6.
The A45R protein is located in the viral core. (A) Immunogold labeling and negative staining electron microscopy of purified IMV particles. IMV purified by sucrose gradient centrifugation from RK13 cells infected with either vWTA45R or vΔA45R was adsorbed to carbon-coated nickel 400-mesh grids, permeabilized (+ Triton X-100) or unpermeabilized (− Triton X-100), and incubated with MAb 2.B.11. Bound antibody was detected with 5-nm gold particle-conjugated goat α-mouse Ig. Representative viral particles are shown. Bar, 100 nm. (B) Immunogold labeling of thin sections of purified IMV particles. Purified vWTA45R IMV particles were pelleted by high-speed centrifugation, fixed with 8% PFA, and embedded in LR White (London Resin Company). Ultrathin sections were immunolabeled with MAb 2.B.11, followed by 10-nm gold particle-conjugated goat α-mouse Ig, and were examined by electron microscopy. Some gold particles are indicated by arrows. Bar, 100 nm. (C) Fractionation of purified IMV by detergent treatment. Purified vWTA45R IMV particles (IMV) were incubated in lysis buffer containing 1% NP-40 in the absence or presence of 50 mM DTT, and soluble envelopes (M1 and M2, respectively) were removed from viral cores (C) by centrifugation. Cores were partitioned into soluble (SC) and insoluble (IC) fractions by treatment with 0.5% sodium deoxycholate and 0.1% SDS. Fractions were then subjected to SDS-PAGE (15% gel), transferred to nitrocellulose, blotted with MAb 2.B.11, and developed with ECL reagents. Molecular size markers are shown.
The localization of the A45R protein was analyzed further by processing purified vWTA45R IMV for electron microscopy and subsequent immunogold labeling of ultrathin sections with MAb 2.B.11 (Fig. 6B). Careful examination of the distribution of the gold particles in IMV particles indicated that the great majority (80%) were located either on the surface of the core or in the space between the surrounding envelope and the core. No staining of vΔA45R IMV was observed.
Finally, to confirm the subviral localization of A45R protein, virions were treated with NP-40 in the presence or absence of DTT to release detergent-soluble membrane proteins from the cores, and these were partitioned further into soluble and insoluble fractions by treatment with deoxycholate and SDS. Fractions were then analyzed by SDS-PAGE and immunoblotting with antibodies against D8L, L4R, and A45R. The detection of the VV D8L protein in the membrane fraction and L4R in the core fraction demonstrated the effectiveness of the fractionation (data not shown). Both the 13.5- and 65-kDa A45R proteins were retained in the core after NP-40–DTT fractionation and were recovered in the deoxycholate-SDS-soluble fraction of the core (Fig. 6C).
Analysis of SOD activity in VV-infected cells and purified IMV particles.
Although the A45R protein expressed in E. coli did not exhibit either SOD activity or inhibitory activity toward bovine Cu-Zn SOD, it was possible that this protein did not have the correct folding to allow functional activity. Therefore, SOD activity assays were performed with extracts of BSC-1, RK13, HeLa, P388D1, and U937 cells 14 h after infection with vWTA45R, vΔA45R, or vRA45R. No differences in SOD activity were detected in mock-infected or virus-infected cells (data not shown). Since the A45R protein is incorporated into virions, similar experiments were carried out with purified IMV particles, and neither SOD activity nor inhibitory activity was detected (data not shown). These data indicate that the WR A45R protein does not have SOD activity or any effect on the cellular SOD, at least in the cells and under the conditions analyzed.
Virulence of VV lacking the A45R gene.
