Abstract
Previous studies have shown that the 5′ arm of the influenza A virus virion RNA promoter requires a hairpin loop structure for efficient endonuclease activity of influenza virus RNA polymerase, an activity that is required for the cap-snatching activity of primers from host pre-mRNA. Here we examine whether a hairpin loop is also required in the 3′ arm of the viral RNA promoter. We study point mutations at each nucleotide position (1 to 12) within the 3′ arm of the promoter as well as complementary “rescue” mutations which restored base pairing in the stem of a potential hairpin loop. Our results suggest that endonuclease activity is absolutely dependent on the presence of a 3′ hairpin loop structure. This is the first direct evidence for RNA secondary structure within the 3′ arm being required for a specific stage, i.e., endonuclease cleavage, in the influenza virus replicative cycle.
Influenza A virus is a segmented, negative-sense RNA virus. The virion RNA of influenza virus serves several functions. During transcription it acts as the template and as a cofactor in endonuclease cleavage of host pre-mRNA (17, 31), leading to the synthesis of viral mRNA which is capped at its 5′ end and polyadenylated at its 3′ end (18). Virion RNA (vRNA) also acts as the template for full-length cRNA, which in turn serves as a template for the synthesis of new vRNA molecules. Influenza virus RNA synthesis takes place within the nucleus of infected cells, consistent with the requirement for host cell pre-mRNAs and for splicing (26), suggesting that influenza virus transcription may be coupled to host cell RNA polymerase II transcription (14). Inhibitors selective for either influenza virus endonuclease or RNA polymerase activities suggest that the active site for endonuclease cleavage is separate from the transcription active site (50, 51).
The influenza virus RNA polymerase is a heterotrimer formed by the PB1, PB2, and PA subunits (25, 26). All three subunits are required for transcription and replication, although the role of PA is poorly understood (37, 44, 45). In the virion the polymerase proteins are associated with vRNA and the nucleoprotein to form ribonucleoprotein complexes. It is likely that base pairing between the conserved 3′ and 5′ termini of the influenza virus vRNA leads to the formation of an RNA panhandle structure; however, it is probable that the polymerase itself helps to stabilize the interactions between the 5′ and 3′ termini (7, 19, 20, 23, 31, 33, 35).
PB1 interacts with both PB2 and PA (20, 22, 52) and is also involved in binding vRNA (11, 12, 15, 31). PB1 also binds cRNA, although the binding sites are different from those for vRNA (16). PB1 contains amino acid motifs present in other RNA-dependent RNA polymerases (38) and is the subunit responsible for polymerization (24). Although it has been reported that the endonuclease site residues in PB2 (32), recent evidence favors its location being in PB1 (31a).
PB2 is the cap-binding protein. Cap-binding regions have been mapped, and capped RNA can be cross-linked to PB2 (3, 4, 21, 31a, 36, 46, 53). Defective PB2 mutants do not cleave cap structures from mRNA (54) and there are potential sequence similarities between PB2 and known cap-binding proteins (8). In addition, capped RNA-directed RNA synthesis in vitro is inhibited by antisera against PB2 (5). The extreme N-terminal region of PB2 interacts with PB1 (21).
The cap-binding activity of the polymerase is stimulated by the presence of the 5′-terminal nucleotides of the influenza virus vRNA (6, 31). However, although both vRNA and cRNA bind the polymerase and are efficiently transcribed, only vRNA activates endonuclease cleavage (6). This is presumably because of subtle differences in the promoters (30) causing separate regions of PB1 to bind to vRNA and cRNA (16).
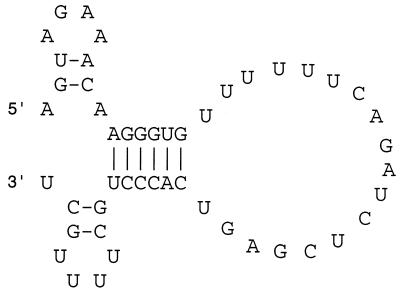
Several models have been proposed for the secondary structure of the influenza virus vRNA promoter in what is a somewhat controversial field (2, 9–13, 26, 34, 41). It is now accepted that both the vRNA 5′ and 3′ ends are required for replication and transcription (6, 12, 13, 17, 41, 49). Chemical probing methods indicate that the promoter structure is made up of a panhandle structure with an internal bulge (2). However, other work suggests that the 10 5′-terminal vRNA nucleotides and 9 3′ nucleotides do not base pair with each other (9, 12). This observation led to the proposal of the “RNA fork” model, based on in vitro transcription assays, in which nucleotides within the 5′ promoter arm base pair with cognate nucleotides on the 3′ promoter arm, leaving the terminal nucleotides unpaired (12, 13). The corkscrew model, based on in vivo reporter gene experiments, extends this model further by proposing local secondary structures, or hairpin loops, within the “single-stranded” 3′- and 5′-terminal nucleotides (Fig. 1) (9). In all cases the base pairs within the promoter, rather than the identity of the nucleotides, were found to be important for polymerase activity.
FIG. 1.
Promoter structure of the 49-nt-long model influenza virus vRNA used in this study drawn in a corkscrew-type conformation (9).
In vitro transcription and polyadenylation of orthomyxovirus vRNA templates are dependent on a 5′ hairpin loop (27, 43), but it is known that the formation of a 3′ hairpin loop is not necessary for replication and transcription primed with a dinucleotide primer (13, 29, 42, 43). Previously, the role of the vRNA 5′ promoter sequence on endonuclease activity was studied in isolation from those sequences that regulate transcription (30). Here we extend that study and now report the mutagenic analysis of the 3′-terminal nucleotides of the influenza A virus vRNA with regard to endonuclease activity. We show that a 3′ hairpin loop is required, thus formally assigning a role for this structure and reconciling the perceived contradictions between existing in vivo and in vitro data (9, 10, 12, 13, 39, 43).
