Abstract
G207 is an oncolytic herpes simplex virus (HSV) which is attenuated by inactivation of viral ribonucleotide reductase (RR) and deletion of both γ134.5 genes. The cellular counterparts that can functionally substitute for viral RR and the carboxyl-terminal domain of ICP34.5 are cellular RR and the corresponding homologous domain of the growth arrest and DNA damage protein 34 (GADD34), respectively. Because the thymidylate synthetase (TS) inhibitor fluorodeoxyuridine (FUdR) can alter expression of cellular RR and GADD34, we examined the effect of FUdR on G207 bioactivity with the hypothesis that FUdR-induced cellular changes will alter viral proliferation and cytotoxicity. Replication of wild-type HSV-1 was impaired in the presence of 10 nM FUdR, whereas G207 demonstrated increased replication under the same conditions. Combined use of FUdR and G207 resulted in synergistic cytotoxicity. FUdR exposure caused elevation of RR activity at 10 and 100 nM, whereas GADD34 was induced only at 100 nM. The effect of enhanced viral replication by FUdR was suppressed by hydroxyurea, a known inhibitor of RR. These results demonstrate that the growth advantage of G207 in FUdR-treated cells is primarily based on an RR-dependent mechanism. Although our findings show that TS inhibition impairs viral replication, the FUdR-induced RR elevation may overcome this disadvantage, resulting in enhanced replication of G207. These data provide the cellular basis for the combined use of RR-negative HSV mutants and TS inhibitors in the treatment of cancer.
G207 is a ribonucleotide reductase (RR)-negative herpes simplex virus type 1 (HSV-1) which was designed for brain tumor therapy and is presently under clinical evaluation as a new treatment for malignant glioma (29, 31). Recently, it was demonstrated that this HSV mutant also demonstrates a high oncolytic potency against colorectal cancer cells (28).
The strategy of G207 typifies the strategy used by many candidate oncolytic viruses that specifically attempt to target tumor cells by deletion of viral RR and γ134.5. First, viral RR is inactivated by inserting the Escherichia coli lacZ gene into the infected cell protein 6 (ICP6) locus that codes for the large subunit of RR. RR catalyzes the reduction of ribonucleotides to the corresponding deoxyribonucleotides, thereby providing sufficient precursors for the de novo synthesis of DNA. In mammalian cells, RR is highly expressed during S phase and under DNA damage and repair conditions (4, 6, 17, 19, 39). Most herpesviruses encode their own RR, and their replication is therefore independent of the host cell cycle (5, 35). The inactivation of ICP6 in G207 makes viral DNA replication completely dependent on the cellular enzyme, and consequently replication of this mutant becomes largely dependent on host cell conditions. It is therefore reasonable to conceive that cell cycle alterations or DNA damage and repair conditions might modulate the replication of this herpes vector. The second mutation in G207 is the deletion of both γ134.5 loci. The γ134.5 gene encodes a protein (ICP34.5) with at least two functions. One allows HSV to replicate and spread within the central nervous system (10, 41). The second function confers HSV the ability to escape from a host defense mechanism against viral infections by preventing the cellular shutoff of protein synthesis (11, 24). This function can be substituted by the ICP34.5 homologous domain of the cellular growth arrest and DNA damage protein 34 (GADD34), a protein which is induced by DNA damage (24).
Chemotherapy is an established modality in the treatment of malignancies. Fluorodeoxyuridine (FUdR) is a widely used chemotherapeutic drug to treat colorectal cancer. It is rapidly converted to the active metabolite 2′-deoxy-5-fluorouridine 5′- monophosphate (FdUMP) by phosphorylation via thymidine kinase. FdUMP inhibits the enzyme thymidylate synthetase (TS) by forming a covalent complex with both the sulfhydryl residue of TS and methylenetetrahydrofolate. Inhibition of TS causes a depletion of dTTP with subsequent imbalance of intracellular deoxynucleotide triphosphate pools (13, 27, 42). This inhibition induces cytotoxicity through several mechanisms. Nucleotide pool imbalances have been shown to induce a specific endonuclease with double-strand breakage activity in FM3A cells (42). Other studies have demonstrated that excessive dUTP/dTTP ratios result in uracil misincorporation and misrepair leading to DNA strand breaks (3, 22, 26). The ability of FUdR to be incorporated into nascent DNA has been suggested as another mechanism of cytotoxic action (13). Furthermore, FUdR has profound effects on cell cycle and DNA replication by causing early S-phase blockade, loss of histone H1, and retarded DNA elongation (15).
The cellular counterparts which can functionally replace the viral RR and the carboxyl-terminal domain of ICP34.5 are cellular RR and the corresponding homologous domain of GADD34, respectively. The diverse effects of FUdR on cell cycle and DNA suggest that this drug may influence the expression of these two cellular proteins and thus the replication of G207. Therefore, we investigated the interaction between FUdR and G207 with special emphasis on cellular RR and GADD34. In these studies we examined two colorectal cell lines expressing different degrees of sensitivity to FUdR. Here, we report that FUdR-induced cellular alterations provide an environment favorable for replication of G207. Based on the two major mutations in G207, we also demonstrate mechanisms by which FUdR induces enhanced replication of this HSV mutant.
MATERIALS AND METHODS
Cell lines and culture.
HCT8 cells with two different degrees of sensitivity to 5-fluorouracil (5-FU) and FUdR were used for this study. HCT8 cells were obtained from the American Type Culture Collection (CCL-224; Rockville, Md.). The resistant cell line was cloned from HCT8 cells after exposure to 15 μM 5-FU for 7 days (HCT8/FU7dR) as previously described (1). Both cell lines were maintained in RPMI 1640 supplemented with 10% fetal calf serum (FCS), 100 μg of penicillin/ml, and 100 μg of streptomycin/ml. Vero cells (African green monkey kidney) were grown in Eagle's minimal essential medium supplemented with 10% FCS.
Viruses.
