Two FDA-approved compounds, AKT inhibitors and pitavastatin, synergize to induce cell death in triple-negative breast cancer, motivating evaluation of the efficacy of this combination in clinical trials.
Abstract
Triple-negative breast cancer (TNBC) is responsible for a disproportionate number of breast cancer patient deaths due to extensive molecular heterogeneity, high recurrence rates, and lack of targeted therapies. Dysregulation of the phosphoinositide 3-kinase (PI3K)/AKT pathway occurs in approximately 50% of TNBC patients. Here, we performed a genome-wide CRISPR/Cas9 screen with PI3Kα and AKT inhibitors to find targetable synthetic lethalities in TNBC. Cholesterol homeostasis was identified as a collateral vulnerability with AKT inhibition. Disruption of cholesterol homeostasis with pitavastatin synergized with AKT inhibition to induce TNBC cytotoxicity in vitro in mouse TNBC xenografts and in patient-derived estrogen receptor (ER)–negative breast cancer organoids. Neither ER-positive breast cancer cell lines nor ER-positive organoids were sensitive to combined AKT inhibitor and pitavastatin. Mechanistically, TNBC cells showed impaired sterol regulatory element-binding protein 2 (SREBP-2) activation in response to single-agent or combination treatment with AKT inhibitor and pitavastatin, which was rescued by inhibition of the cholesterol-trafficking protein Niemann-Pick C1 (NPC1). NPC1 loss caused lysosomal cholesterol accumulation, decreased endoplasmic reticulum cholesterol levels, and promoted SREBP-2 activation. Taken together, these data identify a TNBC-specific vulnerability to the combination of AKT inhibitors and pitavastatin mediated by dysregulated cholesterol trafficking. These findings support combining AKT inhibitors with pitavastatin as a therapeutic modality in TNBC.
Significance: Two FDA-approved compounds, AKT inhibitors and pitavastatin, synergize to induce cell death in triple-negative breast cancer, motivating evaluation of the efficacy of this combination in clinical trials.
Introduction
Triple-negative breast cancer (TNBC) is an aggressive disease with the worst 5-year survival rate of all breast cancer subtypes (1). TNBC is characterized by the lack of expression of the estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (HER2). TNBC often recurs on standard of care chemotherapy, and targeted therapy options are limited to PARP inhibitors and immunotherapy, which show efficacy in only subsets of patients (2, 3). As such, there is a pressing need to identify targetable vulnerabilities in TNBC. The phosphoinositide 3-kinase (PI3K)/AKT signaling pathway is hyperactivated in nearly 50% of TNBC cases and promotes cell growth, survival, and metabolism (2–5). Because of the high prevalence of mutations in the PI3K/AKT pathway across cancer types, inhibitors of multiple nodes of the pathway have been developed, including PI3K, AKT, and mTOR inhibitors (6, 7). In 2023, the catalytic AKT inhibitor AZD5363 (capivasertib, Truqap) was FDA-approved in combination with endocrine therapy (fulvestrant, Faslodex) to treat subsets of patients with hormone receptor (HR)–positive breast cancer (8–10). Yet, the clinical use of PI3K/AKT inhibitors across distinct cancer lineages remains limited, and their efficacy depends on identifying combination therapies to limit on-target toxicities and acquired resistance (6, 11). Thus far, attempts to combine PI3K/AKT inhibitors with other drugs in the clinic have been mostly limited to standard-of-care regimens (6, 11, 12). Here, we present an unbiased approach to characterize synergistic drug combinations with PI3K/AKT inhibitors in TNBC.
We identify cholesterol homeostasis as a collateral vulnerability with AKT inhibition in TNBC. Cholesterol is synthesized through a series of energy-consuming reactions that occur primarily in the endoplasmic reticulum (13, 14). Cholesterol biosynthesis is regulated by end-product feedback inhibition to ensure sufficient cholesterol for dividing cells, thereby preventing unnecessary energy expenditure in sterol-replete conditions (14–16). When endoplasmic reticulum cholesterol levels are low, the transcription factor sterol regulatory element-binding protein 2 (SREBP-2) is escorted out of the endoplasmic reticulum and into the Golgi by SREBP cleavage-activating protein (SCAP). In the Golgi, SREBP-2 is sequentially cleaved and activated by two proteases, and active SREBP-2 enters the nucleus to promote the transcription of target genes, including LDLR, HMGCR, and INSIG1 (14, 15, 17). Statins, a class of FDA-approved drugs, inhibit 3-hydroxy-3-methylglutaryl coenzyme A reductase (HMGCR) at the first rate-limiting step of cholesterol synthesis. Statin-mediated inhibition of HMGCR lowers endoplasmic reticulum cholesterol levels and subsequently activates SREBP-2 (18–20). SREBP-2 activation results in upregulation of low-density lipoprotein receptor (LDLR) to lower plasma cholesterol levels and paradoxical upregulation of HMGCR (21). Here, we characterize synergy with combination AKT inhibitor and statin treatment in TNBC.
Materials and Methods
Cell lines
The following commercially available cell lines were used: SUM159 (Asterand Bioscience/BioIVT, SUM159PT), MDA-MB-468 (ATCC, HTB-132), BT20 (ATCC, HTB-19), T47D (ATCC, HTB-133), MCF7 (ATCC, HTB-22), BT474 (ATCC, HTB-20), HCC70 (ATCC, CRL-2315), MCF10A (ATCC, CRL-10317), HepG2 (ATCC, HB-8065), BT549 (ATCC, HTB-122), HCC1937 (ATCC, CRL-2336), ZR-75-1 (ATCC, CRL-1500), MDA-MB-361 (ATCC, HTB-27), HEK293T (ATCC, CRL-11268), parental T47D (Myles Brown Lab), fulvestrant-resistant T47D clones 1 to 3 (Myles Brown Lab), PC3 (ATCC, CRL-1435), and LNCaP (ATCC, CRL-1740). SUM159, MDA-MB-468, T47D, MCF7, BT474, HCC70, BT549, HCC1937, ZR-75-1, parental T47D, PC3 and LNCaP cells were cultured in RPMI 1640 medium (Gibco, 11875093) supplemented with 10% fetal bovine serum (FBS; GeminiBio, 100-106). Fulvestrant-resistant T47D cells were cultured in RPMI 1640 medium (Gibco, 11875093) supplemented with 10% FBS (Gemini Bio, 100-106) and 100 nmol/L fulvestrant (Selleckchem, S1191). BT20 and HepG2 cells were cultured in Eagle's Minimum Essential Medium (EMEM; Corning, 10-009-CV) supplemented with 10% FBS (GeminiBio, 100-106). MDA-MB-361 and HEK293T cells were cultured in DMEM with L-glutamine, 4.5 g/L glucose and sodium pyruvate (Fisher Scientific, MT10013CV) supplemented with 10% FBS (GeminiBio, 100-106). MCF10A cells were cultured in standard MCF10A growth medium without antibiotics [DMEM/F12 medium (Wisent Bioproducts, 319-075-CL), 5% horse serum (GeminiBio, 100508), 10 µg/mL insulin (Thermo Fisher Scientific/Gibco, A11382II), 0.5 mg/mL hydrocortisone (Sigma-Aldrich, H4001), 20 ng/mL EGF (R&D Systems, 236-EG-01M), and 100 ng/mL cholera toxin (List Biological Laboratories, 100B)]. The cell lines were maintained at 37°C in a 5% CO2 cell culture incubator and passaged at 70% to 90% confluency. To passage, cells were washed once with 1× PBS and incubated for 5 to 10 minutes at 37°C with 0.25% trypsin, 0.1% EDTA (Fisher Scientific, MT25053CI). Cells were passaged up to five times in the same dish and were maintained in culture for up to 1 month. Cells routinely tested negative for Mycoplasma contamination (Lonza, LT07-218).
Genome-wide CRISPR/Cas9 screen
Viral transduction, cell seeding, drug treatments, and harvest
SUM159 cells were transduced at a multiplicity of infection of ∼0.3 with CRISPR/Cas9 lentivirus (Steve Elledge Lab) containing 94,495 sgRNAs, with 3 to 4 sgRNAs per gene, and at least 500-fold representation. Cells were selected with 1 µg/mL puromycin (Corning, 61-385-RA) for 3 days. After selection, day-0 cell pellets were collected, and the remaining infected cells were expanded for 3 days and then plated into treatment arms at 100,000 cells/mL (2 × 106 cells/plate) in tissue culture–treated 15-cm plates. The cells were treated for 72 hours with DMSO (vehicle) or cytostatic doses of the PI3Kα-selective inhibitor BYL719 (0.4 µmol/L; Active Biochem, A-1214) or the catalytic AKT inhibitor GDC-0068 (3 µmol/L; Selleckchem, S2808). At the endpoint, cell pellets were harvested and processed as described below.
Genomic DNA isolation for sequencing
Cell pellets were resuspended in 10 mmol/L Tris (Fisher Scientific, BP152-500) pH, 8.0 and 10 mmol/L EDTA (Fisher Scientific, E478-500; TE buffer) to a final concentration of 2 to 10 million cells per 1 mL of TE buffer. Cell pellets were disrupted by pipetting. SDS (0.5% final concentration; AmericanBio, AB01920-00500) and proteinase K (0.5 mg/mL final concentration; Invitrogen, 25530049) were added, and the cells were incubated in a 55°C water bath overnight, with a few inversions to promote cell lysis. When digestion was complete (homogeneous, clear solution), NaCl (0.2 mol/L final concentration; Fisher Scientific, BP358-10) was added. Phenol–chloroform/chloroform extraction was then performed using MaXtract High Density tubes (QIAGEN, 129073). The tubes were pre-spun according to the QIAGEN MaXtract High Density tube manual. The samples were mixed with equal parts of phenol:chloroform (Invitrogen, 15593031) in the MaXtract High Density tubes, shaken for 1 minute for extraction, and spun at 1,500g for 5 minutes; the aqueous DNA phase separated on top. This extraction was repeated with chloroform (Fisher Scientific, C298-500), and the aqueous phase was transferred to a 50-mL conical tube. The conical tubes were incubated at 50°C for 1 hour to evaporate residual chloroform. RNase A (25 µg/mL final concentration; QIAGEN, 19101) was added, and the samples were incubated at 37°C overnight. The phenol:chloroform/chloroform extraction was repeated. DNA was precipitated by adding 1/10 v/v 3 mol/L sodium acetate (Sigma-Aldrich, S2889; pH, 5.2) and two volumes 100% ethanol (Pharmco, 111000200CSGL) and incubated at −20°C overnight. The samples were spun for 30 to 45 minutes at 4,500 rpm at 4°C. DNA was washed three times with 1 to 1.5 mL 70% ethanol and allowed to dry open cap at 37°C for 10 to 20 minutes. DNA was resuspended in 1 mL of TE buffer by gently pipetting and incubating overnight at 55°C.
PCR amplification of genomic DNA
Genomic DNA was PCR amplified in three consecutive steps. All PCRs were performed using Q5 Hot Start High-Fidelity DNA Polymerase (New England Biolabs, M0493L) and the Q5 Reaction Buffer Pack (New England Biolabs, B9027S). For PCR #1, 400 µg of DNA was added to the reaction for each condition with 6 µg of DNA per 50 µL reaction. PCR #1 primers amplified sgRNA sequences from the genomic DNA:
LC353F (forward): 5′-AAT GGA CTA TCA TAT GCT TAC CGT- AAC TTG AAA GTA TTT CG-3′
LCR2L (reverse): 5′-TCT ACT ATT CTT TCC CCT GCA CTG- TTG TGG GCG ATG TGC GCT CTG-3′
For PCR #1, the thermocycling parameters were 98°C for 2 minutes, 24 cycles of 98°C for 10 seconds, 65°C for 30 seconds, 72°C for 45 seconds, and 72°C for 10 minutes. Following PCR #1, reactions for each condition were pooled and 10 µL of each reaction was run on agarose gel to confirm a 287-bp product. Isopropanol (Pharmco, 231000099) and 3 mol/L sodium acetate (Sigma-Aldrich, S2889; pH, 5.2) were added to each sample, and the samples were precipitated at −20°C for 1 hour. They were spun at 10,000 rpm for 30 minutes, and the pellets were washed two times with fresh 70% ethanol. The samples were then air-dried for 10 to 15 minutes and resuspended in 50 to 100 µL of TE buffer. The pellets were dissolved at 55°C for 1 hour and mixed by pipetting a few times that hour.