MATERIALS AND METHODS
Construction of influenza virus-like model RNAs from a plasmid.
The methods used were essentially the same as those published previously (30) and are given here only in brief. Plasmid pBXB49 (30) contains 49 nucleotides (nt) of influenza virus vRNA sequence derived from the 5′ and 3′ ends of segment 8 of influenza virus A/PR/8/34. Point mutants of pBXB49 were made by standard methods. Influenza virus vRNA-like RNA synthesized by T7 RNA polymerase runoff transcription reactions was worked up and quantitated as before except that unincorporated ribonucleoside triphosphates were removed by a QIAquick nucleotide removal kit.
Preparation of recombinant influenza A virus polymerase/endonuclease and the synthesis of the 67-nt-long cap donor.
Recombinant vaccinia virus vectors which express influenza A virus PB1, PB2, or PA protein were used to infect HeLa cells, and nuclear extracts were prepared as described before (30, 48). 32P-labeled cap zero donors (67 nt long) were prepared by a slight modification of a method described previously (17, 30) using QIAquick nucleotide removal columns both for the workup of the transcription reaction and for removing unincorporated [32P]GTP.
Capped RNA endonuclease assays.
The endonuclease assay was similar to one described before (30) except that usually 1 μl of recombinant polymerase was used in a 5-μl reaction which typically contained 20 to 30 nM capped 32P-, end-labeled 67-nt-long pGEM-7Zf(+)-derived RNA probe and an excess (about 1 pmol) of wild-type or mutant 49-mer model RNA. The reaction was usually incubated at 30°C for 20 min. Products were analyzed by 15% polyacrylamide gel electrophoresis in 7 M urea and quantified by phosphorimage analysis. The yield of substrate and product was measured, and endonuclease activity was expressed as the percentage of substrate cleaved.
RESULTS
Systematic study of endonuclease activity using vRNA with point mutations in the 3′ promoter arm.
Flick et al. (9) suggested that a corkscrew model of the vRNA promoter (Fig. 1) was a valid secondary structure model based on in vivo reporter gene assays. In contrast, no evidence for the presence of the 3′ hairpin loop component of the corkscrew was obtained in studies of in vitro transcription (13) or in studies of the cis-acting requirements for polyadenylation (43). Here we tested whether the 3′ hairpin loop of the corkscrew is needed for endonuclease activity by studying endonuclease cleavage in isolation from transcription and replication. We synthesized 49-nt-long influenza virus-like model RNAs (Fig. 1) by T7 transcription of plasmid pBXB49 which were added to the endonuclease reactions in excess. Point mutants of pBXB49 were then used to study the cis-acting vRNA requirements for endonuclease activity. This methodology was similar to that used previously (30) to study the 5′ arm of the vRNA promoter.
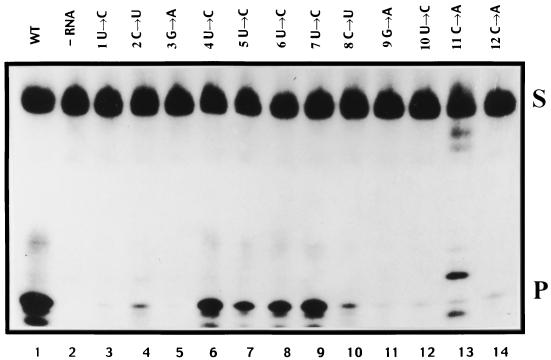
Initially, a complete set of point mutants was constructed at each nucleotide, from nt 1 to 12 of the 3′ influenza virus vRNA promoter arm, with each nucleotide being mutated to two alternative nucleotides. These mutants were used to determine which nucleotides or possible secondary structures within the 3′ arm of the influenza virus vRNA have a role in endonuclease function using recombinant influenza virus polymerase prepared by vaccinia virus expression vectors for the PB1, PB2, and PA proteins. Endonuclease assays (see Materials and Methods) were conducted using a 32P-labeled 67-nt-long cap donor (see Materials and Methods) with each of the mutant constructs, and the percent cleavage activity, which gives rise to a 12-nt-long cleavage product, was expressed relative to the control wild-type construct. A representative autoradiograph is shown in Fig. 2. The results of three independent sets of reactions with each virion RNA construct, quantified by phosphorimage analysis, are summarized in Fig. 3.
FIG. 2.
Effect of mutations in each nucleotide of the influenza virus vRNA 3′ promoter arm analyzed by 15% polyacrylamide gel electrophoresis in 7 M urea. Lane 1, wild-type (WT) vRNA (49-mer); lane 2, negative control with no added model vRNA; lanes 3 to 14, positions of mutations and nucleotides changed. S, 67-nt-long capped RNA substrate; P, 12-nt-long cleavage product. Note that the nonspecific bands in lane 13 were caused by RNase contamination.
FIG. 3.
