Abstract
Recently, remarkable progress has been made in developing effective combination drug therapies that can control but not cure retroviral replication. Even when effective, these drug regimens are toxic, they require demanding administration schedules, and resistant viruses can emerge. Thus the need for new gene-based therapies continues. In one such approach, capsid-targeted viral inactivation (CTVI), nucleases fused to viral coat proteins are expressed in infected cells and become incorporated during virion assembly. CTVI can eliminate infectious murine retrovirus titer in tissue culture. Here we describe transgenic mice expressing fusions of the Moloney murine leukemia virus (Mo-MuLV) Gag protein to staphylococcal nuclease. This work tests the protective effect and demonstrates in vivo proof-of-principle of CTVI in transgenic mice expressing endogenous proviral copies of Mo-MuLV. The antiviral protein-expressing mice are phenotypically normal, attesting to the lack of toxicity of the fusion protein. The Mo-MuLV infection was much less virulent in transgenic littermates than in nontransgenic littermates. Gag-nuclease expression reduced infectious titers in blood up to 10-fold, decreased splenomegaly and leukemic infiltration, and increased life spans up to 2.5-fold in transgenic relative to nontransgenic infected animals. These results suggest that gene therapies based on similar fusion proteins, designed to attack human immunodeficiency virus or other retroviruses, could provide substantial therapeutic benefits.
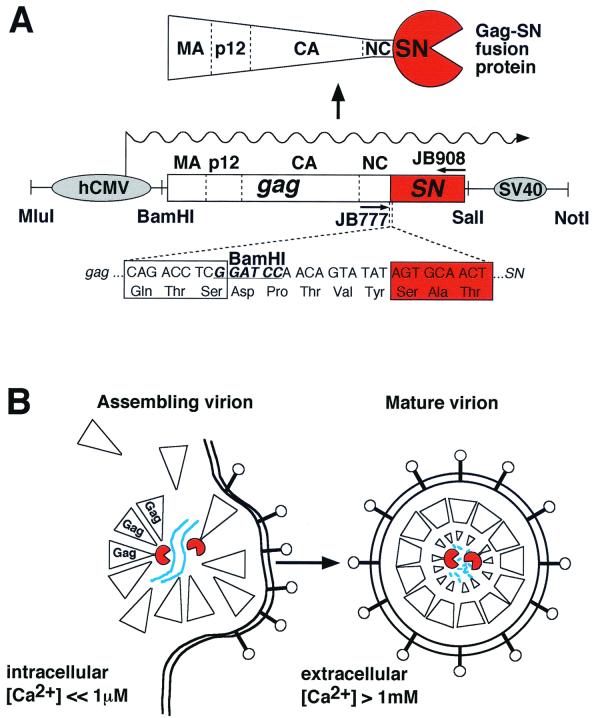
A number of genetic strategies to interfere with retrovirus replication are being explored. In a general strategy called intracellular immunization (1), genes encoding macromolecules that interfere with viral multiplication are introduced into virus-susceptible cells. Antiviral transgenes include antisense RNAs, ribozymes, RNA decoys, dominant-negative versions of viral proteins, and intracellular antibodies (9). We and others have explored the antiretroviral effects of expressing fusions between structural proteins of virions and several nucleases, including Barnase (a general RNase) (31), Escherichia coli RNase HI (35, 42, 43), and the calcium-dependent staphylococcal nuclease (SN) (32, 36, 42, 43). Our previous work with retroviruses explored antiviral effects against Moloney murine leukemia virus (Mo-MuLV) and human immunodeficiency virus (HIV) (5). In order to inactivate Mo-MuLV, we examined the antiviral effect of a construct in which the full-length Mo-MuLV gag gene is fused in frame to the N terminus of the SN gene (Fig. 1A). These Gag-SN fusion proteins are enzymatically active, nontoxic to tissue culture cells, and have antiviral activity. They are nontoxic to cells, presumably because intracellular calcium concentrations are very tightly regulated at submicromolar concentrations, whereas SN requires millimolar calcium ion for activity. The Gag-SN fusion proteins are efficiently encapsidated into virions, where they undergo proteolytic processing. When the virions are shed into the extracellular milieu, Gag-SN encounters millimolar concentrations of calcium ion, leading to viral RNA degradation and loss of infectivity (Fig. 1B). Mo-MuLV Gag-SN polyproteins were previously demonstrated to have a long-term prophylactic and therapeutic effect that can virtually eliminate the production of infectious Mo-MuLV in tissue culture (32, 36, 42).
FIG. 1.
Organization of the antiviral transgene and mode of action. (A) Structure of the hCMV-gag-SN transgene. The gag-SN transgene was constructed as described in Materials and Methods. hCMV, hCMV-P/E element; box, gag-SN open reading frame (SN sequences depicted in red); SV40, simian virus 40 polyadenylation signal; wavy line, mRNA. (B) Mechanism of viral inactivation (see text for further detail). Gag-SN protein (left) is incorporated into assembling virions via Gag-Gag interactions. Assembled virions are shed from the cell and undergo proteolytic processing and exposure to high Ca2+ concentrations, leading to viral RNA degradation (right). Blue line, viral RNA; lollipops, viral Env proteins; large triangles, Gag precursor proteins; red portions, SN.
We have previously studied the virus-inactivating mechanism, general feasibility, and promising efficacy of the capsid-targeted viral inactivation (CTVI) strategy in cell culture in an MuLV model (32, 35, 36, 42). In this publication, we describe an animal model for evaluating this antiretroviral therapy. It was important to determine whether Gag-SN fusion protein expression interferes with retroviral multiplication in an infected animal. Furthermore, it was crucial to ascertain whether the Gag-SN fusion protein could be tolerated in animals or was toxic. This is the first transgenic mouse model for a protein-based antiviral strategy. (A murine in vivo nucleic-acid-based antiviral strategy was described previously [15].)
MATERIALS AND METHODS
DNA constructs and transgenic mice.
The 1,570-bp human cytomegalovirus-promoter/enhancer (hCMV-P/E)-simian virus 40 (SV40) polyadenylation site expression cassette was obtained by PCR with primers JB353 (5′-GGGCCCACGCGTTTCGAGCTCGCCCGACAT-3′) and JB354 (5′-GGAACCGCGGCCGCAAAACGACGGCCAGTGCC-3′) from plasmid pRK5 (46). The MluI-NotI fragment in the plasmid TFAneo (12) containing two Rous sarcoma virus long terminal repeats (LTRs) was replaced by the 1,570-bp hCMV-P/E-SV40 poly(A) cassette after its digestion with MluI and NotI, generating pGN1717. The gag-SN gene cassette from pGN1595, flanked by XbaI and SalI restriction sites, was inserted into the multiple cloning site of pGN1717, which separates the hCMV-P/E element from the SV40 poly(A) signal, leading to pGN1743. The construct was successfully tested for expression of the 85-kDa Gag-SN fusion protein in transient transfection experiments. The 3,692-bp MluI-NotI fragment from pGN1743 carrying the Mo-MuLV gag-SN gene fusion under control of the hCMV-P/E element and the SV40 poly(A) signal was purified by electroelution and Qiagen-Tip20 column before being microinjected into fertilized mouse oocytes. Transgenic founder mice (SJL × C57BL/6) were generated by DNX, Inc. (Princeton, N.J.). Two of three independent founder mice (SJL × C57BL/6) transmitted the transgene, generating lines E and F. The founders were backcrossed to C57BL/6 mice (93.8% isogenic to C57BL/6). C57BL/6 mice and Mov13 mice were purchased from Jackson Laboratories (Bar Harbor, Maine). Mov14 mice were kindly provided by Ruth Curry in the laboratory of Rudolf Jaenisch (Whitehead Institute, Boston, Mass). The origin and characteristics of the Mov13 and Mov14 strains (on a C57BL/6 background) have been described previously (20, 30, 34). All mice were bred under standard pathogen-free conditions at the animal facility of the Johns Hopkins University School of Medicine.
Nuclear magnetic resonance imaging.
The development of splenomegaly in Mov14/+; gagSN/+ mice and their nontransgenic littermates was monitored by nuclear magnetic resonance imaging at the Department of Radiology Division of Magnetic Resonance Imaging at the Johns Hopkins University. After the mice were 2 months old, the spleens of six litters were imaged every 2 to 3 weeks until at least one member of each litter had developed a significantly enlarged spleen. Then the entire litter was sacrificed, and the spleens were weighed.
PCR.