PCRs #2 and #3 were performed to attach Illumina adaptors and barcodes to samples. For PCR #2, 500 ng of DNA per sample of the PCR #1 product was added to each reaction. PCR #2 included primers of variable sequence lengths to increase library complexity:
Forward primers were an equimolar cocktail of the following staggered primers:
KMN_stagger_PCR2_F01: 5′-ACACTCTTTCCCTACACGACGCTCTTCCGATCTTCTTGTGGAAAGGACGAACACCG-3′
KMN_stagger_PCR2_F02: 5′-ACACTCTTTCCCTACACGACGCTCTTCCGATCTcTCTTGTGGAAAGGACGAACACCG-3′
KMN_stagger_PCR2_F03: 5′-ACACTCTTTCCCTACACGACGCTCTTCCGATCTagTCTTGTGGAAAGGACGAACACCG-3′
KMN_stagger_PCR2_F04: 5′-ACACTCTTTCCCTACACGACGCTCTTCCGATCTgagTCTTGTGGAAAGGACGAACACCG-3′
KMN_stagger_PCR2_F05: 5′-ACACTCTTTCCCTACACGACGCTCTTCCGATCTcgagTCTTGTGGAAAGGACGAACACCG-3′
KMN_stagger_PCR2_F06: 5′-ACACTCTTTCCCTACACGACGCTCTTCCGATCTtcgacTCTTGTGGAAAGGACGAACACCG-3′
KMN_stagger_PCR2_F07: 5′-ACACTCTTTCCCTACACGACGCTCTTCCGATCTatcaacTCTTGTGGAAAGGACGAACACCG-3′
KMN_stagger_PCR2_F08: 5′-ACACTCTTTCCCTACACGACGCTCTTCCGATCTgaacgaaTCTTGTGGAAAGGACGAACACCG-3′
Reverse primer (reverse): 5′-GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTTCTACTATTCTTTCCCCTGCACTGT-3′
For PCR #2, the thermocycling parameters were 98°C for 30 seconds; six cycles of 98°C for 10 seconds, 55°C for 30 seconds, and 72°C for 45 seconds; and 72°C for 10 minutes. Following PCR #2, 3 to 5 µL of each reaction was run on agarose gel to confirm a 302-bp product. For PCR #3, 2 µL per sample of the PCR #2 product was added to each reaction. PCR #3 primer sequences were as follows:
KMN_LCV2_PCR3F (forward): 5′-AATGATACGGCGACC-ACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCG-ATCT-3′
P7-indexing primer (reverse, xxxxxxxx denotes the eight-nucleotide barcode): 5′-CAAGCAGAAGACGGCATACGAGATxxxxxxxxGTGACTGGAGTTCAGACGTGT-3′
For PCR #3, the thermocycling parameters were 98°C for 30 seconds; six cycles of 98°C for 10 seconds, 55°C for 30 seconds, and 72°C for 45 seconds; and 72°C for 10 minutes. Following PCR #3, 5 µL of each reaction was run on agarose gel to confirm a 358-bp product. The samples were mixed proportionally, and the mixed sample was run on agarose gel. DNA was isolated using the QIAquick Gel Extraction Kit (QIAGEN, 28704). DNA was sequenced by next-generation sequencing at the Biopolymers Facility at Harvard Medical School (Illumina NextSeq 500) in two separate runs with 400 million plus reads per run and single-indexed reads. Sequencing data were pooled for analysis.
Model-based Analysis of Genome-wide CRISPR/Cas9 Knockout analysis of screen data
Next-generation sequencing data were processed for Model-based Analysis of Genome-wide CRISPR/Cas9 Knockout (MAGeCK) analysis. The reads were trimmed, aligned to the reference genome, and counted. Read count tables were analyzed by MAGeCK (22).
Custom CRISPR/Cas9 minipool screen
SUM159, MDA-MB-468, and BT20 cells were transduced at a multiplicity of infection of ∼0.3 with a CRISPR/Cas9 lentivirus (the Genetic Perturbation Platform, Broad Institute) containing 3,011 sgRNAs, with 10 to 13 sgRNAs per gene and at least 1000-fold representation. This library also contained 50 sgRNAs targeting five essential genes (13 sgRNAs/gene) and 400 negative controls, including 100 no site and 300 intergenic sgRNAs. Cells were selected with 2 µg/mL puromycin for 3 days. After selection, day-0 cell pellets were collected, and the remaining infected cells were expanded for 4 days and then plated into treatment arms in tissue culture–treated 15-cm plates (SUM159: 94,375 cells/mL; MDA-MB-468: 377,500 cells/mL; BT20: 566,250 cells/mL). The cells were treated for 72 hours with DMSO (vehicle) or GR50 doses of the PI3Kα-selective inhibitor BYL719 (SUM159: 2.28 µmol/L; MDA-MB-468: 13.9 µmol/L; BT20: 2 µmol/L) or the catalytic AKT inhibitor GDC-0068 (SUM159: 4.34 µmol/L; MDA-MB-468: 8.49 µmol/L; BT20: 0.9 µmol/L). At the endpoint, cell pellets were harvested and the genomic DNA was extracted using the NucleoSpin Blood L, Midi genomic DNA extraction kit (MACHEREY-NAGEL, 740954.20). Genomic DNA was PCR amplified by the Genetic Perturbation Platform at the Broad Institute using the following primers:
P5 ARGON (forward): 5′-TTGTGGAAAGGACGAAACACCG-3′
P7 BEAKER (reverse): 5′-CCAATTCCCACTCCTTTCAAG-ACCT-3′
The thermocycling parameters were 95°C for 5 minutes; 28 cycles of 95°C for 30 seconds, 53°C for 30 seconds, and 72°C for 20 seconds; and 72°C for 10 minutes. Next-generation sequencing was also performed by the Genetic Perturbation Platform at the Broad Institute (HiSeq 2500/50 cycles) with approximately 150 million reads per sequencing lane. Data analysis was performed using the Broad Institute’s CRISPR Gene Scoring Tool.
Cell density (sulforhodamine B) assays
The cells were seeded in tissue culture–treated 96-well plates in 80 to 100 µL of appropriate growth media (SUM159: 1,000–2,000 cells/well; MDA-MB-468: 4,000 cells/well; BT20: 6,000 cells/well; T47D: 6,000 cells/well; MCF7: 4,000 cells/well; BT474: 6,000 cells/well; HCC70: 6,000 cells/well; MCF10A: 2,000 cells/well; HepG2: 4,000 cells/well; BT549: 4,000 cells/well; HCC1937: 4,000 cells/well; ZR-75-1: 4,000 cells/well; MDA-MB-361: 7,000 cells/well; parental T47D: 6,000 cells/well; fulvestrant-resistant T47D clones 1 to 3: 6,000 cells/well; PC3: 3,000 cells/well; LNCaP: 5,000 cells/well). The cells were incubated for 24 hours, and the cell density was assayed using sulforhodamine B (SRB; Sigma-Aldrich, 230162) staining, as previously described (23), to determine the number of cells at the start of the experiment (day 0). The cells were then treated with 5 to 20 µL of drug for indicated periods of time to bring the final volume in each well to 100 µL. At the endpoint, the relative cell density was assayed using SRB staining. The cell density at each time point was normalized to the day-0 control. For dose curve and double-dose curve experiments, these values were normalized from 0 to 100 using GraphPad Prism, wherein an empty well (background) served as the 0% reference and untreated cells served as the 100% reference. Normalized cell densities were plotted versus log10 drug concentration, and a nonlinear curve was fit using the log(inhibitor) versus normalized response–variable slope function in GraphPad Prism. IC50 values were calculated by GraphPad Prism on the basis of the nonlinear curve fit. The cell density data for experiments that were not dose curves or double-dose curves were transformed to log2(Y) values using GraphPad Prism and plotted such that values less than 0 indicated the loss of cell density or cell death.
GR50 calculator
GR50 values were calculated for the CRISPR/Cas9 minipool screen using the online GR calculator (http://www.grcalculator.org/grcalculator/). Cells were seeded, and the cell density was measured as described for the cell density assays. Cell densities at day 0 (24 hours after seeding) and 72 hours after drug treatment were used to calculate the GR50 values.
Synergy calculations
For proliferation assays with two inhibitors, synergy scores were calculated using the synergyfinder R package (24). Highest single-agent synergy scores were reported for each drug dose combination tested and displayed as a heatmap.
Incucyte cell death assays
Cells were seeded in black-walled, tissue culture–treated 96-well plates as described for the cell density assays. The media were changed 24 hours later to 90 µL of growth media containing 1:1,000 Incucyte Nuclight Rapid Red Dye (Sartorius, 4717) and 1:1,000 NucView 488 caspase-3/7 substrate (Biotium, 30029). The cells were treated with 10 µL of drug-containing media. Plates were then placed in the Incucyte instrument, and images were taken every 2 hours for 72 hours. The Incucyte software was used to train a model to count cells as those expressing the nuclear dye, Incucyte Nuclight Rapid Red Dye. A model was also trained to count green cells or those with cleaved caspase-3/7 signal. Four images were taken per well and averaged, and quadruplicate wells were assayed for each condition. The percentage of cell death was measured as the number of cells with an overlapping signal (red and green) divided by the number of red cells.
Mouse studies
All animal experiments were performed at the Beth Israel Deaconess Medical Center and approved by and in accordance with the guidelines of the Beth Israel Deaconess Medical Center Institutional Animal Care and Use Committee. HCC70 cells were maintained in RPMI 1640 supplemented with 10% FBS, and the cells had tested negative for Mycoplasma before injection. On the day of injection, HCC70 cells were washed two times with 1× PBS, trypsinized, and counted. A total of 5 × 106 cells per mouse were resuspended in 100 µL of serum-free RPMI 1640 and placed on wet ice. They were mixed with Matrigel (Corning, 356230) in a 1:1 (v/v) ratio and injected subcutaneously into the flanks of 50 NSG mice.
Tumors were allowed to grow for 18 days before switching mice from standard chow to a low geranylgeraniol diet for 3 days (Research Diets, D22092101l—modified open standard diet with 15 kcal % fat canola oil). Then, 10 to 14 mice were assigned to each treatment group (vehicle, AZD5363, pitavastatin, and combination), and treatments were administered daily by oral gavage. AZD5363 (AstraZeneca) was prepared as a suspension in 0.5% carboxymethylcellulose and was dosed daily for 4 days of the week at 100 mg/kg, followed by a 3-day AKT inhibitor holiday. Pitavastatin (Selleckchem, S1759) was prepared by diluting 100 mmol/L pitavastatin in DMSO in 0.5% carboxymethylcellulose and was dosed daily at 100 mg/kg. Tumors were measured with calipers two times per week (length and width) for 35 days (24 days after the start of treatments). After 24 days of treatment, all mice were euthanized. Four mice in each group were treated with AZD5363 (2 hours) or pitavastatin (6 hours) prior to euthanasia and sections of tumor and liver were snap-frozen and fixed in 10% formalin for immunoblotting and immunohistochemistry, respectively.
Tissues were homogenized in cold RIPA lysis buffer [150 mmol/L Tris-HCl, 150 mmol/L NaCl, 0.5% (w/v) sodium deoxycholate (Sigma-Aldrich, D6750), 1% (v/v) NP-40 (Sigma-Aldrich, I3021); pH, 7.5] containing 0.2% SDS, 2 mmol/L sodium pyrophosphate, 10 mmol/L sodium fluoride, 0.5% (v/v) protease inhibitor cocktail, 100 nmol/L calyculin A, 4 mmol/L sodium orthovanadate (Sigma-Aldrich, S6508), and 2× Halt protease and phosphatase inhibitor cocktail (Fisher Scientific, PI78443), added just before use. Tissue homogenization was facilitated by using gentleMACS Dissociator (Miltenyi Biotec, 130-093-235). Samples were then centrifuged at 4,000g for 5 minutes at 4°C. The supernatants were transferred to new tubes, and samples were spun at maximum speed for 15 minutes at 4°C. The supernatants were again transferred to new tubes, and this process of centrifugation and supernatant collection was repeated. Once the supernatants were no longer cloudy, protein concentrations were measured, and immunoblotting was performed as described in the Immunoblotting section.
Patient-derived organoid cultures
For characteristics of patient-derived organoid cultures (PDO), see Supplementary Table S1. Propagation and culturing of PDOs were performed as previously described (25). Briefly, PDOs were incubated in 1× Dispase-II solution with 2 mg/mL collagenase for 30 to 45 minutes at 37°C and mechanically disrupted by passing through a 26G needle. PDOs were washed once with Advanced DMEM/F12 supplemented with 5% FBS and pelleted by centrifugation at 400g for 5 minutes. PDO fragments were embedded in Cultrex Reduced Growth Factor Basement Membrane Extract type 2 (Trevigen; R&D Systems, 3533-001-02), 50 µL drops were plated into a 24-well plate, and 500 µL PDO medium was added 30 minutes later (25). To assess drug sensitivity, 200 to 600 PDO fragments were plated into eight-well chamber slides, and 1 µmol/L AZD5363 (Cayman Chemical, 15406) and/or 5 µmol/L pitavastatin (Selleckchem, S1759) was added the following day. After 96 hours of drug treatment, PDOs were pulsed with 10 µmol/L 5-ethynyl-2′-deoxyuridine (EdU) for 4 hours and fixed with 4% paraformaldehyde for 30 minutes. CellProfiler software was used to measure PDO area for 10 to 20 PDOs at the endpoint.
To assess cell proliferation and apoptosis, fixed PDOs were permeabilized with wash buffer (0.3% Triton X-100 in PBS) for 20 minutes. EdU labeling was performed for 40 minutes using the Click-iT EdU Cell Proliferation Kit for Imaging (Invitrogen, C10337) according to the manufacturer’s description. PDOs were washed three times with wash buffer, blocked for 1 hour with blocking buffer (5% goat serum, 0.2% BSA, 0.3% Triton X-100 in PBS), and incubated overnight with anti-cleaved caspase-3 (Cell Signaling Technology, 9661) in blocking buffer at 4°C. The following day, the PDOs were washed extensively with wash buffer, incubated with secondary antibody (Alexa Fluor 488) for 2 hours at room temperature, washed with wash buffer, and mounted using VECTASHIELD Antifade Mounting Medium with DAPI (VectorLabs, H-1200-10). PDOs were imaged with Zeiss LSM 880 confocal microscope. To assess proliferation, 10 to 20 PDOs per treatment condition were imaged and the ratio of EdU-positive cells per total number of cells was quantified. To assess apoptosis, 10 to 20 PDOs per treatment condition were scored on the basis of the presence of cleaved caspase-3 staining.