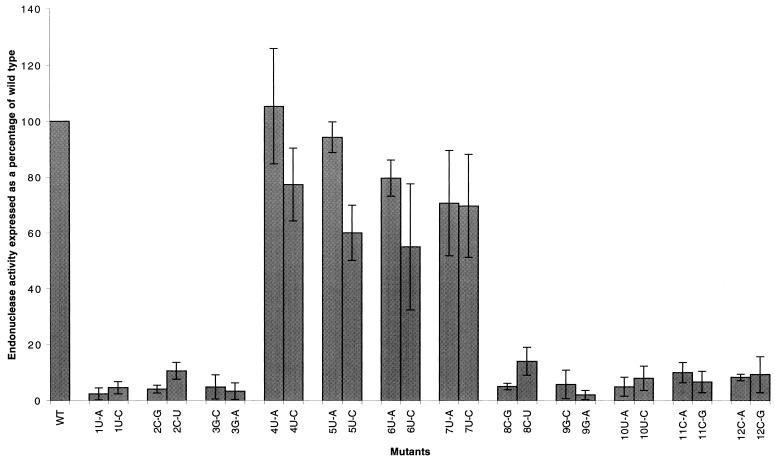
Quantitation of endonuclease cleavage by phosphorimage analysis. Standard deviations of the means were calculated from at least three independent experiments. Results are corrected for minor nonspecific background cleavage in the absence of added RNA. Results for each mutant are expressed as a percentage of wild-type vRNA, which was set at 100%. The percent substrate cleaved in the presence of wild-type vRNA varied from 22 to 35% in different experiments.
It can be seen that significant endonuclease activity compared to that of the wild type was still evident in RNA with mutations at nt 4, 5, 6, and 7 (Fig. 3). However, cleavage was essentially at background levels (but see later comments on nt 2 [C→U] and 8 [C→U]) in the presence of RNA with mutations in positions 1 to 3 and 8 to 12, suggesting that these nucleotides were essential for endonuclease activity per se or that they were involved in base pairs with other residues, thus forming essential secondary structures.
Hairpin loop in the 3′ promoter arm is required for endonuclease activity.
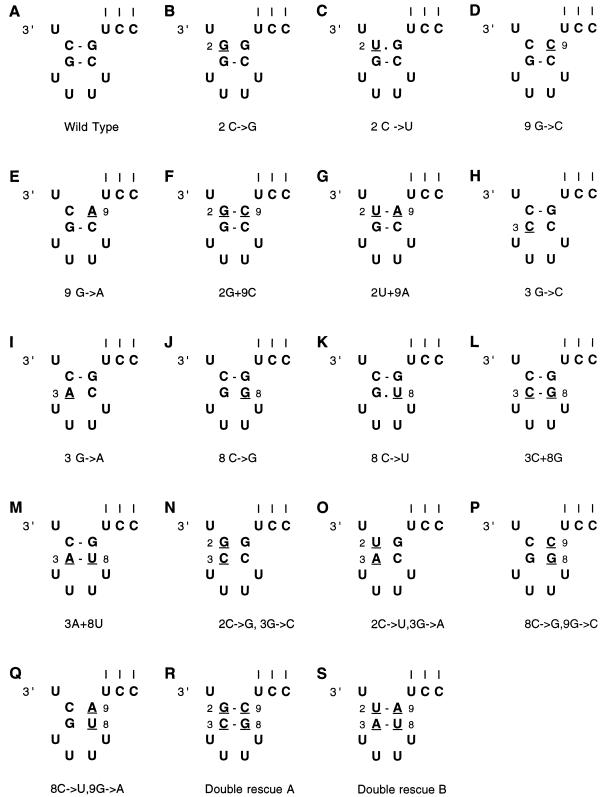
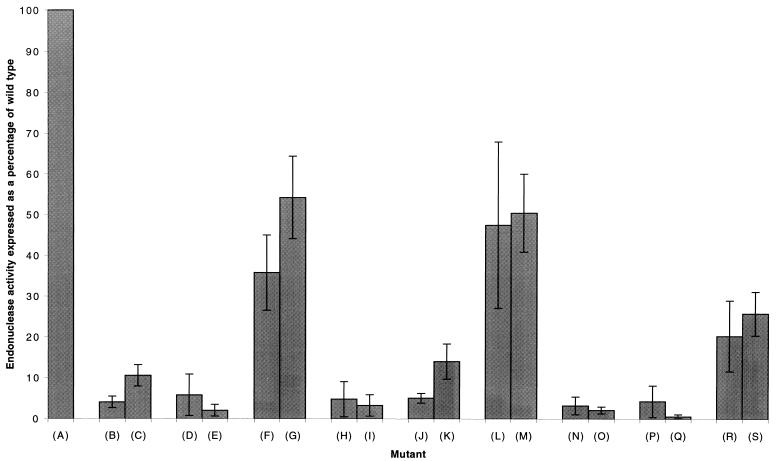
Nucleotide substitutions at positions 2, 3, 8, and 9 reduced endonuclease activity to the background level of detection (Fig. 2 and 3). To test whether this effect was due to the specific identity of these nucleotides or to a possible hairpin loop structure formed by the base pairing of nt 2 and 3 with 9 and 8, respectively, a series of rescue mutations was constructed (Fig. 4). The results of endonuclease assays (averages of three or more experiments) are summarized in Fig. 5. It was found that a mutation from C to G at position 2 (Fig. 4B), which reduced endonuclease activity to background levels, could be rescued to 36% of the wild-type activity (Fig. 5, average of three experiments) by making the complementary mutation (G→C) at position 9 (Fig. 4F). Likewise, a mutation from C to U at position 2 (Fig. 4C), which reduced endonuclease activity to approximately 9% of the wild-type activity, could be rescued to 54% of the wild-type activity (average of three experiments) by making the complementary mutation (G→A) at position 9 (Fig. 4G).
FIG. 4.
The 3′ end of the 49-nt-long RNA constructs used to investigate the effects of mutations in the stem of the 3′ hairpin loop on endonuclease activity. The mutations are underlined and numbered in the different constructs (A to S). The three vertical lines indicate base pairing to the 5′ end of the RNA.
FIG. 5.