Heterozygous Mov13/+ littermates were identified by PCR (11). The common 5′-primer JB1362 (5′-TCAGCTTTGTGGACCTCCGG-3′) is specific for exon 1 of the mCol1a1 gene (nucleotides [nt] 122 to 141); one 3′ primer, JB1363 (5′-GACCCCTCTATACAGAACGC-3′), is reverse complementary to sequences in the first intron of the collagen gene (nt 257 to 238). The Mo-MuLV-specific primer JB1364 (5′-CTTCTGCTCCCCGAGCTCAA-3′) recognizes nt 8236 to 8217 of the Mo-MuLV proviral DNA (39). PCR generates a 136-bp product from the wild-type mCol1a1 allele template and an additional 283-bp product if the Mov13 allele is present.
Approximately 2 μg of tail DNA was used per PCR; the primer sequences used to amplify a 546-bp fragment of the gag-SN transgene (Fig. 1) were JB777 (5′-CTACTGCAAAGAAAAGGGGCAC-3′) and JB908 (5′-GACCTGAATCAGCGTTGTCTTCG-3′). Primers used to amplify a 202-bp fragment of the internal control gene Xist (7) were JB1959 (5′-GAAGTGAATTGAAGTTTTGGTCTAG-3′) and JB1960 (5′-GGGACCTAACTGTTGGCTTTATCAG-3′). PCR was performed under the following conditions. There were two initiation cycles with denaturation at 94°C for 4 min, annealing at 65°C for 1 min, and elongation at 72°C for 2 min. This was followed by 30 cycles of amplification with denaturation at 94°C for 1 min, annealing at 65°C for 1 min, and elongation at 72°C for 1 min and concluded with incubation at 72°C for 3 min followed by cooling to 4°C.
RT-PCR.
Reverse transcription (RT)-PCR was performed as follows. CD90+ T lymphocytes were isolated with MACS CD90 (Thy 1.2) MicroBeads (Miltenyi Biotec, Auburn, Calif.; order no. 491–01), to separate T- and non-T-cell (predominantly B cell) fractions. A total of 106 cells were lysed in 1 ml of lysis-binding buffer containing LiDS-LiCl (Dynal, Oslo, Norway). Isolation of mRNA was carried out with magnetic Dynabeads oligo(dT) 25 (Dynal; product no. 610.11) according to the manufacturer's protocol. Isolated mRNA was treated with 10 U of RNase-free DNase I (Boehringer Mannheim, Mannheim, Germany; catalog no. 776785), in the presence of 50 U of RNase inhibitor (Boehringer Mannheim; catalog no. 799017) and 2.5 mM MgCl2 plus 10 mM Tris-HCl (pH 8.0) for 30 min at 37°C. DNase was inactivated for 10 min at 75oC. First-strand cDNA synthesis was performed with avian myeloblastosis virus (AMV) reverse transcriptase (Boehringer Mannheim; catalog no. 1483188) as described by the manufacturer's protocol. Immediately before cDNA synthesis, mRNA was incubated for 5 min at 65°C and cooled down on ice. RNA was reverse transcribed in the presence of antisense oligonucleotide primer JB908 (Fig. 1). The reaction was carried out in a total volume of 20 μl containing 10 mM Tris, 50 mM KCl (pH 8.3), 5 mM MgCl2, 1 mM (each) deoxynucleoside triphosphates (dNTPs), 0.01 mg of gelatin per ml, 50 U of RNase inhibitor, 20 U of AMV reverse transcriptase, and 2 mM antisense oligonucleotide primer. The reaction mixture was incubated for 60 min at 42oC and subsequently for 5 min at 99oC for heat inactivation of reverse transcriptase. The AmpliWax PCR Gem-facilitated hot start process (Perkin-Elmer; catalog no. N808–0100) was employed for PCR amplification of transgene-specific cDNAs to decrease degenerate priming and multiplex PCR during the assembly of the reaction. Each PCR amplification consisted of heat activation of AmpliTaq polymerase (Perkin-Elmer) at 94°C for 4 min. This was followed by 32 cycles of 94°C for 1 min, 65°C for 1 min, and 72°C for 1 min, with a final extension at 72°C for 10 min with the Gag-SN-specific primer pair JB908 and JB777. The presence of gag-SN gene-specific transcripts gave rise to 546-bp PCR products. A 1-kb DNA ladder served as the molecular weight marker (Gibco/BRL catalog no. 15615–016).
Virus or VLP preparation, immunoblot analysis, and focus formation assay.
Virions used as positive controls for immunoblotting and zymograms were isolated from supernatants of Gag-SN-expressing chicken embryo fibroblast (CEF) cells infected with amphitropic Mo-MuLV derivative Mo(4070A) (32, 36). One to 2 weeks postinfection, virions from 30 ml of supernatant were prepared as described previously (36). In order to isolate virus-like particles (VLP), 23 mice of each line (E and F) that were homozygous for the gag-SN transgene and 23 C57BL6 mice were sacrificed at 20 to172 days of age. Sera obtained from each line were pooled and adjusted to a volume of 50 ml with virion buffer (50 mM Tris-HCl [pH 6.8], 100 mM NaCl). After removal of residual cells, VLP were pelleted by centrifugation of the diluted sera for 30 min at 25,000 rpm at 4°C in a Beckman SW28 rotor, washed twice in 500 μl of virion buffer, and resuspended in 30 μl of the same buffer.
Immune complexes were detected with the Amersham ECL enhanced chemiluminescence kit. For ECL immunoblot analysis of protein extracts from mouse tissues with purified anti-SN antibody, each tissue sample (160 mg of tissue), except for lymph nodes (LN), was ground with a Brinkmann Polytron homogenizer (model PT10/35) twice for 1 min in 1 ml of 3× Laemmli buffer containing 2 mM phenylmethylsulfonyl fluoride at room temperature. LN were dissected and minced with a surgical scalpel. All LN of one animal were pooled and resuspended in 100 μl of 3× Laemmli buffer. The total volume of Laemmli buffer added depended on the size and texture of tissue, so that the final extract solution was not too viscous for loading. Then 20 μl (2.4 to 3.2 mg) of each tissue was loaded in each lane of sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels (8% polyacrylamide) gels. Virus particles were prepared as described previously by infection of CEF cells with Mo(4070A) (32, 36).
Following electrophoresis, proteins were transferred by electroblotting according to standard methods (41) to an Immobilon-P membrane (Millipore) prewet with 100% methanol and incubated in transfer buffer for 10 min. Protein transfer was performed in a Genie apparatus (Idea Scientific) at 24 V for 60 min. After transfer, filters were preabsorbed for 1 h at room temperature with blocking buffer (4% bovine serum albumin in phosphate-buffered saline [PBS]), incubated with a 1:1,000 dilution of affinity-purified anti-SN polyclonal antibody, and washed with PBS–0.05%Tween 20.
Infectious Mo-MuLV titers were determined by the S+L− focus formation assay as described previously (3, 36).
Zymograms.
Organs and tissues were homogenized in 3× Laemmli buffer. Homogenized tissue samples were boiled for 5 min and centrifuged at 14,000 rpm in an Eppendorf centrifuge for 5 min. The supernatant was boiled for additional 2 min and loaded on an SDS-PAGE gel (15% polyacrylamide) containing 10 μg of sheared herring sperm DNA per ml. Following electrophoresis at 20 mA for 4 h in a Hoefer SE 260 slab gel electrophoresis unit, the gel was placed in water for 3 h and gently shaken in a mixture containing 40 mM Tris-HCl (pH 9.5), 0.1 mM CaCl2, 100 mM glycine, and 0.5 mg of ethidium bromide per ml at room temperature for 3 h to 2 days. The incubation allows protein refolding and enzymatic action, and the digested regions of the gel are detected under UV light as dark bands on a fluorescent background of ethidium bromide bound to the DNA. Gels were photographed with an Eagle Eye detection system (Stratagene) at 40 integrations.
Immunohistochemistry.
Hematoxylin- and eosin-stained tissue sections of spleen and liver were examined in a masked fashion to evaluate presence and severity of leukemic cells. To quantify leukemic infiltrates in the liver, a monoclonal antibody directed against CD45 (clone 30-F11; Pharmingen, San Diego, Calif.) (26, 44) was used for immunohistochemical staining. To ensure uniformity of staining essential for quantitative image analysis, samples were stained by an Optimax Plus automated cell stainer (BioGenex, San Ramon, Calif.). Briefly, formalin-fixed, paraffin-embedded tissue sections were deparaffinized and rehydrated and then postfixed in Streck tissue fixative for 20 min. For antigen retrieval, tissues were rinsed in water and heated in a microwave in Na-citrate (0.01 M [pH 6.0]) for 8 min. Endogenous peroxidase was quenched with 3% H2O2 in water for 10 min, and then sections were blocked with buffered casein for 5 min. Primary antibody was applied to the tissues for 60 min at room temperature, the tissues were washed in wash buffer, and secondary biotinylated multilink antibody (BioGenex) was applied for 20 min. The tissues were washed again, and streptavidin-horseradish peroxidase conjugate was added for another 20 min. The sections were then washed, and diaminobenzidine tetrahydrochloride in buffer containing H2O2 was applied to the sections for 10 min. The sections were washed, dehydrated, and mounted. To permit accurate digital quantitation of signal, sections were not counterstained.