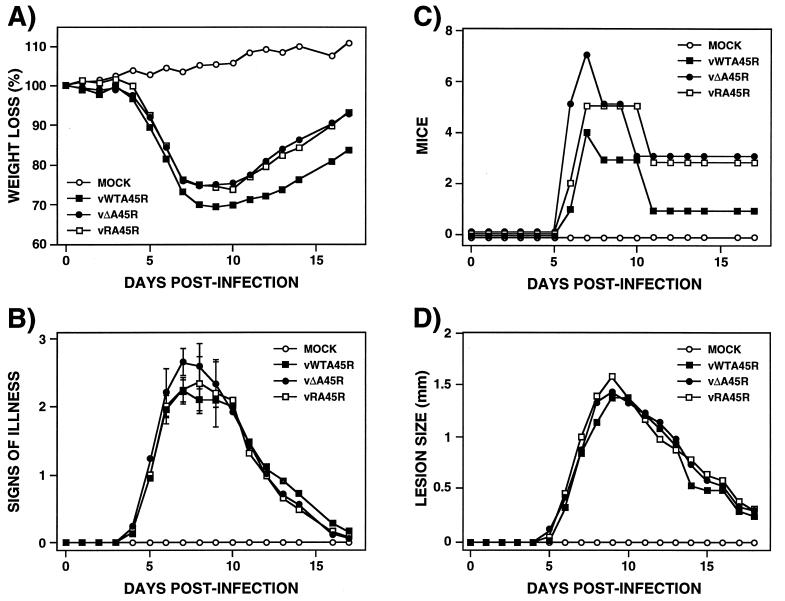
To determine if the A45R gene affects virulence, the outcome of infection with vΔA45R was compared to that with vWTA45R and vRA45R in a murine intranasal model (61). Virulence was assessed by weight loss and signs of illness (piloerection, arched back, and reduced mobility). Animals infected with each virus at doses of 105 or 106 PFU per animal all suffered severe weight loss and signs of illness and were sacrificed by day 6 p.i. (data not shown). At 104 PFU there were no significant differences in the amount of weight loss or the rate of recovery between mice infected with the different viruses (Fig. 7A), but mice infected with vΔA45R showed slightly higher signs of illness on days 6 to 8 compared with mice infected with either vWTA45R or vRA45R (Fig. 7B). Statistical analysis using the unpaired Student's t test found a significant difference (P = 0.048) between the mean values for the vΔA45R-infected group and each other group on day 7 only. Since the difference is significant only on 1 day and the P value is just within the limit, it is very difficult to conclude, at least in this model, that A45R plays any role in virulence. However, when the number of animals presenting severe signs of illness at different times p.i. was plotted (Fig. 7C), an early onset of severe signs in vΔA45R-infected animals was observed, suggesting a possible effect of A45R on the outcome of infection.
FIG. 7.
Virulence of vΔA45R in mice. (A, B, and C) Murine intranasal model. Groups of 10 female BALB/c mice, 5 to 6 weeks old, were mock infected or infected intranasally with 104 PFU of the indicated virus per animal. Percent weight changes (A), signs of illness (B), and the number of animals with illness scores of ≥2.5 (C) were analyzed daily. Animals that had lost more than 30% of their body weight were sacrificed. The mean weight of each group of mice was expressed as a percentage of the mean weight of that group of animals immediately prior to infection. Signs of illness were scored from zero to four, and the mean value for each group is shown. The number of animals that presented strong signs of illness (scores of ≥2.5) is represented at different days p.i. Standard deviations in panel B are given as error bars for days 6 to 9. (D) Murine intradermal model. Groups of six 8 to 9-week-old female BALB/c mice were mock infected or infected intradermally in the left ear pinnae with 104 PFU of the indicated virus. The diameters of lesions produced were determined daily using a micrometer. Each data point represents the mean for a group.
The virulence of vΔA45R was then analyzed in a murine intradermal model (57). In this model, inoculation of VV strain WR into the ear pinnae leads to a localized and limited infection where virus spread is minimal and the mice suffer no signs of systemic illness. Groups of six animals were injected intradermally in the left ear pinnae with doses of 104, 105, or 106 PFU of either vWTA45R, vΔA45R, or vRA45R/ear, and the diameters of resulting lesions were measured daily. Figure 7D shows that animals infected with these viruses at 104 PFU/ear did not present significant differences in lesion size. Identical results were obtained at doses of 105 and 106 PFU/ear (data not shown). In a second experiment, the titers of infectious virus in ears at different times p.i. and the cellular and humoral immune responses were determined as described previously (57). There was no significant difference in virus titer or in cellular and humoral immune response after infections with the different viruses (data not shown). Finally, the role of the A45R gene in virulence was assessed in a rabbit skin model. For this purpose, a New Zealand White rabbit was inoculated subcutaneously along the left and right flanks with either vWTA45R, vΔA45R, or vRA45R at 104, 105, or 106 PFU/site, and the diameters of lesions produced were determined daily. As described for the murine intradermal model, there was no difference in size among the lesions produced by the three viruses (data not shown). A histological examination of virus-induced lesions in both models was performed, and again no differences were detected among the different viruses (data not shown). These results show that, at least in the last two models, the A45R gene does not contribute to virus virulence.