Quantitation of endonuclease cleavage in the presence of model vRNAs mutated to destroy the 3′ hairpin loop structure and model RNAs containing a second rescue mutation designed to restore base pairing within the stem of the hairpin loop of mutated constructs. Results are corrected for minor nonspecific background cleavage in the absence of added RNA. Results for each mutant are expressed as a percentage of wild-type activity, which was set at 100%. The percent substrate cleaved in the presence of wild-type vRNA varied from 22 to 35% in different experiments. Letters in parentheses refer to the constructs (A to S) shown in Fig. 4. Note that the point mutants are identical to those in Fig. 3 and are repeated here for ease of comparison with the rescue mutants.
When position 3 was mutated from G to C (Fig. 4H), endonuclease activity was reduced to background levels but was rescued to 48% of the wild-type activity (average of three experiments) by introducing the complementary mutation (C→G) at position 8 (Fig. 4L). Similarly, when nt 3 was changed from G to A (Fig. 4I), endonuclease activity was reduced to background levels, but on addition of a complementary mutation at position 8 (C→U) (Fig. 4M), activity was again increased to 51% that of the wild type. In agreement with these results, the point mutants at nt 8 and 9 which disrupt base pairs are in all cases inactive (Fig. 3 and 4). When nt 8 was mutated from C to U, an activity of 16% of the wild-type activity was observed, which was higher than the background, although still significantly lower than that of the rescue mutants (Fig. 4K and 5). These results indicate that a 3′ hairpin loop structure involving base pairs between nt 2 and 9 and nt 3 and 8 is required for endonuclease activity. It is interesting to note that when the C→U mutation was made at position 2 or 8 endonuclease activity was higher than for other mutations to the stem of the possible hairpin loop (Fig. 5), possibly due to the formation of a G-U wobble pair (see Discussion).
To further test the hypothesis that a 3′ hairpin loop, rather than specific nucleotides within the influenza virus vRNA 3′ promoter arm, is required for endonuclease activity, double mutants which would disrupt both base pairs in the stem of the loop were constructed. When a C→G mutation was made at position 2 and a G→C mutation was made at position 3, endonuclease activity was reduced to background levels (Fig. 5). Likewise, when a C→U mutation was made at position 2 and a G→A mutation was made at position 3, endonuclease activity was abrogated. Again, when a C→G mutation at position 8 was combined with a G→C mutation at position 9 or a C→U mutation at position 8 was combined with a G→A mutation at position 9, endonuclease activity was reduced to background levels (Fig. 5). When rescue mutations were made which allowed the formation of base pairs between nt 2 and 9 and nt 3 and 8 (Fig. 4R and S), endonuclease activity was rescued to 19 and 23%, respectively. In both cases this was significantly higher than background levels, although still significantly lower than any of the single base pair rescue mutants (Fig. 5).
DISCUSSION
The RNA polymerase complex of influenza virus, composed of the PB1, PB2, and PA subunits, carries out all of the essential replicative functions of the virus (26). Not only is the polymerase required for the synthesis of mRNA, vRNA, and cRNA, but it also possesses an endonuclease activity which is strictly dependent on the presence of the vRNA promoter whereby the complex cleaves cap structures together with 9 to 15 or so heterologous nucleotides from host mRNA.
A thorough mutagenic analysis of the 5′ arm of the vRNA promoter had previously shown that a 5′ hairpin loop formed by base pairing between nt 2 and 9 and nt 3 and 8, respectively, of the 5′ promoter arm was essential for cleavage activity (30). These same positions had also been found to be important in previous in vitro and in vivo studies of transcription and polyadenylation (9, 12, 39, 43). In the present study, mutation of nt 2, 3, 8, and 9 of the 3′ promoter arm destroyed endonuclease activity (Fig. 2 and 3), suggesting that the 3′ arm of the influenza virus vRNA promoter may possess a similar hairpin loop structure to the 5′ arm (Fig. 1).
To determine whether the 3′ hairpin loop was required for endonuclease activity, mutations were made at 3′ positions 2 and 3, thus disrupting the stem of the hairpin loop (Fig. 4). Mutations which would destroy a putative 3′ hairpin loop were also made at positions 8 and 9. In all cases the residues were changed to at least two alternative nucleotides (usually a transition and a transversion) to rule out possible sequence-specific effects. All mutants which destroyed possible base pairing within the stem of a putative hairpin loop reduced endonuclease activity markedly, usually to background levels of detection. Complementary mutations made at positions 9 and 8 reformed the base pairs with nt 2 and 3, respectively (Fig. 5). These complementary mutations partially rescued endonuclease activity in all cases, indicating the importance of a 3′ hairpin loop in endonuclease activity. However, the fact that the rescue was not 100%, with typical values lying between 35 and 55%, indicated that there were some sequence-specific effects. Interestingly, when a C→U mutation was made at position 2 or position 8, residual endonuclease activity was higher than when other mutations were made to the stem of the possible hairpin loop. The fact that these wobble base pairs were only weakly active argues that two standard Watson-Crick base pairs are required to ensure sufficient stability (Fig. 4C and K). In agreement with previous studies, the mutation of 3′ nt 10, 11, and 12, which are involved in base pairs with the 5′ promoter arm, destroyed endonuclease activity (Fig. 3) (30). It is possible that insertions or deletions within the hairpin loop or other mutations beyond the scope of this paper may affect endonuclease activity. However, the data presented here show that the 3′ hairpin loop appears to be the major determinant of endonuclease activity.
Binding of recombinant influenza virus polymerase, devoid of influenza virus vRNA, to a capped RNA is dependent on the addition of the 5′ arm of the influenza virus vRNA (6). However, the cap structure is not cleaved from the mRNA unless the 3′ arm of the vRNA is also present (6, 17, 31). The 5′ component of the influenza virus vRNA promoter was not mutated in the present study and so cap binding should not have been affected. Therefore, the step that was abrogated by the mutants which disrupted the 3′ hairpin loop was probably the actual cleavage of the capped substrate. Loss of activity could have occurred either because the polymerase complex failed to bind the 3′ promoter arm lacking the hairpin loop structure or because when bound it did not cause the presumed allosteric changes required to activate the endonuclease.
