Abstract
Purpose/objective(s)
Biomarkers for extracranial oligometastatic disease remain elusive and few studies have attempted to correlate genomic data to the presence of true oligometastatic disease.
Methods
Patients with non-small cell lung cancer (NSCLC) and brain metastases were identified in our departmental database. Electronic medical records were used to identify patients for whom liquid biopsy-based comprehensive genomic profiling (Guardant Health) was available. Extracranial oligometastatic disease was defined as patients having ≤5 non-brain metastases without diffuse involvement of a single organ. Widespread disease was any spread beyond oligometastatic. Fisher’s exact tests were used to screen for mutations statistically associated (p<0.1) with either oligometastatic or widespread extracranial disease. A risk score for the likelihood of oligometastatic disease was generated and correlated to the likelihood of having oligometastatic disease vs widespread disease. For oligometastatic patients, a competing risk analysis was done to assess for cumulative incidence of oligometastatic progression. Cox regression was used to determine association between oligometastatic risk score and oligoprogression.
Results
130 patients met study criteria and were included in the analysis. 51 patients (39%) had extracranial oligometastatic disease. Genetic mutations included in the Guardant panel that were associated (p<0.1) with the presence of oligometastatic disease included ATM, JAK2, MAP2K2, and NTRK1, while ARID1A and CCNE1 were associated with widespread disease. Patients with a positive, neutral and negative risk score for oligometastatic disease had a 78%, 41% and 11.5% likelihood of having oligometastatic disease, respectively (p<0.0001). Overall survival for patients with positive, neutral and negative risk scores for oligometastatic disease was 86% vs 82% vs 64% at 6 months (p=0.2). Oligometastatic risk score was significantly associated with the likelihood of oligoprogression based on the Wald chi-square test. Patients with positive, neutral and negative risk scores for oligometastatic disease had a cumulative incidence of oligometastatic progression of 77% vs 35% vs 33% at 6 months (p=0.03).
Conclusions
Elucidation of a genomic signature for extracranial oligometastatic disease derived from non-invasive liquid biopsy appears feasible for NSCLC patients. Patients with this signature exhibited higher rates of early oligoprogression. External validation could lead to a biomarker that has the potential to direct local therapies in oligometastatic patients.
Keywords: NSCLC - lung adenocarcinoma - EGFR - ALK - BRAF - KRAS - RET - MET - PD-L1 - ROS1, genomic study, metastatic NSCLC, oligometastatic disease, brain metastases
Introduction
The concept of oligometastatic cancer was originally presented in the 1990’s by Hellman and Weichselbaum (1). This theory posits that a population of patients with truly limited sites of metastatic disease exists in which aggressive local therapy may improve outcomes and, in some cases, even effect a cure. The evidence for the potential curability of some patients with oligometastatic disease derived originally from series of liver metastasectomy in colorectal cancer (2) as well as lung metastasectomy with several types of primary cancer (3).
Stereotactic body radiation therapy (SBRT), or stereotactic ablative radiotherapy (SABR), represents a popular option for treatment of oligometastatic disease given its non-invasive nature compared to metastasectomy, and its delivery of ablative doses of radiation. SBRT is characterized by hypofractionated (limited number of treatments) external delivery of radical irradiation dose with highly conformal treatment planning targeted at a well-defined target volume, employing modern techniques of stereotactic or image-guided patient setup (4). Multiple prospective studies have now demonstrated that SBRT may benefit the population with oligometastatic disease (5, 6). Presently open studies (including the STOP and HALT studies) intend on assessing the effect of the addition of SBRT to standard systemic therapy for oligoprogressive cancer (7). Future directions include the assessment of the role of SBRT in the population of oligometastatic patients receiving immunotherapy given the potential for release of tumor antigens and stimulation of an immune response to tumor (8).
A controversy that has emerged for the management of oligometastatic disease is how to truly define it clinically. While a common definition utilized historically is five or fewer metastases (9), various definitions have been employed both for clinical trial entrance criteria in recent studies and for defining this entity to assess for appropriateness of SBRT (10). Multiple recent studies have employed a more liberal definition with allowance for >5 total metastases including brain metastases, consistent with the general observation that protocols have generally focused on extent of extracranial disease in their definitions of oligometastatic disease (11, 12). Newer clinical trials of metastatic NSCLC have employed a cutoff of up to 6 extracranial metastatic sites for classification of oligometastatic disease; this was irrespective of extent of intracranial metastatic disease, so long as patients (a) did not have leptomeningeal carcinomatosis (aggressive metastatic involvement of meningeal regions) and (b) underwent brain-specific treatment via whole-brain radiation therapy (WBRT) or stereotactic radiosurgery (10, 13, 14).
Despite efforts to properly define oligometastatic disease, there is likely a dichotomy between patients with truly a limited metastatic disease burden, and those who will progress to diffuse widespread disease in a short period of time. This dichotomy is likely driven by biology, and several attempts have been made to identify biologically the patients with truly limited metastatic disease. Thus far, these attempts have focused predominantly on measurement of serum levels of circulating markers, and this has been complicated by the sensitivity of detection (15).
A recently published series demonstrated that genomic profiling acquired from serum samples could predict the responses to systemic therapy in advanced NSCLC (16). This profiling was able to successfully segregate patients into genomic clusters that were prognostic for treatment response. Given the predictive power of this genomic profiling technique and the non-invasiveness of a liquid biopsy platform, we decided to use this technique in an attempt to identify a genomic profile for oligometastatic extracranial disease. We sought to do so using a population of patients with primary lung cancer, focusing on NSCLC We focused on patients with metastatic disease secondary to NSCLC, the most common type of lung cancer and a leading cause of brain metastases worldwide (17).