DISCUSSION
VV and other poxviruses express a wide variety of proteins that are nonessential for virus replication in vitro but help the virus to evade the host response to infection. In this paper we have characterized the VV A45R protein, which shares 39% amino acid identity with eukaryotic Cu-Zn SOD. Data presented show that the A45R protein is expressed late during infection and is incorporated into IMV particles but lacks SOD activity. A virus with a disrupted A45R gene replicated normally in vitro and had unaltered virulence in mice and rabbits.
The A45R gene was selected for study because VV virulence might be influenced by the presence of SOD, an enzyme that provides defense against damage caused by superoxide radicals (18). This enzyme could be used by poxviruses to avoid the toxic effects of UV light and protect against the DNA-, lipid-, and protein-damaging effects of reactive oxygen species. Furthermore, vertebrate macrophages and neutrophils produce superoxide radicals as a means of killing invading microorganisms (7, 58). Therefore, a virus-encoded SOD activity might induce virus resistance to destruction by an oxidative burst when the virus enters macrophages either after phagocytosis or as part of its replicative cycle.
Published sequence data for four strains of VV and four strains of variola virus showed that the A45R protein is highly conserved and shares more than 96% amino acid identity. However, the protein has lost some surface peptide loops and the Zn-binding domain that are critical for SOD activity (51), suggesting that it was unlikely to have SOD activity. Nonetheless, it was possible that the A45R protein might be active in other VV strains or other orthopoxviruses. There are precedents for a VV gene being functional in some VV strains but not others. For instance, an active tumor necrosis factor receptor is expressed by 3 out of 16 VV strains (4). Therefore, the A45R gene was sequenced in 12 additional VV strains, camelpox virus, and cowpox virus. The protein was highly conserved, but in all cases the two major deletions affecting residues critical for SOD activity were present. In agreement with sequence data, no SOD activity was detected for the protein expressed in E. coli (Fig. 1) or in extracts of virus-infected cells in comparison with extracts of uninfected or deletion mutant virus-infected cells, or in purified virus particles.
Within the Poxviridae, proteins related to SOD are present in the leporipoxviruses myxoma virus (12) and Shope fibroma virus (60), the molluscipoxvirus molluscum contagiosum virus (48), and the entomopoxvirus B Amsacta moorei virus (9). Sequence analysis revealed that all have deletions or substitutions within the coding region that are likely to render the protein inactive, with the exception of Amsacta moorei virus, where the SOD protein appears to be intact. However, no SOD activity has been reported for the entomopoxvirus protein. Potential SODs have also been identified in other complex DNA viruses including baculovirus (55) and the chlorella virus PBCV-1, a member of the Phycodnaviridae family (30). However, SOD activity in these proteins remains to be demonstrated. It is interesting that members of the Baculoviridae, Poxviridae, and Phycodnaviridae families, which have significantly different replication strategies, carry SOD-like proteins.
The presence of a complete SOD in entomopoxvirus and baculovirus might reflect a function for SOD in viruses replicating in insect hosts, and perhaps this activity might have been lost during the evolution of vertebrate poxviruses. Alternatively, it is possible that poxviruses use a nonfunctional SOD-like protein to inhibit or regulate cellular SOD function or for some other purpose. For instance, the regulation of cellular SOD in poxvirus-infected cells might disrupt the balance of oxidants and antioxidants. Further, since oxidative stress can induce apoptosis (11), this may aid virus dissemination. Additionally, an increase in the cellular oxidant status results in activation of transcriptional factors, such as NF-κB (46), that may be necessary for replication of some viruses. For instance, the Tat protein from human immunodeficiency virus type 1 induces the activation of NF-κB, which is important for virus replication, by down-regulation of cellular MnSOD (59). Another example is the Epstein-Barr virus, which induces autoantibodies against cellular MnSOD, thus contributing to viral pathogenesis (43).