Quantitation of immunohistochemical staining on tissues was performed on 20 adjacent fields of tissue examined at a ×200 magnification encompassing a 2.8-mm2 area of liver (left lobe). Images were captured with a Sensys 2 digital camera (Photometrics, Tucson, Ariz.) and analyzed with IP Lab imaging software (Scanalytics, Vienna, Va.). Images were binarized (each pixel converted to a value of 1 [positive] or 0 [negative]), and the total percent area occupied by positive pixels was calculated. This provides a quantitative measure of the total area occupied by positively stained cells or portions of cells in the area evaluated.
Statistical analysis.
Survival curves were derived by the Kaplan-Meier (KM) method with SAS version 6.12 for Windows (SAS Institute, Cary, N.C.). P values for the comparisons of the equality of survival time across transgene strata were based on the log-rank test. Analysis of spleen masses was done with a mixed linear model (SAS version 6.12) with fixed parameters of age and transgene group and with litter treated as a random effect (covariance parameter) to control for correlations within litters (25).
RESULTS
Mouse lines expressing Gag-SN.
In order to express a gag-SN transgene, we sought a relatively strong promoter active in many tissues, especially in the hematopoietic compartment, in which the target virus, Mo-MuLV, multiplies. Thus we could evaluate overall toxicity in a wide variety of tissues, as well as antiviral efficacy. The hCMV-E/P (8) appeared most useful for our experiments, because this control element facilitated transgene expression in 28 tissues analyzed (13). Moreover, hCMV-driven transgenes were reproducibly shown to be highly expressed in spleen, thymus, and other lymphatic tissues (13, 15, 45).
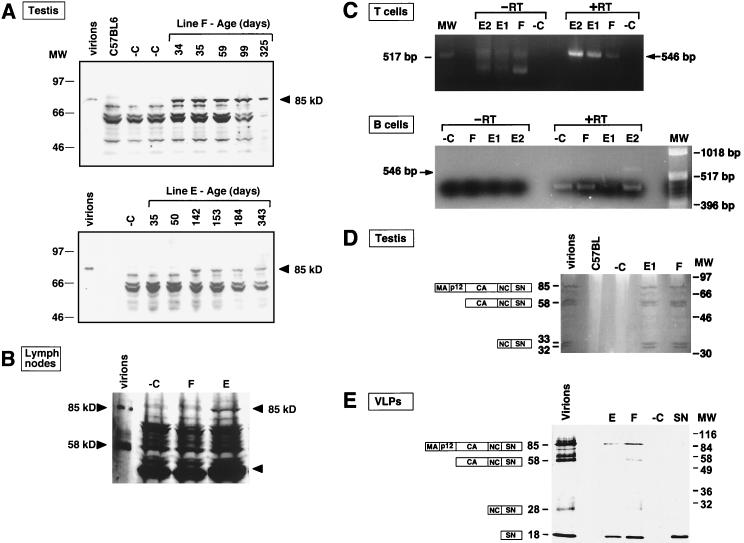
The hCMV-E/P was joined to DNA encoding Gag-SN (Fig. 1A), and Gag-SN protein expression was confirmed in transient transfection experiments (data not shown). Previous studies have proven that expression of this Gag-SN protein in tissue culture cells dramatically reduces the output of infectious progeny retrovirus particles and that this antiviral effect depends on the enzymatic activity of the SN protein (32, 36). Using a PCR screen for the presence of the Gag-SN coding region, we identified three founder mice, two of which transmitted the transgene to progeny, which allowed us to establish transgenic lines E and F. We examined whether the Gag-SN fusion protein was expressed in a variety of tissues in these lines. Both transgenic lines were backcrossed to C57BL/6 three times before further experiments were performed. Figure 2 and the results summarized in Table 1 demonstrate Gag-SN expression by RT-PCR and/or immunoblotting in testis and hematopoietic tissues in lines E and F.
FIG. 2.
Regulation of expression of the enzymatically active antiviral Gag-SN fusion protein in transgenic lines (A) Time course of expression of the antiviral transgene in testis. Testis tissues from 34- to 343-day-old line E and F mice were characterized by immunoblot analysis with anti-SN antibody. Results were reproduced with two or three mice for each age and line. Virus particles containing Gag-SN fusion proteins (virions) (36) served as positive controls. Testis tissue from nontransgenic littermates of line E and F mice, respectively, served as a negative control (−C). The other bands migrating faster than the 85-kDa Gag-SN fusion protein are caused by nonspecific cross-reaction. (B) Antiviral gene products are detectable in lymphatic tissue of line E mice. LN tissue from line F (age 145 days) and line E (age 75 days) mice as well as C57BL/6 (negative control [−C]) mice was analyzed. One additional band in line E tissue in the 40-kDa range (arrow) may result from proteolytic processing of the 85-kDa Gag-SN fusion protein. (C) RT-PCR analysis of poly(A) RNA from B and CD90+ T cells isolated from LN confirms the presence of gag-SN transcripts (546-bp RT-PCR product) in lymphatic cells of line E and F mice. The animals were 412 (E1), 371 (E2), and 460 (F [−C]) days old. PCR without cDNA synthesis (−RT) served as a control for contamination of the poly(A) RNA preparation with genomic DNA. Poly(A) RNA from a nontransgenic littermate (−C) was used as negative control. (D) Zymogram analysis of testis tissue expressing Gag-SN fusion proteins demonstrates nucleolytic activity of the fusion protein. The gel was loaded with 2 mg of testis tissue extract from transgenic lines E and F, a nontransgenic littermate (−C), and C57BL/6. Gag-SN incorporating MuLV particles that were released from tissue culture cells (virions) served as a positive control for nucleolytic activity. Gag-SN precursor protein and products of proteolytic processing incorporated into virus particles are depicted as boxes. (E) Immunoblot analysis of viral pellet fractions from sera of line E and F mice with anti-SN antibody. VLP preparations from 23 homozygous mice each of lines E and F are shown. VLP preparations from 23 nontransgenic C57BL/6 mice served as negative control (−C). Gag-SN incorporating MuLV particles (virions) released from tissue culture cells (36) and 5 ng of purified SN were loaded as positive controls.
TABLE 1.
Gag-SN expression in tissues of transgenic mouse lines E and Fa
| Tissue Type |
n
|
Result by:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Northern blotting
|
Immunoblotting
|
Zymogram
|
RT-PCR
|
|||||||
| E (total, 20) | F (total, 24) | E | F | E | F | E | F | E | F | |
| Circulatory | ||||||||||
| Heart | 1 | 1 | − | − | ||||||
| Lung | 1 | 2 | − | − | − | |||||
| Neural | ||||||||||
| Brain (whole) | 2 | 2 | − | − | − | |||||
| Gastrointestinal | ||||||||||
| Intestine | 1 | 1 | − | − | − | − | ||||
| Liver | 2 | 2 | + | + | ||||||
| Genitourinary | ||||||||||
| Kidney | 1 | 2 | + | − | − | |||||
| Testis | 6 | 6 | + | + | + | + | + | |||
| Ovary | 1 | 1 | − | + | ||||||
| Lymphoid | ||||||||||
| LN | 2 | 1 | + | − | ||||||
| Spleen | 1 | 1 | − | − | − | − | + | |||
| Thymus | 1 | 2 | − | + | − | + | + | |||
| B lymphocytes | 1 | 1 | + | − | ||||||
| T lymphocytes | 2 | 1 | + | + | ||||||
| Other | ||||||||||
| Bone marrow | 1 | 1 | + | − | + | |||||
| Skeletal muscle | 1 | 1 | − | − | ||||||
| Skin | 1 | 1 | + | + | ||||||
| Tail tissue | 9 | 11 | + | + | ||||||
Twenty line E and 24 line F mice 34 to 548 days old were analyzed for expression of the antiviral gene product by RT-PCR, RNA and immunoblotting analyses, and Zymogram assays. All tissues that tested positive for the antiviral gene product by immunoblot analysis exhibited the 85-kDa Gag-SN fusion protein. Proteolytic cleavage products were not detectable in immunoblot analyses. Zymogram analyses revealed 30- and 18-kDa proteolytic cleavage products in bone marrow, thymus, spleen, and testis. A 58-kDa intermediate product could be detected exclusively in testis (Fig. 2D). −, no expression; +, expression.