Creation of the multimutated, replication-competent type 1 herpes virus G207 has been described previously (31). G207 was constructed from the R3616 mutant based on wild-type HSV-1 strain F. This mutant contains a 1-kb deletion from the coding domains of both γ134.5 loci and an insertion of the E. coli lacZ gene into the ICP6 gene, which encodes the large subunit of RR. HSV-1(F) is the parental wild-type virus of G207, whereas KOS is a different strain of wild-type HSV-1. Viruses were propagated on Vero cells. G207 was a gift of S. D. Rabkin and R. L. Martuza. HSV-1(F) and KOS were provided by NeuroVir, Inc. (Vancouver, Canada).
p53 mutational analysis.
Genomic DNA was extracted from HCT8 and HCT8/7dR cells. Exons 5 through 9 of the p53 gene were amplified by PCR and analyzed for mutations by single-strand confirmation polymorphism.
Cytotoxicity assay.
The cytotoxicity of G207 and FUdR (Floxuridine; Roche Laboratories Inc., Nutley, N.J.) was assessed by measuring cytoplasmic lactate dehydrogenase (LDH) activity (CytoTox 96 nonradioactive cytotoxicity assay; Promega, Madison, Wis.). All cytotoxicity assays were performed in 24-well plates starting with 2 × 104 cells per well. At various time points following the start of treatment, adherent cells were washed with phosphate-buffered saline (PBS) and cytoplasmic LDH was released by lysis buffer (PBS, 1.2% [vol/vol] Triton X-100). Activity of the lysate was measured with a coupled enzymatic reaction, which converts a tetrazolium salt into a red formazan product. Absorbance was measured at 450 nm using a microplate reader (EL 312e; Bio-Tek Instruments, Winooski, Vt.). Cytotoxicity was expressed as the percentage of maximal LDH release of treated cells compared to that of untreated cells (control).
Viral titration.
Vero cultures were carried for at least one subculture in a solution containing Eagle's minimal essential medium, 2 mM l-glutamine, 10% FCS. Cultures were plated at a density of 106 cells per well on 6-well plates and incubated at 37°C in 5% CO2 in air in a humidified incubator. The following day, cultures were washed twice with PBS and serial dilutions of cell lysates (0.8 ml/well) were adsorbed onto triplicate dishes for 4 h at 37°C. Cell lysates were prepared by four freeze-thaw cycles. Following adsorption, inoculum was removed and cultures were overlaid with agar-containing medium. Cultures were stained with neutral red 2 days postinoculation, and plaque formation was assessed the next day.
β-Galactosidase activity.
Activity of β-galactosidase was determined by monitoring the conversion of o-nitrophenyl galactoside (ONPG) to o-nitrophenol and galactose (β-Galactosidase Reporter Assay; Pierce, Rockford, Ill.). Cells were lysed and incubated with ONPG at 37°C for 30 min. The reaction rate was determined by spectrophotometric measurement of o-nitrophenol (λ = 405 nm). Using the molar extinction coefficient for o-nitrophenol (ɛλ = 4.5 × 103 M−1 cm−1), 1 U of β-galactosidase activity was defined as the cleavage of 1 nmol of ONPG to o-nitrophenol and galactose in 1 min at 37°C.
Histochemical staining for β-galactosidase.
Cells were trypsinized, resuspended in media, and washed with PBS. Cytospin slides were prepared by centrifuging 1 ml of a cell suspension containing 105 cells at 1,000 × g for 6 min. Slides were stained with X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) and incubated for 4 h at 37°C. After being washed with PBS, slides were counterstained with 0.1% nuclear fast red.
Cell cycle analysis.
Cell cycle analysis was performed on nuclear preparations by flow cytometry as previously described (32). Briefly, cell monolayers were carefully washed with PBS to remove cellular debris. Following trypsinization, cells were washed in PBS and resuspended in NP-40 solution (10 mM NaCl, 3.4 mM sodium citrate, 0.03% NP-40, 63 μM ethidium bromide, 10 μg of RNase A/ml). After 1 h of incubation at room temperature, an equal volume of high-sucrose solution (0.25 M sucrose, 78 mM citric acid, 100 μM ethidium bromide) was added. DNA content of ethidium bromide-stained nuclei was determined on a FACScalibur. Data were analyzed with FACStation running CellQuest software (Becton Dickinson, San Jose, Calif.).
Cell extraction of RR.
Cells grown in 250-cm2 flasks were trypsinized and washed twice in ice-cold PBS. Cells were centrifuged at 300 × g for 5 min at 4°C and resuspended in 3 volumes of low-salt extraction buffer (10 mM HEPES [pH 7.2], 2 mM dithiothreitol). Viable cell count of the resuspension was determined by trypan blue exclusion. After 30 min of incubation on ice, the cell suspension was drawn through a 28G½ needle 10 times. The crude homogenate was centrifuged at 100,000 × g for 60 min at 4°C to remove cellular debris. The supernatant fraction was dialyzed against 1,000 volumes of the low-salt extraction buffer for 4 h with one buffer change after 2 h using dialysis cassettes with a molecular weight cutoff of 10,000 (Slide-A-Lyzer Dialysis Cassettes; Pierce). The dialyzed extract was snap-frozen in liquid nitrogen and stored at −80°C until analysis. All extraction procedures were performed at 4°C.
Assay for CDP reductase activity.
