Abstract
Background
Septic shock is associated with high mortality and there is significant heterogeneity in the host response. The aim of this study was to understand the genome-wide expression transcriptomic signatures in children with septic shock and correlate them with outcomes.
Methods
This was a prospective study conducted on children (aged 1 month to 18 years) admitted to the PICU (June–December 2021) with septic shock. Demographic details, clinical details, and administered treatment were collected. Differential gene expression analysis was performed to understand the genes and pathways affecting in different subjects.
Results
Fifteen patients were recruited (Septic shock survivors (n = 5), nonsurvivors (n = 5), and non-sepsis controls (n = 5). The median age of the patients in survivors and nonsurvivors was 15 (13, 24) months and 180 (180, 184) months, respectively. The sepsis-survivors vs nonsepsis possessed 983 upregulated and 624 downregulated genes while comparing sepsis nonsurvivors (SNS) with nonsepsis yielded 1,854 upregulated and 1,761 downregulated genes. Further, the lowest number of deregulated genes (383 upregulated and 486 downregulated) were present in SNS compared to sepsis survivors. The major Reactome pathways, found upregulated in SNSs relative to survivors included CD22 mediated B cell receptor (BCR) regulation, scavenging of heme from plasma, and creation of C4 and C2 activators while T cell receptor (TCR) signaling, the common pathway of fibrin clot formation and generation of second messenger molecules were found to be downregulated.
Conclusion
Mortality-related gene signatures are promising diagnostic biomarkers for pediatric sepsis.
How to cite this article
Lalitha AV, Vasudevan A, Moorthy M, Ramaswamy G. Profiling Molecular Changes of Host Response to Predict Outcome in Children with Septic Shock. Indian J Crit Care Med 2024;28(9):879–886.
Keywords: Children, Mortality, RNA sequence, Sepsis
Highlights
Septic shock is a heterogenous syndrome with a dysregulated host response to infection leading to life-threatening organ dysfunction which if non-responsive to treatment results in death.
Whole transcriptome analysis is a powerful tool that can be used for early recognition, risk stratification, and specific targeted therapies.
Mortality-related gene signatures identified from the current study may be used as a prognostic marker.
Introduction
Sepsis is characterized by an array of abnormalities in both innate and adaptive immune systems. Sepsis accounts for 60–80% of pediatric mortality per year according to WHO.1–3 In addition, it is responsible for approximately 20% of PICU admissions and remains the leading cause of morbidity and mortality worldwide.3,4 The presence of multiorgan dysfunction syndrome, the high severity of illness at admission, and the need for multiple inotropes, are important risk factors for mortality.5,6
Central to sepsis pathophysiology is a dysregulated host inflammatory response7–9 suggesting that host-directed immunomodulators could be of therapeutic benefit.10 Despite the prevalence and impact of septic shock in the pediatric population, our understanding of the human response to sepsis remains inadequate and cripples our ability to modify outcomes. Therefore, bridging this knowledge gap is important. The characterization of patients at the molecular level is a promising approach to identifying more specific targets for new therapeutic interventions in selected groups of patients.11 Transcriptome profiling is a powerful tool to investigate and characterize the molecular mechanisms underlying specific physiological or pathological conditions. Changes in host gene expression in response to infection may occur in any part of the body, with the continuous interaction between blood and tissues allowing blood cells to act as biosensors for the changes. In patients with septic shock, major transcriptomic changes occur within the first 48 hours of shock development and are associated with mortality in the ICU.8 In the present project, we wish to study the transcriptomic changes in children with septic shock and attempt to identify molecular signatures to predict the patient outcome.
Methods
This prospective study was conducted between June 2021 and August 2021 in the PICU of an academic and referral hospital in South India. The trial was registered at the Clinical Trials Registry of India (CTRI number: CTRI/2019/07/020322), and approved by the Institutional Ethical Committee (Ref no:164/2019). Written informed consent was obtained from the parents/guardians of all children enrolled in the study for human experimentation and according to the Declaration of Helsinki of 1975.