Materials and methods
Data acquisition, inclusion, and exclusion
This study was approved by the Institutional Review Board at the Wake Forest School of Medicine. We retrospectively analyzed our departmental brain metastasis database of patients who underwent stereotactic radiosurgery (SRS) for definitive treatment of intracranial metastatic disease. SRS refers to a modality of high-precision external beam radiotherapy akin to SBRT that can be employed in the brain (18). SRS was herein performed through the Gamma Knife (GK) platform via the Leksell GammaPlan treatment planning system (Elekta AB, Stockholm). Patients met the following additional inclusion criteria: (a) initial diagnosis of brain metastasis between August 2012 and September 2021, (b) primary NSCLC cancer diagnosis, and (c) liquid biopsy-based comprehensive genomic profiling (Guardant Health). Patients with a diagnosis of small cell lung cancer (SCLC) were excluded from the analysis. Moreover, patients who had genomic profiling through other platforms were excluded.
Electronic medical records (EMR) were used to determine clinical and demographic characteristics of patients, including gender, age, race, Karnofsky performance scale (KPS) (19), number of metastases at first GK, number of metastases at first distant brain failure (development of new intracranial metastases not previously treated with GK) (11), systemic disease burden (extracranial oligometastatic disease status), and brain metastasis velocity (20). EMR were subsequently used to identify patients for whom liquid biopsy-based comprehensive genomic profiling (Guardant Health) was available. Patients who had genomic profiling through other platforms were excluded from the analysis.
Genomic profiling
Comprehensive genomic profiling was performed using the Guardant 360 Platform (Guardant Heath, Redwood City, CA). This platform is a minimally invasive blood test assessing cell free tumor DNA for 55+ known mutations (21). Blood samples were acquired prior to commencing systemic therapy using a CLIA certified next generation sequencing test as published by Leighl et al. (22).
Clinical definitions
Extracranial oligometastatic disease was herein defined as patients having ≤5 non-brain metastases without diffuse involvement of a single organ (12, 23, 24). This definition is consistent with that used in a previous randomized trial, wherein more than a third of patients underwent CNS-directed therapy (stereotactic radiosurgery or whole-brain radiotherapy) for non-leptomeningeal intracranial metastatic disease (13). As in that study, this present study did not include patients with leptomeningeal carcinomatosis. Widespread disease was any spread beyond oligometastatic. Oligoprogression was defined as a limited progression in ≤5 metastatic sites (25). Widespread progression was defined as any progression beyond oligoprogression.
Development of oligometastatic genomic signature
Our group has previously published a precursor genomic study using this patient database, serving as the methodological basis of this current study (23). The oligometastatic gene signature was herein derived via a four-step process starting with a systematic screen for genes associated with nine clinical outcomes of interest. Fisher’s exact tests were used to identify any mutations that had a modest association (p<0.1) with the binary outcome of extracranial metastatic disease burden (oligometastatic vs. widespread). Herein, the goal of screening for predictive genes was to identify a set of genes that could together predict the outcome of interest (oligometastatic extracranial disease) rather than to identify any specific gene that alone would be a significant predictor. Thus, a p-value threshold of 0.1 was set a priori; while above the threshold for statistical significance (0.05), this represents a common threshold for detecting statistically meaningful signals, as is seen in the testing for interactions in statistical models (26, 27). Next, a score of +1 was assigned for every mutation present associated with oligometastatic disease, and -1 was assigned for every mutation associated with widespread disease. Scores were summed for each patient to create a composite risk score for the likelihood of oligometastatic disease, with scores subsequently correlated to the likelihood of having oligometastatic disease vs widespread disease.
Oligometastatic and survival analyses
For all included patients, survival analyses were performed to generate Kaplan-Meier overall survival curves. Patients with oligometastatic extracranial disease were then analyzed separately for patterns of progression, as this represents the subset of patients which could be particularly benefited by the oligometastatic genomic signature to predict for true oligoprogression vs widespread progression (in contrast, patients with widespread disease, by definition, could only progressive in a widespread manner). A Fine Gray competing risk analysis was done to assess for cumulative incidence of oligometastatic progression accounting for the potential competing risks of widespread progression of extracranial disease or death as previously described by Fine and Gray (28). Cox regression was used to determine association between oligometastatic risk score and oligometastatic progression (29). The Wald chi-square test was subsequently used to assess strength of association with different patterns of progression (oligogression vs. widespread progression) in the oligometastatic subset (30).
Results
Patient demographics
130 patients met study criteria and were included in the overall analysis ( Figure 1 ). Among these patients, 51 (39%) presented with oligometastatic extracranial metastatic disease at the time of initial GK, whereas 64 (49%) had widespread extracranial disease and 15 (12%) had no extracranial disease. The median number of brain metastases for the oligometastatic subset was 2 (range: 1-14). Histological subtyping of the 51 patients were as follows: 37 (73%) adenocarcinoma, 7 (14%) squamous cell carcinoma, and 7 (14%) other [3 with mixed histology, 2 with poorly differentiated with epithelioid or spindle cell features, and 2 with poorly differentiated NSCLC not otherwise specified (NOS)]. A similar distribution was observed in the overall 130 patient population, with 97 of 130 (75%) with a biopsy-proven adenocarcinoma. Other characteristics of the overall patient population are summarized in Table 1 .
Figure 1.