Activity of RR was determined by a modified Steeper and Stuart method (38). Conversion of CDP to dCDP was monitored using [14C]CDP (53 mCi/mmol; Movarek Biochemicals, Inc.) as substrate and rattlesnake venom from Crotalus adamanteus (Sigma) to hydrolyze nucleotides to nucleosides. The reaction mixture contained the following concentrations of ingredients in a final volume of 150 μl: 40 μM CDP, 10 μM [14C]CDP (0.08 μCi), 6 mM dithiothreitol, 4 mM magnesium acetate, 2 mM ATP, 50 mM HEPES (pH 7.2), and 100 μl of extract (0.2 to 0.7 mg of protein). The enzyme reaction was carried out for 30 min at 37°C and stopped by incubation at 100°C for 4 min. Nucleotides were hydrolyzed by adding 50 μl of carrier dCMP (6 mM dCMP, 2 mM MgCl2, 6 mM Tris-HCl [pH 8.8]) and a 25-μl snake venom suspension (50 mg/ml). After 3 h of incubation at 37°C, the reaction mixture was heat inactivated by boiling for 4 min. Heat-precipitated material was removed by centrifugation at 14,000 × g for 10 min at room temperature. [14C]deoxycytosine was separated from [14C]cytosine by covalent chromatography using phenylboronic acid columns (BondElut PBA; Varian, Harbor City, Calif.). Triethenolamine buffer (pH 10) was added to the supernatant fraction to a final concentration of 0.4 M, and 1 ml of this mixture was applied to the column. Fractions were collected and measured for radioactivity by liquid scintillation spectrometry (LS 6000IC Liquid Scintillation System; Beckman Instruments, Inc., Fullerton, Calif.). One unit of enzyme activity was defined as conversion of 1 nmol of CDP to the product dCDP in 1 h at 37°C.
Northern hybridization.
Total cellular RNA was isolated by guanidine thiocyanate-phenol-chloroform extraction (8). RNA was denatured, electrophoresed through a 1.2% formaldehyde-agarose gel, and blotted to a nitrocellulose membrane by standard techniques. Following prehybridization, membranes were hybridized in 50% formamide at 40°C to a full-length GADD34 and β-actin cDNA probe labeled with [32P]dCTP by the random primer method. Membranes were washed and exposed to Hyperfilm (Amersham Pharmacia Biotech) at −80°C. Densitometric analysis was carried out on scanned films using NIH Image software. Relative GADD34 levels were calculated as the ratio GADD34/β-actin.
RESULTS
Synergistic cytotoxicity of G207 and FUdR.
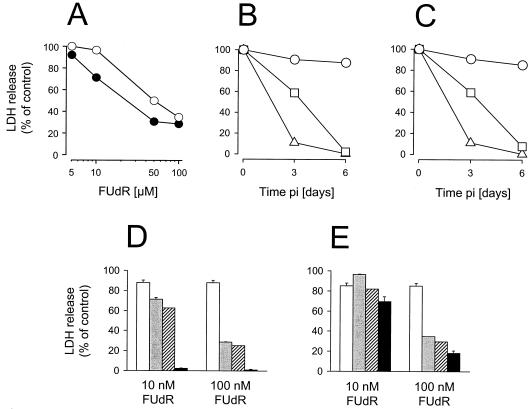
HCT8 cells were more sensitive to FUdR than were HCT8/7dR cells, as demonstrated by lower LDH release and a higher percentage of sub-G1 fraction (Fig. 1A and see Fig. 5A and B). Both cell lines showed similar viral cytotoxicity profiles. Viral infection at a multiplicity of infection (MOI) of 1.0 or 0.1 resulted in complete cell killing at day 6, while G207 at an MOI of 0.01 had only marginal cytotoxic effects (Fig. 1B and C). To test the hypothesis that FUdR can enhance viral cytotoxicity, we decided to use G207 at an MOI of 0.01, since viral cytotoxicity at MOIs of 1.0 and 0.1 was excessively high. G207 (MOI 0.01) combined with either 10 or 100 nM FUdR resulted in nearly complete killing of HCT8 cells by day 6 (Fig. 1D). This effect was higher than the calculated additive effect from each single treatment, indicating synergistic effects of combined treatment. Furthermore, the degree of synergy was more pronounced in HCT8 cells than in the less FUdR-sensitive cell line HCT8/7dR (Fig. 1D and E).
FIG. 1.
Cytotoxic effect of G207 and FUdR. Cell viability was assessed as a function of maximal release of intracellular LDH. (A through C) Cytotoxicity of G207 and FUdR. HCT8 (solid circle) and HCT8/7dR (open circle) cells were exposed to cumulative FUdR concentrations (5, 10, 50, and 100 nM), and viability was determined at day 6 following the start of treatment (A). Viral cytotoxicities in HCT8 (B) and HCT8/7dR (C) at day 3 and 6 p.i. are also shown. Cells were infected with G207 at MOIs of 1.0 (triangles), 0.1 (squares), and 0.01 (circles). (D and E) Cell viability for single (G207 or FUdR) and combined treatment in HCT8 (D) or HCT8/7dR (E) cells at day 6. Cells were infected with G207 at an MOI of 0.01 (open bars), exposed to FUdR (gray bars), or treated with G207 (MOI, 0.01) and FUdR (10 nM or 100 nM) in combination (black bars). Additive effects were calculated as the product from each single treatment (striped bars). All assays were performed in quadruplicate for each condition (averages ± standard errors of the means).
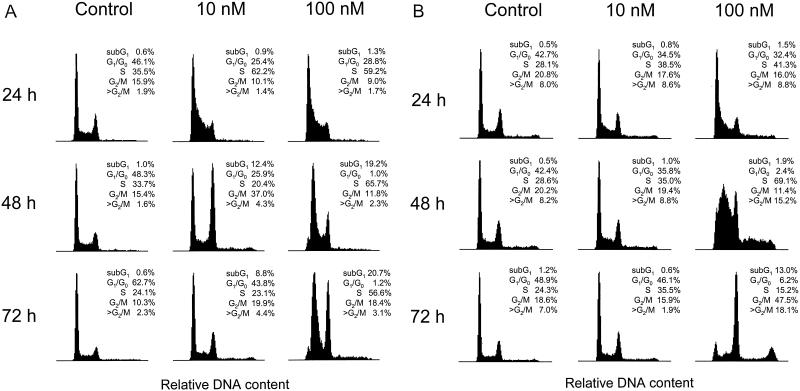
FIG. 5.
Effect of FUdR on cell cycle. Asynchronously growing cells (106) were plated in 20 ml of medium onto 75-cm2 flasks. Twelve hours later FUdR was added to the medium to final concentrations of 10 and 100 nM. Untreated cells served as the control. DNA content was measured on ethidium bromide-stained nuclei by fluorescence-activated cell sorter analysis at 24, 48, and 72 h following the start of treatment. Cell cycle analysis of HCT8 (A) and HCT8/7dR (B) cells was performed based on the shown side-scatter histograms. Histograms were gated for the sub-G1 fraction (<G1/G0 and >G2/M).