An A45R-specific MAb was isolated and was used to identify the A45R protein in VV-infected cells as a major 13.5-kDa polypeptide that was expressed late during infection, consistent with the presence of the TAAAT motif (15) close to the translational start site. Additionally, a minor higher-molecular-size polypeptide of about 65 kDa was detected. The presence of this high-molecular-weight protein might be explained by postranslational modifications of the 13.5-kDa protein, or by the formation of homo- or hetero-oligomeric structures. Although no disulfide-bonded oligomers were detected, many of the glycine residues involved in the formation of cellular SOD homodimers (40) are conserved in A45R. Further study is required to address the nature of this 65-kDa protein.
Immunofluorescence microscopy showed that the A45R protein was distributed widely in the nuclei and cytosol of infected cells. Interestingly, a similar pattern was described for eukaryotic Cu-Zn SODs (13). Small proteins (up to 50 kDa) can diffuse passively through the nuclear pore, whereas proteins destined for active nuclear import possess specific nuclear localization signals (NLSs) that are recognized by protein import receptors (34). A45R and Cu-Zn SODs do not have NLSs, suggesting that either they diffuse through the nuclear pore due to their small size, they contain unknown NLSs, or they interact with other proteins that contain NLSs.
The presence of the A45R protein in the core fraction of viral particles was demonstrated by Western blotting and immunogold labeling of purified virus particles. This was evident from labeling of thin sections and permeabilized particles, and the absence of labeling of the surface of intact IMV. These data were consistent with the biochemical data that showed that the protein was present in the soluble fraction of the core. Interestingly, the 65-kDa protein detected in infected cells was also incorporated into the IMV core. This polypeptide is recognized specifically by the MAb, and therefore it is likely to be the result of the interaction of A45R with itself or with other cellular or viral proteins. A study of VV protein-protein interactions by two-hybrid analysis has been reported recently (36). This analysis indicated that A45R interacts with itself, with A4L, a virion protein that localizes between the core and IMV membrane (14), and with J1R, another virion protein that forms homodimers (5). Further study of these interactions in vivo and their functions in the virus life cycle is needed.
The high conservation of the A45R gene in all orthopoxviruses analyzed and the association of the protein with viral particles suggest an important role during VV infection. To address the essentiality and function of A45R, a recombinant virus lacking the majority of the A45R gene was constructed and compared with wild-type and revertant viruses. The growth properties, plaque phenotype, and infectious intra- and extracellular virus yields of the deletion mutant virus were indistinguishable from those of the wild-type and revertant viruses in all cell lines tested, including monocyte-macrophage cell lines. Additionally, A45R was found to play no role in virus virulence in either a murine or a rabbit intradermal model. In a murine intranasal model, a possible early onset of signs of illness was detected after infection with the deletion mutant, suggesting a possible function of A45R in the progression of the infection.
This very limited, if any, role of A45R in virulence makes A45R the first example of a VV structural protein that is nonessential for virus replication in vitro and in vivo. There are other structural VV proteins that are nonessential in vitro, but in all cases a role in virulence has been reported. For instance, D8L and A56R (virus hemagglutinin), membrane proteins of IMV and EEV particles, respectively, are nonessential for virus replication in vitro, but the deletion or disruption of these genes produces virus attenuation in vivo (17, 31, 39, 45). It is possible that the A45R gene appears to be nonessential because its function is compensated for by other viral or host proteins. Despite the A45R gene being unnecessary for virus replication in monocyte-macrophage cell lines, the protein could be important for viral replication in resting macrophages in vivo. We have shown that the WR strain of VV does not replicate in primary macrophage cultures, consistent with previous reports (10, 38). This abortive infection might be due to the disruption in VV strain WR of the gene related to p28 of ectromelia virus, which is critical for in vitro replication of ectromelia virus in murine resident peritoneal macrophages (47). Further studies with VV strains that encode a complete p28 might elucidate the possible function of A45R during VV replication in macrophages. Alternatively, the protein may play a role in some circumstances that we have not studied, such as in an alternate host, or after exposure to extremes of temperature or UV light. Experiments to investigate some of these possibilities are in progress.
