Abstract
Purpose:
Sarcoma encompasses a diverse group of cancers that are typically resistant to current therapies, including immune checkpoint blockade (ICB), and underlying mechanisms are poorly understood. The contexture of sarcomas limits generation of high-quality data using cutting-edge molecular profiling methods, such as single-cell RNA-sequencing, thus hampering progress in understanding these understudied cancers.
Experimental Design:
Here, we demonstrate feasibility of producing multimodal single-cell genomics and whole-genome sequencing data from frozen tissues, profiling 75,716 cell transcriptomes of five undifferentiated pleomorphic sarcoma and three intimal sarcoma samples, including paired specimens from two patients treated with ICB.
Results:
We find that genomic diversity decreases in patients with response to ICB, and, in unbiased analyses, identify cancer cell programs associated with therapy resistance. Although interactions of tumor-infiltrating T lymphocytes within the tumor ecosystem increase in ICB responders, clonal expansion of CD8+ T cells alone was insufficient to predict drug responses.
Conclusions:
This study provides a framework for studying rare tumors and identifies salient and treatment-associated cancer cell intrinsic and tumor microenvironmental features in sarcomas.
Translational Relevance.
Understanding genomic and transcriptional heterogeneity across and within sarcomas at baseline and during therapy may provide insights into development of future therapies.
Introduction
Single-cell genomics, and specifically single-cell RNA-sequencing (scRNA-seq), has propelled characterization of the immune microenvironment in many solid tumors (1–3). However, it faces significant challenges in aggressive malignancy of mesenchymal origin with limited treatment options and poor clinical outcomes in the advanced and metastatic settings. Although immune checkpoint inhibitors (ICI) have transformed the therapeutic landscape for many cancers, different sarcoma subtypes have variable response rates to ICIs, and only few exhibit durable, clinically significant responses to anti-PD1 checkpoint blockade (4, 5). The development of novel immunotherapies is hindered by a relatively limited understanding of the underlying contribution of niche-specific immunity in different subtypes of sarcoma, particularly given the varied clinical and molecular features exhibited by each of the more than 100 subtypes. Single-cell genomics has the potential to provide valuable insights into the drivers of response and resistance to ICIs by identifying changes in rare cellular subpopulations or unique cellular states that are otherwise lost in bulk genomic and transcriptomic analysis. Such methods may be the key to discovering novel immunotherapeutic targets in sarcoma.
The successful implementation of scRNA-seq techniques in sarcoma, however, is limited by specimen and technical requirements. In particular, scRNA-seq requires immediate processing of a relatively large amount of fresh tissue (milligrams to grams), which is not conducive to clinical workflow. Particularly with rare tumors, collaborative studies between multiple institutions designed to incorporate larger numbers of specimens become impractical. The processing of fresh tissue also demands tremendous infrastructure in the form of skilled technicians and specialized instruments, making this a high-cost endeavor with limited clinical utility. Additionally, sarcoma originates from muscle, adipose tissue, bone, and cartilage, making these tissues physically difficult to dissociate for single-cell analysis; aggressive disaggregation methods are required, which typically involve enzymes, varied temperatures, and flow cytometry (6). Studies have identified artifactual signals caused by harsh dissociation protocols needed for fresh tissue processing, and in one study of sarcoma dissociation protocols in patient-derived sarcoma xenograft models, biases in the transcriptome were identified in cellular stress-related inflammatory pathways (6, 7). Such artifactual cellular stress signals are particularly confounding in studies looking to elucidate changes in the sarcoma immune microenvironment.
Due to these technical challenges, few studies have employed single-cell sequencing methods to investigate sarcomas (8–11). In these studies, the fragility of tumor cells leaves the majority nonviable after undergoing the harsh dissociation process required for sarcoma scRNA-seq; the resulting analyses focused on immune cells, which provides only partial insights into the complex ecosystems of these tumors. Interpretation of these studies is also challenging given relatively small cohorts with heterogenous patient populations exposed to varying treatments.
To address some of these challenges and improve clinical utility, we leveraged methods proven to be useful for the generation of single-cell genomics data from archival melanoma and lung cancer tissues (12–14). These methods can be applied to relatively small input material (nanograms to micrograms) that can be derived from clinical biopsy specimens and allow single-nucleus RNA-sequencing (snRNA-seq). The resulting single-cell transcriptome, matched T-cell receptor sequencing (TCR-seq), and population-matched low pass whole-genome sequencing (lp-WGS) are of high quality. We describe a pilot study utilizing these methods on archival tumor tissues of two sarcoma subtypes, including intimal sarcoma (INS; ref. 15) and undifferentiated pleomorphic sarcoma (UPS; refs. 4, 16), including two matched pair samples from immune checkpoint blockade (ICB)–treated patients.
Materials and Methods
Patient tissue collections and ethical approval
Fresh and frozen tissue specimens were collected under IRB approved protocols at New York Presbyterian Hospital/Columbia University Medical Center (AAAT5388). For the comparison of scRNA-seq and single-nucleus RNA-sequencing (snRNA-seq) protocols, surgical specimens were reviewed by qualified pathologists according to institutional guidelines and immediately placed in ice-cold RPMI 1640 (Thermo Fisher Scientific; #21875034) without supplements and transported to the laboratory space for immediate processing (scRNA-seq). Other samples were flash frozen for subsequent snRNA-seq.
Tissue processing
Processing of frozen tissues was performed as previously described. Briefly, tissue blocks embedded in optimal cutting temperature (Tissue-Tek, Sankura; #4583) were sectioned on a Leica CM1950 cryostat (Leica) into 20-µm-thick curls (four to five for each samples) and placed in 5-mL tubes (Eppendorf), washed with ice-cold PBS (Thermo Fisher Scientific, #10010023), and spun at 400 g for 2 minutes, and supernatants were discarded. The tissue was then resuspended in 1-mL Salt Tris (ST) buffer (146-mmol/L NaCl, 10-mmol/L Tris-HCL pH7.5, 1-mmol/L CaCl2, and 21-mmol/L MgCl2 in ultrapure water) with 0.03% Tween-20 Sigma Aldrich, #p7949 (=TST buffer) with 0.1% BSA (New England Biolabs, B9000S) and supplemented with or without 40-U/mL RNAse inhibitor (RNAse OUT, Thermo Fisher Scientific). The suspension was thoroughly pipetted 15 × using a 1-mL pipette to mechanically dissociate the tissue and left to incubate for 5 minutes on ice. After 5 minutes, the pipetting step was repeated, and the reaction was quenched using 4-mL ST buffer with or without 40-U/mL RNAse inhibitor (RNAse OUT, Thermo Fisher Scientific, #10777019; Supplementary Table S1). The sample was filtered through prewetted 70-µm nylon mesh filters (Thermo Fisher Scientific) into a 50-mL conical tube and the filter was washed with 5-mL ST buffer. The tube was then centrifuged at 500 × g for 5 minutes to collect the dissociated nuclei, and supernatants were discarded. Nuclei were resuspended in 100- to 400-µL ST buffer and filtered with a 40-µm mesh filter attached (Thermo Fisher Scientific) and counted in a Neubauer counting chambers (Bulldog Bio, Inc.) after staining of nuclear DNA with 50-µg/mL Hoechst 33342 (Thermo Fisher Scientific, H3570). The exemplary fresh tissue specimen was processed as previously described.
snRNA/scRNA and TCR library preparation
A range of 0.9 to 1.5 × 103 nuclei were loaded in ST buffer without RNAse inhibitor using a Chromium controller and Chromium reagents (10x Genomics) or 5′V2 capture (#1000006 and #1000263). After reverse transcription and cleanup, cDNA libraries were generated according to manufacturer instructions with one additional cycle of cDNA amplification to account for the relatively lower amount of RNA in nuclei compared with whole cells. TCR libraries were prepared from amplified cDNA libraries according to manufacturer instructions using the following reagents (all 10x Genomics): Chromium Single Cell V(D)J Enrichment Kit for human T cells (#1000005) was used for cDNA generated with Chromium Single Cell V(D)J reagents (#1000006), and final sequencing libraries were prepared using Chromium i7 multiplexing kit (#120262). Single Cell Human TCR Amplification Kit (#1000252) was used for cDNA generated with Chromium Next GEM Single Cell 5′ V2 reagents (#1000263), and final sequencing libraries were prepared using library construction kit (#1000190) and Dual Index Kit TT set A (#1000215).