Subject Recruitment
Children (1 month to 18 years) admitted to the PICU and meeting pediatric-specific criteria for septic shock were eligible.12 Patients treated with corticosteroids or immunosuppressive agents and patients with co-morbidities including severe acute malnutrition, chronic liver diseases, chronic kidney diseases (CKD), and acquired immunodeficiency syndrome infection were excluded. Demographic details, clinical details at onset, and administered treatment were collected. The severity of illness was calculated using the Pediatric Risk of Mortality score-III.13 All children were treated according to the 2017 American College of Critical Care Medicine pediatric septic shock guidelines.14 All-cause mortality was tracked up to 28 days. Included cases were categorized as septic shock survivors and nonsurvivors. In addition, we identified the children admitted to the pediatric ward with the following exclusion criteria as non-septic controls (NS): A recent febrile illness (within 2 weeks), recent use of anti-inflammatory medications (within 2 weeks), or any history of chronic or acute disease even remotely associated with inflammation.
Bio-sample Collection, Processing, and RNA Isolation
Blood sample (3–5 mL) was collected (EDTA and PAXgene™ reagent tubes) on day 1 of admission to PICU after obtaining informed consent from parents or legal guardians. RNA was isolated from blood collected in PAXgene tubes using PAXgene blood miRNA kit (Cat#763134). The rest of the protocol was followed as per the manufacturer's guidelines. RNA was eluted in 30 µl of Elution Buffer BR5 and stored at –80°C till further use.
Library Preparation and mRNA Sequencing
NEB Ultra II directional RNA-Seq Library Prep kit protocol was used to prepare libraries for total RNA sequencing (NEB, Cat# E7760L). Prepared libraries were quantified using Qubit High Sensitivity Assay (Invitrogen, Cat# Q32851). The obtained libraries were pooled and diluted to the final optimal loading concentration before cluster amplification on the Illumina NovaSeq 6000 flow cell to generate 151 bp paired-end reads.
mRNA-Seq Data Analysis
The high-quality paired-end reads after appropriate trimming were mapped to the human reference genome (hg38), using HISAT2 v.2.2.1 and quantified further using Feature Count v.2.0.1. Differential gene expression analysis was performed using DESeq2 R package with thresholds considered for defining upregulated and downregulated genes (p-value < 0.05 for statistical significance, log2 (foldchange) >1 and log2 (foldchange) <1). Gene set enrichment analysis (GSEA) was used to identify regulated Reactome pathways, gene ontology (biological process and molecular functions), and hallmark genesets using the fgsea package in R. Venn diagram was generated using the online webserver, Venny (https://bioinfogp.cnb.csic.es/tools/venny). The function of the markers found unique to non-survivor vs nonsepsis was analyzed using both Reactome pathway annotation as well as for enrichment, by DAVID webserver (https://david.ncifcrf.gov/home.jsp).
Receiver Operating Characteristic (ROC) Curve and Survival Analysis of Selected Markers
The GSE185263 (adult cohort) and GSE26440 (pediatric cohort) datasets, obtained using the NCBI GEO (https://www.ncbi.nlm.nih.gov/geo/) database were used for the external validation of the diagnostic value of selected genes. The pROC R package was used for analyzing the diagnostic value in terms of area under the curve (AUC) to differentiate between survivors and nonsurvivors of sepsis. The GSE65682 dataset was chosen for survival analysis which has been performed using the survival and survminer R package. The log rank test was used for statistical analysis and p < 0.05 was considered to indicate statistical significance. Further details of the statistical analysis can be found in the online supplement.
Results
Patients Characteristics
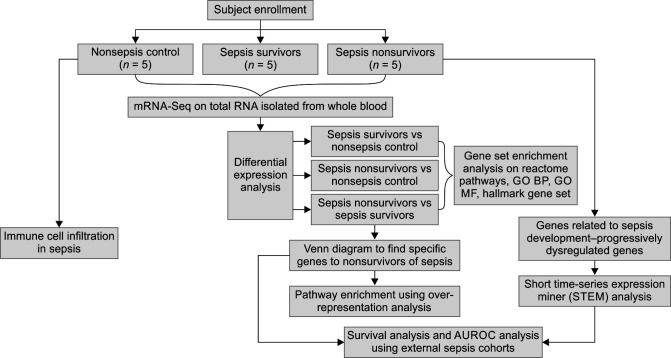
A total of 28 children with septic shock were screened during the study period. Eighteen children were enrolled after excluding 10 children (4 dengue fever, 3 rickettsial fever, 1 CKD, and 2 acute leukemia). All 18 patients could not be included in sequencing due to cost constraints and one sample acted as an outlier (data not shown), which was later removed from the downstream analysis. Ten children with septic shock and 5 nonsepsis children were selected for RNA sequencing. The experimental flow chart of this study is shown in Figure 1. The median age of the patients in survivors (n-5) and nonsurvivors (n-5) was 15 (13, 24) months and 180 (180, 184) months, respectively. The clinical characteristics of the study cohort are shown in Supplementary Table 1.