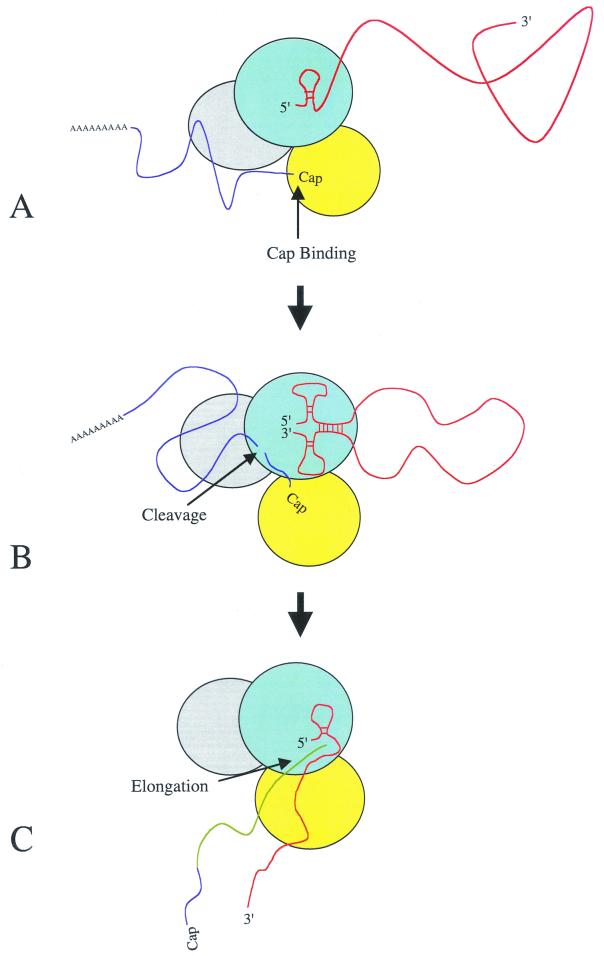
Although the results reported here support the RNA corkscrew promoter model (Fig. 1), the secondary structure seen is likely to be transient. The process of transcription and the accompanying conformational changes that take place within both the vRNA and the PB1, PB2, and PA subunits are complex and not well understood. Our current understanding of the role of the 3′ hairpin loop and the ordered events leading to transcription is outlined schematically in Fig. 6. First the PB1 polymerase subunit binds the 5′ hairpin loop of the influenza virus vRNA promoter (Fig. 6A) (6, 31). This brings about unspecified allosteric changes, activating a cap-binding site on the PB2 subunit and a vRNA 3′ binding site on PB1 (31). As the PB1 subunit binds the vRNA 3′ promoter arm, base pairs form between the 5′ and 3′ promoter arms, thus forming the RNA corkscrew conformation (Fig. 6B). In addition, there is compelling evidence for interactions of the vRNA 3′ and 5′ termini with distinct regions of PB1 that are separated by 300 amino acids in the primary sequence (31). Binding of the 3′ hairpin loop stimulates endonuclease cleavage, possibly on the PB1 subunit, leading to the cleavage of host pre-mRNA (6, 31). Finally, the capped RNA is used to prime transcription of the vRNA and elongation ensues (Fig. 6C). It is obvious that either coincident with or after endonuclease cleavage, melting of the 3′ hairpin loop of vRNA must occur to allow transcription to initiate. During elongation the RNA polymerase remains bound to the 5′ promoter arm, ultimately leading to reiterative incorporation of A nucleotides directed by a track of uridine nucleotides, thus forming a poly(A) tail of mRNA (39, 40, 42, 43).
FIG. 6.
Model for the initiation of influenza virus mRNA transcription. (A) Influenza virus vRNA binds to the PB1 subunit of the polymerase complex via the 5′ hairpin loop, thus activating the cap-binding domain of the PB2 subunit. This leads to cell mRNA being bound by the polymerase PB2 subunit. (B) vRNA 3′ binding site of PB1, together with base pair interactions between the virion RNA 5′ and 3′ termini, leads to the binding of the vRNA promoter 3′ arm, thus activating endonuclease cleavage. (C) Polymerase complex initiates transcription of the virion RNA template using the capped cleavage product as a primer following a conformational change within the promoter leading to the melting of the vRNA 3′ hairpin loop (for full details, see text). Proteins: light blue, PB1; grey, PA; yellow, PB2. RNA: red, vRNA; dark blue, host mRNA; green, influenza virus mRNA.
It is known that influenza virus vRNA activates the polymerase complex, allowing it to cleave mRNA, but influenza virus cRNA, which is superficially similar in sequence, does not (6); however, to date no mechanism has been proposed to explain this. A related orthomyxovirus, Thogoto virus (THOV), appears to have many similarities to influenza virus with respect to transcription and replication and also cleaves host cell pre-mRNA for use as primers (1, 27, 47, 55). In contrast to the present study, however, in the THOV vRNA promoter only the 5′ hairpin loop and not a 3′ hairpin loop was required for endonuclease activity (28). As the THOV cRNA molecule is a complement of the vRNA, it follows that the cRNA lacks a 5′ hairpin loop. Based on these observations an endonuclease switching mechanism was proposed whereby vRNA molecules containing 5′ hairpin loops stimulated endonuclease activity and cRNA molecules lacking the 5′ hairpin loop structure did not (29). Hairpin loops have now been shown to be present and necessary for endonuclease activity in both the 3′ and 5′ promoter arms of the influenza virus vRNA. Because hairpin loops are present in both influenza virus vRNA termini, it follows that they are present in cRNA molecules. Therefore, it seems likely that influenza virus distinguishes between vRNA and cRNA with regard to endonuclease cleavage by a different method from that of THOV.