Sequencing of single nuclei libraries
Final sequencing libraries were quantified using Tapestation D1000 and D5000 reagents (Agilent) and a 2200 TapeStation system. Samples were then mixed and sequenced to target >20,000 reads per cell for gene expression libraries and >5,000 reads per cell for TCR libraries using NovaSeq S4 or HiSeq 4000 (Illumina) with at least 2 × 100 bp read length.
Whole-genome library construction and sequencing
Excess nuclei (>1 × 105) from the sample preparations for sn-RNAseq were collected by centrifugation (500 × g, 5 minutes) and snap frozen after removing all but ∼10-µL ST buffer and stored until further processing at −20°C. If insufficient numbers of nuclei were available after loading, additional curls were processed using the same methods as described above for single nuclei extraction. To extract genomic DNA from nuclei, the nuclei were briefly thawed on wet ice and genomic DNA was extracted using DNeasy Blood and Tissue kit (Qiagen; #69504) according to manufacturer instructions and eluted in RNAse and DNAse free water at 37°C for 5 minutes. The DNA concentration was then quantified using a Nanodrop. Indexed WGS libraries were mixed equimolarly and sequenced on an Illumina MiSeq instrument with 0.1 to 1× coverage using the V2-300 cycle kit (Illumina). We used an R-based package ichorCNA v0.2.0 (17) for estimating tumor fractions in ultra-low pass whole-genome sequencing data, followed by prediction of large-scale copy number variation (CNV). We first converted the raw fastq data to sam using the bowtie2 v2.4.5 (18), followed by converting the.sam file to.bam using samtools v1.16.1 (19). We then generated the read count coverage data using the HMMcopy v1.38.0 R package (20). This creates a WIG file with 1-Mb bins across all chromosomes including reads with a mapping quality more than20. This is provided as an input for ichorCNA v0.2.0 (17), generating genome-wide plots representing the log2 ratio copy number for each bin in the genome. Finally, GISTIC 2.0 (21) was used to assign a copy number to each gene.
Multiplex tissue imaging and spatial analysis
Tissues were heated in the hybridization chamber to allow for adhesion onto the glass side prior to the dewax and staining done in the Leica Bond RX Fully Automounted Research Stainer. Slides were set to stain under the approved Leica 7-Color Double Dispensing protocol in which they were treated with six primary antibodies and 10× Spectral 4′,6-diamidino-2-phenylindole (Akoya, Cat #FP1490). Primary antibodies were detected by an horseradish peroxidase–conjugated secondary antibody (Akoya, ARH1001EA) before opals were applied. Antibodies were diluted with Akoya 1× Antibody Diluent Block (Akoya, Cat# ARD1001EA). The following antibody-opal order and pairings were used to image samples pre- and post-treatment: anti-CD19 Antibody (Leica, Cat # CD19-163-L-CE, Clone BT51E, 1/50) and Opal 570 (Akoya, Cat # FP1488001KT), anti-CD8 Antibody (Leica, Cat #PA0183, Clone 4B11, 1/50) and Opal 650 (Akoya, Cat # FP1496001KT), anti-Ecadherin Antibody (Cell Signaling, Cat #3195, Clone 24E10, 1/200) and Opal 540 (Akoya, Cat # FP1494001KT), anti-CD163 Antibody (Abcam, Cat #ab74604, Clone 10D6, 1/200) and Opal 690 (Akoya, Cat # FP1497001KT), anti-Vimentin Antibody (Leica, Cat #PA0640, Clone V9, 1/2) and Opal 620 (Akoya, Cat # FP1495001KT), and anti-CD103 Antibody (Abcam, Cat #ab129202, Clone EPR4166(2), 1/1,000) and Opal 780 (Akoya, Cat # FP1501001KT).
The analysis was preformed utilizing inForm 3.0 following multispectral imaging of the slides on the Vectra Polaris PhenoImager at 20× magnification. Multispectral fields underwent unmixing to differentiate overlapping signals, autofluorescence, and background. Tissue segmentation was preformed to outline areas exhibiting elevated anti-Vimentin and E-cadherin signal, as well as areas with no signals of interest. Individual cells were segmented with aid from 4,6-diamidino-2-phenylindole to identify nuclei signal. Cellular phenotypes for each cell were also manually identified as belonging to one of the six aforementioned markers. Opals that were not entirely spectrally isolated from each other required further unmixing during cellular phenotyping. The data from each multispectral image was merged to identify the quantity of each cellular phenotype per tissue segmented region.
Computational methods
Generating gene expression matrices
The fresh and frozen sample fastq files provided from raw snRNA sequencing reads were aligned to the reference GRCh38 genome (provided by 10x Genomics Cell Ranger pipeline as “refdata-gex-GRCh38-2020-A”). Gene count quantification was computed using CellRanger v6.1.2 (22), with the expected cell count set to 10,000.
Filtering background noise across gene expression matrices
For the fresh and frozen samples, the remove-background function provided by CellBender v0.2.0 (23) was utilized, using the raw feature matrix (“raw_feature_bc_matrix.h5”) from CellRanger v6.1.2 (22). The expected cells were set to 5,000, and the “total-droplets-included” parameter was set to a value between 10,000 and 20,000 based on the plateau observed in the barcode-rank plot. In doing so technical ambient RNA counts and empty droplets were removed from the downstream analysis.
Quality control and filtering
The expression matrices generated by CellRanger v6.1.2 (22) in which then processed individually in R v4.1.2 using Seurat v4.1.1 (24). Each Seurat object for a corresponding sample was filtered to only keep cells with 300 to 7,500 genes, 600 to 40,000 Unique Molecular Identifiers (UMIs), and <10% of mitochondrial reads. Scrublet v0.2.1 (25) was applied as well to remove doublets, using an expected rate of 11% for each sample, set based on the loading rate. The filtered gene-barcode matrices for each sample was then normalized and log-scaled as per the Seurat pipeline, using the “NormalizeData,” followed by the “LogNormalize” function (24). Gene expression matrices were then scaled using “ScaleData” on a per-sample basis. Following this, principal component (PC) analysis and Uniform Manifold Approximation and Projection for Dimension Reduction (UMAP) dimension were performed using the top 30 computed PCs. The stress signature was computed across each cell in the provided samples (eight frozen one fresh) using the “AddModuleScore” function provided by Seurat. This data was compared with droplet-based sarcoma data accessed via Gene Expression Omnibus from Jerby-Arnon and colleagues (10) and Slyper and colleagues (13) after applying similar filters, keeping cells with 300 to 7,500 genes, and <10% of mitochondrial reads. When plotting stress signatures, expression thresholds between 15 and −5 were applied for visualization purposes (Supplementary Fig. S1).
Integration of individual samples
All individual frozen samples were integrated using the Seurat v4.1.1 (24) canonical correlation analysis pipeline to remove batch effects from each individual sample. This was followed with using the “SelectIntegrationFeatures,” and the “FindIntegrationAnchors” function to select 2,000 anchors using the top 50 dimensions from the from canonical correlation analysis to specify the neighbor search space (24). “IntegrateData” was then called to integrate the eight frozen samples from the precomputed anchors. These data were then normalized and scaled using the “NormalizeData” and “ScaleData” function within Seurat.
Identification of malignant cells and clones using copy number alterations
To identify the chromosomal CNA for individual cells, Numbat v1.1.0 was utilized with default parameters (26). For each sample, immune cells identified via SingleR v1.8.1 using the built in “BlueprintEndcodeData” as a reference for inferring the CNAs in predicted nonimmune cell populations (27). Numbat infers the clonal history via maximum likelihood phylogeny of the profiled samples and was utilized to assign malignant cells to specific clones. Tumor cells were identified with a cutoff of 0.5 using the joint output of posterior aneuploidy probability. We found that across all samples, the malignant and nonmalignant assignments was discordant and aligned with specific clones outputted by numbat. Visualization of segments of inferred amplifications and deletions across each malignant clone aligned with large-scale CNV prediction done with ultralow-pass (ulp) WGS computed with ichorCNA v0.2.0 (17).
DGE and gene set enrichment across malignant clones
Seurat’s “FindAllMarkers” function was used to perform differential gene expression (DGE) based on the normalized data for each numbat clone on a per-sample basis. Using EnrichR, gene set enrichment was performed across MSigDB Hallmarks v7.4.1 (24, 28).