Fig. 1.
Study flowchart
Table 1.
Summary of distribution of differentially expressed genes
| Comparison | No. of significant genes* | No. of upregulated genes* | No. of downregulated genes* |
|---|---|---|---|
| Sepsis survivor vs nonsepsis | 2,457 | 983 | 624 |
| Sepsis nonsurvivor vs nonsepsis | 5,562 | 1,853 | 1,761 |
| Sepsis nonsurvivor vs sepsis survivor | 1,294 | 383 | 486 |
*Significant – p-value < 0.05 and Log2Fc ≥1 and ≤–1; 52% of the genes were found to be expressed with >10 average read count across all samples in the study
Principal Component Analysis and Sample Clustering
Supplementary Table 2 summarizes the overall data generated and the average read quality observed across the reads. Data generation for each sample was in the range of 7–19 GB and the average % Q30 was near 98%. Sample alignment to the Hg38 reference genome was in the range of 96–98%. Unaligned reads were in the range of 2–3%.
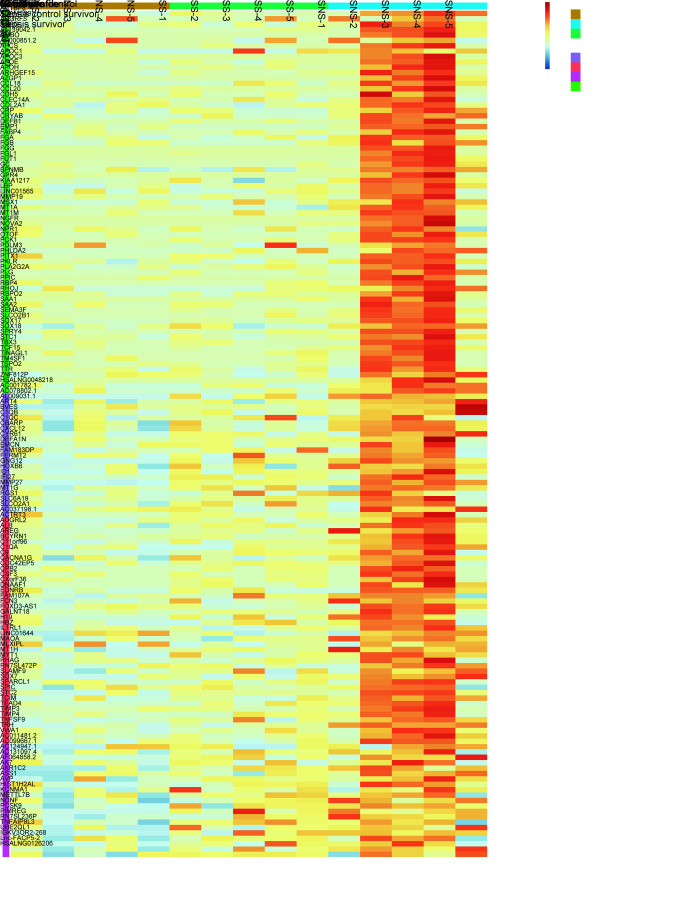
As part of the data quality check, the initial clustering of the samples was observed through a Principal Component Analysis (PCA) plot. The normalized gene expression of all the genes quantified through Feature Counts and later transformed through vast function was used as input. The PCA plot representing the grouping of the samples after the outlier removal is shown in Supplementary Figure 1. Later, the effect of multiple covariates like gender and age (Supplementary Figure 2A) was checked for their effect on gene expression using principle component analysis as well as using hierarchical clustering of highly variant top 500 genes which was visualized as a heatmap (Supplementary Figure 2B). It was observed that based on the top variable 500 genes, it was capable of clustering the samples based on their groups. A set of genes was found with high expression in sepsis nonsurvivor (SNS) while the same found with mild expression in SS, which were analyzed further through differential gene expression analysis. Gender did not have an effect on the clustering of the samples, however, and age did have an effect on the sample clustering. Hence, age was considered as a covariate during the differential expression analysis of the genes.
Fig. 2.