Transgene expression was observed by assaying for Gag-SN protein (Fig. 2A and B), RNA (Fig. 2C), and enzymatic activity (Fig. 2D). Expression was detected in both transgenic lines E and F in 12 of 17 tissues tested, including bone marrow, liver, kidney, skin, testis, ovary, LN, spleen, thymus, and B and T lymphocytes (Table 1). The latter are of particular relevance, because it is within these tissues that the lymphotropic target retrovirus, Mo-MuLV, multiplies. We observed a protein of ∼85 kDa in transgenic protein extracts that reacts with anti-SN antibody (Fig. 2A and B). This transgene-specific protein comigrates with Gag-SN fusion protein produced in tissue culture cells and incorporated into virions (32, 36). We observed very high levels of expression of this Gag-SN protein in testis (Fig. 2A). Remarkably, the transgenic animals expressing Gag-SN protein have normal fertility, indicating that expression of Gag-SN protein is not deleterious to germ cells. Pathological examination confirmed that males of both transgenic lines have active spermatogenesis. This is interesting, because seminal fluid contains high concentrations of calcium. We noted a different time dependence of expression in the two transgenic lines. Whereas line F mice expressed the Gag-SN protein at all ages tested, expression was not detected until day 142 in line E mice (Fig. 2A).
A lower level of expression was seen in LN from line F mice (Fig. 2B). The presence of the antiviral Gag-SN gene product in hematopoietic tissue was confirmed by RT-PCR analysis of mRNA from B and CD90+ T lymphocytes (Fig. 2C).
Finally, we demonstrated by using zymograms that fusion proteins expressed in the animals were enzymatically active in vitro (Fig. 2D). The 85-kDa Gag-SN fusion protein and lower-molecular-weight derivatives produced by proteolytic processing, including the ∼58-kDa capsid-nucleocapsid-SN protein and a 32- to 33-kDa nucleocapsid-SN protein, were observed at high levels in transgenic testis. Interestingly, the enzymatically active proteolytic cleavage products detectable in testis tissue are identical in mobility to those incorporated into MuLV virions in tissue culture (Fig. 2D). It is reasonable to presume that Gag-SN precursor proteins are processed by proteases of endogenous retroviruses. We conclude that intracellular calcium concentrations in testis tissue are maintained at sufficiently low levels that the Gag-SN protein is not active or toxic inside cells, even though the protein is fully active in vitro in the presence of calcium ions. Furthermore, active Gag-SN protein was specifically detected in spleen samples from transgenic animals (not shown), confirming expression of Gag-SN at the protein-enzyme activity level in a tissue relevant to viral infection.
We sacrificed several animals of the E and F lines to seek evidence of toxicity as revealed by abnormal tissue pathology or other unusual phenotypes. No unusual findings specific to the presence of the transgene were noted upon standard pathological analysis of the following tissues: lung, brain, spleen, ovary, testis, kidney, bladder, pancreas, thymus, heart, stomach, small or large intestine, colon, skin, eye, skeletal muscle, spinal cord, harderian gland, tongue, esophagus, trachea, thyroid, reproductive tract, adrenal gland, LN, bone, salivary gland, liver, and nasal cavity. Similarly, complete blood counts and blood chemistry for the transgenic lines were comparable to those of normal littermates. We also searched for evidence of a possible immune response to Gag-SN by screening the sera for anti-SN activity and by extensive pathological examinations, but found none.
The mean longevities of E and F line mice hemizygous for the gag-SN transgene (754 [n = 37] and 742 [n = 61] days, respectively) were essentially identical to that of C57BL/6 control mice (737 days [n = 24]). These data suggest that Gag-SN expression is nontoxic to mice and does not shorten life span.
Gag-SN fusion proteins are incorporated into VLPs.
Since retroviral Gag proteins contain all of the necessary signals for particle formation and Gag-SN fusion proteins have been shown to be competent for particle formation (32), we assayed particulate fractions of sera from line E and F mice, homozygous for the antiviral transgene, for fusion proteins. The sera were assayed for incorporation of fusion proteins into VLPs by immunoblotting with anti-SN antibodies. Figure 2E shows that the isolated particles predominantly contain proteins that comigrate with the 85-kDa Gag-SN fusion proteins and the proteolytic cleavage product SN (18 kDa) incorporated into virions released from Gag-SN-expressing tissue culture cells (36). In the case of line F, 58-kDa CA-NC-SN and 28-kDa NC-SN intermediates are detected in the viral pellet fractions additionally. We conclude that Gag-SN fusion proteins expressed in both transgenic lines are assembled into VLPs and released into the blood.
Reduction of viral titers in Mov lines expressing Gag-SN.
We examined the effects of infection of gag-SN transgenic and nontransgenic littermates by genetic crosses and observed a protective effect of the gag-SN transgene against Mo-MuLV infection. However, Mo-MuLV was still able to multiply in gag-SN transgenic animals, indicating that protection was incomplete.
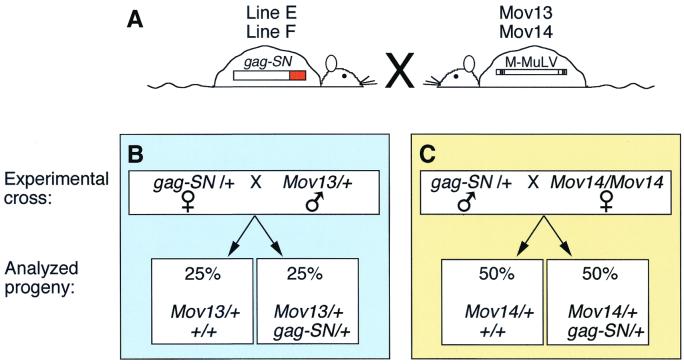
To simplify and to standardize the infection procedure, we employed Mov lines of mice containing endogenous proviral copies of Mo-MuLV. In these strains, virus expression leads to viremia, viral spread, and, ultimately, T-cell leukemia or lymphoma. The infection is early and severe: retrovirus activation is prenatal, and the mice die of virus-induced leukemia at 3 to 6 months. Their disease course and pathogenesis have been studied extensively (2, 4, 10, 16, 19–21, 29, 34). Furthermore, the Mov13 and Mov14 lines have served as quantitative models of in utero infection for evaluating pharmacologic strategies of antiretroviral therapy (27, 37, 38). We set up crosses between Mov13 and Mov14 mice and the two gag-SN transgenic lines E and F (Fig. 3), producing gag-SN transgenic Mov mice (doubly transgenic or Mov13 [Mov14]/+; gag-SN/+) and control Mov littermates negative for the gag-SN transgene (singly transgenic or Mov13 [14]/+;+/+). Since the four primary mouse lines Mov13, Mov14, E, and F have the same genetic background (C57BL/6), this experimental design ensured a high degree of genetic and environmental uniformity for the experimental and control animals.
FIG. 3.
Genetic crosses introducing Mo-MuLV into gag-SN transgenic mice (A) To evaluate the antiviral efficacy of Gag-SN fusion proteins in vivo, heterozygous gag-SN/+ mice of lines E and F were crossed with heterozygous Mov13/+ males or homozygous Mov14/Mov14 females. (B) Mov13/+ mice resulting from crosses of gag-SN/+ females with Mov13/+ males lacking the antiviral transgene (Mov13/+; +/+) were compared with their Mov13/+; gag-SN/+ littermates. Only half of the progeny are informative, because the other half do not inherit Mov13. (C) All progeny of the Mov14 crosses are informative. Comparisons are made between Mov14/+; +/+ and Mov14/+; gag-SN/+ siblings.