Increased β-galactosidase expression in the presence of FUdR.
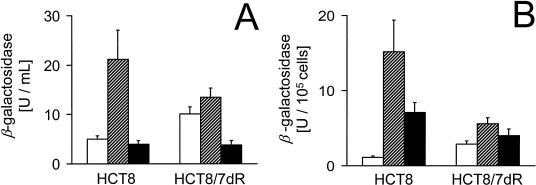
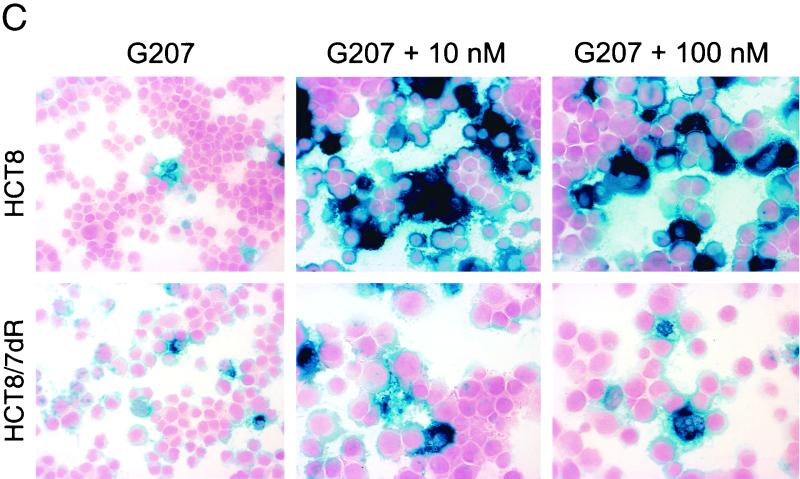
To assess the effect of FUdR on viral infectivity, the activity of β-galactosidase was measured as the product of the lacZ reporter gene in G207. Cells infected with G207 at an MOI of 0.01 followed by treatment with 10 nM FUdR showed the highest total expression of β-galactosidase at day 3 postinfection (p.i.); however, when normalized to viable cell count, exposure to 100 nM FUdR resulted in higher β-galactosidase activity than treatment with G207 only (Fig. 2A and B). Histochemical staining for β-galactosidase of FUdR-exposed cells showed greater staining intensity and a higher proportion of cells positive for staining with X-Gal (Fig. 2C). The degree of enhanced infection by FUdR was more pronounced for HCT8 than for HCT8/7dR cells.
FIG. 2.
Influence of FUdR on β-galactosidase expression following infection with G207. Cells (2 × 104) were plated in 24-well plates, infected with G207 at an MOI of 0.01 in the presence (striped bars, 10 nM; black bars, 100 nM) or absence (open bars) of FUdR. (A) At day 3 following infection cells were lysed and total β-galactosidase activity of cell lysates was measured. (B) Cell counts for each condition were determined in additional wells by trypan blue exclusion, and specific activity was calculated by referring total activity to viable cell number. All assays were performed in triplicate (averages ± standard errors of the means). For lacZ staining, 3 × 105 cells were plated onto 25-cm2 flasks. Twelve hours later cells were infected with G207 in the presence or absence of FUdR under the same conditions as described above. At day 3 p.i., cytospin slides were prepared and stained with X-Gal for lacZ expression. (C) Representative fields for different treatment conditions. Magnification, ×200.
Replication is enhanced for G207 but decreased for wild-type HSV-1 in the presence of FUdR.
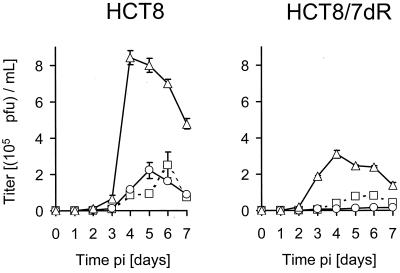
Single-step growth analysis demonstrated a 2-log-higher viral yield for wild-type HSV-1 [HSV-1(F) and KOS] compared to that of G207 in HCT8 cells. Interestingly, burst size of G207 in the presence of FUdR was threefold higher at 36 h p.i. than that of G207 alone, whereas replication of the parental wild-type virus HSV-1(F) was somewhat inhibited under the same conditions. We tested another wild-type HSV-1 (strain KOS) and found a similar reduction of replication in the presence of 10 nM FUdR (Table 1). Multiple-step growth analysis revealed significantly higher viral production in the presence of 10 nM FUdR in both cell lines compared to that of G207 infection alone. Peak titers and overall production of G207 were higher for the parental cell line HCT8 than for HCT8/7dR cells (Fig. 3).
TABLE 1.
Replication of wild-type HSV-1 and G207 in the presence or absence of FUdR
| Virus medium | Viral yield (105 PFU) for a:
|
||
|---|---|---|---|
| HSV-1(F) | KOS | G207 | |
| Virus alone | 325 ± 25 | 119 ± 10 | 1.16 ± 0.22 |
| Virus plus 10 nM FUdR | 215 ± 10 | 57 ± 2 | 3.42 ± 0.28 |
HCT8 cells were infected with HSV-1(F), KOS, or G207 at an MOI of 2. After adsorption for 1 h at 37°C, inoculum was removed, cells were washed with PBS, and medium containing 10 nM FUdR or control medium without FUdR was added. At 36 h p.i., cells and supernatant were harvested and titers of lysates were determined on Vero cells by standard plaque assay. Data are presented as averages ± standard errors of the means of three independent determinations. The fold increase of viral yield between virus alone and virus plus 10 nM FUdR was 0.66 for HSV-1(F), 0.48 for KOS, and 2.95 for G207.
FIG. 3.