In conclusion, our results show that in contrast to transcription and polyadenylation, which are also catalyzed by the influenza virus RNA-dependent RNA polymerase, a hairpin loop in the 3′ arm of the influenza A virus vRNA promoter is stringently required for endonuclease activity studied in vitro. This resolves the apparent contradiction between reporter gene experiments (9) and work carried out in vitro (12, 40, 43). In vivo work has previously stressed the significance of a so-called corkscrew structure involving both 5′ and 3′ hairpin loops, whereas ApG primed transcription, polyadenylation, and polymerase binding function in vitro without the need for such a structure in the 3′ arm. Here we have shown that it is probable that the crucial step in reporter gene work (9) requiring a hairpin loop in the 3′ promoter arm is the activation of endonuclease cleavage.
ACKNOWLEDGMENTS
M. B. L. was supported by the MRC (program grant G9523972 to G.G.B.).
We thank Peter Palese and Jane Sharps for plasmids and Alice Taylor for DNA sequencing.
REFERENCES
- 1.Albo C, Martin J, Portella A. The 5′ ends of Thogoto virus (Orthomyxoviridae) mRNAs are homogeneous in both length and sequence. J Virol. 1996;70:9013–9017. doi: 10.1128/jvi.70.12.9013-9017.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baudin F, Bach C, Cussack S, Ruigrok R W H. Structure of influenza virus RNP. I. Influenza virus nucleoprotein melts secondary structure in panhandle RNA and exposes the bases to the solvent. EMBO J. 1994;13:3158–3165. doi: 10.1002/j.1460-2075.1994.tb06614.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blaas D, Patzelt E, Kuechler E. Cap-recognising protein of influenza virus. Virology. 1982;116:339–348. doi: 10.1016/0042-6822(82)90425-1. [DOI] [PubMed] [Google Scholar]
- 4.Blaas D, Patzelt E, Kuechler E. Identification of the cap binding protein of influenza virus. Nucleic Acids Res. 1982;10:4803–4812. doi: 10.1093/nar/10.15.4803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blok V, Cianci C, Tibbles K W, Inglis S C, Krystal M, Digard P. Inhibition of the influenza virus RNA-dependent RNA polymerase by antisera directed against the carboxy-terminal region of the PB2 subunit. J Gen Virol. 1996;77:1025–1033. doi: 10.1099/0022-1317-77-5-1025. [DOI] [PubMed] [Google Scholar]
- 6.Cianci C, Tiley L, Krystal M. Differential activation of the influenza virus polymerase via template RNA binding. J Virol. 1995;69:3995–3999. doi: 10.1128/jvi.69.7.3995-3999.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Compans R W, Content J, Duesberg P H. Structure of the ribonucleoprotein of influenza virus. J Virol. 1972;4:795–800. doi: 10.1128/jvi.10.4.795-800.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de la Luna S, Martinez C, Ortin J. Molecular cloning and sequencing of influenza virus A/Victoria/3/75 polymerase genes: sequence evolution and prediction of possible functional domains. Virus Res. 1989;13:143–156. doi: 10.1016/0168-1702(89)90012-9. [DOI] [PubMed] [Google Scholar]
- 9.Flick R, Neumann G, Hoffmann E, Hobom G. Promoter elements in the influenza vRNA terminal structure. RNA. 1996;2:1046–1057. [PMC free article] [PubMed] [Google Scholar]
- 10.Flick R, Hobom G. Interaction of influenza virus polymerase with viral RNA in the corkscrew conformation. J Gen Virol. 1999;80:2565–2572. doi: 10.1099/0022-1317-80-10-2565. [DOI] [PubMed] [Google Scholar]
- 11.Fodor E, Seong B L, Brownlee G G. Photochemical cross-linking of influenza A polymerase to its virion RNA promoter defines a polymerase binding site at residues 9 to 12 of the promoter. J Gen Virol. 1993;74:1327–1333. doi: 10.1099/0022-1317-74-7-1327. [DOI] [PubMed] [Google Scholar]
- 12.Fodor E, Pritlove D C, Brownlee G G. The influenza virus panhandle is involved in the initiation of transcription. J Virol. 1994;68:4092–4096. doi: 10.1128/jvi.68.6.4092-4096.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fodor E, Pritlove D C, Brownlee G G. Characterization of the RNA-fork model of virion RNA in the initiation of transcription in influenza A virus. J Virol. 1995;69:4012–4019. doi: 10.1128/jvi.69.7.4012-4019.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fodor E, Mikulasova A, Mingay L J, Poon L L M, Brownlee G G. Messenger RNAs that are not synthesized by RNA polymerase II can be 3′ end cleaved and polyadenylated. EMBO Rep. 2000;1:513–518. doi: 10.1093/embo-reports/kvd111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gonzalez S, Ortin J. Characterization of the influenza virus PB1 protein binding to vRNA: two separate regions of the protein contribute to the interaction domain. J Virol. 1999;73:631–637. doi: 10.1128/jvi.73.1.631-637.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gonzalez S, Ortin J. Distinct regions of influenza virus PB1 polymerase sub-unit recognize vRNA and cRNA templates. EMBO J. 1999;18:3767–3775. doi: 10.1093/emboj/18.13.3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hagen M, Chung T D Y, Butcher J A, Krystal M. Recombinant influenza virus polymerase: requirement of both 5′ and 3′ viral ends for endonuclease activity. J Virol. 1994;68:1509–1515. doi: 10.1128/jvi.68.3.1509-1515.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hay A J. Characterization of influenza virus RNA complete transcripts. Virology. 1982;116:517–522. doi: 10.1016/0042-6822(82)90144-1. [DOI] [PubMed] [Google Scholar]