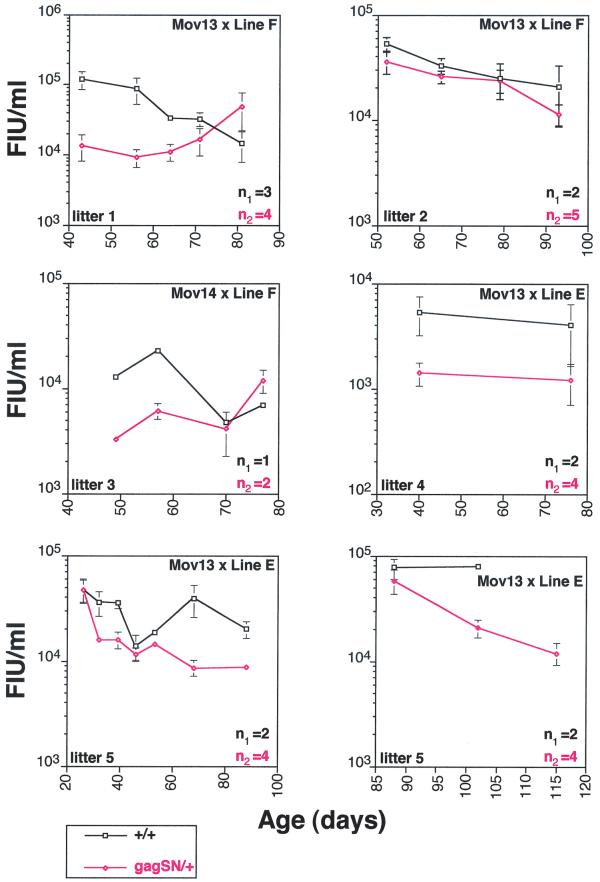
We examined the levels of infectious Mo-MuLV in the serum by bleeding them regularly for several months and then performing S+L− focus formation assays on the serum samples (Fig. 4). We observed consistent reductions in the titers of infectious virus in both gag-SN transgenic lines, ranging from 2- to 10-fold below those of their singly transgenic Mov littermates. Importantly, significant reductions in titer were observed in the Mov13/+ mice. Singly transgenic Mov13/+ mice undergo an especially virulent infection representing a “worst case scenario” that even resists zidovudine (AZT) therapy (38). The pattern of inhibition of virus multiplication differed between lines E and F. In line E, relatively little decrease in titer was observed at young ages (25 to 60 days), but more significant protection was observed at later times. In line F, the opposite trend was observed: namely good protection at early time points (40 to 65 days) and relatively less protection at later time points. The observed antiviral effects parallel the timing of maximal testis gene expression in lines E and F, suggesting that antiviral transgene expression is initiated in line F at an earlier time point than in line E.
FIG. 4.
Infectious titers of sera from Gag-SN-expressing Mov mice are reduced by 2- to 10-fold. Infectious Mo-MuLV titers of sera from Gag-SN-expressing Mov mice were compared with those of their Mov siblings lacking gag-SN. Sera of five different litters were collected every 7 to 15 days up to several months, and infectious titers were determined by a focus formation assay. The litters analyzed result from crosses illustrated in Fig. 3 (upper right corner of each graph). n1 and n2 reflect number of animals compared in each graph. Each graph reflects the mean infectious titers in sera from singly transgenic Mov13 mice (black curve) and their doubly transgenic siblings (red curve). In the case of litter 5, sera were harvested over 85 days. Because of the large number of samples, sera of this litter were assayed in two independent focus formation assays.
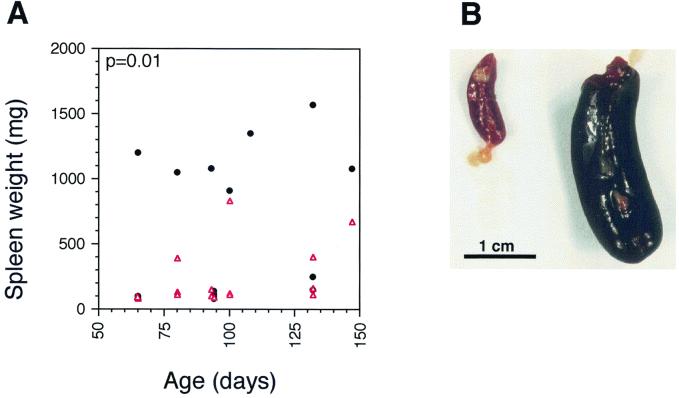
A surrogate assay for viral multiplication and for virus-induced pathology is the development of splenomegaly in infected animals. Proviral integration of Mo-MuLV near proto-oncogenes in cells of the lymphatic system leads to hematopoietic neoplasms, including B- and T-cell lymphomas, with subsequent splenomegaly. A reduction in viral load over the life of the organism, such as in the Gag-SN transgenic mice, is predicted to decrease the probability of these insertional activations of oncogenes. Thus, the antiviral transgene should delay preleukemic physiological changes, most notably splenic enlargement and thymic atrophy, the development of splenomegaly, and the onset of leukemia. Essentially 100% of Mov13/+ animals die with splenomegaly (defined as a spleen weight of ≥400 mg [S. Ruscetti, personal communication]), whereas control C57BL/6 mice have mean spleen weights of 106 mg. However, the mean spleen weights of Mov13/+ mice at death differed: doubly transgenic Mov13/+ mice had reduced spleen weights (574 mg [line E+F; n = 39]) relative to singly transgenic Mov13/+ littermates (762 mg[Mov13/+; n = 48],) although the differences were not statistically significant (P = 0.2). In contrast, the mean spleen weights of Mov14/+ animals intentionally sacrificed at various ages showed a dramatic difference between doubly transgenic (232 ± 60 mg) and singly transgenic (599 ± 150 mg) littermates (Fig. 5) that was highly significant (P = 0.012). Thus a clear attenuation of virus-induced splenomegaly is conferred by the gag-SN transgenes.
FIG. 5.
(A). Splenic enlargement (tumor development) typical of leukemic Mov14/+ mice is delayed in Gag-SN-expressing Mov14/+ animals. The spleen sizes of mice from seven litters resulting from a Mov14/Mov14 × gag-SN/+ cross were monitored by magnetic resonance imaging. As soon as one or more animals in the litter developed enlarged spleens, the relevant litter was sacrificed, and all spleens were weighed. Spleen weights of gag-SN/+, Mov14/+ mice (red triangles, n = 15) were compared with those of nontransgenic Mov14/+ littermates (black circles, n = 16) in a scatter plot. Mean spleen weights of gag-SN/+, Mov14/+ mice (232 mg) are reduced by 61% (P = 0.012) relative to their singly transgenic littermates (599 mg). (B) Marked splenomegaly in a singly transgenic Mov14/+ mouse that developed a T-cell lymphoma (1,300 mg [right]), compared with that in the spleen of an age-matched Mov14/+ littermate expressing the antiviral fusion protein (87 mg [left]).
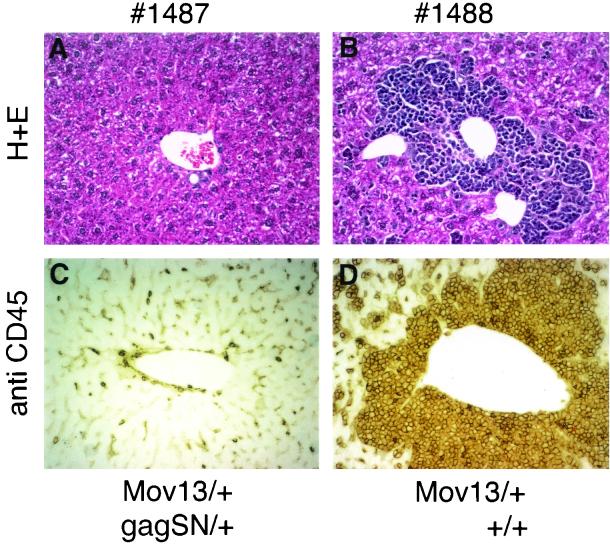
The development of T-cell leukemia in Mov mice is associated with leukemic infiltration of the liver by lymphoid cells. The number of cells infiltrating the liver at a certain time point is another parameter reflecting the stage of leukemia development. We characterized 15 singly or doubly transgenic littermates (four litters) with respect to infiltration of their livers at different ages (Table 2). Since the presence of CD45 distinguishes leukocytes from nonhematopoietic cells (22), we used it as a marker to quantitate leukemic infiltration of nonhematopoietic tissue, like liver, by lymphoid cells. The animals included in this study came from crosses Mov13/+ × gag-SN/+ (line E) and Mov14/Mov14 × gag-SN/+ (line E). None of the seven Gag-SN-expressing Mov mice exhibited CD45+ cells above background level. In contrast, two out of eight singly transgenic Mov13 mice were characterized by significant leukemic infiltration of CD45+ lymphoid cells (Table 2 and Fig. 6).
TABLE 2.