Heat map: Upregulated genes (157) across nonsepsis children and septic shock patients (survivors and nonsurvivors)
Markers Distinguishing Nonsurvival Patients from Survival Patients for Sepsis and Related Pathways
More than half (52%) of the genes were found to be expressed with >10 average read count across all samples in the study. The summary of the distribution of differentially expressed genes (DEGs) in the different comparisons performed is given in Table 1.
By making use of all the statistically significant (p-value < 0.05) genes from the 3 comparisons, a Venn diagram analysis (Supplementary Figure 3) was performed that provided us with the unique list of de-regulated genes within SNS in comparison to SS. 421 genes (184 upregulated and 237 downregulated) were found exclusively dysregulated within SNS relative to SS. The complete list of deregulated genes specific to SNS in comparison to SS along with their Reactome pathway annotation and list of overrepresented pathways leveraged by the DAVID online resource has been listed in Supplementary Table 3. The pathways undertaken by the 421 genes specific to SNSs have been provided in Supplementary Figure 4.
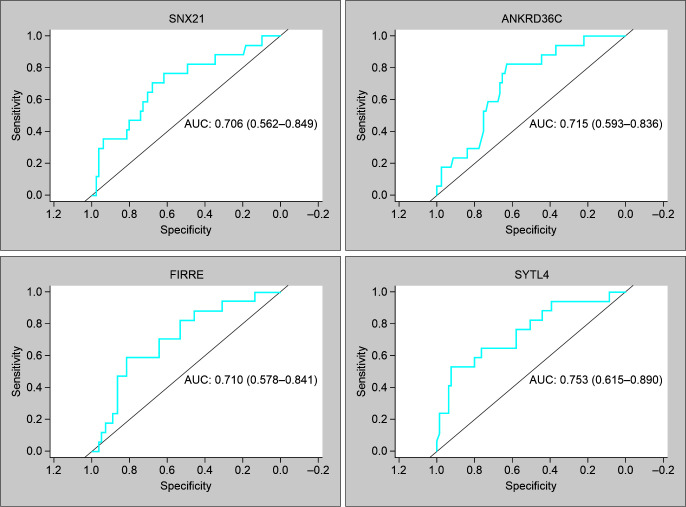
Fig. 3.
ROC curves of genes possessing AUC > 0.7 in GSE26440 to differentiate between septic shock survivors and nonsurvivors
Gene Set Enrichment Analysis of the Dysregulated Genes in Survivors, Nonsurvivors, and Nonsepsis
The Hallmark gene sets, Gene ontology Biological process, Molecular Function, and Reactome pathways were used for the GSEA using the dysregulated genes resulted for each of the three comparisons performed (Supplementary Table 4).
Sepsis Survivors and Nonsepsis Children
Biological processes like Ribonucleo-protein complex biogenesis, positive regulation of cytokine production, and cellular nitrogen compound catabolic process were found to be upregulated while those like regulation of post-synaptic membrane potential and response to lectin were found to be downregulated within the SS in comparison to the NS. Molecular functions like cadherin binding and signaling receptor regulator activity were found to be upregulated while those like functions related to ion channel activity and neuro-transmitter receptor activity were found to be downregulated within the SS in comparison to the NS. The majority of the Hallmark genesets were found to be upregulated including Myc targets and E2F targets while those like Kras signaling and Wnt Beta catenin signaling were found downregulated within the SS relative to the NS. Pathways like Rho-GTPase effectors, Neutrophil degranulation, and Wnt signaling were found upregulated while many pathways related to neuronal transmission were found downregulated within the SS relative to the NS (Supplementary Figure 5).
Sepsis Nonsurvivors and NS Children
Biological processes like negative regulation of protein modification process and negative regulation of intracellular signal transduction were found to be upregulated while those like cilium-dependent cell motility, positive regulation of cell killing, and definitive hemopoiesis were found to be downregulated within the SNS in comparison to the NS. Molecular functions like those related to transcription factor binding and signaling receptor regulator activity were found to be upregulated while those like functions related to ion channel activity and neuro-transmitter receptor activity were found to be downregulated within the SNS in comparison to the NS as similar to observed in SS. The majority of the Hallmark gene sets were found to be upregulated within the SNS as well as observed in SS including E2F targets and TNFA signaling via NFKB while Hedgehog signaling was found downregulated in addition to Wnt beta-catenin signaling, in the SNS relative to the NS, which was found opposite to what was observed within the SS. Also, the Kras signaling function was found upregulated in the nonsurvivors opposite to the expression observed in survivors of sepsis. Pathways like signaling by receptor tyrosine kinases were found upregulated in addition to those found upregulated within the survivors while PD-1 signaling, DAP12 interactions, and generation of secondary messenger molecules were found downregulated within the SNS relative to the NS (Supplementary Figure 6).