Effect of FUdR on viral replication. Viral titers were determined to evaluate the influence of FUdR on viral replication. Cells (5 × 104 per well) were plated in 12-well plates. Twelve hours later cells were infected at an MOI of 0.01 in the presence (▵, 10 nM; ○, 100 nM) or absence (□) of FUdR. Supernatants and cells were harvested daily for the following 7 days p.i., and titers of lysates were determined on Vero cells by standard plaque assay. All assays were performed in triplicate for each condition (averages ± standard errors of the means).
Effect of HU on viral replication and β-galactosidase expression.
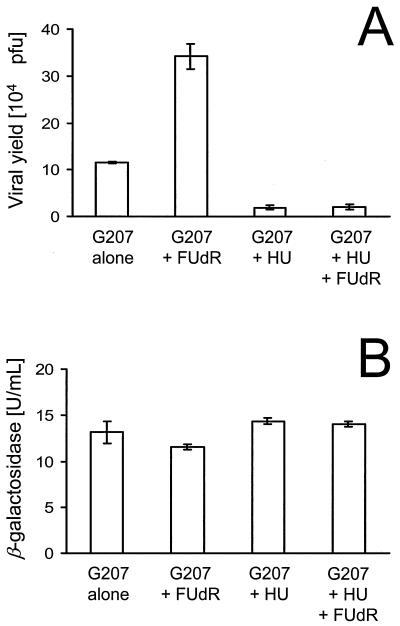
The RR inhibitor hydroxyurea (HU) suppressed viral replication in HCT8 cells by 90%. Furthermore, HU was able to extinguish the FUdR-induced enhanced replication of G207. The degree of inhibition was the same for cells treated with HU alone (1.9 × 104 ± 0.5 × 104 PFU) and for cells treated with HU and FUdR (2.1 × 104 ± 0.5 ×104 PFU). In contrast to viral production, neither FUdR nor HU had significant effects on β-galactosidase expression (Fig. 4).
FIG. 4.
Effects of FUdR and HU on viral replication. HCT8 cells were infected with 2 PFU of G207 per cell. After adsorption for 1 h at 37°C, inoculum was removed, cells were washed with PBS, and medium containing 10 nM FUdR or control medium without FUdR was added. At 8 h p.i., infected cells in the presence or absence of FUdR were exposed to 1 mM HU. At 36 h p.i., cells and supernatant were harvested and lysates were prepared by three cycles of freezing and thawing. Viral titers (A) and β-galactosidase activity (B) of the lysate were determined. All assays were performed in triplicate for each condition (averages ± standard errors of the means).
Cell cycle alteration by FUdR.
Exposure of asynchronously growing cells to FUdR resulted in an increase of the S-phase fraction and a decrease of the G1/G0 fraction; however, this effect was dependent on drug concentration and cell line. Low FUdR concentrations of 10 nM increased the S-phase fraction by 75% in HCT8 and 37% in HCT8/7dR by 24 h. By 48 h both cell lines showed a majority of cells in S phase following treatment with 100 nM FUdR. In contrast to HCT8 cells, which were completely blocked at 100 nM FUdR at S-phase level, HCT8/7dR cells showed only a transient S-phase increase and were able to transit to an apparent G2/M phase. Additionally, we observed that 10 to 20% of HCT8/7dR cells undergo DNA endoreduplication in S phase in the presence of 100 nM FUdR instead of moving to G2/M. Accumulation of these cells in G2/M phase may be due to there being a 4N DNA content resulting from this endoreduplication (Fig. 5A and B).
Elevated activities of RR in the presence of FUdR.
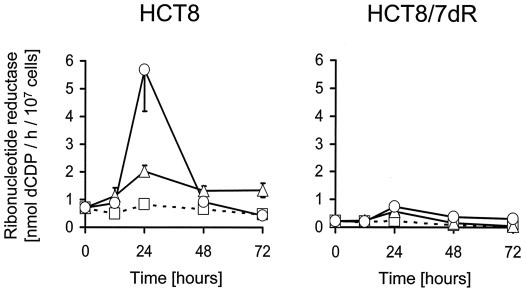
Since replication of G207 is dependent on cellular RR, we tested whether the TS inhibitor FUdR has any effects on RR. Baseline RR activity of exponentially growing cells was approximately 3.2-fold higher for HCT8 cells than for the chemoresistant cell line HCT8/7dR. Figure 6 shows the time-dependent course of RR activity during FUdR exposure. FUdR treatment resulted in an increase of RR activity in both cell lines. This increase was transient, and peak activities were observed simultaneously with the FUdR-induced S-phase elevation at 24 h after the start of treatment (Fig. 5A and B). The degree of activity induction was, however, more pronounced in HCT8 than in HCT8/7dR cells. RR activity in HCT8 treated with 10 nM FUdR remained elevated following peak activity.
FIG. 6.
Effect of FUdR on cellular RR activity. Cells (107) were plated onto 225-cm2 flasks. After 9 h FUdR was added to the medium to a final concentration of 10 or 100 nM. Untreated cells served as a control. RR activity was measured in cellular extracts at various time points in the presence (▵, 10 nM; ○, 100 nM) or absence (□) of FUdR. Activities referred to the cell count. All assays were performed in triplicate for each time point and condition (averages ± standard errors of the means).
Effect of FdUMP on the activity of RR.
FdUMP is the active metabolite of FUdR and inhibits TS. Activity of mammalian RR is highly regulated by feedback inhibition of deoxynucleotides; we therefore tested the idea that FdUMP, the fluorinated form of dUMP, inhibits the activity of RR, which could interfere with replication of G207. Table 2 shows a dose-dependent decline of enzyme activity. Concentrations of FdUMP of 0.001 to 0.1 mM caused only a moderate inhibition of RR, with approximately 80 to 70% RR activity remaining, respectively. Substantial enzyme inhibition was measured in the presence of 1 mM FdUMP, a 10,000-fold higher concentration than the maximal FUdR concentration used in this study.
TABLE 2.