- 19.Heggeness M H, Smith P R, Ulmanen I, Krug R M, Choppin P W. Studies on the helical nucleocapsid of influenza virus. Virology. 1982;118:466–470. doi: 10.1016/0042-6822(82)90367-1. [DOI] [PubMed] [Google Scholar]
- 20.Honda A, Ishihama A. The molecular anatomy of influenza virus RNA polymerase. Biol Chem. 1997;378:483–488. [PubMed] [Google Scholar]
- 21.Honda A, Mizumoto K, Ishihama A. Two separate sequences of PB2 subunit constitute the RNA cap-binding site of influenza virus RNA polymerase. Genes Cells. 1999;4:475–485. doi: 10.1046/j.1365-2443.1999.00275.x. [DOI] [PubMed] [Google Scholar]
- 22.Ishihama A. A multifunctional enzyme with RNA polymerase and RNase activities. Biochimie. 1996;78:1097–1102. doi: 10.1016/s0300-9084(97)86735-1. [DOI] [PubMed] [Google Scholar]
- 23.Jennings P A, Finch J T, Winter G, Robertson J S. Does the higher order structure of the influenza virus nucleoprotein guide sequence rearrangements in influenza viral RNA? Cell. 1983;34:619–622. doi: 10.1016/0092-8674(83)90394-x. [DOI] [PubMed] [Google Scholar]
- 24.Kobayashi M, Toyoda T, Ishihama A. Influenza virus PB1 protein is the minimal and essential sub-unit of RNA polymerase. Arch Virol. 1996;141:525–539. doi: 10.1007/BF01718315. [DOI] [PubMed] [Google Scholar]
- 25.Lamb R A. Genes and proteins of the influenza viruses, p 89–121. In: Krug R M, editor. The influenza viruses. New York, N.Y: Plenum Press; 1989. [Google Scholar]
- 26.Lamb R A, Krug R M. Orthomyxoviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 1353–1395. [Google Scholar]
- 27.Leahy M B, Dessens J T, Nuttall P A. Striking conformational similarities between the transcription promoters of Thogoto and influenza A viruses: evidence for intrastrand base pairing in the 5′ promoter arm. J Virol. 1997;71:8352–8356. doi: 10.1128/jvi.71.11.8352-8356.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leahy M B, Dessens J T, Nuttall P A. In vitro polymerase activity of Thogoto virus: evidence for a unique cap snatching mechanism in a tick-borne orthomyxovirus. J Virol. 1997;71:8347–8351. doi: 10.1128/jvi.71.11.8347-8351.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leahy M B, Dessens J T, Pritlove D C, Nuttall P A. An endonuclease switching mechanism in the virion RNA and cRNA promoters of Thogoto orthomyxovirus. J Virol. 1998;72:2305–2309. doi: 10.1128/jvi.72.3.2305-2309.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leahy M B, Pritlove D C, Poon L M, Brownlee G G. Mutagenic analysis of the 5′ arm of the influenza A virus virion RNA promoter defines the sequence requirements for endonuclease activity. J Virol. 2001;75:134–142. doi: 10.1128/JVI.75.1.134-142.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li M-L, Ramirez B C, Krug R M. RNA-dependent activation of primer RNA production by influenza polymerase: different regions of the same protein subunit constitute the two required RNA-binding sites. EMBO J. 1998;17:5844–5852. doi: 10.1093/emboj/17.19.5844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31a.Li M-L, Rao P, Krug R M. The active sites of the influenza cap-dependent endonuclease are on different polymerase subunits. EMBO J. 2001;20:2078–2086. doi: 10.1093/emboj/20.8.2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Licheng S, Summers D F, Peng Q, Galarza J M. Influenza A virus polymerase sub-unit PB2 is the endonuclease which cleaves host cell mRNA and functions only as the trimeric enzyme. Virology. 1995;208:38–47. doi: 10.1006/viro.1995.1127. [DOI] [PubMed] [Google Scholar]
- 33.Murti K G, Webster R G, Jones I M. Localization of RNA polymerases of influenza viral ribonucleoproteins by immunogold labeling. Virology. 1988;164:562–566. doi: 10.1016/0042-6822(88)90574-0. [DOI] [PubMed] [Google Scholar]
- 34.Neumann G, Zobel A, Hobom G. RNA polymerase I-mediated expression of influenza viral RNA molecules. Virology. 1995;202:477–479. doi: 10.1006/viro.1994.1365. [DOI] [PubMed] [Google Scholar]
- 35.Ortega J, Martin-Benito J, Zurcher T, Valpuesta J M, Carrascosa J L, Ortin J. Ultrastructural and functional analyses of recombinant influenza virus ribonucleoproteins suggest dimerization of nucleoprotein during virus amplification. J Virol. 2000;74:156–163. doi: 10.1128/jvi.74.1.156-163.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Penn C, Blaas D, Mahy E, Mahy B W J. Extragenic suppression of a ts phenotype during recombination between ts mutants of two fowl plague virus strains with a ts mutation in gene 1. J Gen Virol. 1982;62:177–180. doi: 10.1099/0022-1317-62-2-239. [DOI] [PubMed] [Google Scholar]