Sepsis Nonsurvivor and Survivor
Biological processes like phagocytosis recognition, humoral immune response, and cellular response to cortico-steroid stimulus were found to be upregulated while those like leukocyte-mediated cytotoxicity and positive regulation of lymphocyte-mediated immunity were found to be downregulated within the SNS in comparison to the SS. Molecular functions like those related to metallopeptidase activity and immuno-globulin receptor binding were found to be upregulated while those like functions related to chemokine activity and MHC class II protein complex binding were found to be downregulated within the SNS in comparison to the SS. The majority of the Hallmark gene sets were found to be upregulated within the SNS including hypoxia, Heme metabolism, and Wnt beta-catenin signaling while Interferon alpha, gamma response, allograft rejection, androgen response, and Myc targets were found downregulated in the SNS relative to the SS. Pathways like the creation of C4, C2 activators, scavenging of heme from plasma, and CD22 mediated B cell receptor (BCR) regulation were found upregulated while pathways like platelet activation signaling and aggregation, pathways related to interferon signaling and fibrin clot formation were found downregulated within the SNS relative to the SS (Supplementary Figure 7).
Genes Associated with Sepsis Development – Short Time-series Expression Miner (STEM) Analysis
Further, the normalized set of genes was screened for expression patterns within the samples present in the study, using a short time-series expression miner (STEM). The trend of gene expression from three groups of samples were considered further based on the results of STEM analysis among NS, SS and SNS samples. After Bonferroni correction of the p-value, 4 profiles of genes were obtained with significant p-value all belonging to a single cluster. Four of the gene profiles totaling to 157 genes were increasing their expression progressively from SS to SNS (Fig. 2). Most of these progressive upregulated genes were involved in the complement and coagulation cascade which supports the fact that severe sepsis is almost invariably associated with systemic activation of coagulation. Other functions enriched within these 157 genes were the regulation of IGF transport and uptake by IGFBPs, folate metabolism, and negative regulation of blood coagulation. It is to be noted that though majority of the 157 genes belonged to the category of protein coding genes, some of them belonged to lncRNA as well (Supplementary Table 5).
Diagnostic Values of Mortality Signatures to Differentiate between Nonsurvivors and Survivors of Sepsis
The 421 genes which were detected specifically within the SNS relative to SS were further analyzed for their diagnostic value to differentiate between survivors and nonsurvivors using external datasets from the NCBI GEO database. It was observed that within the external pediatric cohort (GSE26440), four genes namely, SNX21, FIRRE, ANKRD36C, and SYTL4 possessed AUC values > 0.7 (Fig. 3). When the same set of 421 genes was validated within an adult cohort (GSE185263), it was seen that none of them had AUC values >0.7 and the maximum AUC value the genes possessed was 0.66. The 157 sepsis development genes obtained through the STEM analysis were also screened for their mortality-based diagnostic value in the external cohorts mentioned above. It was seen that out of 157 genes, 6 of them namely TIMP3, AREG, APOC1, SLAMF9, ASS1, and RGS1 possessed AUC > 0.7 within the pediatric cohort while within the adult cohort, the maximum AUC value of the genes possessed was 0.68. Out of the 2 best sepsis development genes having AUC > 0.7, TIMP3 and AREG were found to have good diagnostic value within the adult cohort as well (AUC > 0.6) (Supplementary Table 6).
Mortality Signatures and Sepsis Development Gene Sets are Prognostic in Nature
We checked the prognostic value of the selected genes in with external Adult cohort (GSE65682). It was observed that from both the 421 genes mortality signature and from within the 157 sepsis development genes, there existed genes whose expression was significantly correlated with the patient outcome.
Out of the mortality gene signatures (n = 421), higher expression of the genes APOL6, TAP2, GBP3, RTP4, GPR18, TRAV20, and DDX60 was associated with higher survival rates of patients with sepsis (p < 0.05). While higher expression of the genes EFEMP2, CA1, PRODH, NAPRT1, TMEM119, and CHKA was associated with a poor outcome in septic patients (p < 0.05) (Supplementary Figure 8A). Out of the Sepsis development gene signatures (n = 157), higher expression of the genes MAOA, RHAG, ID1, PCSK9, GALNTL4, FAM107A, MT1G, LRRC50, ADAMTS9, and SLC6A19 was associated with a poor outcome in septic patients (p < 0.05) (Supplementary Figure 8B).