To assess signaling changes in response to drug treatments, PDOs were incubated with 1 µmol/L AZD5363 (Cayman Chemicals, 15406) and/or 5 µmol/L pitavastatin (Selleckchem, S1759) for 24 hours. PDOs were pooled from six wells and incubated for 1.5 hours on ice in Cell Recovery Buffer supplemented with phosphatase inhibitors (Roche, 4906845001). PDOs were lysed using RIPA buffer (Boston BioProducts, BP-115) containing protease and phosphatase inhibitors (Thermo Fisher Scientific, A32959) and then centrifuged at 13,000 rpm for 10 minutes at 4°C. Protein concentrations were measured using the Pierce BCA Protein Assay Kit. Sample concentrations were normalized using 6× SDS sample buffer [62.5 mmol/L Tris, pH, 6.8; 5% SDS; 18% glycerol (Fisher Scientific G33-500); bromophenol blue (Fisher Scientific, BP115-25); 302 mmol/L dithiothreitol (Fisher Scientific, BP172-25); and RIPA lysis buffer]. Cell lysates were boiled at 95°C for 5 minutes and stored at −20°C. For detection of HMGCR, cell lysates were not boiled or frozen but processed and run on the same day to avoid temperature changes that cause endoplasmic reticulum–associated protein aggregation. Immunoblotting was performed as described in Immunoblotting.
Immunoblotting
Cells were washed once in cold 1× PBS (Boston BioProducts, BM-220) and collected on wet ice in 4°C RIPA lysis buffer [150 mmol/L Tris-HCl, 150 mmol/L NaCl, 0.5% (w/v) sodium deoxycholate (Sigma-Aldrich, D6750), and 1% (v/v) NP-40 (Sigma-Aldrich, I3021); pH, 7.5] containing 0.1% SDS (AmericanBio, AB01920-00500), 1 mmol/L sodium pyrophosphate (Sigma-Aldrich, S390-500), 20 mmol/L sodium fluoride (Fisher Scientific, S25547), 0.5% (v/v) protease inhibitor cocktail (104 mmol/L 4-(2-aminoethyl)benzenesulfonyl fluoride hydrochloride (AEBSF), 80 µmol/L aprotinin, 4 mmol/L bestatin, 1.4 mmol/L E-64, 2 mmol/L leupeptin, and 1.5 mmol/L pepstatin A; Sigma-Aldrich, P8340), and 50 nmol/L calyculin A (LC Laboratories, C-3987), added just before use. Plates were scraped into 1.5-mL microcentrifuge tubes, vortexed, and incubated on wet ice for 15 minutes. The samples were then centrifuged at 14,000 rpm for 10 minutes at 4°C. The supernatants were transferred to new tubes, and protein concentrations were measured by the Bio-Rad DC protein assay (Bio-Rad, Reagent A: 5000113, Reagent B: 5000114). Sample concentrations were normalized using 2× SDS sample buffer [62.5 mmol/L Tris (pH 6.8), 2% SDS, 10% glycerol (Fisher Scientific G33-500), bromophenol blue (Fisher Scientific, BP115-25), and 5% (v/v) β-mercaptoethanol (Sigma-Aldrich, M3148)]. Cell lysates were boiled at 95°C for 5 minutes and stored at −20°C. For detection of HMGCR, cell lysates were not boiled or frozen but processed and run on the same day to avoid temperature changes that cause endoplasmic reticulum–associated protein aggregation. Cell lysates were run by SDS-PAGE on 7.6% to 15% acrylamide gels in 1× running buffer (Boston BioProducts, BP-150). Proteins were transferred to nitrocellulose membranes at 100 V for 90 minutes in cold 1× transfer buffer (Cell Signaling Technology, 12539S). Membranes were blocked with 5% (w/v) bovine serum albumin (BSA; Gold Biotechnology, A-420-100) in tris-buffered saline (TBS; Boston BioProducts, BM-301), rocking for at least 1 hour at room temperature. Membranes were washed briefly in TBS-tween (TBST; Boston Bioproducts, IBB-181X), incubated in primary antibodies diluted in 5% (w/v) BSA in TBST with 0.01% (w/v) sodium azide (Fisher Scientific, S227I-25), and rocked at 4°C overnight. Membranes were washed three times for 10 minutes in TBST, rocking at room temperature. Then, membranes were incubated for 1 hour, rocking at room temperature, with fluorophore-conjugated secondary antibodies (LI-COR Biosciences). Membranes were washed two times for 10 minutes each in TBST and one time for 10 minutes in TBS and imaged with the LI-COR Odyssey CLx or Odyssey M Imaging System (LI-COR Biosciences).
Sterol and nonsterol rescue experiments
Cells were seeded as described for cell proliferation assays. Cells were incubated for 24 hours, and cell density was assayed using SRB staining to determine the number of cells at the start of the experiment (day 0). Cells were then treated with 5 µL of drug and 5 µL of vehicle (7:3 MeOH:NH4OH) or sterol and nonsterol intermediates for 72 hours to bring the final volume in each well to 100 µL: 1 mmol/L mevalonate (Sigma-Aldrich, 90469), 5 µg/mL cholesterol (Sigma-Aldrich, C4951) or 5 µg/mL geranylgeranyl pyrophosphate (GGPP; Cayman Chemicals, 63330). At the endpoint, relative cell density was assayed using SRB staining. Cell density at 72 hours was normalized to the day-0 control. Normalized cell densities were log2-transformed and plotted in GraphPad PRISM such that values greater than 0 indicate increased cell density, while values less than 0 indicate decreased cell density at 72 hours compared with the day-0 control.
RNA sequencing
Cells were plated at 250,000 to 500,000 cells/mL in RPMI 1640 medium supplemented with 10% FBS at 2 mL per well in tissue culture–treated six-well plates to achieve 75% density by the endpoint. The next day, stocks of AZD5363 (Cayman Chemicals, 15406) and pitavastatin (Selleckchem, S1759) were prepared in DMSO, and cells were treated for 24 or 48 hours. A master mix of each drug stock was prepared and used to treat all conditions across biological replicates and was stored at −80°C between treatments. After 24 or 48 hours of treatment, each well was washed once with 1 mL of cold 1× PBS and aspirated completely. Plates were snap-frozen on dry ice and stored at −80°C until all biological replicates were collected. Four biological replicates were seeded on sequential days at the same time of day. Samples were collected for protein harvest in parallel to confirm expected drug effects on cell signaling. Snap-frozen plates were thawed on ice, and RNA was extracted using the NucleoSpin RNA Plus kit (Takara, 740984.50). RNA quantity and purity were assessed by NanoDrop 1000. Samples were submitted to Novogene for integrity assessment (Agilent 2100 analysis), mRNA library preparation (unstranded), and paired-end (150-bp) sequencing on a NovaSeq 6000, S4 flow cell. Data were analyzed as described previously (26).
Database for Annotation, Visualization and Integrated Discovery gene ontology analysis
Genes that were uniquely up- or downregulated with AZD5363, pitavastatin or combination treatment in MDA-MB-468 or T47D cells in the RNA sequencing (RNA-seq) data were analyzed using the Database for Annotation, Visualization and Integrated Discovery (DAVID) 2021 Bioinformatics Resource functional annotation tool. Differentially regulated genes were uploaded as lists with the background genome set to all genes detected in the RNA-seq analysis of that cell line, and biological processes were plotted by −log10 (P value), where the P value is an EASE score, a modified Fisher exact P value for gene enrichment analysis (27, 28).
siRNA transfections
Cells were seeded in 96-well plates in 90 µL of appropriate growth media (SUM159: 1,000–2,000 cells/well; MDA-MB-468: 4,000 cells/well; BT20: 6,000 cells/well; T47D: 6,000 cells/well; MCF7: 4,000 cells/well; BT474: 6,000 cells/well). Cells were incubated for 24 hours before transfection. They were transfected with Lipofectamine RNAiMAX Transfection Reagent (Thermo Fisher Scientific, 13778150) as follows. All reagents were brought to room temperature before proceeding with the transfection. Lipofectamine RNAiMAX was diluted in Opti-MEM reduced-serum media (Thermo Fisher Scientific, 31985070) with 1.5 µL of Lipofectamine RNAiMAX for every 25 µL of Opti-MEM media, vortexed, and incubated at room temperature for 5 minutes. siRNAs were diluted to 500 nmol/L in Opti-MEM media, vortexed, and incubated at room temperature for 5 minutes. Equal volumes of Lipofectamine RNAiMAX in Opti-MEM media and siRNA in Opti-MEM media were combined, vortexed, and incubated for 20 minutes at room temperature. For a lipofectamine-only control, equal volumes of Lipofectamine RNAiMAX in Opti-MEM and Opti-MEM media were combined, vortexed, and incubated at room temperature for 20 minutes. To bring the final volume in each well to 100 µL and the final concentration of siRNA to 25 nmol/L, 10 µL of the Lipofectamine RNAiMAX, siRNA, and Opti-MEM mixtures were then added to appropriate wells. After 24 hours, the media were changed to 90 µL of fresh media using gentle pipetting to avoid disrupting cells, and the cell density of lipofectamine-only control cells was assayed using SRB staining to determine the number of cells at the start of drug treatment (day 0). Cells were then treated with 10 µL of the drug for 72 hours to bring the final volume in each well to 100 µL. Cell density at 72 hours was normalized to the cell density at the start of drug treatment (day 0). Normalized cell densities were log2 transformed and plotted in GraphPad PRISM such that values greater than 0 indicate increased cell density, whereas values less than 0 indicate decreased cell density at 72 hours compared with the day-0 control.
qRT-PCR
RNA was isolated from cells with the NucleoSpin RNA Plus kit (MACHEREY-NAGEL, 740984) according to the manufacturer’s protocol. Reverse transcription was performed using the TaqMan Reverse Transcription kit (Thermo Fisher Scientific, N8080234). cDNA was detected using PowerUp SYBR Green Master Mix for qPCR (Thermo Fisher Scientific, A25776). A master mix of 250 µmol/L forward primer, 250 µmol/L reverse primer, 1× SYBR Green Master Mix, and nuclease-free water up to 10 µL per reaction was prepared. Then, 10 µL of master mix was added to each well of a 384-well plate, and 2 µL of 2.5 ng/µL cDNA was added to each well for 5 ng of cDNA per reaction. The plate was spun briefly at 1,000 rpm. qRT-PCR was performed using a CFX384 Touch Real-Time PCR Detection System (Bio-Rad). The thermocycling parameters were 50°C for 2 minutes; 95°C for 2 minutes; 40 cycles of 95°C for 15 seconds and 60°C for 1 minute; 65°C for 5 seconds; and 95°C for 5 seconds. qRT-PCR was performed in technical triplicate, and quantification of mRNA expression was calculated by the ΔΔCT method with 18S ribosomal RNA as the reference gene. For qRT-PCR primers, which were designed using the NCBI Primer-BLAST tool, see Supplementary Table S2. Primer efficiency was confirmed to be within 90% to 110% for each primer pair in each cell line.
Plasmids
For detailed plasmid information, see Supplementary Table S3. pHAGE-ESR1 was purchased from Addgene (116737). To generate pLenti6/V5-HMGCR, HMGCR was PCR amplified out of pCMV-SPORT6-hHMGCR1 (Addgene, 86085) using the forward primer 5′-GTATACTGGATCCGCCGCCACCATGTTGTCAAGACTT-3′ and reverse primer 5′-TCGGAGCTCGAGGTGGCTGTCTTCTTGGT-3′. PCR-amplified HMGCR was cloned into pLenti6/V5-p53_wt p53 (gift from the Muranen lab) by double restriction enzyme digestion with BamHI (New England Biolabs, R0136) and XhoI (New England Biolabs, R0146) at 37°C for 1 hour, followed by ligation with T4 DNA Ligase (New England Biolabs, M0202) by following the manufacturer’s protocol. To generate mutations in the catalytic residues of HMGCR, four sequential site-directed mutagenesis reactions were performed following the QuikChange II XL Site-Directed Mutagenesis Kit protocol (Agilent, 200521). The following primers were generated using Agilent’s QuikChange Primer Design tool: H866A: 5′-CAGGACATCTTGTCAAAAGTGCCATGATTCACAACAGGTCGA-3′ (forward), 5′-TCGACCTGTTGTGAATCATGGCACTTTTGACAAGATGTCCTG-3′ (reverse), E559A: 5′-CCAATGGCAACAACAGCAGGTTGTCTTGTGGCC-3′ (forward), 5′-GGCCACAAGACAACCTGCTGTTGTTGCCATTGG-3′ (reverse), K691A: 5′-GTTAGTGGTAACTATTGTACTGACGCGAAACCTGCTGCTATAAATTGGAT-3′ (forward), 5′-ATCCAATTTATAGCAGCAGGTTTCGCGTCAGTACAATAGTTACCACTAAC-3′ (reverse), D767A: 5′-CATTGCCTGTGGACAGGCTGCAGCACAGAATGTTG-3′ (forward), 5′-CAACATTCTGTGCTGCAGCCTGTCCACAGGCAATG-3′ (reverse). To generate pLenti6-ER-mRFP, mRFP with a C-terminal KDEL sequence (ER-mRFP) was PCR amplified out of ER-mRFP (Addgene, 62236) using the forward primer 5′-GGTCGGATCCGCCGCCACCATGGACAGCAAAGG-3′ and the reverse primer 5′-GAGGCACCGGTGTTTAGAGCTCATCTTT-3′. PCR-amplified ER-mRFP was cloned into pLenti6/V5-p53_wt p53 (gift from the Muranen lab) by double restriction enzyme digestion with BamHI-HF (New England Biolabs, R0136) and AgeI-HF (New England Biolabs, R3552) at 37°C for 1 hour, followed by ligation with T4 DNA ligase (New England Biolabs, M0202) by following the manufacturer’s protocol.