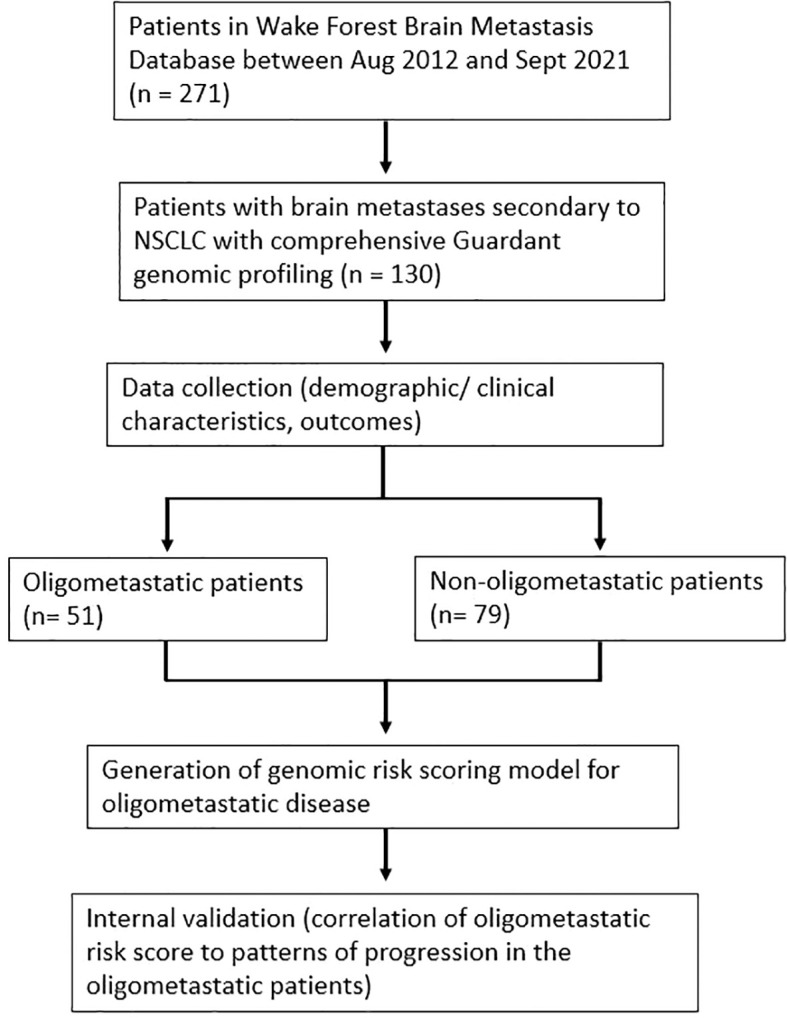
Flow diagram depicting study design.
Table 1.
Patient characteristics.
| Total = 130 patients | Count (%) | Median (Range) |
|---|---|---|
| Sex | ||
| Female | 68 (52%) | |
| Male | 62 (48%) | |
| Age (Years) | 68 (41-85) | |
| Race (White) | 108 (83%) | |
| KPS* | 78 (60-90) | |
| Smoking status | ||
| Former | 83 (64%) | |
| Current | 26 (20%) | |
| Never | 21 (16%) | |
| Number of brain metastases at first GK† | 4 (1-18) | |
| Oligometastatic disease burden | 51 (39%) | |
| BMV‡ (n = 54/130) | ||
| Low (≤4) | 21 (39%) | |
| Intermediate (4-13) | 21 (39%) | |
| High (>13) | 12 (22%) | |
| Concurrent diagnosis | 88 (68%) | |
| Mutated genes (N=77) | ||
| TP53 | 70% | |
| KRAS | 32% | |
| EGFR | 26% | |
| ARID1A, ERBB2, NF1, KTI, STK11, PIK3CA, PDGFRA, AR, MET, BRAF(V600), BRACA1, APC. | 10% - 20% | |
| Other genes | < 10% | |
*KPS, Karnofsky Performance Scale.
†GK, Gamma Knife.
‡BMV, Brain Metastasis Velocity (number of new metastasis after first GK/time in years).
Genomic findings
Genetic mutations included in the Guardant panel that were associated (p<0.1) with the presence of oligometastatic extracranial disease included ATM, JAK2, MAP2K2, and NTRK1, while ARID1A and CCNE1 were associated with widespread disease, based on the overall 130 patient sample ( Table 2 ). Overall, patients with a positive, neutral and negative composite risk score for oligometastatic disease had a 78%, 41% and 11.5% likelihood of having oligometastatic disease, respectively (p<0.0001). Table 3 presents the percentages of risk scores categorized as positive, neutral, and negative among oligometastatic and non-oligometastatic patients, as well as the overall percentage distribution among all patients. Of the 51 patients with oligometastatic disease, 11 (22%) had a positive genomic risk score, 37 (73%) had a neutral score, and 3 (6%) had a negative score.
Table 2.
Genes positively associated with oligometastatic vs. widespread metastatic extracranial disease burden in the 130 patients.
| Name | Class of alteration | P-value | |
|---|---|---|---|
| Genes Predictive of Oligometastatic Metastatic Disease | ATM JAK2 MAP2K2 NTRK1 |
SNV/INV* SNV/INV SNV/INV Fusion |
0.048 0.058 0.058 0.058 |
| Genes Predictive of Widespread Metastatic Disease | ARID1A CCNE1 |
SNV/INV SNV/INV, Amplification |
0.097 0.088 |
*SNV, Single Nucleotide Variant; INV, Insertion and Deletion Variant.
Table 3.
Distribution of risk scores across patients stratified by status of extracranial metastatic disease burden.