Effect of FdUMP on RR activity
| FdUMP concn (mM) | RR activitya (nmol of dCDP/h) | Activity (%) | Inhibition (%) |
|---|---|---|---|
| 0 | 0.61 ± 0.05 | 100 | 0 |
| 0.001 | 0.47 ± 0.02 | 77.0 | 23.0 |
| 0.01 | 0.45 ± 0.02 | 73.8 | 26.2 |
| 0.1 | 0.43 ± 0.03 | 70.5 | 29.5 |
| 1 | 0.24 ± 0.02 | 39.3 | 60.7 |
| 10 | 0.14 ± 0.01 | 22.9 | 77.1 |
RR was extracted from exponentially growing HCT8 cells as described in Materials and Methods. Extracts were incubated with FdUMP in cumulative concentrations, and RR activity was measured. Data are presented as the averages ± standard errors of the means of three independent determinations of RR activity. One hundred microliters of dialyzed cell extract contained 0.65 mg of protein.
Expression of GADD34 in response to FUdR.
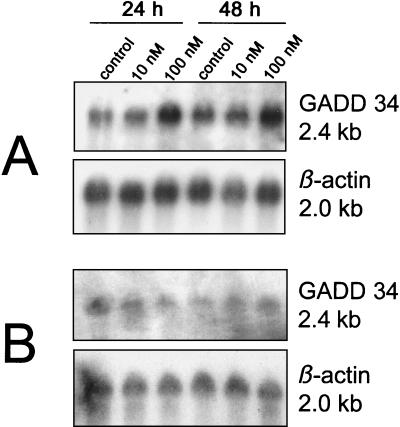
The GADD34 protein is expressed in response to DNA damage. GADD34 and the viral γ134.5 protein contain similar carboxyl-terminal domains that can functionally sustain protein synthesis under stress conditions. We therefore investigated whether FUdR as a DNA damaging agent can induce expression of GADD34, which can complement the γ134.5 deletions in G207. FUdR at 100 nM induced GADD34 in both cell lines, whereas 10 nM had almost no effect. Densitometric reading revealed a 1.9- and 1.6-fold higher mRNA level at 24 and 48 h, respectively, for HCT8 cells and a 1.9-fold higher level at 48 h for HCT8/7dR cells compared to levels for untreated cells (Fig. 7).
FIG. 7.
GADD34 expression in response to FUdR. Northern blots of GADD34 mRNA in HCT8 (A) and HCT8/7dR (B) cells grown in the absence or presence of 10 or 100 nM FUdR for 24 or 48 h. Cells were plated and treated with FUdR according to the methods of the experiment of RR measurement (see the legend to Fig. 5). β-actin served as a loading control for GADD34.
DISCUSSION
These results show that FUdR-induced cellular alterations create an environment that increases the cytotoxic potential of the conditionally replicating HSV G207. The two major mutations in G207 are the deletion of both γ134.5 loci and a lacZ insertion into the ICP6 gene disabling viral RR. The cellular counterparts which can theoretically substitute for the functions of viral RR and the carboxyl-terminal domain of ICP34.5 are host RR and the corresponding homologous domain of GADD34, respectively. We investigated whether FUdR-induced alterations in these cellular proteins may be potential mechanisms by which FUdR produces enhanced replication of G207 and synergistic cytotoxicity of the viral and chemotherapeutic agent.
The inability of G207 to express functional viral RR makes its DNA replication highly dependent on the cellular enzyme. It is therefore reasonable to conclude that alterations in cellular RR activity influence the growth of G207. In light of the observed enhanced replication of G207 by FUdR, we investigated the effect of FUdR on cellular RR. We found elevated RR activity levels in FUdR-treated cells, peaking at 24 h. There are two explanations as to why cells elevate their RR activity by an RR protein increase in response to FUdR. First, RR is cell-cycle regulated and highly expressed in S phase (4, 6, 17). RR consists of homodimeric large (R1) and small (R2) subunits in an α2β2 configuration. Regulation occurs at the level of R2 that is maximally expressed in S phase and undergoes controlled degradation when cells enter mitosis (6). In contrast, levels of R1 are constant and excessive during the cell cycle. A previous study showed that R2 is highly stable in S phase and not degraded in cells blocked in S phase (6). S-phase blockade by FUdR as observed in our study could therefore result in elevated RR activity by delaying R2 degradation. Second, RR activity is also increased under conditions such as DNA damage and repair (19, 39). Recently, another RR small subunit (p53R2) was identified which is induced by DNA damage in a wild-type p53-dependent manner (39). Although the measurement of RR activity alone cannot distinguish how much each mechanism contributes to the overall activity, the DNA-damaging effect of FUdR and the p53 wild-type status of both cell lines suggest that the p53-dependent induction of p53R2 may play an important role in the FUdR-induced increase of cellular RR activity. Another mechanism by which FUdR can increase RR activity independently of the RR protein level involves the allosteric regulation of this enzyme. In contrast to the viral form, the mammalian enzyme is highly regulated by feedback inhibition of deoxynucleotides (2, 12). Thereby, dTTP inhibits CDP and UDP reduction of RR (12). TS inhibition by FdUMP, the major active metabolite of FUdR, results in dTTP depletion and, thus, in activation of RR (27). This phenomenon is not discernible, since RR activity was measured in dialyzed cell extracts when nucleotides were removed. Therefore, the actual difference between increased RR activity in FUdR-treated cells and activity in untreated cells may be even higher intracellularly than that demonstrated in our study.
The TS inhibitor FdUMP represents the fluorinated form of dUMP. A nuclear magnetic resonance spectroscopic study demonstrated binding activity of the diphosphate analogue FdUDP to R1 (36), but no data exist on the inhibitory effect of fluorinated deoxyuridine nucleotides on enzyme activity. We therefore investigated the effect of FdUMP on RR activity in the light of a possible RR inhibition which could interfere with replication of G207. Our inhibition experiment with FdUMP demonstrates that RR is not significantly inhibited at the concentrations of FUdR utilized. Thus, we conclude that FdUMP does not impair viral replication of G207 by compromising RR activity.