Transfection and lentiviral transduction of plasmid DNA
HEK293T cells were transfected with pHAGE-ESR1, pLenti6/V5-HMGCR, or pLenti6-ER-mRFP plasmid DNA as follows. A master mix of 11.1 µg psPAX2 (Gal/Pol), 0.6 µg VSV G, 6.3 µg of plasmid DNA, and 1,200 µL of serum-free, antibiotic-free DMEM was prepared. Then, 54 µL of 1 mg/mL polyethylenamine (Sigma-Aldrich, 408727) was added, and the master mix was vortexed for 10 seconds to mix and then incubated at room temperature for 15 minutes. Tissue culture–treated 10-cm dishes were coated with poly-L-lysine (PLL; Sigma-Aldrich, P1274) for 5 minutes and then washed three times with 1× PBS. HEK293T cells were trypsinized and counted, and 9 × 106 cells were resuspended in 7.8 mL of DMEM supplemented with 10% FBS. Then, approximately 1,200 µL of reaction mixture was added to the HEK293T cell suspension, mixed, and plated on the PLL-coated 10-cm plates. GlutaMAX Supplement (Thermo Fisher Scientific, 35050061) was added to a final concentration of 1×. The cells were incubated for 48 hours. After 24 hours since transfection of HEK293T cells, the target cell lines were seeded in six-well plates at 62,500 to 250,000 cells/mL in complete media (RPMI 1640 supplemented with 10% FBS). After 48 hours since transfection of HEK293T cells, virus was harvested. Media from the HEK293T cells were passed through a 10-mL syringe with a 0.45-µm filter attached. The media were aspirated from the target cells, and virus was added to each well (pHAGE-ESR1: 125 µL). For pLenti6/V5-HMGCR and pLenti6-ER-mRFP, virus was concentrated following the PEG-it Virus Precipitation protocol (LV810A-1). Viral particles were resuspended in 80 µL of PBS, and 30 µL (pLenti6/V5-HMGCR) or 10 µL (pLenti6-ER-mRFP) was added to each well. The volume in each well was brought up to 1 mL with RPMI 1640 supplemented with 10% FBS. Polybrene (Sigma-Aldrich, 107689) was added at a final concentration of 5 µg/mL, and cells were incubated at 37°C for 4 hours. After 4 hours, 3 mL of media was added to each well, and cells were incubated at 37°C overnight. The next day, media were changed on the transduced cells to antibiotic-containing media (1 µg/mL puromycin, 6 µg/mL blasticidin). Cells were maintained and expanded in antibiotic-containing media.
Endoplasmic reticulum cholesterol depletion
Cells cultured in RPMI 1640 supplemented with 10% FBS were trypsinized and seeded into 90 µL of RPMI 1640 supplemented with 10% lipid-depleted FBS (Biowest, S148L) in 96-well plates. Cells were incubated for 24 hours and then treated with 10 µL of DMSO or a high dose (10 µmol/L) of pitavastatin for 1 hour. After 1 hour, cell density was assayed using SRB staining to determine the number of cells at the start of the experiment (day 0). The media were changed to 100 µL of fresh RPMI 1640 supplemented with either 10% lipid-depleted FBS or 10% FBS (complete serum) containing DMSO or a low dose (2 µmol/L) of pitavastatin for 72 hours. At the endpoint, relative cell density was assayed using SRB staining. Cell density at 72 hours was normalized to the day-0 control. These values were normalized from 0 to 100 using GraphPad PRISM, where an empty well (background) served as the 0% reference, and untreated cells within the ±10 µmol/L pitavastatin pretreatment groups served as the 100% reference.
Immunofluorescence
Black-walled, glass bottom, tissue culture–treated plates were coated with PLL (Sigma-Aldrich, P1274) for 5 minutes and then washed three times with 1× PBS. Cells were seeded at 100,000 cells/well in 80 µL of RPMI 1640 supplemented with 10% FBS. Cells were incubated for 24 hours and then treated with 10 µL of DMSO or 1 µmol/L U18666A (Cayman Chemicals, Cholesterol Cell-Based Detection Assay Kit, 10009779) and 10 µL of DMSO or 2 µmol/L pitavastatin (Selleckchem, S1759) for 24 hours. Cells were then fixed with 4% formaldehyde solution (MilliporeSigma, 1004965000) for 1 hour at room temperature and then washed three times with 1× PBS. Cells were then stained with Filipin III for 1 hour, rocking at room temperature in the dark, as indicated by the Cholesterol Cell-Based Detection Assay Kit protocol (Cayman Chemicals, 10009779). After Filipin III staining, the cells were maintained in the dark for the remaining steps. Cells were washed three times with 1× PBS and then blocked with 0.5% BSA (Gold Biotechnology, A-420-100) in 1× PBS for 1 hour, rocking at room temperature. After blocking, the cells were incubated with primary antibody for 1 hour, rocking at room temperature (LAMP1; Cell Signaling Technology, 9091, 1:100). Cells were washed three times with 1× PBS and then incubated with secondary antibody for 30 minutes, rocking at room temperature (goat anti-rabbit IgG Alexa Fluor 488; Thermo Fisher Scientific, A-11008). Cells were then washed two times with 1× PBS and imaged on the Keyence BZ-X800 microscope.
Data availability
The source data and annotated analysis workflows are available on the following Open Science Framework (OSF) project website: https://osf.io/6pw9d/. RNA-seq data have been deposited with Gene Expression Omnibus (GEO) under series accession number GSE252944. A complete list of reagents is provided in Supplementary Tables S2–S6.
Results
Genome-wide CRISPR/Cas9 screen identifies synergy with combined AKT inhibition and cholesterol homeostasis genes
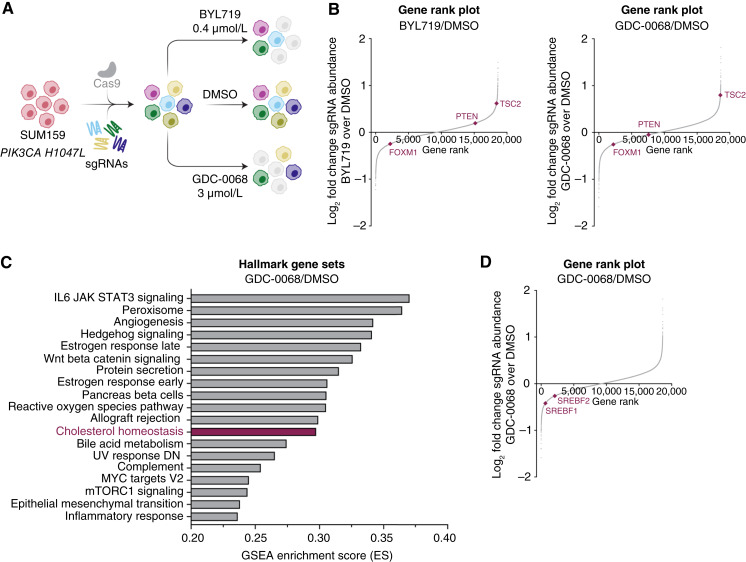
To identify synthetic lethal combinations with PI3K/AKT inhibitors in TNBC, we performed a genome-wide CRISPR/Cas9 screen in PIK3CA-mutant (H1047L) TNBC cells (SUM159). Cells were treated for 72 hours with DMSO (vehicle) or cytostatic doses of the PI3Kα-selective inhibitor BYL719 (0.4 µmol/L) or the catalytic AKT inhibitor GDC-0068 (3 µmol/L; Fig. 1A). As expected, PTEN knockout provided a growth advantage to cells treated with BYL719 but not to cells treated with GDC-0068. TSC2 knockout conferred a growth advantage to cells treated with either BYL719 or GDC-0068. Conversely, FOXM1 knockout synergized to impair cell growth in both drug arms (Fig. 1B). Gene set enrichment analysis identified cholesterol homeostasis as one of the top perturbed pathways in the AKT inhibitor arm of the screen (Fig. 1C). We performed a second CRISPR/Cas9 screen utilizing a minipool library targeting 197 hits from the top perturbed pathways in the AKT inhibitor arm of the genome-wide screen. We identified synergy with AKT inhibition and knockout of cholesterol homeostasis genes across three TNBC cell lines (Supplementary Fig. S1A–S1C). Specifically, knockout of the master lipogenic transcriptional regulators, SREBF1 and SREBF2, synergized with GDC-0068, suggesting that AKT inhibition sensitizes TNBC cells to inhibition of cholesterol biosynthesis (Fig. 1D).
Figure 1.
Genome-wide CRISPR/Cas9 screen identifies synergy with combined AKT inhibition and cholesterol homeostasis gene knockout in TNBC cells. A, Schematic of CRISPR/Cas9 screen. SUM159 cells were transduced with a Cas9-expressing lentivirus containing 94,495 sgRNAs with 3 to 4 sgRNAs per gene. Infected cells were allowed to grow for approximately 1 week before seeding the treatment arms. Cells were treated with DMSO, BYL719 (PI3Kα-selective inhibitor, 0.4 µmol/L), or GDC-0068 (catalytic AKT inhibitor, 3 µmol/L) for 72 hours (N = 3 technical replicates for each treatment arm). B, Rank plots showing the log2-fold change of each gene plotted against the dropout gene rank for the BYL719 and GDC-0068 treatments arms compared with the DMSO arm. Expected changes in PI3K/AKT signaling genes are highlighted, including TSC2, PTEN, and FOXM1. Plots were generated using MAGeCK with a read count cutoff of 50 (N = 3 technical replicates). C, Plot of the top pathways selectively perturbed in the GDC-0068 arm of the CRISPR/Cas9 screen. Analysis was performed via gene set enrichment analysis. D, Rank plot showing the log2-fold change of each gene plotted against the dropout gene rank for the GDC-0068 treatment arm of the CRISPR/Cas9 screen compared with the DMSO arm. The transcription factors SREBF1 and SREBF2 are highlighted. The plot was generated using MAGeCK with a read count cutoff of 50.
Disruption of cholesterol homeostasis synergizes with AKT inhibition in TNBC
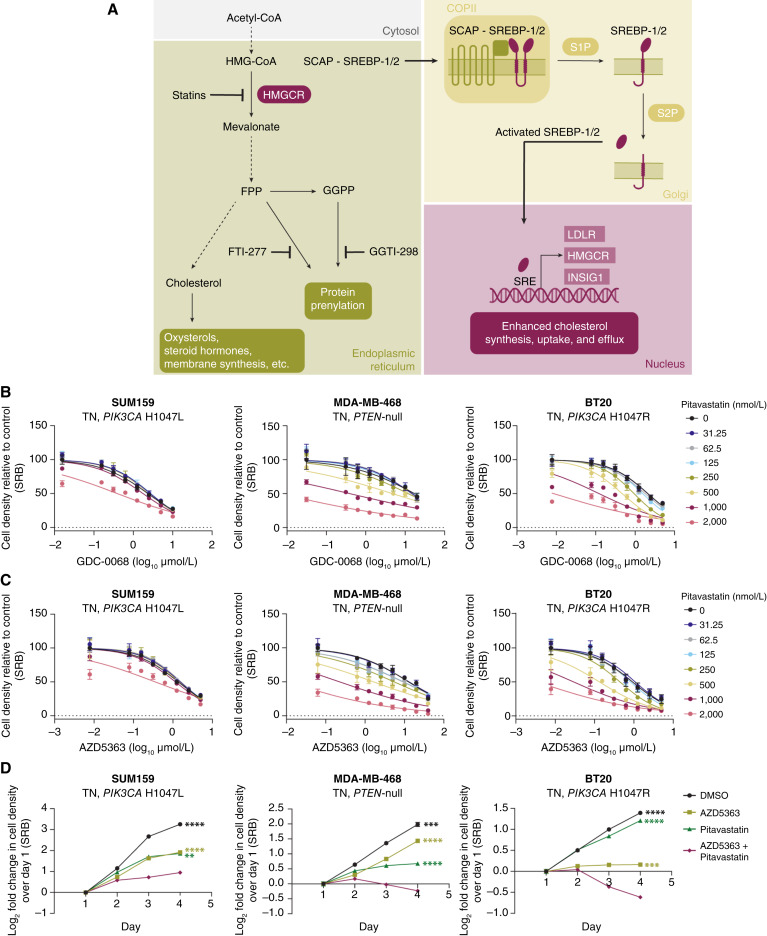
Statins are a family of drugs that inhibit HMGCR, the first rate-limiting enzyme in cholesterol biosynthesis (Fig. 2A; ref. 20). In a panel of TNBC cell lines, the lipophilic statins pitavastatin and lovastatin and the hydrophilic statin rosuvastatin synergized with AKT inhibition (Fig. 2B and C; Supplementary Fig. S2A–S2D). Highest single-agent (HSA) synergy score analysis for the combination of AKT inhibitors (GDC-0068, AZD5363) and statins showed significant synergy (Supplementary Figs. S2B and S2D, S3A and S3B). Pitavastatin was more potent than lovastatin or rosuvastatin, synergizing with AKT inhibitors at 500 to 2,000 nmol/L concentrations, but all three statins showed similar degrees of synergy (Supplementary Figs. S2B and S2D, S3A and S3B). In a 4-day growth curve, the combination of AZD5363 and pitavastatin outperformed DMSO or single agent at impairing proliferation and inducing cell death (Fig. 2D). Nontumorigenic mammary epithelial cells (MCF10A) and liver adenocarcinoma cells (HepG2) were less sensitive to the combination of AKT inhibitor and pitavastatin than TNBC cells, suggesting that this combination is not toxic to all cells (Supplementary Fig. S3C and S3D).