ACKNOWLEDGMENTS
We thank Francisco Iborra and Ana Pombo for advice and assistance with electron and confocal microscopy, Pike Pucklevic for help with generation of MAbs, and Sara Thompson for tissue culture.
This work was supported by grants from The Wellcome Trust and the United Kingdom Medical Research Council. F.A. was the recipient of a long-term EMBO fellowship, D.C.T. was a Wellcome Trust Traveling Fellow, and G.L.S. is a Wellcome Trust Principal Research Fellow.
REFERENCES
- 1.Aguado B, Selmes I P, Smith G L. Nucleotide sequence of 21.8 kbp of variola major virus strain Harvey and comparison with vaccinia virus. J Gen Virol. 1992;73:2887–2902. doi: 10.1099/0022-1317-73-11-2887. [DOI] [PubMed] [Google Scholar]
- 2.Alcamí A, Smith G L. Vaccinia, cowpox, and camelpox viruses encode soluble gamma interferon receptors with novel broad species specificity. J Virol. 1995;69:4633–4639. doi: 10.1128/jvi.69.8.4633-4639.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alcamí A, Symons J A, Collins P D, Williams T J, Smith G L. Blockade of chemokine activity by a soluble chemokine binding protein from vaccinia virus. J Immunol. 1998;160:624–633. [PubMed] [Google Scholar]
- 4.Alcamí A, Khanna A, Paul N L, Smith G L. Vaccinia virus strains Lister, USSR and Evans express soluble and cell-surface tumour necrosis factor receptors. J Gen Virol. 1999;80:949–959. doi: 10.1099/0022-1317-80-4-949. [DOI] [PubMed] [Google Scholar]
- 5.Antoine G, Scheiflinger F, Dorner F, Falkner F G. The complete genomic sequence of the modified vaccinia Ankara strain: comparison with other orthopoxviruses. Virology. 1998;244:365–396. doi: 10.1006/viro.1998.9123. [DOI] [PubMed] [Google Scholar]
- 6.Appleyard G, Hapel A J, Boulter E A. An antigenic difference between intracellular and extracellular rabbitpox virus. J Gen Virol. 1971;13:9–17. doi: 10.1099/0022-1317-13-1-9. [DOI] [PubMed] [Google Scholar]
- 7.Babior B M. The respiratory burst oxidase. Trends Biochem Sci. 1987;12:241–243. [Google Scholar]
- 8.Banham A H, Smith G L. Vaccinia virus gene B1R encodes a 34-kDa serine/threonine protein kinase that localizes in cytoplasmic factories and is packaged into virions. Virology. 1992;191:803–812. doi: 10.1016/0042-6822(92)90256-o. [DOI] [PubMed] [Google Scholar]
- 9.Bawden A L, Glassberg K J, Diggans J, Shaw R, Farmerie W, Moyer R W. Complete genomic sequence of the Amsacta mooreientomopoxvirus: analysis and comparison with other poxviruses. Virology. 2000;274:120–139. doi: 10.1006/viro.2000.0449. [DOI] [PubMed] [Google Scholar]
- 10.Broder C C, Kennedy P E, Michaels F, Berger E A. Expression of foreign genes in cultured human primary macrophages using recombinant vaccinia virus vectors. Gene. 1994;142:167–174. doi: 10.1016/0378-1119(94)90257-7. [DOI] [PubMed] [Google Scholar]
- 11.Buttke T M, Sandstrom P A. Oxidative stress as a mediator of apoptosis. Immunol Today. 1994;15:7–10. doi: 10.1016/0167-5699(94)90018-3. [DOI] [PubMed] [Google Scholar]
- 12.Cameron C, Hota-Mitchell S, Chen L, Barrett J, Cao J X, Macaulay C, Willer D, Evans D, McFadden G. The complete DNA sequence of myxoma virus. Virology. 1999;264:298–318. doi: 10.1006/viro.1999.0001. [DOI] [PubMed] [Google Scholar]