Immune Cell Infiltration in Sepsis
CIBERSORTx was used to calculate the cell type fractions of four categories of cell types, namely, T cells, adaptive immune cells, innate immune cells, and macrophages across all samples from survivors, nonsurvivors of sepsis, and nonsepsis controls. It was observed that the cell type fractions of Treg, macrophages, and mast cell resting were significantly different between NS and SNS (p-value = 0.02, 0.027, and 0.032, respectively) (Additional Results 1).
Discussion
In our study on transcriptomic profiling of pediatric sepsis, we have three main findings: (a) the biological cues on the difference between septic shock survivors and nonsurvivors in terms of DEGs; (b) sepsis development gene signature shows higher expression in nonsurvivors; and (c) the pathways related to coagulation and platelet degranulation is dysregulated in the nonsurvivor group.
Biological processes like lymphocyte mediated immunity was found low in nonsurvivors of sepsis in comparison to sepsis survivors, where blood lymphocyte dysfunction. Similar findings has been studied in sepsis where it would be accompanied by decreased lymphocyte T CD4+, CD8+, and natural killer cells in patients with septic shock.15 The suppression of the mentioned pathway in nonsurvivors could be explained by the reduction observed in the abundance of lymphocyte cell population in nonsurvivors, as quantified by the in silico flow cytometry algorithm of CIBERSORTx.
Platelet activation signaling and aggregation and fibrin clot formation were found to be downregulated within the nonsurvivors while coagulation and complement pathway were found to overexpressed as the sepsis condition worsened towards septic shock. As the severity of sepsis increases, it has been reported to result in Disseminated Intravascular Coagulation (DIC),16 which is characterized by diffuse activation of the coagulation system. Reduced fibrinogen accompanied by a reduction in fibrin clot formation has been reported to be present only during the late course of sepsis.17
The mortality-related signatures detected from the current study would help in predicting the outcome of the septic shock patients irrespective of the age, including the FIRRE, TIMP3, and AREG genes. The FIRRE intergenic repeating RNA element belongs to the long noncoding RNA class and is studied to be responsive to the NF-κB signaling in macrophages which in turn positively regulates the expression of inflammatory genes taking part in innate immunity.18 A recent study has provided evidence on FIRRE locus having physiological roles in hematopoiesis,19 which does explain their possibility of acting as a potential biomarker in differentiating between nonsurvivors and survivors. Many of the 421 genes found specifically in the nonsurvivors were long non-coding in nature (lncRNA), which brings up the importance of profiling the expression of non-coding RNAs from sepsis patients and analyzing their role within the pathogenesis of this disease. Other potential mortality markers TIMP3 and AREG, exhibiting notable AUROC values in both pediatric as well as adult cohorts of sepsis, are known to contribute to septic microvascular endothelial cells (MVEC) barrier20 and disrupted tolerance towards tissue or organ damage associated with acute or chronic inflammation, respectively. AREG was originally identified as a potential early marker for inflammation in preterm babies diagnosed with sepsis through single-cell RNA-Seq assay.21
One of the important finding in our study was able to identify prognostic markers for predicting outcome in septic shock patients. Some of the genes like TAP2, GPR18,22 PCSK923 have already been identified as prognostic markers while others like APOL6, GBP3, RTP4, TRAV20, DDX60, EFEMP2, CA1, PRODH, NAPRT1, TMEM119, CHKA, MAOA, RHAG, ID1, GALNTL4, FAM107A, MT1G, LRRC50, ADAMTS9, and SLC6A19 can be putative prognostic markers for sepsis identified from the current study which requires further validation within larger pediatric cohort.
The EFEMP2 gene was found as a mortality biomarker differentiating between survivors and nonsurvivors of sepsis and as a prognostic marker (p-value < 0.05). This gene though have not been studied in depth about its molecular action of pathogenesis in sepsis, have been established for its role in over expression in macrophages under lipopolysaccharide treatment caused due to bacterial infections.24
Limitations of the Study
Our study on transcriptomic profiling in a specific subset of critically ill patients with septic shock adds to the strength of the study. Moreover, this is the first study in children with septic shock from India. We acknowledge certain limitations. First, the study is based on a single center with a small sample size. Multicenter studies with a larger sample size are needed. Second, our patient population was heterogeneous in age, which may be confounding factors. Nevertheless, we have performed the diagnostic value validation in two cohorts, across different age groups (<10 years and >18 years). Third, major limitation is that there was no healthy control involved in the study and hence, based on the finding the diagnostic performance to differentiate between healthy and septic shock patients cannot be recommended. Despite these limitations, the findings have strong biological significance and enhance our understanding of the differences in the host response to septic shock in children.