Figure 2.
Disruption of cholesterol homeostasis synergizes with AKT inhibition in TNBC cells. A, Cholesterol is synthesized in multiple steps from acetyl-CoA. HMGCR catalyzes the first rate-limiting step of cholesterol biosynthesis. This pathway also generates the prenylation substrates FPP and GGPP. SREBP-1/2 sense low endoplasmic reticulum cholesterol levels and translocate from the endoplasmic reticulum to the Golgi where they are cleaved and activated. N-terminal active SREBP-1/2 enter the nucleus to regulate the transcription of target genes. Drugs targeting this pathway are highlighted, including HMGCR inhibitors (statins) and inhibitors of protein farnesylation (FTI-277) and geranylgeranylation (GGTI-298). B and C, TNBC cell lines (SUM159, MDA-MB-468, and BT20) were treated with increasing doses of GDC-0068 (SUM159, 0–10 µmol/L; MDA-MB-468, 0–20 µmol/L; and BT20, 0–5 µmol/L; B) or AZD5363 (SUM159, 0–5 µmol/L; MDA-MB-468, 0–40 µmol/L; BT20, 0–5 µmol/L; C) and pitavastatin (0–2,000 nmol/L) for 72 hours, and cell density was measured by SRB assay. Data are represented as mean ± SD (N = 3 technical replicates). D, TNBC cell lines (SUM159, MDA-MB-468, and BT20) were treated with DMSO, AZD5363 (SUM159, 2.5 µmol/L; MDA-MB-468, 10 µmol/L; BT20, 1.25 µmol/L), pitavastatin (SUM159, 4 µmol/L; MDA-MB-468, 2 µmol/L; BT20, 0.5 µmol/L), or a combination of AZD5363 and pitavastatin for 72 hours, and cell density was measured daily by SRB assay. Data are represented as mean ± SD (N = 3 technical replicates). Statistical analysis was performed using two-way ANOVA with Dunnett multiple comparison test. *, significant differences compared with the AZD5363 and pitavastatin combination treatment on day 4. **, P = 0.0021; ***, P = 0.0002; ****, P < 0.0001.
We hypothesized that statins would also synergize with PI3K inhibitors, since PI3K acts upstream of AKT, and as expected, the knockout of SREBF1 and SREBF2 synergized with BYL719 in the genome-wide CRISPR/Cas9 screen (Supplementary Fig. S4A). In a panel of TNBC cell lines, BYL719 synergized with pitavastatin (Supplementary Fig. S4B and S4C). Furthermore, Torin1–mediated inhibition of mTORC1/2, which regulates SREBP-1/2 activation downstream of PI3K/AKT, synergized with pitavastatin, suggesting that synergy between PI3K/AKT inhibitors and statins is mTORC1/2-dependent (Supplementary Fig. S4D and S4E). Upstream of PI3K/AKT/mTORC1, epidermal growth factor receptor inhibition with erlotinib also synergized with pitavastatin in a panel of TNBC cell lines (Supplementary Fig. S5A and S5B). Together, these data show that PI3K/AKT pathway inhibition synergizes with inhibition of cholesterol biosynthesis in TNBC.
AKT inhibition synergizes with pitavastatin to induce TNBC cytotoxicity in vitro in xenografts and in PDOs
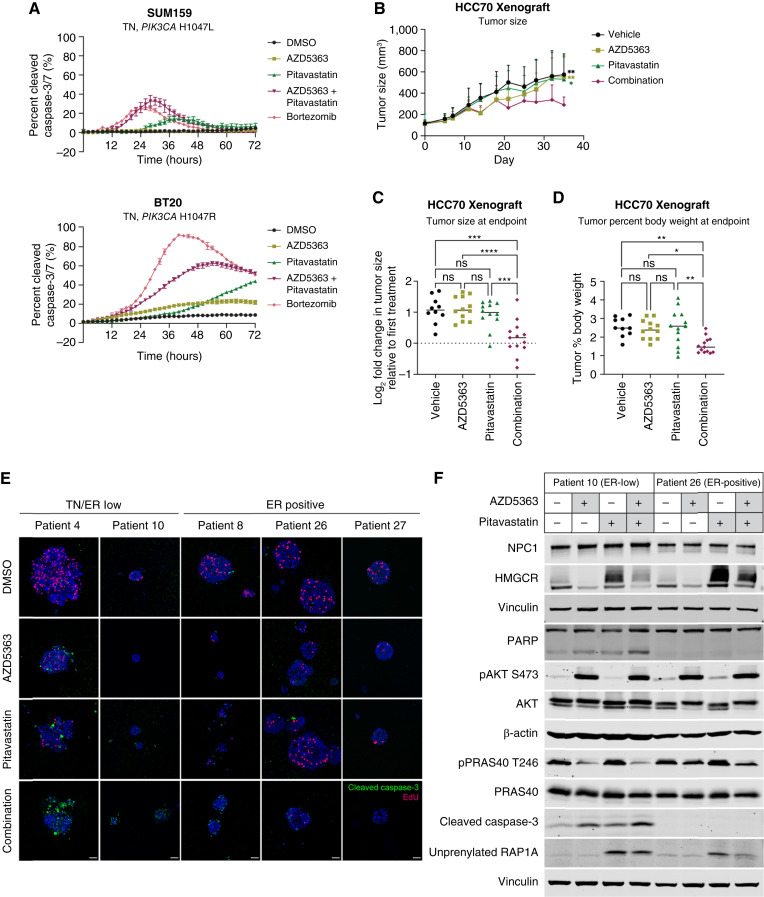
Combination AZD5363 and pitavastatin treatment for 72 hours induced significant TNBC cell death compared with either single agent alone (Fig. 3A). To evaluate the efficacy of this combination in preclinical models, we used HCC70 TNBC xenografts. In vitro, AKT inhibitors synergized with pitavastatin in HCC70 cells (Supplementary Fig. S6A). HCC70 tumor–bearing mice were maintained on a low geranylgeraniol chow diet to limit the rescue of GGPP levels after pitavastatin treatment (29, 30). In a pilot experiment, HCC70 tumor growth was impaired by pitavastatin treatment in mice that were maintained on a low geranylgeraniol chow diet, but not in mice on the standard chow diet (Supplementary Fig. S6B). Mice were treated with 100 mg/kg AZD5363 (4 days on, 3 days off) and 100 mg/kg pitavastatin (daily) by oral gavage. Single agent AZD5363 or pitavastatin did not significantly affect tumor growth. Combination AZD5363 and pitavastatin significantly impaired tumor growth and decreased tumor size and tumor percent body weight at endpoint (Fig. 3B–D; Supplementary Fig. S6C and S6D). Both drugs showed on-target efficacy as detected using markers of PI3K/AKT pathway activation (pAKTSer473, pPRAS40Thr246, pAKTThr308, pGSK3βSer9, and pS6Ser240/244), protein prenylation (RHEB, unprenylated RAP1A, and HDJ2) and HMGCR (Supplementary Fig. S6E). In a panel of breast cancer PDOs, the combination of AZD5363 and pitavastatin impaired the growth of all PDOs but induced cytotoxicity primarily in triple-negative (TN)/ER-low PDOs (Fig. 3E; Supplementary Fig. S7A–S7D). Consistent with these results, pitavastatin-induced HMGCR upregulation was impaired in an ER-low PDO compared with an ER-positive PDO, and this was associated with a greater accumulation of unprenylated RAP1A (Fig. 3F). These data show that the combination of AZD5363 and pitavastatin induces cytotoxicity in preclinical models of TNBC.
Figure 3.
AKT inhibitors synergize with pitavastatin to induce TNBC cytotoxicity. A, TNBC cell lines (SUM159 and BT20) were treated with DMSO, AZD5363 (SUM159, 5 µmol/L; BT20, 1.25 µmol/L), pitavastatin (SUM159, 4 µmol/L; BT20, 2 µmol/L), or a combination of AZD5363 and pitavastatin, and the total cell number (rapid red nuclear dye) and number of dead cells (cleaved caspase-3/7 dye) were measured every 2 hours for 72 hours by Incucyte Live-Cell Analysis. Data are represented as mean ± SD of percent cleaved caspase-3/7 signal (N = 4 technical replicates). B–D, HCC70 cells were injected subcutaneously into NSG mice, and tumors were allowed to grow for 21 days before starting treatments. Mice were switched to a low geranylgeraniol chow diet 3 days before starting treatments and were treated once daily with vehicle (0.5% carboxymethylcellulose, N = 10), 100 mg/kg AZD5363 (4 days on, 3 days off, N = 12), 100 mg/kg pitavastatin (daily, N = 12), or both (N = 13) for 24 days. B, Tumor size (mm3) was measured every 3 to 4 days, starting 10 days after injection of cells. C, Tumor size (mm3) was measured at the endpoint. D, Tumor weight and mouse body weight were measured at the endpoint, and the tumor percent body weight was calculated by dividing tumor weight by mouse body weight. For B–D, statistical analysis was performed using two-way ANOVA with Tukey multiple comparison test (*, significant differences; in B, significant differences compared with the AZD5363 and pitavastatin combination treatment at the endpoint). E, A panel of breast cancer PDOs were treated with DMSO, 1 µmol/L AZD5363, 5 µmol/L pitavastatin, or the combination of AZD5363 and pitavastatin for 96 hours and then pulsed with EdU and stained with a cleaved caspase-3 antibody. A representative image for each PDO in each treatment condition is shown. Scale bars, 40 µm. F, Immunoblots of NPC1, HMGCR, PARP, pAKTSer473, pPRAS40Thr246, cleaved caspase-3, unprenylated RAP1A, β-actin, and vinculin in an ER-low (patient 10) and ER-positive (patient 26) organoid treated with DMSO, 1 µmol/L AZD5363, 5 µmol/L pitavastatin, or the combination for 24 hours. *, P = 0.0332; **, P = 0.0021; ***, P = 0.0002; ****, P < 0.0001; ns, not significant.
AKT inhibition does not synergize with pitavastatin in ER-positive breast cancer cells
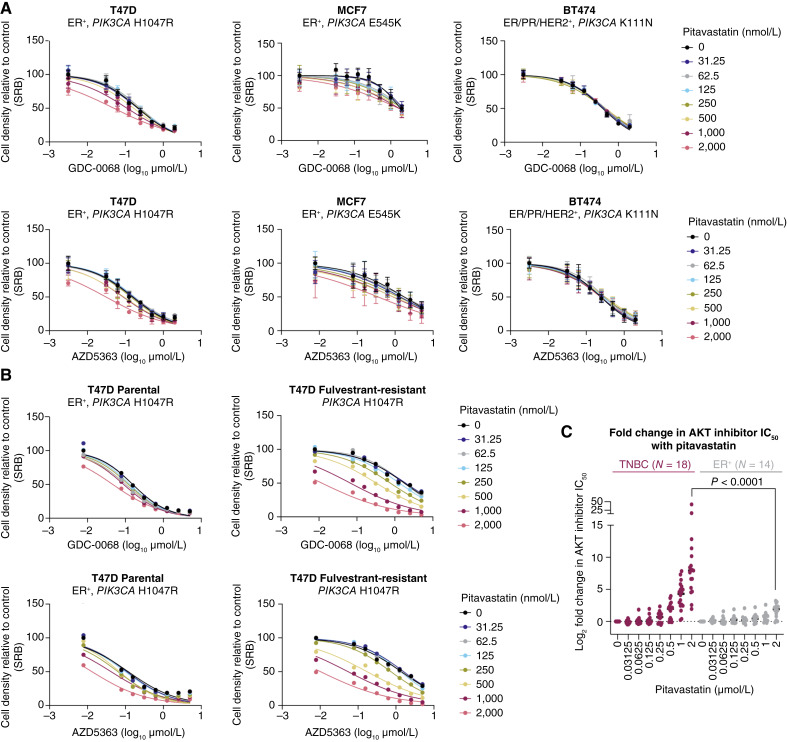
We further tested the combination in a panel of PI3K/AKT pathway–mutant ER-positive breast cancer cell lines. ER-positive breast cancer cells were completely insensitive to the combination of AKT inhibitor and pitavastatin (Fig. 4A, Supplementary Fig. S8A and S8B). Similarly, ER-positive breast cancer cells were insensitive to the combination of BYL719 or Torin 1 plus pitavastatin (Supplementary Fig. S9A–S9D). Endocrine therapy (fulvestrant)–resistant T47D cells were exquisitely sensitive to AKT inhibitor and pitavastatin combination therapy, compared with the matched parental T47D cells, and this coincided with a loss of ER expression in fulvestrant-resistant cells (Fig. 4B; Supplementary Fig. S10A and S10B). In a panel of six TN and five ER-positive breast cancer cell lines, all TNBC cell lines were sensitive to combination catalytic (GDC-0068 and AZD5363) or allosteric AKT (ARQ 092 and MK-2206) inhibitor and pitavastatin, whereas none of the ER-positive breast cancer cell lines were sensitive (Fig. 4C). Furthermore, androgen receptor–negative prostate cancer cells were also sensitive to the combination of AKT inhibitor and pitavastatin, whereas androgen receptor–positive prostate cancer cells were not sensitive (Supplementary Fig. S10C). We hypothesized that HR expression contributes to statin resistance, however overexpression of ER in SUM159 and fulvestrant-resistant T47D cells did not affect sensitivity to pitavastatin or to the combination of AKT inhibitor plus pitavastatin (Supplementary Fig. S11A and S11B). Similarly, degradation of ER with fulvestrant did not sensitize ER-positive breast cancer cells to pitavastatin, indicating that ER expression is not sufficient to mediate resistance to the antiproliferative effects of statins (Supplementary Fig. S11C). This demonstrates that HR-negative breast cancer cells either have a unique dependency on the cholesterol biosynthesis pathway or have dysregulated cholesterol homeostasis.