Inferred CNA comparison between RNA and DNA
Fraction of genome altered (FGA) measurements were utilized to correlate inferred CNA across individual cells within each snRNA-seq sample with the large-scale CNV prediction made using ulp-WGS. Numbat’s outputted consensus strand provided a probability of a genomic event occurring across specific segments using snRNA. This probability, multiplied by the number of SNPs in the given segments, estimated the FGA across each sample (26). This was correlated with the output from ichorCNA using ulp-WGS (17). Here, FGA measured the genome fraction with a segment mean >0.2 or less than −0.2. The Pearson correlation coefficient and P-values between these measures was reported, along with the regression fit line. Residuals between each point and the regression fit were computed, squared, and averaged to get the mean squared error (MSE); 1.96 times the square root of the MSE was used to obtain the 95% confidence interval plotted around the regression line.
Computation of the tumor-diversity index
Tumor diversity was defined using a diversity score index, following the methodology of Ma and colleagues (29). All malignant cells across the eight snRNA samples were merged into a single unintegrated object. The normalized count matrix was subsequently projected onto 50 PCs through PCA. For each sample, the centroid was identified as the arithmetic mean of all 50 PCs for the corresponding malignant cells. The diversity index was defined as the average Euclidean distance between each associated malignant cell and the centroid for a specific sample. Similar as Ma and colleagues (29), outliers were excluded by filtering cells with any PCs beyond ±4 standard deviations for a given particular sample. These extreme cells were omitted from the diversity index calculation (29).
Comparison of treatment-resistant and treatment-responsive malignant clones in patients X and Y
The malignant cells pertaining to patient X (Sample S167 for Pre IO, Sample S410 for post-IO) were normalized and then using Seurat’s “FindIntegrationAnchors” to select 2,000 anchors between the two samples (24). This was repeated for patient Y. Following the two samples were then integrated using the “IntegratedData” and then scaled. The Seurat clustering pipeline was utilized with the Louvain algorithm and a resolution of 0.8 to identify 16 malignant cell clusters between the two samples in patient X. Inspection of these clusters results in malignant cell clusters that contained more than 30% of samples from the post-IO sample were labeled as treatment-resistant malignant cells, and the remainder as treatment-responsive malignant cells.
DGE was done on patient X using Seurat’s “FindAllMarkers” function across the integrated malignant cell population, locating markers for treatment-resistant malignant cells and treatment-responsive malignant cells (2). Using EnrichR, gene set enrichment analysis (GSEA) was performed across Hallmarks of MSigDB v7.4.1 (28) given the identified marker list. For patient Y, DGE was done across the integrated malignant cell population to identify markers that pertained to the pre-IO and the post-IO tumor cell population. GSEA was carried out in the same way. For patient X, the top 12 returned pathways for the treatment-resistant and treatment-responsive malignant clusters were plotted (two of these pathways were shared, visualizing 20 total pathways). The returned combined score was rescaled between 0 and 1. This was repeated for patient Y, comparing pre- and post-treatment malignant cells and visualizing the top 20 shared pathways following gene set enrichment.
Nonmalignant cell type annotation
Nonmalignant cells across each sample were integrated using the Seurat integration pipeline with 2,000 identified integration anchors, following normalization and scaling of the nonmalignant cells for each individual sample to account for batch effects. Fourteen clusters were then identified within this integrated object using the Louvain algorithm with a resolution of 0.2. Broad cell type labeling was done with manual annotation based on the assigned markers for each cluster. This was refined using the expression level of known markers and signatures. Further refined myeloid and T-cell annotations were done following boarder refinements in the manner. The annotated T cells were selected, and the outputted Seurat object was rescaled and re-clustered with a resolution of 0.8. These clusters were then subject to another round of manual annotation using the cluster markers outputted by “FindAllMarkers” (24). This was repeated with the myeloid cell population.
DC analysis
Diffusion component (DC) analysis was performed as a nonlinear dimensionality reduction method to examine the primary components across refined CD8+ T-cell and myeloid populations. Fifty DCs were computed using the Destiny v3.8.1 (30), and the resulting eigenvectors were merged to their respective Seurat objects (30). The top three DCs were then visualized across the refined CD8+ T-cell and myeloid populations. Nonclassical monocytes were removed from myeloid population prior to DC calculation for visualization purposes. Cells that were in the top 10 percentile of CD8+ T cells in the expression of TOX were assigned to be TOX+. The same method was used for the assignment of TC7+ CD8+ T cells.
Integration of TCR data
CellRanger v6.1.2 (22) was used to align TCR fastq files. We used scRepertoire v1.7.2 (31) to filter contig outputs, following the Cell Ranger pipeline. Clonotypes were assigned based on two TCR chains, enabling analysis of clonotype dynamics. We examined unique clonotypes and combined the TCR information with mRNA expression data using the Seurat pipeline.
Analysis shared MPs using NMF
Estimation of MPs using KINOMO
Nonnegative matrix factorization (NMF) performs dimensionality reduction across nonnegative data, such as seen with snRNA-seq. Kernel dIfferentiability correlation-based NMF algorithm using Kullback–Leibler divergence loss Optimization (KINOMO), a semi-supervised NMF model was utilized here using the RNA assay as the input (bioRxiv 2022.05.02.490362). Moreover, KINOMO consists of a factorization error analysis capable of handling outliers with regularization and structure perseveration. Metaprograms (MPs) are estimated using the sample-wise factors/programs returned and are co-correlated using a Spearmen’s correlation. Significant testing and/or correlation across factors (threshold of 0.4–0.9) are computed iteratively to eventually obtain the observed MPs.
Normalized gene contribution per MP
For each MP, a list of unique genes was identified and ranked based on a weighted stouffer integrated expression value. For each of the “H” matrices generated sample wise (returned by KINOMO), we identify the cell barcodes associated with each program. An expectation maximization–Gaussian mixture model (EM-GMM) was run on the normalized single-cell data to identify the modality of gene distribution (bioRxiv 2022.05.02.490362). Based on the modality (could be unimodal, bimodal, or multimodal), the peak was identified that we term as the “normalized gene contribution,” associated with each gene unique to a specific MP.
GSEA across MPs
The selected representative genes with the highest observed gene contribution to a given MP were assigned for GSEA. Using these genes alongside EnrichR (28), GSEA was performed across Hallmarks of MSigDB v7.4.1 (32), given the identified gene set for each MP (1). For comparing MP3 and MP6 to MP1, MP2, MP4, and MP5, the assigned gene sets for MP3 and MP6 were concatenated, and GSEA was performed. This was compared with the GSEA results performed on the concatenated gene sets assigned to MP1, MP2, MP4, and MP5. The returned combined score for both sets of GSEA was rescaled between 0 and 1 individually and the top 20 representative pathways were visualized (24, 28).
Immune resistance signature measurement
The given cells assigned to each MP returned by KINOMO analysis were measured for their associated expression of immune resistance signature using the “AddModuleScore” function provided by Seurat.
Ligand–receptor analysis
ContactTracing was used to identify ligand–receptor interactions within and patient Y that differ prior and following ICB treatment (33). ContactTracing provides a systems approach to predicting conditionally dependent cell interactions and their effects. ContactTracing exploits the variability within single-cell data without reliance on prior knowledge of downstream targets to provide an unbiased estimate of ligand–receptor interactions within the TME (33). ContactTracing was built to account for the fact that in a given TME, it is unlikely all ligand-producing cells and receptor-expressing cells are simultaneously involved in a specific cell-to-cell interaction (33). Thus, for each potential ligand–receptor pair a likelihood ratio test was conducted comparing receptor-expressing to receptor-null cells within a given cell cluster. To distinguish between co-expressing ligands and receptors, ContactTracing measured the variability in these results between pre- and post-treatment conditions in patients X and Y (33). Inputs for ligand–receptor interactions were pulled from curated protein–protein interactions curated by CellPhoneDB v2.1.4 (34). A threshold fraction of minimum expression for receptors and ligands was set to 0.05 for a given cell type was used to filter ligand–receptor pairs. ContactTracing’s “make_circos_plot” function, alongside custom ligand minimum log fold-change filters ranging from 0.15 to 0.35 for visual appeal, was used to generate the visualized circus plot (33).