- 37.Perales B, Ortin J. The influenza A virus PB2 polymerase subunit is required for the replication of viral RNA. J Virol. 1997;71:1381–1385. doi: 10.1128/jvi.71.2.1381-1385.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Poch O, Sauvaget I, Delarue M, Tordo N. Identification of four conserved motifs among the RNA-dependent polymerase encoding elements. EMBO J. 1990;8:3867–3874. doi: 10.1002/j.1460-2075.1989.tb08565.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Poon L M, Pritlove D C, Sharps J, Brownlee G G. The RNA polymerase of influenza virus, bound to the 5′ end of virion RNA, acts in cis to polyadenylate mRNA. J Virol. 1998;72:8214–8219. doi: 10.1128/jvi.72.10.8214-8219.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Poon L M, Pritlove D C, Fodor E, Brownlee G G. Direct evidence that the poly(A) tail of influenza virus is synthesized by reiterative copying of a U track in the virion RNA template. J Virol. 1999;73:3473–3476. doi: 10.1128/jvi.73.4.3473-3476.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pritlove D C, Fodor E, Seong B L, Brownlee G G. In vitro transcription and polymerase binding studies of the termini of influenza A virus cRNA: evidence for a cRNA panhandle. J Gen Virol. 1995;76:2205–2213. doi: 10.1099/0022-1317-76-9-2205. [DOI] [PubMed] [Google Scholar]
- 42.Pritlove D C, Poon L M, Fodor E, Sharps J, Brownlee G G. Polyadenylation of influenza virus mRNA transcribed in vitro from model virion RNA templates: requirement for 5′ conserved sequences. J Virol. 1998;72:1280–1286. doi: 10.1128/jvi.72.2.1280-1286.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pritlove D C, Poon L M, Devenish L J, Leahy M B, Brownlee G G. A hairpin loop at the 5′ end of influenza A virus virion RNA is required for synthesis of poly(A)+ mRNA in vitro. J Virol. 1999;73:2109–2114. doi: 10.1128/jvi.73.3.2109-2114.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sans-Ezquerro J J, de la Luna S, Ortin J, Nieto A. Individual expression of influenza virus PA protein induces degradation of coexpressed proteins. J Virol. 1995;69:2420–2426. doi: 10.1128/jvi.69.4.2420-2426.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sans-Ezquerro J J, Fernandez Santaren J, Sierra T, Aragon T, Ortega J, Ortin J, Smith G L, Nieto A. The PA influenza polymerase sub-unit is a phosphoprotein. J Gen Virol. 1998;79:471–478. doi: 10.1099/0022-1317-79-3-471. [DOI] [PubMed] [Google Scholar]
- 46.Shi L C, Peng Q H, Galarza J M. Influenza virus RNA polymerase subunit PB2 is the endonuclease which cleaves host cell mRNA and functions only as the trimeric enzyme. Virology. 1995;208:38–47. doi: 10.1006/viro.1995.1127. [DOI] [PubMed] [Google Scholar]
- 47.Siebler J, Haller O, Kochs G. Thogoto and Dhori virus replication is blocked by inhibitors of the cellular polymerase II activity but does not cause shutoff of the host cell protein synthesis. Arch Virol. 1996;141:1587–1594. doi: 10.1007/BF01718257. [DOI] [PubMed] [Google Scholar]
- 48.Smith G L, Levin J Z, Palese P, Moss B. Synthesis and cellular location of ten influenza polypeptides individually expressed by recombinant vaccinia viruses. Virology. 1987;160:336–345. doi: 10.1016/0042-6822(87)90004-3. [DOI] [PubMed] [Google Scholar]
- 49.Tiley L S, Hagen M, Matthews J T, Krystal M. Sequence-specific binding of the influenza virus RNA polymerase to sequences located at the 5′ ends of the viral RNAs. J Virol. 1994;68:5108–5116. doi: 10.1128/jvi.68.8.5108-5116.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tisdale M, Ellis M, Klumpp K, Court S, Ford M. Inhibition of influenza-virus transcription by 2′-deoxy-2′-fluoroguanosine. Antimicrob Agents Chemother. 1995;39:2454–2458. doi: 10.1128/aac.39.11.2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tomassini J, Selnick H, Davies M E, Armstrong M E, Baldwin J, Bourgeois M, Hastings J, Hazuda D, Lewis J, McClements W, Ponticello G, Radzilowski E, Smith G, Tebben A, Wolfe A. Inhibition of cap (m7GpppXm)-dependent endonuclease of influenza virus by 4-substituted 2,4-dioxobutanoic acid compounds. Antimicrob Agents Chemother. 1994;38:2827–2837. doi: 10.1128/aac.38.12.2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Toyoda T, Adyshev D M, Kobayashi M, Iwata A, Ishihama A. Molecular assembly of the influenza virus RNA polymerase: determination of the sub-unit–sub-unit contact sites. J Gen Virol. 1996;77:2149–2157. doi: 10.1099/0022-1317-77-9-2149. [DOI] [PubMed] [Google Scholar]
- 53.Ulmanen I, Broni B A, Krug R M. Role of two of the influenza virus core P proteins in recognizing cap 1 structures (m7GpppNm) on RNAs and in initiating viral RNA transcription. Proc Natl Acad Sci USA. 1981;78:7355–7359. doi: 10.1073/pnas.78.12.7355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ulmanen I, Broni B A, Krug R M. Influenza virus temperature-sensitive cap (m7GppNm)-dependent endonuclease. J Gen Virol. 1983;45:27–35. doi: 10.1128/jvi.45.1.27-35.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Weber F, Haller O, Kochs G. Conserved vRNA end sequences of Thogoto-orthomyxovirus suggests a new panhandle structure. Arch Virol. 1997;142:1029–1033. doi: 10.1007/s007050050138. [DOI] [PubMed] [Google Scholar]