Figure 4.
Pitavastatin does not synergize with AKT inhibition in ER-positive breast cancer cells. A, ER-positive breast cancer cell lines (T47D, MCF7, and BT474) were treated with increasing doses of GDC-0068 (0–2 µmol/L) or AZD5363 (T47D, 0–2 µmol/L; MCF7, 0–5 µmol/L; BT474, 0–2 µmol/L) and pitavastatin (0–2,000 nmol/L) for 72 hours, and cell density was measured by SRB assay. Data are represented as mean ± SD (N = 3 technical replicates). B, Parental and fulvestrant-resistant T47D cells were treated with increasing doses of GDC-0068 (0–5 µmol/L) or AZD5363 (0–5 µmol/L) and pitavastatin (0–2,000 nmol/L) for 72 hours, and cell density was measured by SRB assay (N = 1 technical replicate). C, Log2-fold change in AKT inhibitor IC50 (GDC-0068, AZD5363, MK2206, and ARQ 092) with 2 vs. 0 µmol/L pitavastatin was calculated for six TNBC and five ER-positive breast cancer cell lines. Data are represented around the median (N = the number of cell line and AKT inhibitor combinations tested). Statistical analysis was performed for the 2 µmol/L pitavastatin conditions using an unpaired, nonparametric Mann–Whitney test (P < 0.0001).
To measure the dependency of breast cancer cells on the cholesterol biosynthesis pathway, we treated a panel of TN and ER-positive breast cancer cells with the geranylgeranyltransferase-I inhibitor GGTI-298, which inhibits protein geranylgeranylation downstream of statins. Both TN and ER-positive breast cancer cells were sensitive to GGTI-298 and to the combination of AZD5363 and GGTI-298, suggesting that all breast cancer cells depend on the cholesterol biosynthesis pathway for survival through the generation of the prenylation substrate GGPP (Supplementary Fig. S12A and S12B). AZD5363 and pitavastatin-induced TNBC cell death was rescued by the addition of mevalonate, the product of HMGCR, or GGPP, further suggesting that loss of GGPP contributes to AKT inhibitor and pitavastatin-mediated cell death (Supplementary Fig. S12C and S12D). After statin treatment, but not GGTI-298 treatment, ER-positive breast cancer cells maintain sufficient GGPP levels. By contrast, only RAS-altered breast cancer cells were sensitive to the farnesyltransferase inhibitor, FTI-277 (Supplementary Fig. S13A and S13B). Cholesterol did not rescue combination therapy–induced cell death, but rather potentiated the cytotoxicity of AZD5363 and pitavastatin, likely by further inhibiting cholesterol biosynthesis through negative feedback (Supplementary Figs. S12C and S14A; refs. 15, 16). Consistent with this hypothesis, supplementing ER-positive breast cancer cells with exogenous cholesterol sensitized these cells to pitavastatin (Supplementary Fig. S14B). Altogether, these data show that the combination of AKT inhibition and pitavastatin induces apoptotic cell death by critically depleting GGPP in TNBC cells, which display dysregulated cholesterol homeostasis.
Impaired SREBP-2 activation sensitizes TNBC cells to statins
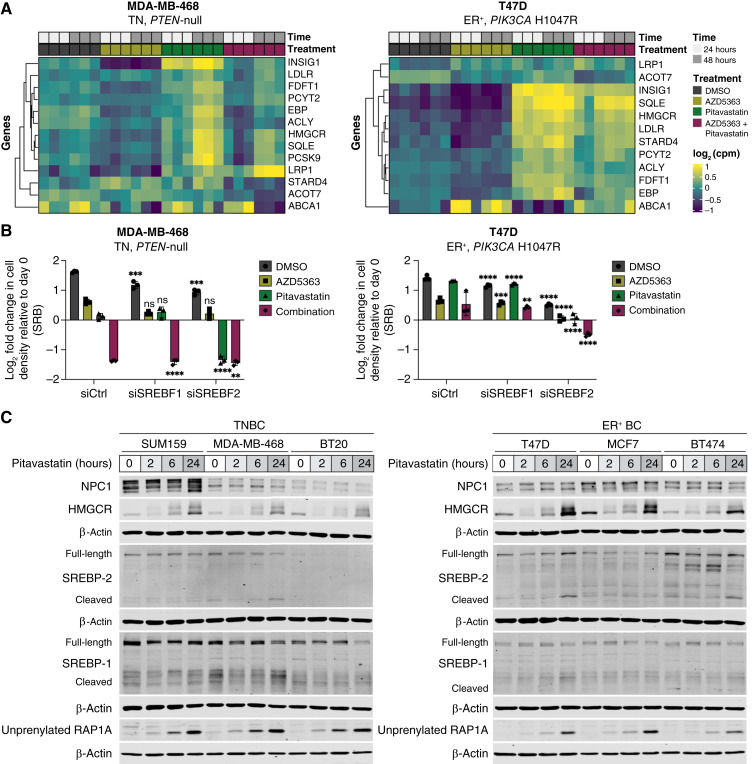
To characterize the differential sensitivity of TN and ER-positive breast cancer cells to combined AKT inhibitor and pitavastatin, we performed bulk RNA-seq in statin-sensitive TNBC cells (MDA-MB-468) and statin-resistant ER-positive breast cancer cells (T47D) treated with cytostatic doses of single-agent AKT inhibitor (AZD5363), pitavastatin, or the combination for 24 or 48 hours. Statin-resistant ER-positive cells rapidly upregulated and sustained expression of SREBF2 target genes, including HMGCR, in response to pitavastatin or combination AZD5363 and pitavastatin (Fig. 5A; Supplementary Figs. S15A–S15D, S16A, and S16B). This is the canonical response to low sterol conditions, which is mediated by the transport of the SCAP–SREBP-2 complex to the Golgi, SREBP-2 cleavage, and subsequent translocation of active SREBP-2 to the nucleus where it promotes the transcription of target genes, including LDLR, INSIG1, and HMGCR (14, 15, 17). By contrast, statin-sensitive TNBC cells were unable to increase the expression of SREBF2 target genes as potently or rapidly as ER-positive cells in response to pitavastatin or combination AZD5363 and pitavastatin (Fig. 5A; Supplementary Figs. S15A–S15D, S16C–S16D). Depletion of SREBF2, but not SREBF1, further sensitized TNBC cells to pitavastatin and sensitized ER-positive breast cancer cells to pitavastatin and combination AKT inhibitor and pitavastatin by impairing pitavastatin-induced HMGCR upregulation (Fig. 5B; Supplementary Fig. S17A). Similarly, SREBP-1/2 inhibition with fatostatin, which binds SCAP and inhibits the transport of SREBP-1/2 from the endoplasmic reticulum to the Golgi, impaired HMGCR upregulation after combination AZD5363 and pitavastatin treatment and sensitized ER-positive breast cancer cells to pitavastatin (Supplementary Fig. S17B–S17C). Over a 24-hour time course of pitavastatin treatment, ER-positive breast cancer cells upregulated HMGCR mRNA and protein levels more significantly than TNBC cells, and higher HMGCR expression was associated with reduced unprenylated RAP1A levels (Fig. 5C; Supplementary Fig. S18A).
Figure 5.
TNBC cells have impaired pitavastatin-induced SREBP-2 activation. A, RNA-seq was performed in TN (MDA-MB-468) and ER-positive (T47D) breast cancer cells treated with DMSO, AZD5363 (MDA-MB-468, 10 µmol/L; T47D, 0.25 µmol/L), pitavastatin (1 µmol/L), or a combination of AZD5363 and pitavastatin for 24 or 48 hours. Data for SREBF2 target genes are represented as log2 counts per million for each replicate (N = 3 biological replicates per condition). B, TN (MDA-MB-468) and ER-positive (T47D) breast cancer cells were transfected with siControl (siCtrl), siSREBF-1, or siSREBF-2 for 24 hours and then treated with DMSO, AZD5363 (MDA-MB-468, 15 µmol/L; T47D, 0.25 µmol/L), pitavastatin (2 µmol/L), or a combination of AZD5363 and pitavastatin for 72 hours, and cell density was measured by SRB assay. Data are represented as mean ± SD (N = 3 technical replicates). Statistical analysis was performed using two-way ANOVA with Tukey multiple comparison test (*, significant differences compared with the matched treatment condition in the siCtrl cells). C, Immunoblots of NPC1, HMGCR, SREBP-1/2, unprenylated RAP1A, and β-actin in TN (SUM159, MDA-MB-468, and BT20) and ER-positive (T47D, MCF7, BT474) breast cancer cell lines treated with DMSO or 2 µmol/L pitavastatin for 2, 6, or 24 hours. **, P = 0.0021; ***, P = 0.0002; ****, P < 0.0001; ns, not significant. cpm, counts per million.
To test whether HMGCR expression is sufficient to confer resistance to pitavastatin, we attempted to overexpress wild-type or catalytically inactive HMGCR in TN or ER-positive breast cancer cells. However, intracellular cholesterol levels are tightly regulated, thereby significantly limiting HMGCR overexpression, to the extent that expression of wild-type HMGCR did not desensitize TNBC cells to pitavastatin (Supplementary Fig. S18B and S18C). To determine whether HMGCR expression is necessary to confer resistance to pitavastatin, we depleted HMGCR in a panel of TN and ER-positive breast cancer cells and treated them with AZD5363, pitavastatin, or the combination. HMGCR depletion further sensitized TNBC cells to single-agent pitavastatin but did not potentiate the cytotoxic effects of combination AZD5363 and pitavastatin (Supplementary Fig. S18D). By contrast, HMGCR depletion significantly sensitized ER-positive breast cancer cells to pitavastatin and induced cytotoxicity to combination AZD5363 and pitavastatin (Supplementary Fig. S18E).
Since AKT is known to regulate SREBP-2 activation, we hypothesized that AKT inhibitors synergize with pitavastatin in TNBC through enhanced suppression of pitavastatin-induced HMGCR upregulation (31–33). In a panel of TNBC cell lines, AZD5363 decreased HMGCR expression, and pitavastatin increased HMGCR expression. The combination of AZD5363 and pitavastatin impaired pitavastatin-induced HMGCR expression, consistent with impaired SREBP-2 activation (Supplementary Fig. S19A). These data suggest that statin-induced HMGCR upregulation occurs through SREBP-2 activation, but HMGCR is also regulated by proteasomal degradation. Pretreatment of TN and ER-positive breast cancer cells with the proteasome inhibitor MG132 before a 24-hour treatment with pitavastatin or combination AZD5363 plus pitavastatin did not increase pitavastatin-induced HMGCR upregulation. Pretreatment with the translation inhibitor cycloheximide abolished pitavastatin-induced HMGCR upregulation, indicating that new synthesis accounts for increased HMGCR expression after statin treatment (Supplementary Fig. S19B and S19C). Collectively, these data show that AKT inhibitors synergize with pitavastatin in TNBC by potently suppressing SREBP-2 activation, resulting in decreased flux through the cholesterol biosynthesis pathway.
NPC1 inhibition causes lysosomal cholesterol accumulation and rescues pitavastatin sensitivity
SREBP-2 activity is primarily regulated by cholesterol levels in the endoplasmic reticulum. We reasoned that perturbing intracellular cholesterol trafficking would alter SREBP-2 activity and pitavastatin sensitivity. Treatment of TN and ER-positive breast cancer cells with chloroquine, an autophagy inhibitor that causes lysosomal cholesterol accumulation, increased HMGCR expression, and SREBF2 depletion abolished this increase (Supplementary Fig. S19D). Increased HMGCR expression induced by chloroquine and pitavastatin rescued the antiproliferative effects of pitavastatin in TNBC cells (Supplementary Fig. S19E).