Statistics and reproducibility
Samples for contrasting scRNA-seq with snRNA-seq were chosen based on the tissue that was accessible and had enough substance for fresh and sequential frozen analysis as collected in the Izar laboratory. No prior statistical method was used to predetermine sample size. Experiments were conducted without randomization, and the investigators were not blinded to allocation during experiments and assessment.
Data availability
Data can be accessed on Gene Expression Omnibus under accession number: GSE243381. Additional data requests should be directed to the corresponding author. Code is available viahttps://github.com/IzarLab/sarcoma-sn.
Results
Multimodal single-cell profiling from archival sarcoma specimens
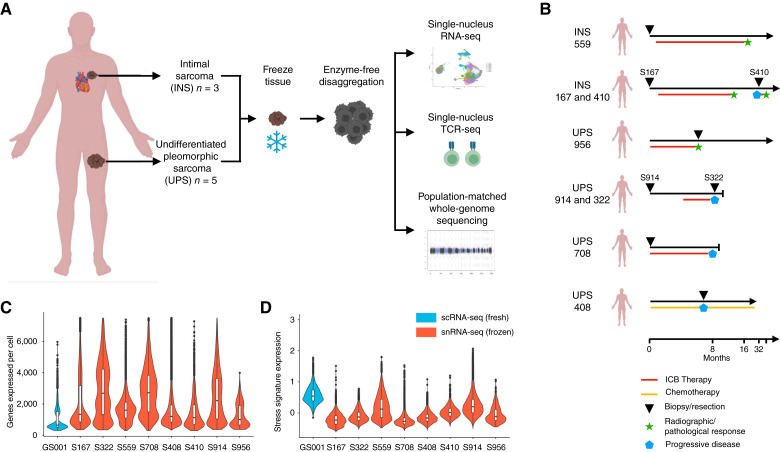
Here, we performed snRNA/TCR-seq coupled with population-matched WGS of UPSs and INSs of the pulmonary artery (Fig. 1A). In total, we profiled eight specimens from six patients, encompassing five UPSs and three INSs, and paired specimens from two patients (one UPS and INS each) treated with ICB (Fig. 1B). These included four pretreatment specimens (with two patients who subsequently had a clinical response to ICB), three post/on-ICB and one recurrent lesion from a patient treated with doxorubin. Following stringent quality control and removal of ambient RNA, a total of 75,716 cell transcriptomes were included in subsequent analysis, including an average of 9,465 cell transcriptomes per specimen (Supplementary Table S1). To emphasize the technical quality achievable from performing profiling of frozen tissues, we also generated single-cell RNA-seq of another sarcoma specimen directly from fresh tissue (Supplementary Table S1). The median number of genes detected per cell, a commonly used metric for data quality, from snRNA-seq was 1,723 (range 1,006–2,720) compared with 648 in scRNA-seq (Fig. 1C). The frequency of mitochondrial reads in snRNA-seq versus scRNA-seq was 1.17% versus 4.78% (Supplementary Table S1). Furthermore, expression of a stress signature, artifactually introduced during tissue processing, was virtually absent in snRNA-seq but was strongly expressed in scRNA-seq (Fig. 1D). We also validated this observation by analyzing previously published data of synovial sarcoma (10), Ewing sarcoma, and rhabdomyosarcoma (13) on which either fresh or frozen single-cell profiling was performed (Supplementary Fig. S1). Together, these results indicate that single-cell profiling from frozen sarcoma tissues yielded high quality, whereas avoiding potential artifactual signals introduced by processing of fresh tissues. Furthermore, the tissue input for snRNA-seq is substantially smaller compared with scRNA-seq, thus, overall favoring profiling from frozen material.
Figure 1.
Experimental design and comparison of sequencing quality metrics between fresh and frozen samples. A, Outline for this study, describing sample preparation, and summary of analytic approaches used. B, Representative schematic describing the origin of each sample and patient treatment at the time of resection. C and D, Violin plots and boxplots displaying the (C) genes detected per cell and (D) stress signature expression per cell. The top and bottom edges of the boxplot display the 75th and 25th percentiles, respectively. Blue violin plots are from scRNA-seq from fresh tissue, and red violin plots are from snRNA-seq on frozen tissue.
Genomic and transcriptional cancer cell heterogeneity
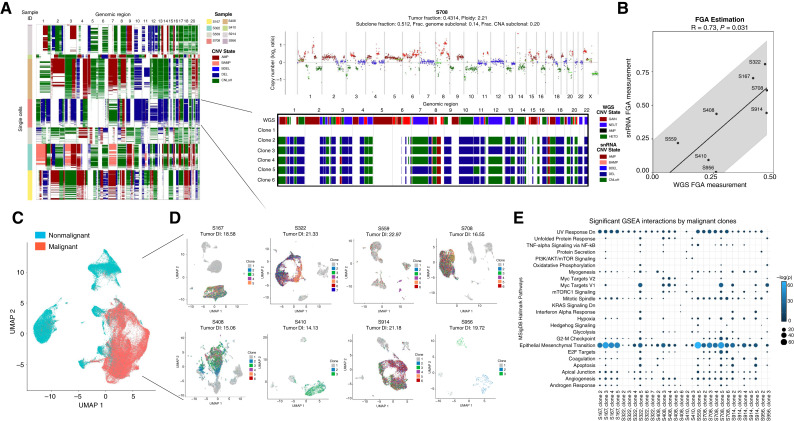
Defining malignant from nonmalignant cells in contexts in which cancer cells share a high degree of transcriptional similarity with stromal cells within the tumor ecosystem, such as in the case of sarcomas, can be challenging. To address this, we first performed inference of CNAs, which should be present in malignant and largely absent in nonmalignant cells in snRNA-seq (Fig. 2A). Concurrently, we performed lp-WGS on DNA isolated from the same cell pool on which we also performed snRNA-seq on (Supplementary Table S1) and determined CNAs using ichorCNA (17). We find strong concordance among the inferred and measured CNAs on RNA and DNA, respectively, and concordant tumor purity (Fig. 2B) overall demonstrating that assignment of cancer cells was robust. Across all eight specimens, we identified 46,670 malignant and 29,046 nonmalignant cells (Fig. 2C).
Figure 2.
A, Representation of inferred CNAs in single cells across the chromosomal landscape of all eight snRNA-seq samples provided by Numbat (left). On the right is a detailed analysis of the selected S708 sample. IchorCNA on low pass whole-genome sequencing (ulp-WGS) revealing CNA (y-axis, log2 ratio), with amplification segments in red, deleted segments in green, gain segments in maroon, and copy neutral segments in blue (top; ref. 1). A breakdown of the copy number segments for each identified clone (bottom). The top bar provides a comparison of the inferred CNAs per clone based on snRNA-seq to CNA segments based on ulp-WGS for the selected sample. B, Pearson correlation and P-value of the FGA based on CNAs computed using ulp-WGS (x-axis) as compared with the estimated FGA based on CNAs computed using snRNA-seq (y-axis). Regression line and 95% confidence interval plotted, calculated using the MSE of the residuals. C, Merged, integrated, UMAP embedding displaying the annotated assignment of malignant (red) and non-malignant (blue) cell types across all eight snRNA-seq samples. D, Sample-wise UMAP embeddings of each snRNA-seq sample colored by respective CNA clones, revealing the impact of CNA differences on transcriptional output. For each sample, the measured tumor-diversity index is listed. E, GSEA of genes differentially expressed in clone across all eight samples that were assigned to malignant cell types. Only significant (P < 0.05) pathways are plotted for each clone. Dot size and color refer to the assigned q-score (−log adjusted P-value).
We first explored cancer cell heterogeneity within individual specimens. Clustering of malignant cells from each specimen revealed variable degree of heterogeneity (measured by a diversity score, “Methods”; Fig. 2D). DGE among cancer cells within each patient revealed different drivers of variability. Using GSEA, we found transcriptional programs of clones within individual patients varied around few pathway axes, including epithelial-to-mesenchymal-transition (EMT), UV response dn (down-signature), mitotic spindle, myogenesis, cell migration, and apical junction, suggesting conserved programs and interactions with immune-mediated effects (Fig. 2E; Supplementary Table S2). Furthermore, these data also indicated that variability underlying aneuploidy patterns across patients introduced an important bias in direct comparisons of patient specific gene expression patterns.