- 13.Crapo J D, Oury T, Rabouille C, Slot J W, Chang L Y. Copper,zinc superoxide dismutase is primarily a cytosolic protein in human cells. Proc Natl Acad Sci USA. 1992;89:10405–10409. doi: 10.1073/pnas.89.21.10405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cudmore S, Blasco R, Vincentelli R, Esteban M, Sodeik B, Griffiths G, Krijnse Locker J. A vaccinia virus core protein, p39, is membrane associated. J Virol. 1996;70:6909–6921. doi: 10.1128/jvi.70.10.6909-6921.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davison A J, Moss B. Structure of vaccinia virus late promoters. J Mol Biol. 1989;210:771–784. doi: 10.1016/0022-2836(89)90108-3. [DOI] [PubMed] [Google Scholar]
- 16.Falkner F G, Moss B. Transient dominant selection of recombinant vaccinia viruses. J Virol. 1990;64:3108–3111. doi: 10.1128/jvi.64.6.3108-3111.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Flexner C, Hugin A, Moss B. Prevention of vaccinia virus infection in immunodeficient mice by vector-directed IL-2 expression. Nature. 1987;330:259–262. doi: 10.1038/330259a0. [DOI] [PubMed] [Google Scholar]
- 18.Fridovich I. Superoxide radical and superoxide dismutases. Annu Rev Biochem. 1995;64:97–112. doi: 10.1146/annurev.bi.64.070195.000525. [DOI] [PubMed] [Google Scholar]
- 19.Goebel S J, Johnson G P, Perkus M E, Davis S W, Winslow J P, Paoletti E. The complete DNA sequence of vaccinia virus. Virology. 1990;179:247–266. doi: 10.1016/0042-6822(90)90294-2. [DOI] [PubMed] [Google Scholar]
- 20.Halliwell B, Gutteridge J M C. Free radicals in biology and medicine. 3rd ed. Oxford, United Kingdom: Oxford University Press; 1999. [Google Scholar]
- 21.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- 22.Hirt P, Hiller G, Wittek R. Localization and fine structure of a vaccinia virus gene encoding an envelope antigen. J Virol. 1986;58:757–764. doi: 10.1128/jvi.58.3.757-764.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hughes S J, Johnston L H, de Carlos A, Smith G L. Vaccinia virus encodes an active thymidylate kinase that complements a cdc8 mutant of Saccharomyces cerevisiae. J Biol Chem. 1991;266:20103–20109. [PubMed] [Google Scholar]
- 24.Ichihashi Y, Matsumoto S, Dales S. Biogenesis of poxviruses: role of A-type inclusions and host cell membranes in virus dissemination. Virology. 1971;46:507–532. doi: 10.1016/0042-6822(71)90056-0. [DOI] [PubMed] [Google Scholar]
- 25.Isaacs S N, Kotwal G J, Moss B. Reverse guanine phosphoribosyltransferase selection of recombinant vaccinia viruses. Virology. 1990;178:626–630. doi: 10.1016/0042-6822(90)90367-z. [DOI] [PubMed] [Google Scholar]
- 26.Jewett S L, Latrenta G S, Beck C M. Metal-deficient copper-zinc superoxide dismutases. Arch Biochem Biophys. 1982;215:116–128. doi: 10.1016/0003-9861(82)90285-5. [DOI] [PubMed] [Google Scholar]
- 27.Johnson G P, Goebel S J, Paoletti E. An update on the vaccinia virus genome. Virology. 1993;196:381–401. doi: 10.1006/viro.1993.1494. [DOI] [PubMed] [Google Scholar]
- 28.Kerr S M, Smith G L. Vaccinia virus DNA ligase is nonessential for virus replication: recovery of plasmids from virus-infected cells. Virology. 1991;180:625–632. doi: 10.1016/0042-6822(91)90076-n. [DOI] [PubMed] [Google Scholar]
- 29.Kettle S, Blake N W, Law K M, Smith G L. Vaccinia virus serpins B13R (SPI-2) and B22R (SPI-1) encode Mr38.5 and 40K, intracellular polypeptides that do not affect virus virulence in a murine intranasal model. Virology. 1995;206:136–147. doi: 10.1016/s0042-6822(95)80028-x. [DOI] [PubMed] [Google Scholar]
- 30.Lu Z, Li Y, Que Q, Kutish G F, Rock D L, Van Etten J L. Analysis of 94 kb of the chlorella virus PBCV-1 330-kb genome: map positions 88 to 182. Virology. 1996;216:102–123. doi: 10.1006/viro.1996.0038. [DOI] [PubMed] [Google Scholar]
- 31.Maa J S, Rodriguez J F, Esteban M. Structural and functional characterization of a cell surface binding protein of vaccinia virus. J Biol Chem. 1990;265:1569–1577. [PubMed] [Google Scholar]