Conclusion
This is the first study from the Indian cohort identifying the gene expression pattern in children with septic shock. The mortality-related signatures detected from the current study may help in predicting the outcome of septic shock. The pathways related to coagulation and platelet degranulation deregulated in the non survivors. Hence, targeting hemostatic disruptors/inhibitors would be an elective way to prevent fatality due to sepsis.
Acknowledgment
The authors would like to thank Theracues Innovations Pvt. Ltd., Bengaluru, India, a service provider for RNA extraction, sequencing, and bioinformatic analysis.
Supplementary Materials
All the supplementary materials are available online on the website of www.ijccm.org.
Orcid
Lalitha AV https://orcid.org/0000-0002-6168-1677
Anil Vasudevan https://orcid.org/0000-0003-4338-0325
Manju Moorthy https://orcid.org/0009-0002-5282-9147
Gopalakrishna Ramaswamy https://orcid.org/0009-0004-4643-4274
Footnotes
Source of support: This work was supported by a grant from the Rajiv Gandhi University of Health Sciences (RGU/ADV-RES/BR/19/2019-20).
Conflict of interest: None
References
- 1.Kissoon N, Carcillo JA, Espinosa V, Argent A, Devictor D, Madden M, et al. World federation of pediatric intensive care and critical care societies: Global sepsis initiative. Pediatr Crit Care Med. 2011;12(5):494–503. doi: 10.1097/PCC.0b013e318207096c. [DOI] [PubMed] [Google Scholar]
- 2.Kissoon N, Argent A, Devictor D, Madden MA, Singhi S, van der Voort E, et al. World federation of pediatric intensive and critical care societies‐its global agenda. Pediatr Crit Care Med. 2009;10(5):597–600. doi: 10.1097/PCC.0b013e3181a704c6. [DOI] [PubMed] [Google Scholar]
- 3.Khan MR, Maheshwari PK, Masood K, Qamar FN, Haque AU. Epidemiology and outcome of sepsis in a tertiary care PICU of Pakistan. Indian J Pediatr. 2012;79(11):1454–1458. doi: 10.1007/s12098-012-0706-z. [DOI] [PubMed] [Google Scholar]
- 4.Hartman ME, Linde‐Zwirble WT, Angus DC, Watson RS. Trends in the epidemiology of pediatric severe sepsis. Pediatr Crit Care Med. 2013;14(7):686–693. doi: 10.1097/PCC.0b013e3182917fad. [DOI] [PubMed] [Google Scholar]
- 5.Kaur G, Vinayak N, Mittal K, Kaushik JS, Aamir M. Clinical outcome and predictors of mortality in children with sepsis, severe sepsis, and septic shock from Rohtak, Haryana: A prospective observational study. Indian J Crit Care Med. 2014;18(7):437–441. doi: 10.4103/0972-5229.136072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anthwal P, Kumar N, Manchanda A, Garg B. Six‐hour sepsis bundle decreases mortality: Truth or illusion – A prospective observational study. Indian J Crit Care Med. 2018;22(12):852–857. doi: 10.4103/ijccm.IJCCM_147_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Singer M, Deutschman CS, Seymour CW, Shankar-Hari M, Annane D, Bauer M, et al. The Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3). JAMA. 2016;315(8):801. doi: 10.1001/jama.2016.0287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wiersinga WJ, Leopold SJ, Cranendonk DR, van der Poll T. Host innate immune responses to sepsis. Virulence. 2014;5(1):36–44. doi: 10.4161/viru.25436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aziz M, Jacob A, Yang WL, Matsuda A, Wang P. Current trends in inflammatory and immunomodulatory mediators in sepsis. J Leukoc Biol. 2013;93(3):329–342. doi: 10.1189/jlb.0912437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Delano MJ, Ward PA. Sepsis-induced immune dysfunction: Can immune therapies reduce mortality? J Clin Invest. 2016;126(1):23–31. doi: 10.1172/JCI82224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferrer R, Martin-Loeches I, Phillips G, Osborn TM, Townsend S, Dellinger RP, et al. Empiric antibiotic treatment reduces mortality in severe sepsis and septic shock from the first hour. Crit Care Med. 2014;42(8):1749–1755. doi: 10.1097/CCM.0000000000000330. [DOI] [PubMed] [Google Scholar]