Recurrent cancer cell programs associated with ICB resistance
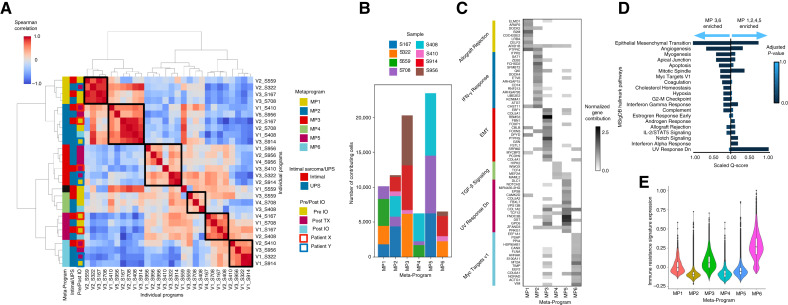
To overcome this bias and enable comparisons across the entire cohort of patients, we used an NMF approach (KINOMO; bioRxiv 2022.05.02.490362). In this approach, we first determined gene expression programs (factors) of variability within each specimen and then determined the co-correlation of these programs across the entire cohort (MPs; Fig. 3A). This yielded 28 programs (average 3.5 per sample) and six recurrent MPs that were composed of programs from three or more patients each (Fig. 3B). Importantly, each specimen harbored cells with cell cycle programs indicating active proliferation, as expected. Most MPs harbored programs from a mixture of clinical contexts (pre-/post-therapy) and sarcoma types (UPS and INSR).
Figure 3.
A, Spearman correlation of programs, merged into six MPs. Boxes to the right of the correlation plot were used to indicate, from left to right, the assigned MP of an individual MP, if the individual program belongs to a patient with intimal or UPS sarcoma, and if the individual program belongs to a patient, extracted pre-/post-ICB treatment. In the rightmost column, paired samples from patient X are outlined in red, and paired samples from patient Y are outlined in blue. B, Stack plot showing the contribution of each individual sample to each meta-program. C, Normalized gene contribution to MPs identified using KINOMO. Filtered and selected representative genes (rows) are seen along with their contribution to specific MPs (columns). The most significant GSEA pathway is listed on the right for the set of genes that have the highest normalized contribution for that set of genes. D, Side-by-side comparison of GSEA of the representative genes with a normalized gene contribution highest in either MP3 or MP6, to genes with a normalized gene contribution highest in either MP1, MP2, MP4, or MP5. Combined scores are rescaled to be between 0 and 1. Top of 20 gene sets are visualized. E, Violin plots and boxplots displaying the immune resistance signature expression per cell associated with each MP.
Given the overlap and redundancy in several of these pathways, we next sought to identify unique gene signatures that specifically determine the function and the normalized gene contribution of each MP (“Methods”; ref. 35). This revealed unique biologic functions of gene programs driving specific MPs (Fig. 3C). MP1 enriched for allograft rejection, UV response dn, Interferon gamma response, UV response dn, interferon alpha, KRAS signaling, and interferon gamma responses, MP2 for interferon gamma response and mitotic spindle, MP3 for EMT, myogenensis, angiogenesis, coagulation, and hypoxia and UV response dn, MP4 for TGBb and Notch signaling, MP5 for EMT, mitotic spindle, and UV response dn, MP6 for MYC targets, cholesterol homeostasis, mTORC1 signaling, and EMT. Notably, MP3 and MP6 programs strongly enriched for ICB-resistance signature were previously described in melanoma (Fig. 3E) and compared with other MPs, more strongly enriched for EMT, angiogenesis, MYC targets, among others (Fig. 3D). Importantly, a patient (S408) with recurrence after doxorubicin therapy did not contribute to MP3 or MP6, suggesting that these programs did not simply represent resistance to any therapy but potentially specifically to ICB. Together, these data indicate that recurrent programs exist in cancer cells among multiple patients with resistance to ICB.
Cancer cell evolution among patients with paired samples
We next interrogated the two patients for which we had paired specimens. Patient X was a patient with INS who had a pretreatment biopsy (S167), received pembrolizumab for approximately 12 months, achieved a complete response, discontinued therapy, and had a metastatic recurrence 21 months later (S410) that was resected and profiled. The patient went on to receive another course of pembrolizumab for 8 months, which was discontinued due to neutrophilic dermatitis but remained disease free for 12 months after treatment discontinuation. Patient Y was a patient with UPS who underwent a pretreatment biopsy (S914), underwent three cycles of treatment with ipilimumab and nivolumab, and underwent another biopsy (S322) at progression of disease (Fig. 1B).
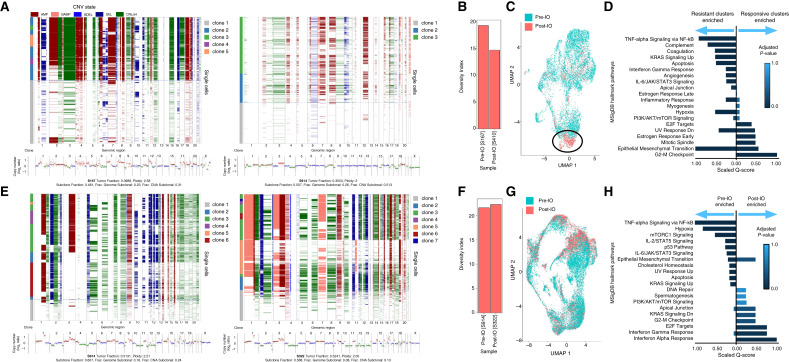
Inference of CNAs of patient X samples revealed that a dominant aneuploidy pattern in the pretreatment specimen (e.g., Chr 4 amplification, Chr 8 amplification, and Chr 16 deletion) and a minor clone (Clone 2) with a divergent pattern (e.g., 19 amplification; Fig. 4A). Notably, the overall diversity decreased in the relapse specimen (Fig. 4B) and clone 2 emerged as dominant clone in the recurrence specimen suggesting that these preexisting cells were resistant to pembrolizumab. Consistently, cancer cell transcriptomes from the post-IO therapy specimens clustered distinctly (Fig. 4C), and were characterized by DGE of EMT, angiogenesis, TNFα signaling via NF-kB (Fig. 4D).
Figure 4.
A, For patient X, for sample S167, taken prior to treatment (left), ichorCNA analysis across ulp-WGS displaying CNA (bottom). The x-axis indicates chromosome number, and the y-axis indicates copy number. Inference of CNAs using snRNA-seq provided by Numbat is plotted across single cells, with the x-axis indicating chromosome number (top). The ichorCNA analysis and the inferred CNA are visualized for sample S410, taken following treatment (right). B, Bar graph displaying the change in tumor-diversity index in sample S167 (prior to treatment) and sample S410 (following treatment). C, UMAP of all integrated malignant cells from samples S167 and S410, affiliated with patient X, colored by sample origin. Circled is a cluster of treatment-resistant malignant cells primarily found in the posttreatment sample. D, Side-by-side comparison of GSEA of the representative gene markers for the annotated treatment-responsive malignant clusters and the treatment-resistant malignant clusters found in patient X. Combined scores are rescaled to be between 0 and 1. Top of 20 gene sets are visualized. E, For patient Y, for sample S914, taken prior to treatment (left), ichorCNA analysis across ulp-WGS displaying CNA (bottom). The x-axis indicates chromosome number, and the y-axis indicates copy number. Inference of CNAs using snRNA-seq provided by Numbat is plotted across single cells, with the x-axis indicating chromosome number (top). The ichorCNA analysis and the inferred CNA are visualized for sample S322, taken following treatment (right). F, Bar graph displaying the change in tumor-diversity index in sample S914 (prior to treatment) and sample S322 (following treatment). G, UMAP of all integrated malignant cells from samples S322 and S914, affiliated with patient Y, colored by sample origin. H, Side-by-side comparison of GSEA of the representative gene markers for the malignant cells that originated in the pretreatment sample compared with the malignant cells originating from the posttreatment sample. Combined scores are rescaled to be between 0 and 1. Top of 20 gene sets are visualized.