| Risk Score | Oligometastatic Patients (n = 51) | Widespread Metastatic Patients (n = 64) | Non-Metastatic Patients (n = 15) | Total Patients (n = 130) |
|---|---|---|---|---|
| Positive | 22% | 5% | 0% | 11% |
| Neutral | 73% | 69% | 60% | 69% |
| Negative | 6% | 27% | 40% | 20% |
Survival and treatment outcomes
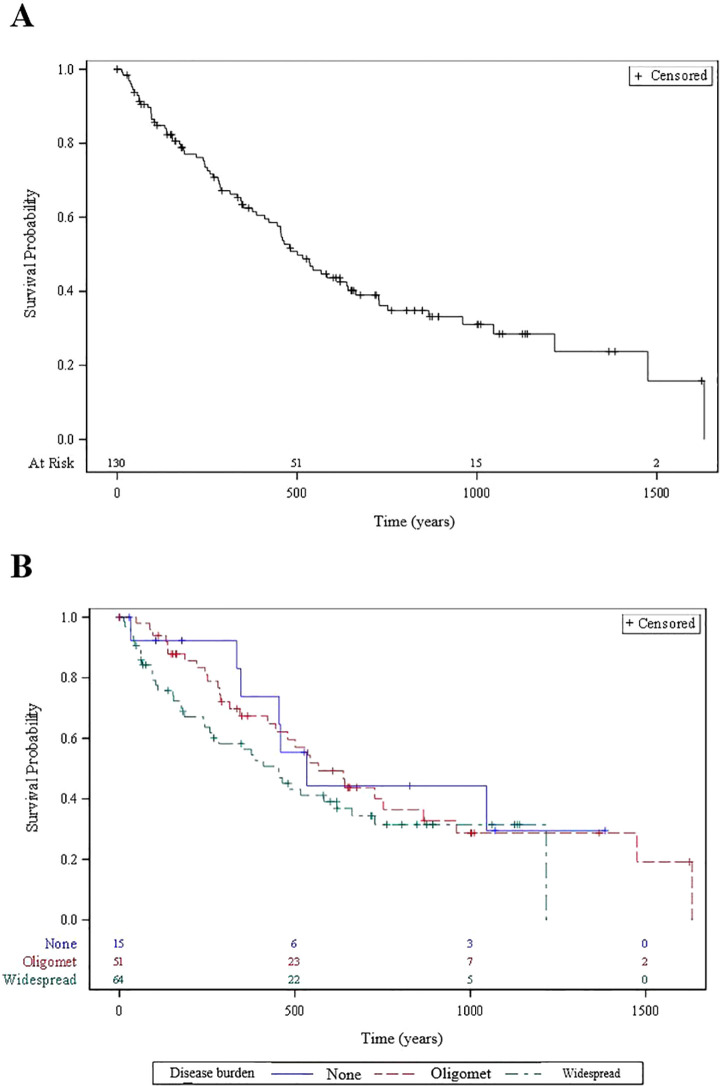
Among the 130 patients, overall survival for patients with positive, neutral and negative risk scores for oligometastatic disease was 86% vs 82% vs 64% at 6 months (p=0.2). Overall survival for patients with oligometastatic vs widespread disease was 88% vs 73% and 67% vs 59% at 6 months and 12 months, respectively. Kaplan-Meier survival curves based on the overall 130 patient population and stratified to extracranial oligometastatic disease burden are shown in Figure 2 . Seven of 51 (14%) oligometastatic patients underwent SBRT to extracranial oligometastatic site(s) as part of their treatment course; all but one patient received this ablative treatment after their first GK.
Figure 2.
Kaplan Meier overall survival curves for (A) 130 patients overall and (B) patients stratified according to extracranial metastatic disease burden status.
Patterns of progression
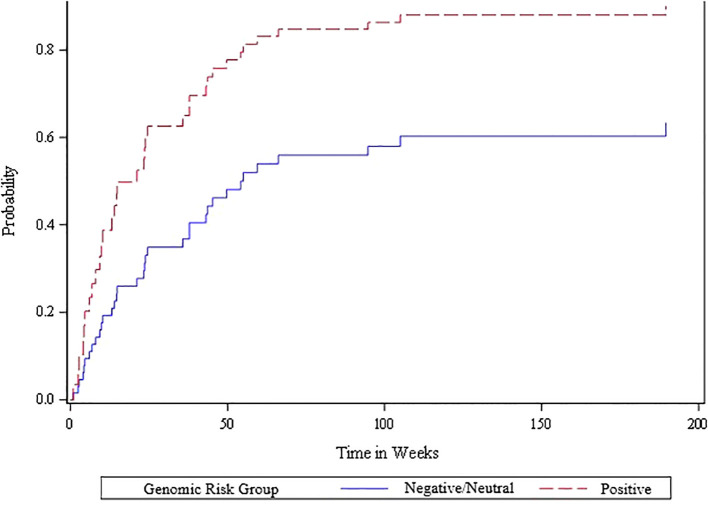
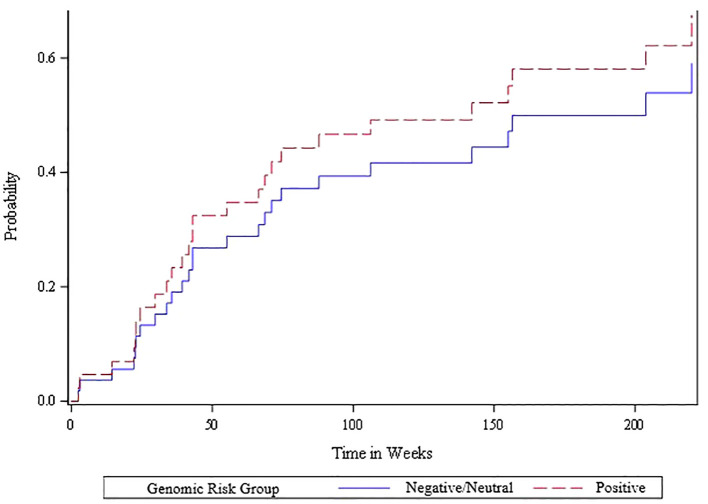
The competing risk analysis based on the 51 oligometastatic patients found that the oligometastatic risk score was significantly associated with the likelihood of oligometastatic progression based on the Wald Chi-square test ( Figure 3 ). Within this oligometastatic subset, 91% of the patients with a positive risk score experienced an oligoprogression event, compared to 62% and 67% of patients with a neutral or negative risk score, respectively. Moreover, 35 (69%) patients subsequently experienced oligoprogression, 13 (25%) either died or experienced widespread progression (thus had competing events), and 3 (6%) had no event. Since there were only three of 51 patients with a negative genomic score we divided the scores into neutral/negative vs positive; we found that in the competing risk analysis those with the positive risk score exhibited a higher rate of oligometastatic progression – hazard ratio of 2.29 (95% CI of 1.1 to 4.8). Additionally, patients with positive, neutral and negative risk scores for oligometastatic disease had a cumulative incidence of oligometastatic progression of 77% vs 35% vs 33% at 6 months (p=0.03 from competing risk model) ( Figure 3 ). In contrast, the Wald chi-square test was not statistically significant (p=0.57) when comparing neutral/negative vs positive scores with the endpoint of widespread extracranial progression ( Figure 4 ).