Based on the observed elevated RR activities induced by FUdR, the question arises as to whether this FUdR effect is the cause for enhanced replication of G207 or whether both phenomena occur coincidentally. Pioneering studies with ICP6 lacZ insertion and ICP6 deletion mutants have shown evidence that cellular RR alone can sustain viral replication (20, 21). Dependence of these mutants on cellular RR activity is demonstrated by the following facts: (i) RR is present only in rapidly dividing cells and is nearly undetectable in differentiated growth-arrested cells (16, 18, 40), and (ii) growth of RR-negative HSV mutants is not restricted in rapidly dividing cells but is severely compromised in growth-arrested cells (20, 21). In our study we have shown that G207 has a replicative advantage in FUdR-treated cells and that both FUdR-induced phenomena, increase in cellular RR and increase in viral replication, were clearly more pronounced in HCT8 than in the less FUdR-sensitive line HCT8/7dR. Although these findings support the concept that cellular RR supports viral DNA synthesis of RR-negative HSV mutants, it is not evident that enhanced replication of G207 is based on the observed FUdR-induced RR effect. To elucidate this point we tried to neutralize the FUdR effect of enhanced replication by specific inhibition of RR with HU, which inactivates the tyrosyl free radical of R2 required for substrate reduction (23). An RR-dependent mechanism is suggested by the suppression of the FUdR-induced enhanced replication of G207 and, more importantly, the fact that there is the same degree of inhibition of G207 replication by HU treatment alone as by HU treatment with FUdR.
A significant finding which yields further insight into the interaction of FUdR and viral replication is the observation that replication of wild-type HSV-1 is impaired in HCT8 cells by FUdR, whereas replication of G207 is enhanced under the same conditions. This inhibition of wild-type HSV-1 replication, which has an intact endogenous RR, likely involves the inhibitory effect of FUdR on de novo thymidylate biosynthesis. Inhibition of TS by the active metabolite FdUMP results in depletion of the intracellular dTTP pool (13, 27, 42). This leads to an inhibition of cellular DNA synthesis, demonstrated by S-phase blockade in our study. Since viral DNA synthesis requires cellular deoxynucleotides, including dTTP, DNA replication of wild-type HSV-1 likely suffers from TS inhibition in the same way that cellular DNA replication does. This mechanism should also affect G207, but the replication benefit from RR elevation by FUdR is sufficiently great for this RR-negative HSV mutant that it seems to overcome the effect of TS inhibition and results in enhanced replication in HCT8 cells under these conditions. The overall replication of G207 is a sum of the net FUdR effect of enhanced replication by RR activity and impaired replication by TS inhibition.
Although we present evidence that enhanced replication of G207 by FUdR is based on an RR-dependent mechanism and is related to ICP6 inactivation in G207, it is conceivable that other mechanisms contribute to enhanced replication in the presence of FUdR. This idea may have special implications regarding the nature of the second major mutation in G207, the deletion of both γ134.5 loci (31). This mutation attenuates the virulence of G207 by the following mechanism. Cellular protein synthesis is regulated at the level of translational eukaryotic initiation factor 2 (eIF-2). The dephosphorylated form of eIF-2 initiates protein synthesis, whereas phosphorylation of eIF-2 terminates protein synthesis (33, 37). One major host defense mechanism against viral infection is the activation of double-stranded RNA-dependent protein kinase that phosphorylates eIF-2 (11, 34). This results in shutoff of cellular protein synthesis and prevents synthesis of viral proteins. Wild-type HSV overcomes this defense mechanism by expression of its γ134.5 genes. The complex of ICP34.5 and cellular protein phosphatase 1α dephosphorylates eIF-2 and precludes the shutoff of protein synthesis (7, 11). The absence of ICP34.5 shuts off protein synthesis in cells infected with γ134.5-null mutants, including G207 (24).
Interestingly, the carboxyl-terminal domain of ICP34.5 has great homology to the carboxyl terminus of the murine and human GADD34 protein (9, 30, 43). The gadd34 gene belongs to a family of five genes which are inducible by stressful growth arrest and DNA damage (25, 43). The carboxyl-terminal domain of GADD34 binds to protein phosphatase 1α and can functionally substitute for the corresponding domain of ICP34.5 by preventing cessation of protein synthesis (24). Since FUdR is a DNA damaging agent, it was hypothesized that FUdR may sustain protein synthesis by inducing gadd34 in the face of infection with G207. Indeed, FUdR at concentrations of 100 nM induced GADD34 in both cell lines, whereas 10 nM FUdR had almost no effect on GADD34. These data would indicate that the main reason for the observed growth advantage of G207 in FUdR-treated cells is based on the FUdR-induced RR effect. The mechanism by which GADD34 can complement the γ134.5 defect seems not to play a significant role, since the highest β-galactosidase expression and viral production were observed in HCT8 cells treated with 10 nM FUdR when gadd34 was not induced; however, this mechanism may play a role at higher FUdR concentrations.
Finally, the question arises as to how the dose of FUdR given to patients corresponds to the in vitro dose that is effective in enhancing replication of G207. FUdR is one of the most commonly used drugs for regional therapy of colorectal liver metastases and is delivered by continuous pump infusion via the hepatic artery at a daily dose of 0.25 mg per kg of body weight. Although the measurement of FUdR concentration inside the tumor is not technically feasible, the concentration of FUdR in hepatic arterial blood would most appropriately correspond to the in vitro FUdR concentration on cultured HCT8 cells. The concentration of FUdR in hepatic arterial blood can be calculated from hepatic arterial blood flow, dose, and the molecular weight (246) of FUdR. Given a mean hepatic arterial blood flow of 375 ml/min for a patient with 70 kg of body weight, a continuous daily dose of 0.25 mg/kg results theoretically in a FUdR concentration of 130 nM in hepatic arterial blood. Thus, the in vitro dose of 10 nM that is most effective in enhancing replication of G207 is approximately 1 log lower than the expected FUdR concentration in hepatic arterial blood. This has encouraging clinical implications for a combined FUdR and G207 therapy, since the in vitro concentrations of 10 nM FUdR are clinically attainable far below the maximal tolerable dose, with minimal side effects of FUdR.