In contrast, patient Y who had intrinsic resistance to ICB had diverse clones (Fig. 4E), but no change in diversity with treatment (Fig. 4F), showed a higher degree of mixing between both specimens in gene expression space (Fig. 4G) and a preserved population of cycling cells (Fig. 4H). Inference of CNAs of patient Y revealed at least three major cancer clones before therapy and minimal change at the time of IO resistance. Notably, clustering revealed highly similar global gene expression and both specimens contributed to MP3 and MP6, which is reflective of an ICB-resistant cell state (Fig. 3E), overall demonstrating that treatment did not alter genomic, transcriptomic, and programmatic features, indicating that these were intrinsically resistant to ICB.
Tumor microenvironment, T-cell clonality, and myeloid polarization
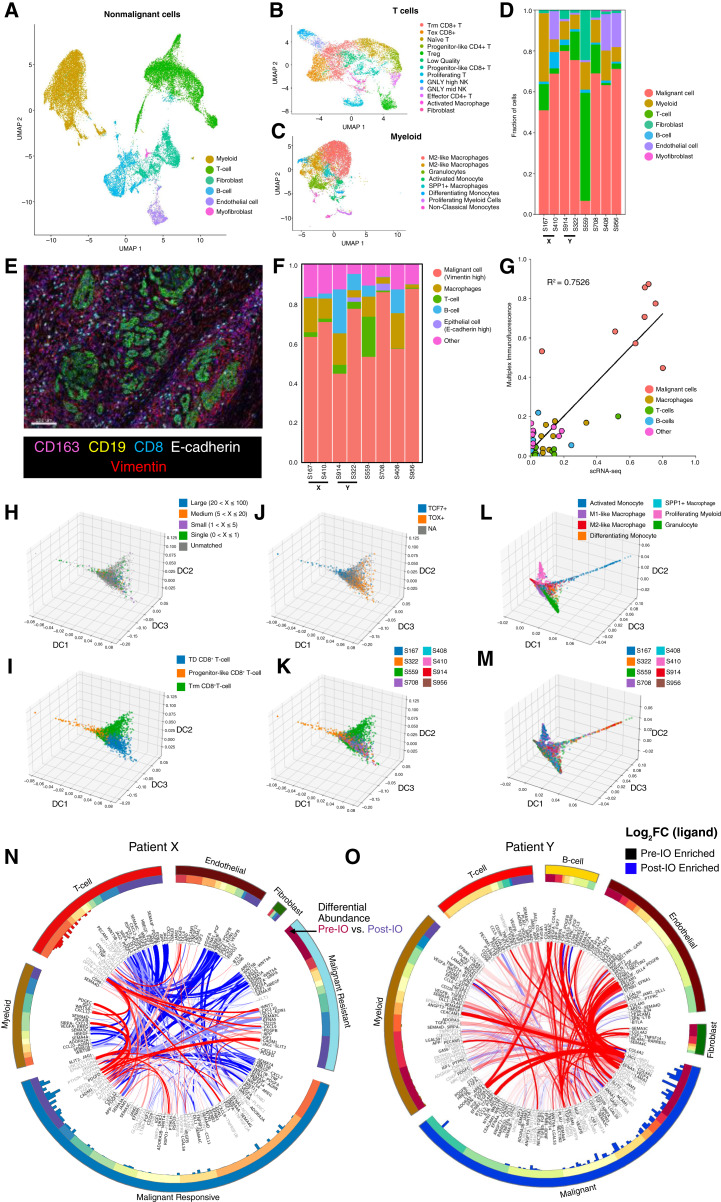
We next focused our analysis on the tumor microenvironment (TME). First, we used a multi-step process to determine cell types and major functional cell states (Fig. 5A; “Methods”) and granular annotation of T/NK and myeloid cells (Fig. 5B and C). Across the entire cohort, major cell types of the TME were T lymphocytes and NK cells (k = 9,300), B lymphocytes (k = 3,320), myeloid (k = 11,185) and endothelial cells (k = 1,213), and fibroblasts (k = 3,907). We found compositional differences across these cell types that may associate with prior-treatment exposures and clinical outcomes (Fig. 5D). To validate this, we performed multiplex ImmunoFluorescence (mIF) and stained for multiple cell markers (CD8, CD19, CD163, E-cadherin, and Vimentin; Fig. 5E). We observed a similar cellular composition distribution across all the samples (Fig. 5F). For example, the tumor of patient 559, who had a durable complete response to ICB, was densely infiltrated with T and B lymphocytes when assessed through snRNA-seq and mIF staining. To this extent, we observed a good correlation comparing the proportion of each cell type across each patient using the snRNA-seq and mIF approaches (Fig. 5G).
Figure 5.
A, Merged, integrated, UMAP embedding displaying the annotated assignment nonmalignant cell types across all eight snRNA-seq samples. B and C, UMAP embedding of the integrated T cells (B) and myeloid cells (C) across all eight snRNA-seq samples, colored by cell type assignment. D, Stacked bar plots denoting the percentage breakdown of each observed cell type across each of the snRNA-seq samples. E, Representative multiplexed mIF image showing all the markers. F, Stacked bar plots denoting the percentage breakdown of each observed cell type across each of the patients using mIF. G, Correlation plot between different cellular proportion in snRNA-seq and mIF across all the patients. H–K, CD8+ T cells projected onto the DC space, colored by (H) clonotypes, (I) manual cell type assignment, (J) TCF+/TOX+ assignment, and (K) derived sample. L and M, Selected myeloid cells projected onto the DC space, colored by (L) manual cell type assignment and (M) derived sample. N and O, ContactTracing circus plot highlighting all pre-IO and post-IO significant ligand–receptor interactions across multiple cell types for patient X (N) and for patient Y (O). Each outer segment represents a specific cell type and across which ligands and receptors are represented. These are ordered based on the first DC (DC1) computed through ContactTracing. DC1 is visualized in the next ring. The histogram in the inner circle shows the magnitude of pretreatment ligand effects with an FDR q-value <0.05 computed by ContactTracing. Each interacting ribbon links a ligand to a receptor across multiple cell types, with ribbon thickness proportional to the number of genes expressing a pretreatment (red) or posttreatment (blue) dependent interaction effect. Ligands (black) and receptors (gray) are labeled at ribbon endpoints. Ribbon opacity is determined by the log fold-change difference of the complementary ligand across the pretreatment and posttreatment samples. Ribbons are filtered to involve a minimum of 12 pre- and posttreatment dependent interaction effects in the target cell type.
The TCR-seq data consist of 1,925 raw clonotypes, which are mapped to 982 T-cell transcriptomes. Within this matched subset, there are 737 distinct TCRs identified, out of which 325 clonotypes exhibited expansion (Supplementary Table S3). DC analysis integrating gene expression and TCR data revealed a trajectory spanning progenitor-like CD8+ T cells, tissue-resident memory–like T cells and terminally differentiated (TD) T cells with enrichment of clonally expanded T cells in the TD compartment (Fig. 5H and I), which also corresponded to increased expression of TOX and reduced expression of TCF7 (Fig. 5J). Surprisingly, T cells along the progenitor-like to TD spectrum arose from all patients, including those with response and resistance to ICB suggesting that clinical outcomes were not sufficiently explained by T-cell phenotypes (Fig. 5K). This suggest that T-cell responses alone are insufficient to predict responses ICB, but additional features, such cancer cell genomic evolution, may contribute. Similarly, although some patients showed compositional enrichment of some myeloid subpopulations (Fig. 5L and M), most samples harbored a spectrum of diverse macrophages.
Homotypic and heterotypic interactions within the tumor ecosystem are shaped by ICB
Given the absence of clear compositional TME differences among ICB responding compared with resistant patients, we reasoned that functional changes in the form of cellular and ligand–receptor interactions among cells within the tumor ecosystem may exist. We focused on these interactions analyses of the matched patient specimens using ContactTracing (33). This novel tool enables conditional inferences of cellular interactions and exploits inherent biologic and technical variability of single-cell data among donor (expressing a ligand) and recipient cells (expressing the respective receptor). The interaction patterns between patient X and patient Y showed significant differences over their treatment course: In patient X, treatment-resistant malignant cells displayed a quantitative decrease in heterotypic interactions (e.g., T cells with malignant cells and endothelial cells with malignant cells), as compared with treatment-responsive malignant cells. Nonetheless, important T-cell/malignant cell interactions were sustained among treatment-resistant malignant cell populations, including CD226-NECTIN2, and CXCL9/CXCL10-DPP4, suggesting sustained effects of prior ICB exposure. Homotypic interactions among cancer cells were strongly enriched for growth factor–receptor interactions, including FGF1-FGFR, IGF1-IGFR, and WNT5A-ROR1, suggesting several paracrine/autocrine loops that maintain or promote cell survival and proliferation (Fig. 5N). In contrast, although overall cellular interactions significantly decreased on a quantitative level in patient Y, there were a sustained number of homotypic malignant cell interactions including FGF2-FGFR1, JAG-NOTCH3, GAS6-AXL, and TGFB1-TGFBR3, following ICB exposure (Fig. 5O). Together, these results suggest that in intrinsic and acquired ICB resistance, a dense network of autocrine/paracrine signaling is present and highlight the need to fully investigate all portions of the tumor ecosystem to dissect clinical phenotypes in response to systemic therapies.