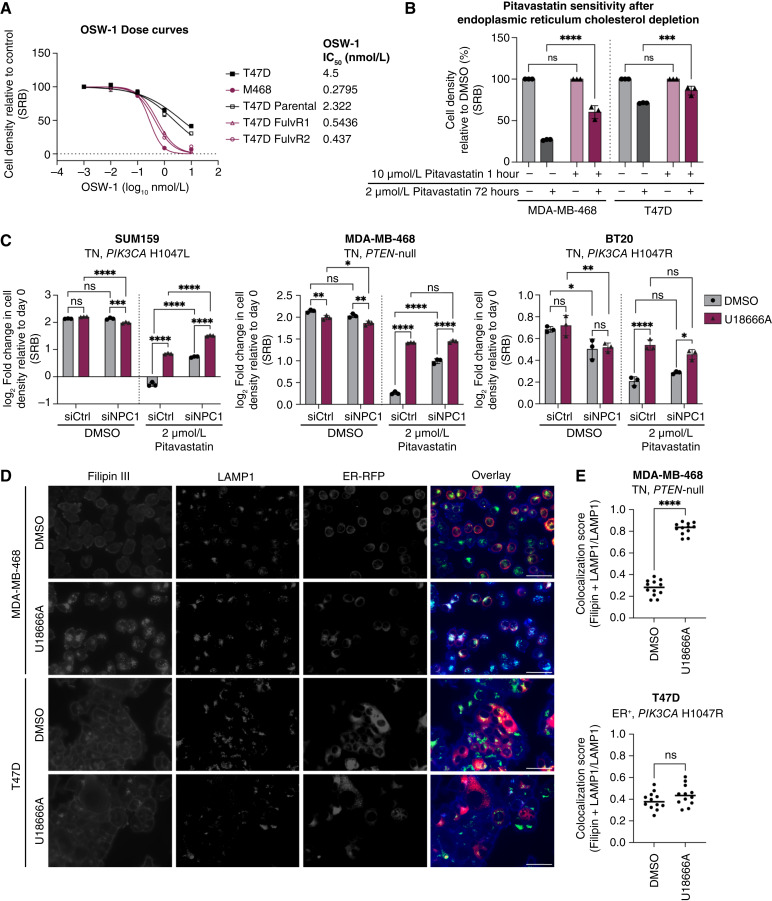
We next treated cells with OSW-1, a potent inhibitor of the oxysterol-binding protein, which transports cholesterol out of the endoplasmic reticulum. Pitavastatin-sensitive breast cancer cells were more sensitive to OSW-1 than ER-positive pitavastatin-resistant cells (Fig. 6A). Moreover, treatment with low-dose OSW-1 (0.1 nmol/L) sensitized ER-positive breast cancer cells to pitavastatin (Supplementary Fig. S20A). OSW-1 does not induce endoplasmic reticulum stress as a single agent or in combination with pitavastatin, suggesting that the antiproliferative effects are specifically due to endoplasmic reticulum cholesterol accumulation (Supplementary Fig. S20B). To standardize baseline endoplasmic reticulum cholesterol levels across TN and ER-positive breast cancer cells, we cultured cells in media with 10% lipid-depleted serum, containing no cholesterol, thereby limiting the cellular source of cholesterol to de novo synthesis. Concomitant addition of high dose of pitavastatin (10 µmol/L) for 1 hour inhibits cholesterol synthesis, resulting in selective depletion of cholesterol from the endoplasmic reticulum. After standardizing endoplasmic reticulum cholesterol levels, the cells were treated with a low dose of pitavastatin for 72 hours in media supplemented with 10% lipid-depleted serum or cholesterol-rich 10% FBS (complete serum). TNBC cells maintained in lipid-depleted serum were desensitized to pitavastatin, indicating that endoplasmic reticulum cholesterol levels determine statin sensitivity (Fig. 6B). By contrast, TNBC cells treated with a low dose of pitavastatin in complete serum remained pitavastatin sensitive, suggesting that these cells took up and trafficked exogenous cholesterol to the endoplasmic reticulum, thereby limiting SREBP-2 activation (Supplementary Fig. S20C).
Figure 6.
NPC1 inhibition causes lysosomal cholesterol accumulation and rescues pitavastatin sensitivity. A, ER-negative (MDA-MB-468 and T47D fulvestrant-resistant clones 1 and 2) and ER-positive (T47D and parental T47D) breast cancer cell lines were treated with a range of concentrations of OSW-1 (0–10 nmol/L) for 72 hours, and cell density was measured by SRB assay. Data are represented as mean ± SD (N = 3 technical replicates). IC50 values for each cell line are reported. B, TN (MDA-MB-468) and ER-positive (T47D) breast cancer cells were seeded into media supplemented with 10% lipid-depleted serum and treated for 1 hour with vehicle or high dose of pitavastatin (10 µmol/L). Media were removed and replaced with media supplemented with 10% lipid-depleted serum and vehicle or low dose pitavastatin (2 µmol/L) for 72 hours, and cell density was measured by SRB assay. Data are represented as mean ± SD (N = 3 technical replicates). Statistical analysis was performed using two-way ANOVA with Šidák multiple comparison test. C, TNBC cells (SUM159, MDA-MB-468, and BT20) were transfected with siControl (siCtrl) or siNPC1 and then treated with DMSO or 1 µmol/L U18666A and DMSO or 2 µmol/L pitavastatin for 72 hours, and cell density was measured by SRB assay. Data are represented as mean ± SD (N = 3 technical replicates). Statistical analysis was performed using two-way ANOVA with Šidák multiple comparison test. D, TN (MDA-MB-468) and ER-positive (T47D) breast cancer cells expressing red fluorescent protein in the endoplasmic reticulum (ER-RFP; red) were treated with DMSO or 1 µmol/L U18666A for 24 hours. Cells were fixed with 4% formaldehyde and stained with Filipin III (blue) and a LAMP1 antibody (green). Representative images are shown. Scale bars, 50 µm. E, Quantification of Filipin III and LAMP1 colocalization normalized to total LAMP1 from 12 nonoverlapping fields. Statistical analysis was performed using an unpaired, two-tailed parametric t test. *, P = 0.0332; **, P = 0.0021; ***, P = 0.0002; ****, P < 0.0001; ns, not significant.
The cholesterol trafficking protein Niemann-Pick C1 (NPC1) transports cholesterol from the lysosome to the endoplasmic reticulum. Depletion or inhibition of NPC1 results in lysosomal cholesterol accumulation and decreased endoplasmic reticulum cholesterol levels. In a panel of TNBC cell lines, siRNA-mediated depletion of NPC1 or treatment with the NPC1 inhibitor U18666A rescued the antiproliferative effects of pitavastatin (Fig. 6C). Since U18666A can inhibit other cholesterol-trafficking proteins beyond NPC1, we depleted a panel of genes reported to be inhibited by U18666A and evaluated pitavastatin sensitivity. Depletion of GRAMD1A/B/C or OSBPL9 did not rescue pitavastatin sensitivity, indicating that the loss of NPC1 alone is sufficient to rescue the antiproliferative effects of pitavastatin (Supplementary Fig. S21A). Combined depletion of NPC1 and treatment with U18666A did not significantly outperform depletion or inhibitor alone, further demonstrating that NPC1 alone can mediate pitavastatin sensitivity in TNBC (Fig. 6C). Depletion of SREBF2 abrogated the NPC1 inhibitor–mediated rescue of pitavastatin in TNBC, suggesting that NPC1 inhibition activates SREBP-2 (Supplementary Fig. S21B). NPC1 depletion or inhibition in combination with pitavastatin increased HMGCR upregulation, and this increase was abolished upon depletion of SREBF2 (Supplementary Fig. S21C). These data indicate that NPC1 inhibition rescues pitavastatin sensitivity by decreasing endoplasmic reticulum cholesterol levels to promote SREBP-2 activation and subsequent HMGCR upregulation.
Finally, we visualized subcellular cholesterol localization in TN and ER-positive breast cancer cells expressing red fluorescent protein in the endoplasmic reticulum and stained with Filipin III (a cholesterol stain) and LAMP1 (a lysosomal marker). At baseline, ER-positive breast cancer cells showed elevated lysosomal cholesterol levels compared with TNBC cells (Fig. 6D and E). TNBC cells treated with U18666A rapidly accumulated lysosomal cholesterol, whereas the fraction of cholesterol-positive lysosomes in ER-positive breast cancer cells did not significantly increase from baseline (Fig. 6D and E). Statin-sensitive fulvestrant-resistant T47D cells treated with U18666A also rapidly accumulated lysosomal cholesterol, whereas the fraction of cholesterol-positive lysosomes in parental ER-positive T47D cells did not significantly increase from baseline (Supplementary Fig. S22A and S22B). NPC1 inhibition decreases endoplasmic reticulum cholesterol levels in statin-sensitive breast cancer cells by trapping cholesterol in the lysosome, resulting in enhanced SREBP-2 activation and loss of sensitivity to pitavastatin. Together, these data support a model in which TNBC cells are uniquely sensitive to statins due to elevated endoplasmic reticulum cholesterol, which results in impaired SREBP-2 activation in response to statin. Combination AKT inhibitor and statin more potently suppresses SREBP-2 activation, resulting in cytotoxicity.
Discussion
Given the prevalence of PI3K/AKT pathway hyperactivation in TNBC and the clinical potential of PI3K/AKT inhibitors, we leveraged an unbiased, genome-scale screen to identify collateral vulnerabilities using PI3K/AKT inhibitors as anchor drugs. We identified synergy between AKT inhibition and disruption of cholesterol homeostasis genes, including SREBF2, a master transcriptional regulator of cholesterol biosynthesis. Inhibition of cholesterol biosynthesis with statins synergized with AKT inhibitors in a panel of TNBC cell lines. The combination of AKT inhibitor and pitavastatin induced apoptotic cell death in TNBC cells, mouse xenografts of TNBC, and PDOs of ER-negative breast cancer. ER-positive breast cancer cells and PDOs were resistant to pitavastatin and the combination of AKT inhibitor and pitavastatin. Previous studies have also shown that HR-negative breast and prostate cancer cells are hypersensitive to statins due to impaired SREBP-2 activation (34–36). Here, we found that ER expression alone is not sufficient to mediate resistance to statins nor is ER degradation sufficient to sensitize ER-positive breast cancer cells to statins. Rather, subcellular cholesterol localization determines statin sensitivity. Higher lysosomal and lower endoplasmic reticulum cholesterol levels in ER-positive breast cancer cells allow for rapid activation of SREBP-2 upon statin treatment. By contrast, TNBC cells have altered cholesterol trafficking with reduced lysosomal and enhanced endoplasmic reticulum cholesterol levels, resulting in statin sensitivity that was rescued by depletion of endoplasmic reticulum cholesterol. Dysregulated cholesterol trafficking in TNBC cells significantly sensitizes them to combination treatment with AKT inhibitor and pitavastatin through robust inhibition of the cholesterol biosynthesis pathway.
While statins have been FDA-approved for the treatment of hypercholesterolemia since the 1980s, epidemiological and clinical data on the efficacy of statins in cancer is inconclusive (37–41). Previous studies have reported that lipophilic statins exert anticancer effects in preclinical models, yet no conclusive mechanisms of sensitivity have been described (42–49). Our findings explain the apparent failure of statins as anticancer agents in the clinic. Most clinical trials of statins in cancer have not evaluated pitavastatin, yet pitavastatin is the only statin that can reach cytotoxic anticancer concentrations in human plasma at clinically administered doses (41, 50). In addition to statin selection, patient diet has not been considered in clinical trials. Several studies in preclinical models have shown that diets rich in the GGPP precursor geranylgeraniol can restore cellular GGPP levels and rescue the antiproliferative effects of statins. Standard mouse chow is rich in oils that contain geranylgeraniol, which has likely confounded most preclinical studies of statins (29, 30). Lastly, our data highlight the importance of patient selection for statin clinical trials. Despite studies in cancer cell lines showing statin efficacy in TNBC, most trials have evaluated the efficacy of statins in HR-positive breast cancer (34, 42). Our studies show a negative correlation between ER expression and statin sensitivity and suggest that lysosomal cholesterol levels and statin-induced HMGCR expression could serve as biomarkers of statin response.
Although millions of people worldwide are prescribed statins for cholesterol-lowering benefit, the epidemiological data are insufficient to definitively assess whether statin use affects incidence or outcomes in TNBC. TNBC accounts for approximately 12% of breast cancer cases and is more common in younger women who are less likely to be statin users (1). Since pitavastatin is infrequently prescribed, there is not a large enough population of patients with TNBC to perform any meaningful retrospective analysis on the role of pitavastatin in TNBC.
In summary, we have identified synergy between AKT inhibitors and pitavastatin in TNBC. By cotargeting TNBC cells with AKT inhibitors and statins, SREBP-2 activation and cholesterol biosynthesis are potently inhibited, which further impairs upregulation of HMGCR expression, resulting in GGPP depletion and cytotoxicity. This combination is selective for TNBC cells because of dysregulated cholesterol homeostasis. Since both AKT inhibitors (AZD5363, capivasertib) and pitavastatin are FDA-approved drugs, this motivates the evaluation of the efficacy of this combination in clinical trials of TNBC.
Supplementary Material
Supplementary figures and information
Acknowledgments
This work was supported by grants from the Ludwig Center at Harvard (to A. Toker, M. Brown, K. Cichowski, S.J. Elledge, and T. Muranen), CA253097 (to A. Toker), NSF Graduate Research Fellowship (to A.L. Hillis), Harvard University Landry Cancer Biology Fellowship (to A.L. Hillis and H.E. Manchester), NIH P01CA250959 (to N. Zhang and M. Brown), Susan G. Komen Foundation CCR18547665 (to T. Muranen), Harvard Stem Cell Institute DP-0194-21-00 (to T. Muranen), Sigrid Jusélius Foundation (to J. Högström), Orion Research Foundation (to J. Högström), Maud Kuistila Memorial Foundation (to J. Högström), AACR fellowship (to J. Högström), Cancer Research UK Grand Challenge and the Mark Foundation for Cancer Research (to K. Cichowski of the SPECIFICANCER team), R01CA234600 (to S.J. Elledge), Howard Hughes Medical Institute (to S.J. Elledge), Department of Defense Impact Award W81XWH-20-1-0867 (to D.A. Fruman), and Sir Henry Wellcome Fellowship 220464/A/20/Z (to R.R. Madsen). We thank members of the Ludwig Center at Harvard University for their constructive comments.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Authors’ Disclosures
J. Högström reports grants from the American Association for Cancer Research, Maud Kuistila Memorial Foundation, Sigrid Jusélius Foundation and Orion Research Foundation during the conduct of the study. D.E. Root reports grants from AbbVie, Bristol Myers Squibb, Janssen, Merck and Vir Biotechnology outside the submitted work. M. Brown reports grants from the Ludwig Center at Harvard and Breast Cancer Research Foundation during the conduct of the study; as well as grants from Novartis and personal fees from Novartis, Kronos Bio, FibroGen and GV20 Therapeutics outside the submitted work. K. Cichowski reports other support from Erasca outside the submitted work. S.T. Barry reports employment and being a shareholder with AstraZeneca. R.R. Madsen reports grants from Wellcome Trust during the conduct of the study; as well as personal fees from Nested Therapeutics outside the submitted work. A. Toker reports grants from NCI and nonfinancial support from AstraZeneca during the conduct of the study; as well as personal fees from American Society for Biochemistry and Molecular Biology (ASBMB) and Novo Holdings of Novo Nordisk Foundation, and grants from BioHybrid Solutions outside the submitted work. No disclosures were reported by the other authors.