- 12.Goldstein B, Giroir B, Randolph A, International Consensus Conference on Pediatric Sepsis. International pediatric sepsis consensus conference: Definitions for sepsis and organ dysfunction in pediatrics. Pediatr Crit Care Med. 2005;6(1):2–8. doi: 10.1097/01. [DOI] [PubMed] [Google Scholar]
- 13.Pollack MM, Patel KM, Ruttimann UE. PRISM III: An updated pediatric risk of mortality score. Crit Care Med. 1996;24(5):743–752. doi: 10.1097/00003246-199605000-00004. [DOI] [PubMed] [Google Scholar]
- 14.Davis AL, Carcillo JA, Aneja RK, Deymann AJ, Lin JC, Nguyen TC, et al. American College of Critical Care Medicine Clinical Practice Parameters for Hemodynamic Support of Pediatric and Neonatal Septic Shock. Crit Care Med. 2017;45(6):1061–1093. doi: 10.1097/CCM.0000000000002425. [DOI] [PubMed] [Google Scholar]
- 15.De Pablo R, Monserrat J, Prieto A, Alvarez-Mon M. Role of circulating lymphocytes in patients with sepsis. BioMed Res Int. 2014;2014:1–11. doi: 10.1155/2014/671087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Simmons J, Pittet JF. The coagulopathy of acute sepsis. Curr Opin Anaesthesiol. 2015;28(2):227–236. doi: 10.1097/ACO.0000000000000163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsantes AG, Parastatidou S, Tsantes EA, Bonova E, Tsante KA, Mantzios PG, et al. Sepsis-induced coagulopathy: An update on pathophysiology, biomarkers, and current guidelines. Life. 2023;13(2):350. doi: 10.3390/life13020350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu Y, Liu X, Xie M, Liu M, Ye M, Li M, et al. The NF-κB–responsive long noncoding RNA FIRRE regulates posttranscriptional regulation of inflammatory gene expression through interacting with hnRNPU. J Immunol. 2017;199(10):3571–3582. doi: 10.4049/jimmunol.1700091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lewandowski JP, Lee JC, Hwang T, Sunwoo H, Goldstein JM, Groff AF, et al. The firre locus produces a trans-acting RNA molecule that functions in hematopoiesis. Nat Commun. 2019;10(1):5137. doi: 10.1038/s41467-019-12970-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arpino V, Mehta S, Wang L, Bird R, Rohan M, Pape C, et al. Tissue inhibitor of metalloproteinases 3-dependent microvascular endothelial cell barrier function is disrupted under septic conditions. Am J Physiol-Heart Circ Physiol. 2016;310(11):H1455–H1467. doi: 10.1152/ajpheart.00796.2015. [DOI] [PubMed] [Google Scholar]
- 21.Das A, Ariyakumar G, Kamdar S, Barugahare A, Deveson D, Gee S, et al. Rapid immune perturbations during sepsis and necrotising enterocolitis in preterm babies. Research Square 2022. [DOI] [Google Scholar]
- 22.Zhao M, Zheng Z, Yin Z, Zhang J, Qin J, Wan J, et al. Resolvin D2 and its receptor GPR18 in cardiovascular and metabolic diseases: A promising biomarker and therapeutic target. Pharmacol Res. 2023;195:106832. doi: 10.1016/j.phrs.2023.106832. [DOI] [PubMed] [Google Scholar]
- 23.Chen Z, Chen R, Ou Y, Lu J, Jiang Q, Liu G, et al. Construction of an HLA classifier for early diagnosis, prognosis, and recognition of immunosuppression in sepsis by multiple transcriptome datasets. Front Physiol. 2022;24(13):870657. doi: 10.3389/fphys.2022.870657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Argraves WS, Greene LM, Cooley MA, Gallagher WM. Fibulins: Physiological and disease perspectives. EMBO Rep. 2003;4(12):1127–1131. doi: 10.1038/sj.embor.7400033. [DOI] [PMC free article] [PubMed] [Google Scholar]