Discussion
Single-cell genomics have informed our understanding of the biology, heterogeneity, and mechanisms of response and resistance to modern cancer therapies in a variety of common tumor types, such as carcinomas of the lung, breast, and colon and melanoma. To date, the application of these methods to rare tumors, such as sarcomas, has been limited by several practical and preanalytic challenges (10, 12), such as difficulties of collecting fresh tissues specimens and artifacts associated with fresh tissue disaggregation to a single-cell suspension of these highly rigid tumors.
Here, we show that multimodal single-cell genomics is feasible from small, clinical, frozen specimens of two types of soft tissue sarcomas, including undifferentiated polymorphic sarcoma and INS, in total comprising an important resource. Moreover, this work provides a framework that can be adapted for in-depth analyses of other rare cancers that are understudied.
Overall, our analysis reveals transcriptional and genomic diversity and tumor microenvironmental and T-cell clonal features of these diseases across real-life clinical contexts. A key insight from this work is that understanding the salient complexity of these rare tumors and molecular underpinnings of response to ICB requires examination of the entire ecosystem rather than interrogation of individual parts. We find, for example, that despite adequate T-cell responses (e.g., activation, differentiation, and clonal expansion), some patients had intrinsic or acquired resistance to ICB, indicating that T-cell responses alone are insufficient to predict clinical outcomes. Analysis of cancer cells, using a novel approach that mitigates inter-patient transcriptional variability, identified a program of ICB resistance found in all patients with either intrinsic or acquired resistance to ICB. We previously described this program in single-cell RNA-sequencing analyses of patients with ICB-resistant melanoma and demonstrated its prognostic role in a pan-cancer analysis of The Cancer Genome Atlas (ref. 1). These results suggest that recurring cell programs of ICB resistance may exist across different cancer lineages.
Analysis of paired biopsies from the same patient afforded additional insights. First, in a patient with initial complete response to ICB and subsequent isolated recurrence, we found that resistance was conferred by a major genomic clone that preexisted and emerged during therapy and that strongly expressed the ICB-resistance transcriptional program. Consistently, we observed significant reduction of cancer cell diversity from the pretreatment to the subsequent biopsy, indicating immune pruning of ICB-sensitive clones. In contrast, in another patient with paired biopsies who was intrinsically resistant to ICB, there was no change in cancer cell diversity, genomic or transcriptomic features. These observations are consistent with a recent study of sequential biopsies of melanoma, in which resistance occurred despite adequate T-cell responses and was driven by emergence of a preexisting cancer cell clone that was defined by a distinct aneuploidy pattern (12). This suggests that in some cases, resistance to ICB may be explained by large genomic changes rather than point mutations (e.g., JAK1 and IFNGR) or loss of specific genes (e.g., B2M) required for proper antigen presentation and interferon gamma responses.
Lastly, through inference of cellular interactions from single-cell data, we find that ICB-resistant cancer clones exhibit a dense network of redundant and nonredundant paracrine/autocrine growth signals that may override productive T-cell responses. One example is the paracrine/autocrine activity of GAS6 and its receptor AXL. AXL expression has previously been associated with resistance to oncogene-directed and immune-based therapies in melanoma and nonsmall cell lung cancer, among others (3, 36). Inhibition of this other interactions may improve the activity of ICB, as shown in melanoma; however, the redundant homotypic interactions may require compounds with broader activity (37).
This study has two limitations: First, the sample size is small, which is largely due to the rarity of the studied diseases and accessible archival tissue. Second, the profiled specimens are from a spectrum of clinical contexts that impact resulting gene expression profiles. Despite these limitations, this study contributes significant conceptual advances, including emerging observations, such as the association of large genomic features in resistance to ICB, and the necessity to analyze the tumor as the complex ecosystem to fully illuminate molecular underpinnings of clinical phenotypes. Lastly, we present a technical and analytic framework that is broadly applicable to studying archival human tumor tissues and is particularly relevant for studying rare cancers such as sarcoma which to date have remained underrepresented in (single-cell) genomics studies.
Supplementary Material
Violin plots and boxplots displaying the (a) genes detected per cell and (b) stress signature expression per cell and (c) mitochondrial fraction of reads in samples profiled from fresh or frozen tissue in this study and in indicated studies by Jerby et. al. (dijon) and Slyper et al. (green), respectively. The upper and lower edges of the boxplot display the 75th and 25th percentiles respectively. Blue violin plots are from scRNA-seq from fresh tissue, and red violin plots are from snRNA-seq on frozen tissue.
Supplemental Table 1: Assays performed and basic quality metrics of included samples.
Supplemental Table 2: Pathway enrichment analyses for indicated samples.
Supplemental Table 3: T-cell receptor sequences and clonality.
Acknowledgments
S. Bose is supported by NCI T32 and the Hearst Foundation. K. Luthria and P. Ho are supported by Training Grant T32GM145440. This work was supported by the Jed Ian Taxel Foundation for Rare Cancer Research. B. Izar is supported by National Institute of Health grants R37CA258829, R01CA280414, and R01CA266446, as well as the Pershing Square Sohn Cancer Research Alliance Award, the Burroughs Wellcome Fund Career Award for Medical Scientists, a Tara Miller Melanoma Research Alliance Young Investigator Award, the Louis V. Gerstner Jr Scholars Program, and the V Foundation Scholars Award.
Footnotes
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
Authors’ Disclosures
A.D. Amin reports personal fees from Adaptimmune outside the submitted work. M. Ingham reports grants from Apexigen, Mirati Therapeutics, PTC Therapeutics, Bioatla, and Intensity outside the submitted work as well as employment with and stock ownership in Regeneron. B. Izar reports consultantship for or honoraria from Volastra Therapeutics, Johnson & Johnson/Janssen, Novartis, Eisai, AstraZeneca, and Merck and research funding to Columbia University from Agenus, Alkermes, Arcus Biosciences, Checkmate Pharmaceuticals, Compugen, Immunocore, Regeneron, and Synthekine. No potential conflicts of interest were disclosed by the other authors.
Authors’ Contributions
K. Luthria: Data curation, formal analysis, investigation, visualization, methodology, writing–original draft. P. Shah: Resources, investigation. B. Caldwell: Formal analysis, visualization. J.C. Melms: Investigation. S. Abuzaid: Investigation, methodology. V. Jakubikova: Investigation, methodology. D.Z. Brodtman: Formal analysis, methodology. S. Bose: Investigation, methodology, writing–original draft. A.D. Amin: Validation. P. Ho: Investigation. J. Biermann: Formal analysis. S. Tagore: Formal analysis. M. Ingham: Conceptualization, resources, funding acquisition. G.K. Schwartz: Conceptualization, resources, funding acquisition. B. Izar: Conceptualization, resources, data curation, formal analysis, supervision, funding acquisition, investigation, visualization, methodology, writing–original draft.