Quantitation of leukemic cells in livers of singly versus doubly transgenic Mov13 micea
| Litter | Mouse | Mov13 | gag-SN (E) | %ROIb | Age (days) |
|---|---|---|---|---|---|
| 1 | 1486 | + | + | 1.99 | |
| 1487 | + | + | 4.78 | ||
| 1488 | + | − | 32.51 | ||
| 1491 | + | + | 2.19 | 303 | |
| 1493 | + | + | 2.24 | ||
| 2 | 1713 | + | − | 1.47 | |
| 1418 | + | + | 1.73 | ||
| 1719 | + | − | 57.69 | 237 | |
| 1720 | + | − | 2.88 |
Only singly transgenic Mov13 mice that do not express the antiviral Gag-SN fusion protein exhibit significant infiltration by leukemic cells (mice 1488 and 1719). Livers from 15 mice (4 litters) resulting from Mov13/+ × gag-SN/+ and Mov14/Mov14 × gag-SN/+ crosses were analyzed for leukemic infiltrates by immunohistochemical staining with a monoclonal antibody against CD45 followed by image analysis. Animals included in the table were sacrificed and analyzed at 303 days of age (litter 1) and 237 days of age (litter 2). The presence of the Mov13 and/or gag-SN transgene of line E [gag-SN (E)] is indicated. The remaining six mice (two Mov14 litters, 179 days old) included in the study did not exhibit any infiltration above background (data not shown). Infiltration was quantified as described in Materials and Methods.
%ROI, mean percent area scores of region of interest of liver staining positive for CD45.
FIG. 6.
Infiltration of the liver by leukemic cells is significantly reduced in Gag-SN-expressing Mov13 mice. The difference in infiltration between a singly transgenic Mov13 mouse (no. 1488) and its doubly transgenic Gag-SN-expressing littermate (no. 1487) is demonstrated by hematoxylin and eosin (H+E) staining (A and B) and immunohistochemical staining with anti-CD45 antibody (C and D). Quantitation of immunohistochemical staining of these two mice is included in Table 2.
The data suggest that leukemic infiltration of the liver by lymphoid cells is significantly reduced in Gag-SN-expressing Mov13 mice (Fig. 6). The young age of the Mov14 mice (179 days) included in the study could account for the fact that leukemic infiltration was not even detectable in singly transgenic Mov14 littermates.
Increased longevity of Gag-SN-expressing infected mice.
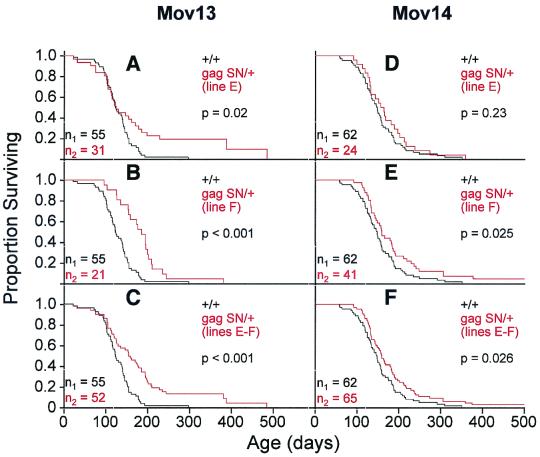
Both doubly transgenic and singly transgenic animals eventually perish of leukemia. However, the gag-SN transgenes reduced infectious viral titer and splenomegaly and delayed leukemic infiltration. Thus it was of interest to determine the effects of the transgene on longevity of infected animals. KM survival analyses of Mov13 and Mov14 animals showed increased longevity of doubly transgenic animals over singly transgenic littermates (Fig. 7). Relatively little effect of transgene status on survival was seen early in life (days 0 to 100). On average, the antiviral transgene conferred a 45% (line E) to 46% (line F) increase in mean longevity of Mov13 animals, corresponding to 57 days. In the case of line E, much of the increased longevity was due to a subset of transgenic animals that showed a long-term survival phenotype not observed in singly transgenic Mov13 animals. One doubly transgenic animal lived 486 days (Fig. 7A to C). Mean life spans of Mov14 mice were increased by 11% (line E) to 23% (line F), corresponding to 17 to 36 days (Fig. 7D to F). One doubly transgenic Mov14 mouse lived for 553 days (Fig. 7D to F).
FIG. 7.
Life spans of doubly transgenic Gag-SN-expressing Mov mice are extended relative to those of their singly transgenic Mov littermates. (A to F) KM survival plots comparing singly transgenic Mov13 (A to C) and Mov14 (D to F) mice (+/+ [black curve]) with their doubly transgenic Mov13 and Mov14 (gag-SN/+ [red curve]) littermates. Life spans of doubly transgenic Mov13 and Mov14 mice (gag-SN/+) of line E (A and D, respectively) and line F (B and E, respectively) were compared independently and collectively (C and F) with those of their singly transgenic (+/+) Mov13 and Mov14 littermates. The mean life span of singly transgenic Mov13 and Mov14 mice, 126 and154 days, respectively, is prolonged to 183 and 171 days in line E and 184 and 187 days in line F, respectively. The colors of the genotype and n correspond to the colors of the KM curves. p, significance of the difference in longevity between the two groups of mice.
gag-SN transgenic Mov mice developed external symptoms of leukemia more slowly than their singly transgenic Mov littermates. These symptoms include premature coat graying, reduced motor activity, and increased time huddling and panting. Thus the gag-SN transgenes increase the longevity and vigor of infected animals, in some cases very dramatically. The increased longevity and vigor are probably direct consequences of decreased infectious retrovirus multiplication.
DISCUSSION
Significant antiviral effect in vivo.
The general feasibility, antiviral efficacy, and retrovirus-inactivating mechanisms of CTVI were previously demonstrated in tissue culture with an MuLV. We developed an animal model for this gene therapeutic antiviral approach and established its efficacy in vivo. Although Gag-SN-expressing Mov mice still produce infectious Mo-MuLV particles, the data presented here clearly show inhibition of initiation of Mo-MuLV leukemia in those mice. Expression of the antiviral fusion protein leads to reduced infectious titers, diminished spleen masses, and extended life spans of doubly transgenic gag-SN/Mov mice.
A strength of CTVI is that it is designed to target a different step in the virus life cycle from those of most other current antiretroviral gene therapy approaches. It has several advantages over other antiviral strategies involving RNA-based inhibitors. Viral escape mutants capable of performing their normal functions nearly as well as the wild type, but resistant to a given antiviral strategy, are a major difficulty in antiretroviral therapy. We hypothesize that mutations in the Mo-MuLV gag gene capable of circumventing the antiviral mechanism would be extremely rare, because the Gag polyprotein plays many roles in virus assembly and infection. Mutants with alterations in gag that fail to interact with Gag-SN should be profoundly impaired with respect to one or more of these functions. Thus, fully functional escape mutants may not arise in the case of CTVI. This hypothesis is supported by our and others' observation that the antiviral effect is sustained in tissue culture during its entire life span up to 96 days (35, 36, 42). Indeed, there are no data so far implying the existence of escape mutants during CTVI.
In our studies, we observed significant antiviral effects of Gag-SN expression in both Mov13 and Mov14 lines. The Gag-SN effects observed in the Mov13 line are remarkable in that this was the only Mov strain in which AZT treatment had no effect (38). Mov13 activates the endogenous virus in many cell types simultaneously (29), and thus AZT treatment cannot prevent the resultant massive viral spread. Obviously, in the case of Mov13, the gag-SN transgene is more potent than the drug AZT and is able to interfere with substantial levels of viral multiplication. We observed significant effects of the transgene on viral infectivity as well as on its pathogenic consequences: splenomegaly, infiltration of the liver by leukemic cells, decreased vigor, and early death.
Limitations.
It is important to understand why protection against Mo-MuLV is incomplete and why a fraction of virions escape inactivation. Three major factors probably limit in vivo antiviral efficacy. (i) Spatial and temporal expression of Gag-SN does not always fully overlap with Mov expression. (ii) The expression level in transgenic animals is low. (iii) During self-assembly, Mo-MuLV might compete with endogenous C-type retroviruses for Gag-SN fusion proteins. All strains of mice contain a few dozen endogenous C-type retroviruses closely related to MuLV (23). Expression of endogenous MuLV varies greatly with inbred mouse strains and with the age of the mice (18, 23, 24). In some cases, expression leads to replication of infectious virus (6). Since Mo-MuLV and endogenous C-type retroviruses are identical in their gag coding regions, incorporation of Mo-MuLV-Gag-fusion proteins is expected. The limited pool of Gag-SN fusion proteins may be competitively incorporated into endogenous retrovirus-like particles.
Further studies will address the question of variation in antiviral transgene expression at the level of individual cells. It is possible that all viral replication occurs in a subset of cells in which antiviral transgene expression is low or nonexistent. This possibility is supported by previous work by VanBrocklin and Federspiel (42). The authors suggested that the major factor determining the level of the therapeutic antiviral effect in the tissue culture model was most likely the population of cells expressing low levels of the Gag-SN polyprotein. After increasing expression of the fusion protein in tissue culture, infectious Mo(4070A) retrovirus production was eliminated.