Figure 3.
Cumulative incidence plot of oligometastatic progression for positive vs. other (neutral/negative) oligometastatic risk scores among the 51 patients with extracranial oligometastatic disease. A Cox proportional hazards regression model was fit to determine the statistical association between the risk score and oligometastatic progression accounting for the competing risk of death or widespread progression, and the Wald chi-square test was statistically significant (p=0.026) when comparing the negative/neutral risk scores versus the positive risk scores.
Figure 4.
Cumulative incidence plot of widespread metastatic progression for positive vs. other (neutral/negative) oligometastatic risk scores among the 51 patients with extracranial oligometastatic disease. A Cox proportional hazards regression model was fit to determine the statistical association between risk score and widespread progression accounting for the competing risk of death. The Wald chi-square test was not statistically significant (p=0.57) when comparing the negative/neutral risk scores versus the positive risk scores.
Discussion
In summary, we present a gene mutational scoring system derived from liquid biopsy that purports to risk stratify patients based on the likelihood of oligometastatic extracranial disease. In this study, the oligometastatic signature predicted not only for the presence of oligometastatic extracranial disease but also for oligoprogression, defined as progression in 5 or fewer extracranial metastatic sites. While the interval validation presented herein is derived from patients with known brain metastases who underwent SRS for definitive treatment of their intracranial metastatic disease, the aim of this study is to develop a genomic risk score to guide treatment decisions pertaining to their extracranial disease.
The potential advantage of identifying an accurate biomarker for oligometastatic extracranial disease is to distinguish the multiple phenotypes within the oligometastatic population, and properly triage their management based on the predicted benefit of treatment. At the present time, clinical trials continue to treat oligometastatic patients as a homogeneous population. However, nearly 50% of patients who present with oligometastatic NSCLC will experience distant progression within 2 years with many of these representing polymetastatic progression (31). Early data suggests that a proportion of patients with oligometastatic disease do benefit from more aggressive treatment (32, 33). If a biomarker could accurately identify the population who would experience early polymetastatic failure, then those patients could be spared aggressive local therapy. Conversely, patients with a true oligometastatic phenotype could be treated with consolidative ablative therapies such as SBRT.
Unfortunately, identification of a clinically useful biomarker for oligometastatic disease in NSCLC has to date proven elusive. The ability of present imaging to determine oligometastatic patients who will have early polymetastatic progression is suboptimal (34). There have been recent efforts to identify cytogenetic biomarkers for true oligometastatic disease. Attempts have included correlation of disease burden with levels of circulating tumor DNA (ctDNA) derived from liquid biopsies to the burden of metastatic disease (35). At this time, however, ctDNA sensitivity for oligometastatic disease is problematic due to the extremely low levels found in the peripheral blood. Another strategy for biomarker identification has been to attempt detection of circulating miRNAs, as these molecules are thought to be regulators of metastatic development and have even been proposed as the molecular basis of prevention of polymetastatic progression (36). The limitations of using circulating miRNAs as a biomarker for oligometastatic disease include issues with variability of miRNA levels from one patient to another given an equivalent burden of disease (37).
The present effort to identify an oligometastatic biomarker uses a genomic signature derived from non-invasive liquid biopsy. Unlike previously described attempts at using liquid biopsy to determine oligometastatic status, the present study uses multiple biomarkers to derive a risk score. The genomic platform used in the study has been validated for use in NSCLC and is widely available clinically (38). The CCNE1 and ARID1A genes were found to be associated with polymetastatic progression. Both ARID1A (39) and CCNE1 (40) play important roles in regulating the cell cycle. Approximately 10% of NSCLC patients have ARID1A gene mutations and these are associated with a poor prognosis (41). CCNE1 is a cyclin that modulates activity of cyclin-dependent kinases for progression through multiple phases of the cell cycle. CCNE1 mutation has been demonstrated in NSCLC to be increasingly overexpressed in higher stage disease, which leads to a worsened disease specific survival (42).
The genes that more associated with oligometastatic phenotype were ATM, JAK2, MAP2K2 and NTRK1, and these play important roles in cellular signaling pathways (43–46). ATM is involved in DNA damage response, JAK2 in cytokine signaling, MAP2K2 in the MAPK pathway, and NTRK1 in neurotrophic signaling. The role of these genes in cellular signaling suggests that dysregulation may contribute to the establishment of an oligometastatic state, wherein cancer cells exhibit a more controlled and localized spread. This distinction highlights the potential dichotomy between the molecular mechanisms driving polymetastatic (cell cycle driven) and oligometastatic disease (cell signaling-driven) in NSCLC.
For any genomic signature for oligometastatic disease, it is important to demonstrate whether the signature is associated with oligometastatic (as opposed to polymetastatic) progression. In the present series, those with a positive risk score for oligometastatic disease were twice as likely as those with neutral or negative risk score for oligometastatic disease to have an oligometastatic pattern of progression. In a randomized phase II study of patients with NSCLC with oligometastatic disease, the median progression free survival was significantly improved in the arm receiving SBRT to oligometastatic disease (33). While the trial by Gomez was not powered to assess the pattern of progression, patients who were not treated with SBRT were more likely to experience failure in known sites of disease (oligometastatic progression). Similar results were found with subsequent phase II prospective studies of oligometastatic NSCLC patients (5, 6). That the present study showed that the oligometastatic signature led to greater oligometastatic failure leads to a potential clinical utility of this genomic biomarker to triage patients to SBRT.