References
- 1. Jerby-Arnon L, Shah P, Cuoco MS, Rodman C, Su MJ, Melms JC, et al. A cancer cell program promotes T cell exclusion and resistance to checkpoint blockade. Cell 2018;175:984–97.e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Yost KE, Satpathy AT, Wells DK, Qi Y, Wang C, Kageyama R, et al. Clonal replacement of tumor-specific T cells following PD-1 blockade. Nat Med 2019;25:1251–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tirosh I, Izar B, Prakadan SM, Wadsworth MH 2nd, Treacy D, Trombetta JJ, et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Sci Apr 2016;352:189–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Tawbi HA, Burgess M, Bolejack V, Van Tine BA, Schuetze SM, Hu J, et al. Pembrolizumab in advanced soft-tissue sarcoma and bone sarcoma (SARC028): a multicentre, two-cohort, single-arm, open-label, phase 2 trial. Lancet Oncol 2017;18:1493–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chen JL, Mahoney MR, George S, Antonescu CR, D’Angelo SP, Van Tine BA, et al. A multicenter phase II study of nivolumab +/− ipilimumab for patients with metastatic sarcoma (Alliance A091401): results of expansion cohorts. J Clin Oncol 2020;38:11511. [Google Scholar]
- 6. Denisenko E, Guo BB, Jones M, Hou R, de Kock L, Lassmann T, et al. Systematic assessment of tissue dissociation and storage biases in single-cell and single-nucleus RNA-seq workflows. Genome Biol 2020;21:130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Truong DD, Lamhamedi-Cherradi S-E, Porter RW. Dissociation Protocols used for Sarcoma Tissues Bias the Transcriptome observed in Single-cell and Single-nucleus RNA sequencing. BMC Cancer 2023;23:488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hong B, Li Y, Yang R, Dai S, Zhan Y, Zhang WB, et al. Single-cell transcriptional profiling reveals heterogeneity and developmental trajectories of Ewing sarcoma. J Cancer Res Clin Oncol 2022;148:3267–80. [DOI] [PubMed] [Google Scholar]
- 9. Wisdom AJ, Mowery YM, Hong CS. Single cell analysis reveals distinct immune landscapes in transplant and primary sarcomas that determine response or resistance to immunotherapy. Nat Commun 2020;12:6410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jerby-Arnon L, Neftel C, Shore ME, Weisman HR, Mathewson ND, McBride MJ, et al. Opposing immune and genetic mechanisms shape oncogenic programs in synovial sarcoma. Nat Med 2021;27:289–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhou Y, Yang D, Yang Q, Lv X, Huang W, Zhou Z, et al. Author Correction: single-cell RNA landscape of intratumoral heterogeneity and immunosuppressive microenvironment in advanced osteosarcoma. Nat Commun 2021;12:2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang Y, Fan JL, Melms JC, Amin AD, Georgis Y, Barrera I, et al. Multimodal single-cell and whole-genome sequencing of small, frozen clinical specimens. Nat Genet 2023;55:19–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Slyper M, Porter CBM, Ashenberg O, Waldman J, Drokhlyansky E, Wakiro I, et al. A single-cell and single-nucleus RNA-Seq toolbox for fresh and frozen human tumors. Nat Med 2020;26:792–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Melms JC, Biermann J, Huang H, Wang Y, Nair A, Tagore S, et al. A molecular single-cell lung atlas of lethal COVID-19. Nature 2021;595:114–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Henick BS, Ingham M, Shirazi M, Marboe C, Turk A, Hsiao S, et al. Assay complementarity to overcome false-negative testing for microsatellite instability/mismatch repair deficiency: a pembrolizumab-sensitive intimal sarcoma. JCO Precis Oncol 2020;4:570–4. [DOI] [PubMed] [Google Scholar]
- 16. D’Angelo SP, Mahoney MR, Van Tine BA, Atkins J, Milhem MM, Jahagirdar BN, et al. Nivolumab with or without ipilimumab treatment for metastatic sarcoma (Alliance A091401): two open-label, non-comparative, randomised, phase 2 trials. Lancet Oncol 2018;19:416–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Adalsteinsson VA, Ha G, Freeman SS, Choudhury AD, Stover DG, Parsons HA, et al. Scalable whole-exome sequencing of cell-free DNA reveals high concordance with metastatic tumors. Nat Commun 2017;8:1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Langmead B, Wilks C, Antonescu V, Charles R. Scaling read aligners to hundreds of threads on general-purpose processors. Bioinformatics 2019;35:421–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Danecek P, Bonfield JK, Liddle J, Marshall J, Ohan V, Pollard MO, et al. Twelve years of SAMtools and BCFtools. GigaScience 2021;10:giab008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lai D, Ha G, Shah S. HMMcopy: copy number prediction with correction for GC and mappability bias for HTS data. Bioconductor version: Release (3.17); 2023. Available from: 10.18129/B9.bioc.HMMcopy. [DOI]
- 21. Mermel CH, Schumacher SE, Hill B, Meyerson ML, Beroukhim R, Getz G. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol 2011;12:R41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zheng GXY, Terry JM, Belgrader P, Ryvkin P, Bent ZW, Wilson R, et al. Massively parallel digital transcriptional profiling of single cells. Nat Commun 2017;8:14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fleming SJ, Chaffin MD, Arduini A, Akkad AD, Banks E, Marioni JC, et al. Unsupervised removal of systematic background noise from droplet-based single-cell experiments using CellBender. Nat Methods 2023;20:1323–35. [DOI] [PubMed] [Google Scholar]
- 24. Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM 3rd, et al. Comprehensive integration of single-cell data. Cell 2019;177:1888–902.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wolock SL, Lopez R, Klein AMS. Scrublet: computational identification of cell doublets in single-cell transcriptomic data. Cell Syst 2019;8:281–91.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gao T, Soldatov R, Sarkar H, Kurkiewicz A, Biederstedt E, Loh PR, et al. Haplotype-aware analysis of somatic copy number variations from single-cell transcriptomes. Nat Biotechnol 2023;41:417–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Aran D, Looney AP, Liu L, Wu E, Fong V, Hsu A, et al. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat Immunol 2019;20:163–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 2013;14:128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ma L, Hernandez MO, Zhao Y, Mehta M, Tran B, Kelly M, et al. Tumor cell biodiversity drives microenvironmental reprogramming in liver cancer. Cancer Cell 2019;36:418–30.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Angerer P, Haghverdi L, Büttner M, Theis FJ, Marr C, Buettner F. destiny: diffusion maps for large-scale single-cell data in R. Bioinformatics 2016;32:1241–3. [DOI] [PubMed] [Google Scholar]
- 31. Borcherding N, Bormann NL. scRepertoire: an R-based toolkit for single-cell immune receptor analysis. F1000Res 2020;9:47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JP, Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst 2015;1:417–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Li J, Hubisz MJ, Earlie EM, Duran MA, Hong C, Varela AA, et al. Non-cell-autonomous cancer progression from chromosomal instability. Nature 2023;620:1080–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Efremova M, Vento-Tormo M, Teichmann SA, Vento-Tormo R. CellPhoneDB: inferring cell-cell communication from combined expression of multi-subunit ligand-receptor complexes. Nat Protoc 2020;15:1484–506. [DOI] [PubMed] [Google Scholar]
- 35. Dempster AP, Laird NM, Rubin DB. Maximum likelihood from incomplete data via the EM algorithm. J R Stat Soc Ser B Methodological 1977;39:1–22. [Google Scholar]
- 36. Noronha A, Belugali Nataraj N, Lee JS, Zhitomirsky B, Oren Y, Oster S, et al. AXL and error-prone DNA replication confer drug resistance and offer strategies to treat EGFR-mutant lung cancer. Cancer Discov 2022;12:2666–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Boshuizen J, Pencheva N, Krijgsman O, Altimari DD, Castro PG, de Bruijn B, et al. Cooperative targeting of immunotherapy-resistant melanoma and lung cancer by an AXL-targeting antibody-drug conjugate and immune checkpoint blockade. Cancer Res 2021;81:1775–87. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Violin plots and boxplots displaying the (a) genes detected per cell and (b) stress signature expression per cell and (c) mitochondrial fraction of reads in samples profiled from fresh or frozen tissue in this study and in indicated studies by Jerby et. al. (dijon) and Slyper et al. (green), respectively. The upper and lower edges of the boxplot display the 75th and 25th percentiles respectively. Blue violin plots are from scRNA-seq from fresh tissue, and red violin plots are from snRNA-seq on frozen tissue.
Supplemental Table 1: Assays performed and basic quality metrics of included samples.
Supplemental Table 2: Pathway enrichment analyses for indicated samples.
Supplemental Table 3: T-cell receptor sequences and clonality.
Data Availability Statement
Data can be accessed on Gene Expression Omnibus under accession number: GSE243381. Additional data requests should be directed to the corresponding author. Code is available viahttps://github.com/IzarLab/sarcoma-sn.