Our studies suggest that high levels of expression correlate with stronger antiviral effects observed in vivo. Expression levels observed in tissue culture experiments in which the identical antiviral fusion proteins were expressed were much higher than those seen in vivo (G.S. and J.D.B., unpublished data). Most of our studies were done with heterozygous transgenes. It should be possible to combine the E and F transgenes in a doubly transgenic line in order to obtain increasingly potent antiviral effects. We were not able to demonstrate expression of the hCMV-controlled Gag-SN protein in every tissue. Soriano and coworkers demonstrated that targeting of a transgene to the ROSA 26 locus in mouse may be a more reliable method to achieve ubiquitous expression during development or in the adult (40, 48). We also expect to obtain improved antiviral effects with more suitable promoters, like the Mo-MuLV LTR promoter itself (15). Putting the antiviral transgene under control of the Mo-MuLV LTR should facilitate coexpression of the MuLV target virus and the antiviral fusion protein in the same tissues. Further studies will also include the characterization of the protective effect of Gag-SN fusion proteins in acutely infected mice. For that purpose, newborn gag-SN transgenic mice will be injected intraperitoneally with different doses of Mo-MuLV, and the protective effect will be quantified over time. Other unknowns that might affect the efficacy of this strategy include the kinetics of calcium seepage into the virion, which is difficult to address experimentally, and the extent to which infection is spread by cell-cell contact. The latter spreading mechanism, which might be especially important in lentiviruses, might fail to expose the nuclease to sufficient calcium ions to kill the virus.
Perspectives.
After optimization of distribution patterns and levels of Gag-nuclease expression in transgenic animals, the methods described here could readily be used to construct improved versions of farm animals that currently suffer from retroviral diseases, including chickens (avian leukosis virus), goats (caprine arthritis-encephalitis virus), sheep (Maedi/Visna virus), cattle (bovine leukemia virus), and horses (equine infectious anemia virus). Transgenic technology has already been developed for many of these animals. In principle, CTVI could also be applied against other important viral pathogens of such animals.
Perhaps the biggest challenges are human retroviruses. Short of germ line gene therapy, targeting a majority of virus-infected cells by a somatic gene therapeutic approach would be necessary for a robust antiviral effect. Our results here indicate that even relatively modest decreases in retroviral titers in vivo can result in significant improvements in clinical outcomes, such as inhibiting the development of leukemia and increasing life span. Previous publications demonstrated that, in the case of HIV, a 2- to 10-fold reduction in infectious particle titer (as observed in our transgenic mouse model) has a significant effect on rate of progression and quality of life of the patients (14, 17, 28, 33). HIV has additional vulnerability to capsid-targeted approaches, in that a variety of viral and cellular proteins are efficiently targeted to virions. For example, fusion of a dominant-negative version of HIV protease to HIV virion proteins showed a significant antiviral effect against HIV-2 in tissue culture (47). Thus capsid-targeted strategies continue to provide a promising approach for therapy against a variety of viruses that affect humans directly and indirectly.
ACKNOWLEDGMENTS
We thank Teresa Zimmers, Ed Hsiao, Alex McPherron, Paul Dunlap, Sarah Richardson, and Se-Jin Lee for useful discussions and technical support. We also thank Dominic Dordai for separation of B and T cells, Michelle Donithan and Scott Zigler for assistance with statistical analyses, V. Chacko for help with magnetic resonance imaging, Tracey Bunton for pathological examinations, Ruth Curry and Rudolf Jaenisch for generous gifts of Mov14 mice, Jürgen Löhler for crucial help with immunohistochemistry, and Alan Rein for Mo-MuLV viral stocks and helpful discussions. We thank Steve Hughes and Mark Federspiel for helpful advice and encouragement during the progress of the project.
This work was supported in part by postdoctoral fellowship Schu1014/1–1 and habilitation grant Schu1014/2–1 of the Deutsche Forschungsgemeinschaft (G.S.) and by NIH grant PO1-AI41215 (J.D.B.).
REFERENCES
- 1.Baltimore D. Intracellular immunization. Nature. 1988;335:395–396. doi: 10.1038/335395a0. [DOI] [PubMed] [Google Scholar]
- 2.Barker D D, Wu H, Hartung S, Breindl M, Jaenisch R. Retrovirus-induced insertional mutagenesis: mechanism of collagen mutation in Mov13 mice. Mol Cell Biol. 1991;11:5154–5163. doi: 10.1128/mcb.11.10.5154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bassin R H, Tuttle N, Fischinger J R. Rapid cell culture assay technique for murine leukaemia viruses. Nature. 1971;229:564–566. doi: 10.1038/229564b0. [DOI] [PubMed] [Google Scholar]
- 4.Berleth T, Nobis P, Jaenisch R, Harbers K. Activation of endogenous retroviral genomes in Mov strains of mice. J Gen Virol. 1987;68:2919–2923. doi: 10.1099/0022-1317-68-11-2919. [DOI] [PubMed] [Google Scholar]
- 5.Boeke J D, Hahn B. Destroying retroviruses from within. Trends Microbiol. 1996;4:421–426. doi: 10.1016/0966-842x(96)10065-2. [DOI] [PubMed] [Google Scholar]
- 6.Boeke J D, Stoye J P. Retrotransposons, endogenous retroviruses, and the evolution of retroelements. In: Varmus H, Hughes S, Coffin J, editors. Retroviruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1997. pp. 343–435. [PubMed] [Google Scholar]
- 7.Borsani G, Tonlorenzi R, Simmler M C, Dandolo L, Arnaud D, Capra V, Grompe M, Pizzuti A, Muzny D, Lawrence C, et al. Characterization of a murine gene expressed from the inactive X chromosome. Nature. 1991;351:325–329. doi: 10.1038/351325a0. [DOI] [PubMed] [Google Scholar]
- 8.Boshart M, Weber F, Jahn G, Dorsch-Häsler K, Fleckenstein B, Schaffner W. A very strong enhancer is located upstream of an immediate early gene of human cytomegalovirus. Cell. 1985;41:521–530. doi: 10.1016/s0092-8674(85)80025-8. [DOI] [PubMed] [Google Scholar]
- 9.Bunnell B A, Morgan R A. Gene therapy for infectious diseases. Clin Microbiol Rev. 1998;11:42–56. doi: 10.1128/cmr.11.1.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doehmer J, Breindl M, Willecke K, Jaenisch R. Genetic transmission of Moloney leukemia virus: mapping of the chromosomal integration site. Hamatol Bluttransfus. 1979;23:561–568. doi: 10.1007/978-3-642-67057-2_72. [DOI] [PubMed] [Google Scholar]
- 11.Dziadek M, Bakker M. Genetic analysis of the preimplantation embryo. Boca Raton, Fla: CRC Press; 1993. [Google Scholar]
- 12.Federspiel M J, Crittenden L B, Hughes S H. Expression of avian reticuloendotheliosis virus envelope confers host resistance. Virology. 1989;173:167–177. doi: 10.1016/0042-6822(89)90232-8. [DOI] [PubMed] [Google Scholar]
- 13.Furth P A, Hennighausen L, Baker C, Beatty B, Woychick R. The variability in activity of the universally expressed human cytomegalovirus immediate early gene 1 enhancer/promoter in transgenic mice. Nucleic Acids Res. 1991;19:6205–6208. doi: 10.1093/nar/19.22.6205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hammer S M, Katzenstein D A, Hughes M D, Gundacker H, Schooley R T, Haubrich R H, Henry W K, Lederman M M, Phair J P, Niu M, Hirsch M S, Merigan T C. A trial comparing nucleoside monotherapy with combination therapy in HIV-infected adults with CD4 cell counts from 200 to 500 per cubic millimeter. N Engl J Med. 1996;335:1081–1090. doi: 10.1056/NEJM199610103351501. [DOI] [PubMed] [Google Scholar]