Authors’ Contributions
A.L. Hillis: Conceptualization, resources, data curation, formal analysis, funding acquisition, validation, investigation, visualization, methodology, writing–original draft, project administration, writing–review and editing. T.D. Martin: Conceptualization, methodology. H.E. Manchester: Investigation, methodology. J. Högström: Conceptualization, investigation, methodology. N. Zhang: Investigation. E. Lecky: Investigation. N. Kozlova: Investigation. J. Lee: Investigation. N.S. Persky: Investigation, methodology. D.E. Root: Resources, formal analysis, methodology. M. Brown: Formal analysis, supervision, project administration. K. Cichowski: Supervision, project administration. S.J. Elledge: Conceptualization, supervision. T. Muranen: Formal analysis, supervision, project administration. D.A. Fruman: Conceptualization, supervision, methodology. S.T. Barry: Resources, supervision. J.G. Clohessy: Conceptualization, investigation, methodology. R.R. Madsen: Conceptualization, formal analysis, supervision, investigation, methodology, project administration. A. Toker: Conceptualization, resources, data curation, formal analysis, supervision, funding acquisition, writing–original draft, project administration, writing–review and editing.
References
- 1. Giaquinto AN, Sung H, Miller KD, Kramer JL, Newman LA, Minihan A, et al. Breast cancer statistics, 2022. CA Cancer J Clin 2022;72:524–41. [DOI] [PubMed] [Google Scholar]
- 2. Garrido-Castro AC, Lin NU, Polyak K. Insights into molecular classifications of triple-negative breast cancer: improving patient selection for treatment. Cancer Discov 2019;9:176–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bianchini G, De Angelis C, Licata L, Gianni L. Treatment landscape of triple-negative breast cancer—expanded options, evolving needs. Nat Rev Clin Oncol 2022;19:91–113. [DOI] [PubMed] [Google Scholar]
- 4. The Cancer Genome Atlas Network . Comprehensive molecular portraits of human breast tumours. Nature 2012;490:61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Manning BD, Toker A. AKT/PKB signaling: navigating the network. Cell 2017;169:381–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Vasan N, Cantley LC. At a crossroads: how to translate the roles of PI3K in oncogenic and metabolic signalling into improvements in cancer therapy. Nat Rev Clin Oncol 2022;19:471–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Vanhaesebroeck B, Perry MWD, Brown JR, André F, Okkenhaug K. PI3K inhibitors are finally coming of age. Nat Rev Drug Discov 2021;20:741–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hamilton E, Schiavon G, Grinsted LM, De Bruin EC, Catanese MT, Rugo HS. 338TiP CAPItello-292: a phase 1b/3 study of capivasertib, palbociclib and fulvestrant vs. placebo, palbociclib and fulvestrant in HR+/HER2− advanced breast cancer. Ann Oncol 2021;32:S514. [Google Scholar]
- 9. Turner N, Howell S, Jhaveri K, Gomez H, Toi M, Hu X, et al. 350TiP A phase III trial of capivasertib and fulvestrant vs. placebo and fulvestrant in patients with HR+/HER2− breast cancer (CAPItello-291). Ann Oncol 2020;31(Suppl 4):S388–9. [Google Scholar]
- 10. Truqap (capivasertib) plus Faslodex approved in the US for patients with advanced HR-positive breast cancer [Internet]. Cambridge, United Kingdom: AstraZeneca; 2023[cited 2023 Dec 14]. Available from:https://www.astrazeneca.com/media-centre/press-releases/2023/truqap-approved-in-us-for-hr-plus-breast-cancer.html. [Google Scholar]
- 11. Hanker AB, Kaklamani V, Arteaga CL. Challenges for the clinical development of PI3K inhibitors: strategies to improve their impact in solid tumors. Cancer Discov 2019;9:482–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Brown JS, Banerji U. Maximising the potential of AKT inhibitors as anti-cancer treatments. Pharmacol Ther 2017;172:101–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bloch K. The biological synthesis of cholesterol. In: Harris RS, Marrian GF, Thimann KV, editors. Vitamins & Hormones [Internet]. Cambridge (MA): Academic Press; 1957[cited 2022 Dec 28]. p. 119–50. Available from:https://www.sciencedirect.com/science/article/pii/S0083672908605099. [DOI] [PubMed] [Google Scholar]
- 14. Goldstein JL, DeBose-Boyd RA, Brown MS. Protein sensors for membrane sterols. Cell 2006;124:35–46. [DOI] [PubMed] [Google Scholar]
- 15. Brown MS, Goldstein JL. Cholesterol feedback: from SchoSchoenheimer’s bottle to Scap’s MELADL. J Lipid Res 2009;50:S15-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Schoenheimer R, Breusch F. Synthesis and destruction of cholesterol in the organism. J Biol Chem 1933;103:439–48. [Google Scholar]
- 17. Brown MS, Goldstein JL. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell 1997;89:331–40. [DOI] [PubMed] [Google Scholar]
- 18. Endo A. A historical perspective on the discovery of statins. Proc Jpn Acad Ser B Phys Biol Sci 2010;86:484–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Endo A, Kuroda M, Tanzawa K. Competitive inhibition of 3-hydroxy-3-methylglutaryl coenzyme a reductase by ML-236A and ML-236B fungal metabolites, having hypocholesterolemic activity. FEBS Lett 1976;72:323–6. [DOI] [PubMed] [Google Scholar]
- 20. Schachter M. Chemical, pharmacokinetic and pharmacodynamic properties of statins: an update. Fundam Clin Pharmacol 2005;19:117–25. [DOI] [PubMed] [Google Scholar]
- 21. Kita T, Brown MS, Goldstein JL. Feedback regulation of 3-hydroxy-3-methylglutaryl coenzyme A reductase in livers of mice treated with mevinolin, a competitive inhibitor of the reductase. J Clin Invest 1980;66:1094–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li W, Xu H, Xiao T, Cong L, Love MI, Zhang F, et al. MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol 2014;15:554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Vichai V, Kirtikara K. Sulforhodamine B colorimetric assay for cytotoxicity screening. Nat Protoc 2006;1:1112–6. [DOI] [PubMed] [Google Scholar]
- 24. Ianevski A, Giri AK, Aittokallio T. SynergyFinder 3.0: an interactive analysis and consensus interpretation of multi-drug synergies across multiple samples. Nucleic Acids Res 2022;50:W739–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hogstrom JM, Cruz KA, Selfors LM, Ward MN, Mehta TS, Kanarek N, et al. Simultaneous isolation of hormone receptor–positive breast cancer organoids and fibroblasts reveals stroma-mediated resistance mechanisms. J Biol Chem 2023;299:105021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Erickson EC, You I, Perry G, Dugourd A, Donovan KA, Crafter C, et al. Multiomic profiling of breast cancer cells uncovers stress MAPK-associated sensitivity to AKT degradation. Sci Signal 2024;17:eadf2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sherman BT, Hao M, Qiu J, Jiao X, Baseler MW, Lane HC, et al. DAVID: a web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res 2022;50:W216–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009;4:44–57. [DOI] [PubMed] [Google Scholar]
- 29. de Wolf E, Abdullah MI, Jones SM, Menezes K, Moss DM, Drijfhout FP, et al. Dietary geranylgeraniol can limit the activity of pitavastatin as a potential treatment for drug-resistant ovarian cancer. Sci Rep 2017;7:5410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jawad MJ, Ibrahim S, Kumar M, Burgert C, Li W-W, Richardson A. Identification of foods that affect the anti-cancer activity of pitavastatin in cells. Oncol Lett 2022;23:73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Porstmann T, Santos CR, Griffiths B, Cully M, Wu M, Leevers S, et al. SREBP activity is regulated by mTORC1 and contributes to Akt-dependent cell growth. Cell Metab 2008;8:224–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Peterson TR, Sengupta SS, Harris TE, Carmack AE, Kang SA, Balderas E, et al. mTOR complex 1 regulates lipin 1 localization to control the SREBP pathway. Cell 2011;146:408–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Han J, Li E, Chen L, Zhang Y, Wei F, Liu J, et al. The CREB coactivator CRTC2 controls hepatic lipid metabolism by regulating SREBP1. Nature 2015;524:243–6. [DOI] [PubMed] [Google Scholar]
- 34. Goard CA, Chan-Seng-Yue M, Mullen PJ, Quiroga AD, Wasylishen AR, Clendening JW, et al. Identifying molecular features that distinguish fluvastatin-sensitive breast tumor cells. Breast Cancer Res Treat 2014;143:301–12. [DOI] [PubMed] [Google Scholar]
- 35. Clendening JW, Pandyra A, Li Z, Boutros PC, Martirosyan A, Lehner R, et al. Exploiting the mevalonate pathway to distinguish statin-sensitive multiple myeloma. Blood 2010;115:4787–97. [DOI] [PubMed] [Google Scholar]
- 36. Longo J, Mullen PJ, Yu R, van Leeuwen JE, Masoomian M, Woon DTS, et al. An actionable sterol-regulated feedback loop modulates statin sensitivity in prostate cancer. Mol Metab 2019;25:119–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Dickerman BA, García-Albéniz X, Logan RW, Denaxas S, Hernán MA. Avoidable flaws in observational analyses: an application to statins and cancer. Nat Med 2019;25:1601–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Beckwitt CH, Brufsky A, Oltvai ZN, Wells A. Statin drugs to reduce breast cancer recurrence and mortality. Breast Cancer Res 2018;20:144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kuoppala J, Lamminpää A, Pukkala E. Statins and cancer: a systematic review and meta-analysis. Eur J Cancer 2008;44:2122–32. [DOI] [PubMed] [Google Scholar]
- 40. Osmak M. Statins and cancer: current and future prospects. Cancer Lett 2012;324:1–12. [DOI] [PubMed] [Google Scholar]
- 41. Abdullah MI, de Wolf E, Jawad MJ, Richardson A. The poor design of clinical trials of statins in oncology may explain their failure—lessons for drug repurposing. Cancer Treat Rev 2018;69:84–9. [DOI] [PubMed] [Google Scholar]
- 42. Campbell MJ, Esserman LJ, Zhou Y, Shoemaker M, Lobo M, Borman E, et al. Breast cancer growth prevention by statins. Cancer Res 2006;66:8707–14. [DOI] [PubMed] [Google Scholar]
- 43. Beckwitt CH, Shiraha K, Wells A. Lipophilic statins limit cancer cell growth and survival, via involvement of Akt signaling. PLoS One 2018;13:e0197422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ehmsen S, Pedersen MH, Wang G, Terp MG, Arslanagic A, Hood BL, et al. Increased cholesterol biosynthesis is a key characteristic of breast cancer stem cells influencing patient outcome. Cell Rep 2019;27:3927–38.e6. [DOI] [PubMed] [Google Scholar]
- 45. Kang S, Kim E-S, Moon A. Simvastatin and lovastatin inhibit breast cell invasion induced by H-Ras. Oncol Rep 2009;21:1317–22. [DOI] [PubMed] [Google Scholar]
- 46. Koyuturk M, Ersoz M, Altiok N. Simvastatin induces apoptosis in human breast cancer cells: p53 and estrogen receptor independent pathway requiring signalling through JNK. Cancer Lett 2007;250:220–8. [DOI] [PubMed] [Google Scholar]
- 47. Wolfe AR, Debeb BG, Lacerda L, Larson R, Bambhroliya A, Huang X, et al. Simvastatin prevents triple-negative breast cancer metastasis in pre-clinical models through regulation of FOXO3a. Breast Cancer Res Treat 2015;154:495–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kotamraju S, Williams CL, Kalyanaraman B. Statin-induced breast cancer cell death: role of inducible nitric oxide and arginase-dependent pathways. Cancer Res 2007;67:7386–94. [DOI] [PubMed] [Google Scholar]
- 49. Sethunath V, Hu H, De Angelis C, Veeraraghavan J, Qin L, Wang N, et al. Targeting the mevalonate pathway to overcome acquired anti-HER2 treatment resistance in breast cancer. Mol Cancer Res 2019;17:2318–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Juarez D, Fruman DA. Targeting the mevalonate pathway in cancer. Trends Cancer 2021;7:525–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures and information
Data Availability Statement
The source data and annotated analysis workflows are available on the following Open Science Framework (OSF) project website: https://osf.io/6pw9d/. RNA-seq data have been deposited with Gene Expression Omnibus (GEO) under series accession number GSE252944. A complete list of reagents is provided in Supplementary Tables S2–S6.