There are several limitations to the present study. First of all, the limited sample size leads to potential that a larger dataset may arrive with a more robust model. The dataset was derived from patients with brain metastases, and it is unclear whether a validation dataset of patients without brain metastases would provide a similar genomic signature. While oligometastatic disease is a prognostic factor for patients with brain metastases in general (11), it is possible that the subset of brain metastasis patients with NSCLC and oligometastatic disease may have their own pattern of behavior and progression. In spite of these limitations, the study did show that patients with the oligometastatic genomic signature were also more likely to fail in an oligometastatic pattern, whereas those oligometastatic patients without the oligometastatic signature had a greater degree of polymetastatic failure - serving as a form of internal validation. In this study, we suggest a trend towards improved survival (at 6 months) with the positive oligometastatic risk score, consistent with our finding that oligometastatic patients on presentation exhibited superior survival outcomes compared to their non-oligometastatic counterparts across time. At the same time, patients with the oligometastatic genomic signature tended to exhibit faster rates of oligoprogression. In a possible interpretation of these seemingly disparate findings, our data may indirectly support the notion of oligoprogression as a distinct entity that may be more amenable to local therapies associated with improved survival (25). As such, future directions include validation with a larger and more diverse dataset, potentially in patients without brain metastases. Ideally this should be done at an independent institution from which the primary data were derived. Should this genomic signature prove valid, it would provide a clinical tool that could be used for management of oligometastatic patients.
Funding Statement
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics statement
The studies involving humans were approved by Institutional Review Board at the Wake Forest School of Medicine. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
Author contributions
AC: Writing – original draft, Writing – review & editing. RD’A: Writing – original draft, Writing – review & editing. MF: Writing – review & editing. MA: Writing – review & editing. JH: Writing – review & editing. YW: Writing – review & editing. MS: Writing – review & editing. JR: Writing – review & editing. TL: Writing – review & editing. WP: Writing – review & editing. CC: Writing – review & editing. ST: Writing – review & editing. AL: Writing – review & editing. JW: Writing – review & editing. WL: Writing – review & editing. JS: Writing – review & editing. CW: Writing – review & editing. FX: Writing – review & editing. MC: Writing – original draft, Writing – review & editing.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- 1. Hellman S, Weichselbaum RR. Oligometastases. J Clin Oncology: Off J Am Soc Clin Oncol. (1995) 13:8–10. doi: 10.1200/JCO.1995.13.1.8 [DOI] [PubMed] [Google Scholar]
- 2. Hughes KS, Rosenstein RB, Songhorabodi S, Adson M, Ilstrup D, Fortner J, et al. Resection of the liver for colorectal carcinoma metastases. A multi-institutional study of long-term survivors. Dis Colon Rectum. (1998) 31(1):1–4. doi: 10.1007/BF02552560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Pastorino U, Buyse M, Friedel G, Ginsberg RJ, Girard P, Goldstraw P, et al. Long-term results of lung metastasectomy: prognostic analyses based on 5206 cases. J Thorac Cardiovasc Surg. (1997) 113:37–49. doi: 10.1016/S0022-5223(97)70397-0 [DOI] [PubMed] [Google Scholar]
- 4. Rieber J, Streblow J, Uhlmann L, Flentjed M, Dumae M, Ernst I, et al. Stereotactic body radiotherapy (SBRT) for medically inoperable lung metastases-A pooled analysis of the German working group "stereotactic radiotherapy". Lung Cancer. (2016) 97:51–8. doi: 10.1016/j.lungcan.2016.04.012 [DOI] [PubMed] [Google Scholar]
- 5. Petty WJ, Urbanic JJ, Ahmed T, Hughes R, Levine B, Rusthoven K, et al. Long-term outcomes of a phase 2 trial of chemotherapy with consolidative radiation therapy for oligometastatic non-small cell lung cancer. Int J Radiat Oncology Biology Phys. (2018) 102:527–35. doi: 10.1016/j.ijrobp.2018.06.400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Palma DA, Olson R, Harrow S, Gaede S, Louie AV, Haasbeek C, et al. Stereotactic ablative radiotherapy versus standard of care palliative treatment in patients with oligometastatic cancers (SABR-COMET): A randomised, phase 2, open-label trial. Lancet. (2019) 393:2051–58. doi: 10.1016/S0140-6736(18)32487-5 [DOI] [PubMed] [Google Scholar]
- 7. Kinj Rémy, Muggeo E, Schiappacasse L, Bourhis J, Herrera. FG. Stereotactic body radiation therapy in patients with oligometastatic disease: clinical state of the art and perspectives. Cancers. (2022) 14. doi: 10.3390/cancers14051152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Onderdonk BE, Chmura SJ. The yin and yang of cytoreductive SBRT in oligometastases and beyond. Front Oncol. (2019) 9:706. doi: 10.3389/fonc.2019.00706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lievens Y, Guckenberger M, Gomez D, Hoyer M, Iyengar P, Kindts I, et al. Defining oligometastatic disease from a radiation oncology perspective: an ESTRO-ASTRO consensus document. Radiotherapy Oncology: J Eur Soc Ther Radiol Oncol. (2020) 148:157–66. doi: 10.1016/j.radonc.2020.04.003 [DOI] [PubMed] [Google Scholar]