- 15.Han L, Yun J S, Wagner T E. Inhibition of Moloney murine leukemia virus-induced leukemia in transgenic mice expressing antisense RNA complementary to retroviral packaging sequences. Proc Natl Acad Sci USA. 1991;88:4313–4317. doi: 10.1073/pnas.88.10.4313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harbers K, Kuehn M, Delius H, Jaenisch R. Insertion of retrovirus into the first intron of alpha 1(I) collagen gene to embryonic lethal mutation in mice. Proc Natl Acad Sci USA. 1984;81:1504–1508. doi: 10.1073/pnas.81.5.1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ho D D. Viral counts count in HIV infection. Science. 1996;272:1124–1125. doi: 10.1126/science.272.5265.1124. [DOI] [PubMed] [Google Scholar]
- 18.Huebner R J, Kellorf G J, Sarma P S, Lane W T, Turner H C, Gilden R V, Orozlan S, Meier H, Myers D D, Peters R L. Group-specific antigen expression during embryogenesis of the genome of the C-type RNA tumor viruses: implications for ontogenesis and oncogenesis. Proc Natl Acad Sci USA. 1970;67:366–376. doi: 10.1073/pnas.67.1.366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jaenisch R, Harbers K, Schnieke A, Lohler J, Chumakov I, Jahner D, Grotkopp D, Hoffmann E. Germline integration of Moloney murine leukemia virus at the Mov13 locus leads to recessive lethal mutation and early embryonic death. Cell. 1983;32:209–216. doi: 10.1016/0092-8674(83)90511-1. [DOI] [PubMed] [Google Scholar]
- 20.Jaenisch R, Jähner D, Nobis P, Simon I, Löhler J, Harbers K, Grotkopp D. Chromosomal position and activation of retroviral genomes inserted into the germ line of mice. Cell. 1981;24:519–529. doi: 10.1016/0092-8674(81)90343-3. [DOI] [PubMed] [Google Scholar]
- 21.Jähner D, Jaenisch R. Integration of Moloney leukaemia virus into the germ line of mice: correlation between genotype and virus activation. Nature. 1980;287:456–458. doi: 10.1038/287456a0. [DOI] [PubMed] [Google Scholar]
- 22.Johnson P, Maiti A, Ng D H W. CD45: a family of leukocyte-specific cell surface glycoproteins. In: Herzenberg L A, Weir D M, Blackwell C, editors. Weir's handbook of experimental immunology. Vol. 2. Cambridge, Mass: Blackwell Science; 1997. pp. 62.1–62.16. [Google Scholar]
- 23.Keshet E, Schiff R, Itin A. Mouse retrotransposons: a cellular reservoir of long terminal repeat (LTR) elements with diverse transcriptional specificities. Adv Cancer Res. 1991;56:215–251. doi: 10.1016/s0065-230x(08)60482-0. [DOI] [PubMed] [Google Scholar]
- 24.Kozak C A, Ruscetti S. The retroviridae. New York, N.Y: Plenum Press; 1992. pp. 405–481. [Google Scholar]
- 25.Laird N M, Ware J H. Random-effects models for longitudinal data. Biometrics. 1982;38:963–974. [PubMed] [Google Scholar]
- 26.Ledbetter J A, Herzenberg L A. Xenogeneic monoclonal antibodies to mouse lymphoid differentiation antigens. Immunol Rev. 1979;47:63–90. doi: 10.1111/j.1600-065x.1979.tb00289.x. [DOI] [PubMed] [Google Scholar]
- 27.Lee J S, Mullaney S, Bronson R, Sharpe A H, Jaenisch R, Balzarini J, De Clerq E, Ruprecht R M. Transplacental antiretroviral therapy with 9-(2-phosphonylmethoxyethyl)adenine is embryotoxic in transgenic mice. J Acquir Immune Defic Syndr. 1991;4:833–838. [PubMed] [Google Scholar]
- 28.Mellors J W, Rinaldo C R, Jr, Gupta P, White R M, Todd J A, Kingsley L A. Prognosis in HIV-1 infection predicted by the quantity of virus in plasma. Science. 1996;272:1167–1170. doi: 10.1126/science.272.5265.1167. [DOI] [PubMed] [Google Scholar]
- 29.Moser T, Harbers K, Kratochwil K. Tissue- and stage-specific activation of an endogenous provirus after transcription through its integration site in the opposite orientation. Mol Cell Biol. 1996;16:384–389. doi: 10.1128/mcb.16.1.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Munke M, Harbers K, Jaenisch R, Francke U. Chromosomal mapping of four different integration sites of Moloney murine leukemia virus including the locus for alpha 1(I) collagen in mouse. Cytogenet Cell Genet. 1986;43:140–149. doi: 10.1159/000132312. [DOI] [PubMed] [Google Scholar]
- 31.Natsoulis G, Boeke J D. New antiviral strategy using capsid-nuclease fusion proteins. Nature. 1991;352:632–635. doi: 10.1038/352632a0. [DOI] [PubMed] [Google Scholar]
- 32.Natsoulis G, Seshaiah P, Federspiel M J, Rein A, Hughes S H, Boeke J D. Targeting of a nuclease to murine leukemia virus capsids inhibits viral multiplication. Proc Natl Acad Sci USA. 1995;92:364–368. doi: 10.1073/pnas.92.2.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O'Brien W A, Hartigan P M, Martin D, Esinhart J, Hill A, Benoit S, Rubin M, Simberkoff M S, Hamilton J D. Changes in plasma HIV-1 RNA and CD4+ lymphocyte counts and their risk of progression to AIDS. N Engl J Med. 1996;334:426–431. doi: 10.1056/NEJM199602153340703. [DOI] [PubMed] [Google Scholar]
- 34.Schnieke A, Stuhlmann H, Harbers K, Chumakov I, Jaenisch R. Endogenous Moloney leukemia virus in nonviremic Mov substrains of mice carries defects in the proviral genome. J Virol. 1983;45:505–513. doi: 10.1128/jvi.45.2.505-513.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schumann G, Cannon K, Ma W-P, Crouch R J, Boeke J D. Anti-retroviral effect of a gag-RNase HI fusion gene. Gene Ther. 1997;4:593–599. doi: 10.1038/sj.gt.3300421. [DOI] [PubMed] [Google Scholar]
- 36.Schumann G, Qin L, Rein A, Natsoulis G, Boeke J D. Therapeutic effect of Gag-nuclease fusion protein on retrovirus-infected cell cultures. J Virol. 1996;70:4329–4337. doi: 10.1128/jvi.70.7.4329-4337.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sharpe A H, Hunter J J, Ruprecht R M, Jaenisch R. Maternal transmission of retroviral disease and strategies for preventing infection of the neonate. J Virol. 1989;63:1049–1053. doi: 10.1128/jvi.63.3.1049-1053.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sharpe A H, Hunter J J, Ruprecht R M, Jaenisch R. Maternal transmission of retroviral disease: transgenic mice as a rapid test system for evaluating perinatal and transplacental antiretroviral therapy. Proc Natl Acad Sci USA. 1988;85:9792–9796. doi: 10.1073/pnas.85.24.9792. . (Erratum, 86:2045, 1989.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shinnick T M, Lerner R A, Sutcliffe J G. Nucleotide sequence of Moloney murine leukemia virus. Nature. 1981;293:543–548. doi: 10.1038/293543a0. [DOI] [PubMed] [Google Scholar]
- 40.Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 41.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.VanBrocklin M, Federspiel M J. Capsid-targeted viral inactivation can eliminate the production of infectious murine leukemia virus in vitro. Virology. 2000;267:111–123. doi: 10.1006/viro.1999.0113. [DOI] [PubMed] [Google Scholar]
- 43.VanBrocklin M, Ferris A L, Hughes S H, Federspiel M J. Expression of a murine leukemia virus Gag-Escherichia coli RNase HI fusion polyprotein significantly inhibits virus spread. J Virol. 1997;71:3312–3318. doi: 10.1128/jvi.71.4.3312-3318.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van Ewijk W, van Soest P L, van den Engh G J. Fluorescence analysis and anatomic distribution of mouse T lymphocyte subsets defined by monoclonal antibodies to antigens Thy-1, Lyt-1, Lyt-2, and T-200. J Immunol. 1981;127:2594–2604. [PubMed] [Google Scholar]
- 45.von Schmidt E, Christoph G, Zeller R, Leder P. The cytomegalovirus enhancer: a pan-active control element in transgenic mice. Mol Cell Biol. 1990;10:4406–4411. doi: 10.1128/mcb.10.8.4406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wood W I, Cachianes G, Henzel W J, Winslow G A, Spencer S A, Hellmiss R, Martin J L, Baxter R C. Cloning and expression of the growth hormone-dependent insulin-like growth factor-binding protein. Mol Endocrinol. 1988;2:1176–1185. doi: 10.1210/mend-2-12-1176. [DOI] [PubMed] [Google Scholar]
- 47.Wu X, Liu H, Xiao H, Conway J A, Kappes J C. Inhibition of human and simian immunodeficiency virus protease function by targeting Vpx-protease-mutant fusion protein into viral particles. J Virol. 1996;70:3378–3384. doi: 10.1128/jvi.70.6.3378-3384.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]