- 32.Mackett M, Smith G L, Moss B. The construction and characterization of vaccinia virus recombinants expressing foreign genes. In: Glover D M, editor. DNA cloning: a practical approach. Vol. 2. Oxford, United Kingdom: IRL Press; 1985. pp. 191–211. [Google Scholar]
- 33.Massung R F, Liu L I, Qi J, Knight J C, Yuran T E, Kerlavage A R, Parsons J M, Venter J C, Esposito J J. Analysis of the complete genome of smallpox variola major virus strain Bangladesh-1975. Virology. 1994;201:215–240. doi: 10.1006/viro.1994.1288. [DOI] [PubMed] [Google Scholar]
- 34.Mattaj I W, Englmeier L. Nucleocytoplasmic transport: the soluble phase. Annu Rev Biochem. 1998;67:265–306. doi: 10.1146/annurev.biochem.67.1.265. [DOI] [PubMed] [Google Scholar]
- 35.McCord J M, Fridovich I. Superoxide dismutase. An enzymic function for erythrocuprein (hemocuprein) J Biol Chem. 1969;244:6049–6055. [PubMed] [Google Scholar]
- 36.McCraith S, Holtzman T, Moss B, Fields S. Genome-wide analysis of vaccinia virus protein-protein interactions. Proc Natl Acad Sci USA. 2000;97:4879–4884. doi: 10.1073/pnas.080078197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moss B. Poxviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Vol. 2. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2637–2671. [Google Scholar]
- 38.Natuk R J, Holowczak J A. Vaccinia virus proteins on the plasma membrane of infected cells. III. Infection of peritoneal macrophages. Virology. 1985;147:354–372. doi: 10.1016/0042-6822(85)90138-2. [DOI] [PubMed] [Google Scholar]
- 39.Niles E G, Seto J. Vaccinia virus gene D8 encodes a virion transmembrane protein. J Virol. 1988;62:3772–3778. doi: 10.1128/jvi.62.10.3772-3778.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Parge H E, Hallewell R A, Tainer J A. Atomic structures of wild-type and thermostable mutant recombinant human Cu,Zn superoxide dismutase. Proc Natl Acad Sci USA. 1992;89:6109–6113. doi: 10.1073/pnas.89.13.6109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Parkinson J E, Smith G L. Vaccinia virus gene A36R encodes a Mr43–50 K protein on the surface of extracellular enveloped virus. Virology. 1994;204:376–390. doi: 10.1006/viro.1994.1542. [DOI] [PubMed] [Google Scholar]
- 42.Petrone W F, English D K, Wong K, McCord J M. Free radicals and inflammation: superoxide-dependent activation of a neutrophil chemotactic factor in plasma. Proc Natl Acad Sci USA. 1980;77:1159–1163. doi: 10.1073/pnas.77.2.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ritter K, Kuhl R J, Semrau F, Eiffert H, Kratzin H D, Thomssen R. Manganese superoxide dismutase as a target of autoantibodies in acute Epstein-Barr virus infection. J Exp Med. 1994;180:1995–1998. doi: 10.1084/jem.180.5.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rodriguez J F, Janeczko R, Esteban M. Isolation and characterization of neutralizing monoclonal antibodies to vaccinia virus. J Virol. 1985;56:482–488. doi: 10.1128/jvi.56.2.482-488.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rodriguez J R, Rodriguez D, Esteban M. Insertional inactivation of the vaccinia virus 32-kilodalton gene is associated with attenuation in mice and reduction of viral gene expression in polarized epithelial cells. J Virol. 1992;66:183–189. doi: 10.1128/jvi.66.1.183-189.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schreck R, Rieber P, Baeuerle P A. Reactive oxygen intermediates as apparently widely used messengers in the activation of the NF-κB transcription factor and HIV-1. EMBO J. 1991;10:2247–2258. doi: 10.1002/j.1460-2075.1991.tb07761.