- 10. Dingemans A-MC, Hendriks LEL, Berghmans T, Levy A, Hasan B, Faivre-Finn C, et al. Definition of synchronous oligometastatic non–small cell lung cancer—A consensus report. J Thorac Oncol. (2019) 14:2109–19. doi: 10.1016/j.jtho.2019.07.025 [DOI] [PubMed] [Google Scholar]
- 11. Ayala-Peacock DN, Peiffer AM, Lucas JT, Isom S, Griff Kuremsky J, Urbanic JJ, et al. A nomogram for predicting distant brain failure in patients treated with gamma knife stereotactic radiosurgery without whole brain radiotherapy. Neuro-Oncology. (2014) 16:1283–88. doi: 10.1093/neuonc/nou018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tsai CJ, Yang JT, Shaverdian N, Patel J, Shepherd AF, Eng J, et al. Standard-of-care systemic therapy with or without stereotactic body radiotherapy in patients with oligoprogressive breast cancer or non-small-cell lung cancer (Consolidative Use of Radiotherapy to Block [CURB] oligoprogression): an open-label, randomised, controlled, phase 2 study. Lancet (London England). (2024) 403:171–82. doi: 10.1016/S0140-6736(23)01857-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Iyengar P, Wardak Z, Gerber DE, Tumati V, Ahn C, Hughes RS, et al. Consolidative radiotherapy for limited metastatic non-small-cell lung cancer: A phase 2 randomized clinical trial. JAMA Oncol. (2018) 4:e173501. doi: 10.1001/jamaoncol.2017.3501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. University of Wisconsin, Madison and AstraZeneca . Comprehensive Stereotactic Body Radiotherapy (SBRT) to All Sites of Oligometastatic Non-small Cell Lung Cancer (NSCLC) Combined With Durvalumab (MEDI4736) and Tremelimumab Dual Immune Checkpoint Inhibition. Madison, Wisconsin, United States: clinicaltrials.gov; (2023). Available at: https://classic.clinicaltrials.gov/ct2/show/NCT03275597. [Google Scholar]
- 15. Cortinovis D, Malapelle U, Pagni F, Russo A, Banna GL, Sala E, et al. Diagnostic and prognostic biomarkers in oligometastatic non-small cell lung cancer: A literature review. Trans Lung Cancer Res. (2021) 10:3385–34005. doi: 10.21037/tlcr [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Smith MR, Wang Y, D’Agostino R, Jr, Liu Y, Ruiz J, Lycan T, et al. Prognostic mutational signatures of NSCLC patients treated with chemotherapy, immunotherapy and chemoimmunotherapy. NPJ Precis Oncol. (2023) 7:34. doi: 10.1038/s41698-023-00373-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Skakodub A, Walch H, Tringale KR, Eichholz J, Imber BS, Vasudevan HN, et al. Genomic analysis and clinical correlations of non-small cell lung cancer brain metastasis. Nat Commun. (2023) 14:4980. doi: 10.1038/s41467-023-40793-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Soliman H, Das S, Larson DA, Sahgal A. Stereotactic Radiosurgery (SRS) in the modern management of patients with brain metastases. Oncotarget. (2016) 7:12318–30. doi: 10.18632/oncotarget.7131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Timmermann C. 'Just give me the best quality of life questionnaire': the Karnofsky scale and the history of quality of life measurements in cancer trials. Chronic Illn. (2013) 9:179–90. doi: 10.1177/1742395312466903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Farris M, McTyre ER, Cramer CK, Hughes R, Randolph DM, Ayala-Peacock DN, et al. Brain metastasis velocity: A novel prognostic metric predictive of overall survival and freedom from whole-brain radiation therapy after distant brain failure following upfront radiosurgery alone. Int J Radiat Oncology Biology Phys. (2017) 98:131–41. doi: 10.1016/j.ijrobp.2017.01.201 [DOI] [PubMed] [Google Scholar]
- 21. Ou S-HI, Nagasaka M, Zhu VW. Liquid biopsy to identify actionable genomic alterations. Am Soc Clin Oncol Educ Book. (2018) 38:978–97. doi: 10.1200/edbk_199765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Leighl NB, Page RD, Raymond VM, Daniel DB, Divers SG, Reckamp KL, et al. Clinical utility of comprehensive cell-free DNA analysis to identify genomic biomarkers in patients with newly diagnosed metastatic non-small cell lung cancer. Clin Cancer Research: Off J Am Assoc Cancer Res. (2019) 25:4691–700. doi: 10.1158/1078-0432.CCR-19-0624 [DOI] [PubMed] [Google Scholar]
- 23. Abdulhaleem M, Hunting JC, Wang Y, Smith MR, Agostino RJ, Lycan T, et al. Use of comprehensive genomic profiling for biomarker discovery for the management of non-small cell lung cancer brain metastases. Front Oncol. (2023) 13:1214126. doi: 10.3389/fonc.2023.1214126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Harris S, Chan MD, Lovato JF, Ellis TL, Tatter SB, Bourland JD, et al. Gamma knife stereotactic radiosurgery as salvage therapy after failure of whole-brain radiotherapy in patients with small-cell lung cancer. Int J Radiat Oncology Biology Phys. (2012) 83:e53–59. doi: 10.1016/j.ijrobp.2011.11.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nguyen KT, Sakthivel G, Milano MT, Qiu H, Singh. DP. Oligoprogression in non-small cell lung cancer: A narrative review. J Thorac Dis. (2022) 14:4998–5011. doi: 10.21037/jtd [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Anderson DR, Burnham KP, Thompson WL. Null hypothesis testing: problems, prevalence, and an alternative. J Wildlife Manage. (2000) 64:912. doi: 10.2307/3803199 [DOI] [Google Scholar]
- 27. Maier M, Lakens D. Justify your alpha: A primer on two practical approaches. Adv Methods Practices psychol Sci. (2022) 5:251524592210803. doi: 10.1177/25152459221080396 [DOI] [Google Scholar]
- 28. Fine JP, Gray RJ. A proportional hazards model for the subdistribution of a competing risk. J Am Stat Assoc. (1999) 94:496–5095. doi: 10.1080/01621459.1999.10474144 [DOI] [Google Scholar]
- 29. Abd ElHafeez S, D’Arrigo G, Leonardis D, Fusaro M, Tripepi G, Roumeliotis S. Methods to analyze time-to-event data: the cox regression analysis. Oxid Med Cell Longev. (2021) 2021(1):1302811. doi: 10.1155/2021/1302811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mann HB, Wald A. On the choice of the number of class intervals in the application of the chi square test. Ann Math Stat. (1942) 13:306–17. doi: 10.1214/aoms/1177731569 [DOI] [Google Scholar]
- 31. Hörner-Rieber J, Bernhardt D, Blanck O, Duma M, Eich HT, Gerum S, et al. Long-term follow-up and patterns of recurrence of patients with oligometastatic NSCLC treated with pulmonary SBRT. Clin Lung Cancer. (2019) 20:e667–77. doi: 10.1016/j.cllc.2019.06.024 [DOI] [PubMed] [Google Scholar]
- 32. Ouyang W, Yu J, Nuerjiang S, Li Z, Wang D, Wang X, et al. Stereotactic body radiotherapy improves the survival of patients with oligometastatic non-small cell lung cancer. Cancer Med. (2019) 8:4605–145. doi: 10.1002/cam4.2366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gomez DR, Blumenschein GR, Jr., Lee JJ, Hernandez M, Ye R, Camidge DR, et al. Local Consolidative Therapy versus Maintenance Therapy or Observation for Patients with Oligometastatic Non-Small-Cell Lung Cancer without Progression after First-Line Systemic Therapy: A Multicentre, Randomised, Controlled, Phase 2 Study. Lancet Oncol. (2016) 17:1672–82. doi: 10.1016/S1470-2045(16)30532-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Solanki AA, Weichselbaum RR, Appelbaum D, Farrey K, Yenice KM, Chmura SJ, et al. The utility of FDG-PET for assessing outcomes in oligometastatic cancer patients treated with stereotactic body radiotherapy: A cohort study. Radiat Oncol. (2012) 7:216. doi: 10.1186/1748-717X-7-216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yi X, Ma J, Guan Y, Chen R, Yang L, Xia X. The feasibility of using mutation detection in ctDNA to assess tumor dynamics. Int J Cancer. (2017) 140:2642–475. doi: 10.1002/ijc.30620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lussier YA, Xing HR, Salama JK, Khodarev NN, Huang Y, Zhang Q, et al. MicroRNA expression characterizes oligometastasis(es). PloS One. (2011) 6:e28650. doi: 10.1371/journal.pone.0028650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bianchi F, Nicassio F, Marzi M, Belloni E, Dall’olio V, Bernard L, et al. A serum circulating miRNA diagnostic test to identify asymptomatic high-risk individuals with early stage lung cancer. EMBO Mol Med. (2011) 3:495–5035. doi: 10.1002/emmm.201100154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Laufer-Geva S, Rozenblum AB, Twito T, Grinberg R, Dvir A, Soussan-Gutman L, et al. The clinical impact of comprehensive genomic testing of circulating cell-free DNA in advanced lung cancer. J Thorac Oncology: Off Publ Int Assoc Study Lung Cancer. (2018) 13:1705–16. doi: 10.1016/j.jtho.2018.07.101 [DOI] [PubMed] [Google Scholar]
- 39. Wang K, Kan J, Yuen ST, Shi ST, Chu KM, Law S, et al. Exome sequencing identifies frequent mutation of ARID1A in molecular subtypes of gastric cancer. Nat Genet. (2011) 43:1219–23. doi: 10.1038/ng.982 [DOI] [PubMed] [Google Scholar]
- 40. Shanahan F, Seghezzi W, Parry D, Mahony D, Lees. E. Cyclin E associates with BAF155 and BRG1, components of the mammalian SWI-SNF complex, and alters the ability of BRG1 to induce growth arrest. Mol Cell Biol. (1999) 19:1460–69. doi: 10.1128/MCB.19.2.1460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Jin F, Yang Z, Shao J, Tao J, Reißfelder C, Loges S, et al. ARID1A mutations in lung cancer: biology, prognostic role, and therapeutic implications. Trends Mol Med. (2023) 29:646–585. doi: 10.1016/j.molmed.2023.04.005 [DOI] [PubMed] [Google Scholar]
- 42. Ullah MA, Farzana M, Islam MS, Moni R, Zohora US, Rahman MS. Identification of the prognostic and therapeutic values of cyclin E1 (CCNE1) gene expression in lung adenocarcinoma and lung squamous cell carcinoma: A database mining approach. Heliyon. (2022) 8:e103675. doi: 10.1016/j.heliyon.2022.e10367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lee J-H, Paull. TT. Activation and regulation of ATM kinase activity in response to DNA double-strand breaks. Oncogene. (2007) 26:7741–48. doi: 10.1038/sj.onc.1210872 [DOI] [PubMed] [Google Scholar]
- 44. Jiao H, Berrada K, Yang W, Tabrizi M, Platanias LC, Yi T. Direct association with and dephosphorylation of Jak2 kinase by the SH2-domain-containing protein tyrosine phosphatase SHP-1. Mol Cell Biol. (1996) 16:6985–92. doi: 10.1128/MCB.16.12.6985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zheng CF, Guan. KL. Properties of MEKs, the kinases that phosphorylate and activate the extracellular signal-regulated kinases. J Biol Chem. (1993) 268:23933–39. doi: 10.1016/S0021-9258(20)80474-8 [DOI] [PubMed] [Google Scholar]
- 46. Martin-Zanca D, Hughes SH, Barbacid. M. A human oncogene formed by the fusion of truncated tropomyosin and protein tyrosine kinase sequences. Nature. (1986) 319:743–48. doi: 10.1038/319743a0 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.