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Senkevich T G, Wolffe E J, Buller R M. Ectromelia virus RING finger protein is localized in virus factories and is required for virus replication in macrophages. J Virol. 1995;69:4103–4111. doi: 10.1128/jvi.69.7.4103-4111.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Senkevich T G, Bugert J J, Sisler J R, Koonin E V, Darai G, Moss B. Genome sequence of a human tumorigenic poxvirus: prediction of specific host response-evasion genes. Science. 1996;273:813–816. doi: 10.1126/science.273.5276.813. [DOI] [PubMed] [Google Scholar]
- 49.Shchelkunov S N, Blinov V M, Sandakhchiev L S. Genes of variola and vaccinia viruses necessary to overcome the host protective mechanisms. FEBS Lett. 1993;319:80–83. doi: 10.1016/0014-5793(93)80041-r. [DOI] [PubMed] [Google Scholar]
- 50.Shchelkunov S N, Totmenin A V, Loparev V N, Safronov P F, Gutorov V V, Chizhikov V E, Knight J C, Parsons J M, Massung R F, Esposito J J. Alastrim smallpox variola minor virus genome DNA sequences. Virology. 2000;266:361–386. doi: 10.1006/viro.1999.0086. [DOI] [PubMed] [Google Scholar]
- 51.Smith G L, Chan Y S, Howard S T. Nucleotide sequence of 42 kbp of vaccinia virus strain WR from near the right inverted terminal repeat. J Gen Virol. 1991;72:1349–1376. doi: 10.1099/0022-1317-72-6-1349. [DOI] [PubMed] [Google Scholar]
- 52.Smith G L, Symons J A, Khanna A, Vanderplasschen A, Alcamí A. Vaccinia virus immune evasion. Immunol Rev. 1997;159:137–154. doi: 10.1111/j.1600-065x.1997.tb01012.x. [DOI] [PubMed] [Google Scholar]
- 53.Sodeik B, Cudmore S, Ericsson M, Esteban M, Niles E G, Griffiths G. Assembly of vaccinia virus: incorporation of p14 and p32 into the membrane of the intracellular mature virus. J Virol. 1995;69:3560–3574. doi: 10.1128/jvi.69.6.3560-3574.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tainer J A, Getzoff E D, Richardson J S, Richardson D C. Structure and mechanism of copper, zinc superoxide dismutase. Nature. 1983;306:284–287. doi: 10.1038/306284a0. [DOI] [PubMed] [Google Scholar]
- 55.Tomalski M D, Eldridge R, Miller L K. A baculovirus homolog of a Cu/Zn superoxide dismutase gene. Virology. 1991;184:149–161. doi: 10.1016/0042-6822(91)90831-u. [DOI] [PubMed] [Google Scholar]
- 56.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tscharke D C, Smith G L. A model for vaccinia virus pathogenesis and immunity based on intradermal injection of mouse ear pinnae. J Gen Virol. 1999;80:2751–2755. doi: 10.1099/0022-1317-80-10-2751. [DOI] [PubMed] [Google Scholar]
- 58.Weiss S J. Oxygen, ischemia and inflammation. Acta Physiol Scand Suppl. 1986;548:9–37. [PubMed] [Google Scholar]
- 59.Westendorp M O, Shatrov V A, Schulze-Osthoff K, Frank R, Kraft M, Los M, Krammer P H, Droge W, Lehmann V. HIV-1 Tat potentiates TNF-induced NF-κB activation and cytotoxicity by altering the cellular redox state. EMBO J. 1995;14:546–554. doi: 10.1002/j.1460-2075.1995.tb07030.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Willer D O, McFadden G, Evans D H. The complete genome sequence of Shope (rabbit) fibroma virus. Virology. 1999;264:319–343. doi: 10.1006/viro.1999.0002. [DOI] [PubMed] [Google Scholar]
- 61.Williamson J D, Reith R W, Jeffrey L J, Arrand J R, Mackett M. Biological characterization of recombinant vaccinia viruses in mice infected by the respiratory route. J Gen Virol. 1990;71:2761–2767. doi: 10.1099/0022-1317-71-11-2761. [DOI] [PubMed] [Google Scholar]