In conclusion, we have demonstrated for the first time that FUdR can create a cellular environment which enhances replication and cytotoxic potential of the oncolytic HSV mutant G207. These effects are primarily dependent on the degree of RR activity induction and TS inhibition by FUdR. Furthermore, we present evidence that RR-negative HSV mutants benefit from FUdR-induced cellular alterations but that HSV with intact endogenous RR does not. Based on these observations, RR-negative HSV mutants appear to be attractive candidates for a combination treatment with FUdR or other chemotherapeutic agents. The understanding of mechanisms by which FUdR interacts with viral replication is relevant to future studies evaluating combined therapy with chemotherapeutic agents and oncolytic HSV for cancer.
ACKNOWLEDGMENTS
This work was supported by grants RO1CA75416, RO1CA72632, and RO1CA61524 from the National Cancer Institute (to Y.F.), grant MBC-99366 (to Y.F.) from the American Cancer Society, and grant D/98/27055 (to H.P.) from the German Academic Exchange Service (DAAD).
We thank S. D. Rabkin and R. L. Martuza (Georgetown University Medical Center, Washington, D.C.) for G207; J. Bertino (Memorial Sloan-Kettering Cancer Center, New York, N.Y.) for the cell line HCT8/FU7dR (HCT8/7dR); A. J. Fornace, Jr. (National Institutes of Health, Bethesda, Md.) for the cDNA for human GADD34; C. Cordon-Cardon (Memorial Sloan-Kettering Cancer Center) for performing the p53 mutational analysis; J. G. Cory (East Carolina University, Greenville, N.C.) for valuable suggestions for the functional RR assay; and Y. You for excellent technical assistance.
REFERENCES
- 1.Aschele C, Sobrero A, Faderan M A, Bertino J R. Novel mechanism(s) of resistance to 5-fluorouracil in human colon cancer (HCT8) sublines following exposure to two different clinically relevant dose schedules. Cancer Res. 1992;52:1855–1864. [PubMed] [Google Scholar]
- 2.Averett D R, Lubbers C, Elion G B, Spector T. Ribonucleotide reductase induced by herpes simplex type-1 virus. J Biol Chem. 1983;258:9831–9838. [PubMed] [Google Scholar]
- 3.Ayusawa D, Shimizu K, Koyama H, Takeishi K, Seno T. Accumulation of DNA strand breaks during thymineless death in thymidylate synthetase-negative mutants of mouse FM3A cells. J Biol Chem. 1983;258:12448–12454. [PubMed] [Google Scholar]
- 4.Björklund S, Skog S, Tribukait B, Thelander L. S-phase-specific expression of mammalian ribonucleotide reductase R1 and R2 subunit mRNAs. Biochemistry. 1990;29:5452–5458. doi: 10.1021/bi00475a007. [DOI] [PubMed] [Google Scholar]
- 5.Boehmer P E, Lehman I R. Herpes simplex virus DNA replication. Annu Rev Biochem. 1997;66:347–384. doi: 10.1146/annurev.biochem.66.1.347. [DOI] [PubMed] [Google Scholar]
- 6.Chabes A, Thelander L. Controlled protein degradation regulates ribonucleotide reductase activity in proliferating mammalian cells during the normal cell cycle and in response to DNA damage and replication blocks. J Biol Chem. 2000;275:17747–17753. doi: 10.1074/jbc.M000799200. [DOI] [PubMed] [Google Scholar]
- 7.Cheng G, Gross M, Brett M-E, He B. AlaArg motif in the carboxyl terminus of the γ134.5 protein of herpes simplex virus type 1 is required for the formation of a high-molecular-weight complex that dephosphorylates eIF-2α. J Virol. 2001;75:3666–3674. doi: 10.1128/JVI.75.8.3666-3674.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 9.Chou J, Roizman B. Herpes simplex virus 1 γ134.5 gene function, which blocks the host response to infection, maps in the homologous domain of the genes expressed during growth arrest and DNA damage. Proc Natl Acad Sci USA. 1994;91:5247–5251. doi: 10.1073/pnas.91.12.5247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chou J, Kern E R, Whitley R J, Roizman B. Mapping of herpes simplex virus-1 neurovirulence to γ134.5, a gene nonessential for growth in culture. Science. 1990;250:1262–1266. doi: 10.1126/science.2173860. [DOI] [PubMed] [Google Scholar]
- 11.Chou J, Chen J J, Gross M, Roizman B. Association of a Mr 90,000 phosphoprotein with protein kinase PKR in cells exhibiting enhanced phosphorylation of translation initiation factor eIF-2 alpha and premature shutoff of protein synthesis after infection with γ134.5-mutants of herpes simplex virus 1. Proc Natl Acad Sci USA. 1995;92:10516–10520. doi: 10.1073/pnas.92.23.10516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cory J G, Rey D A, Carter G L, Bacon P E. Nucleoside 5′-diphosphates as effectors of mammalian ribonucleotide reductase. J Biol Chem. 1985;260:12001–12011. [PubMed] [Google Scholar]
- 13.Danenberg P V, Lockshin A. Thymidylate synthetase-substrate complex formation. Mol Cell Biochem. 1982;43:49–57. doi: 10.1007/BF00229539. [DOI] [PubMed] [Google Scholar]
- 14.Danenberg P V, Heidelberger C, Mulkins M A, Peterson A R. The incorporation of 5-fluoro-2′-deoxyuridine into DNA of mammalian tumor cells. Biochem Biophys Res Commun. 1981;102:654–658. doi: 10.1016/s0006-291x(81)80182-9. [DOI] [PubMed] [Google Scholar]
- 15.D'Anna J A, Crissman H A, Jackson P J, Tobey R. Time-dependent changes in H1 content, H1 turnover, DNA elongation, and the survival of cells blocked in early S phase by hydroxyurea, aphidicolin, or 5-fluorodeoxyuridine. Biochemistry. 1985;24:5020–5026. doi: 10.1021/bi00340a010. [DOI] [PubMed] [Google Scholar]
- 16.Elford H L, Freese M, Passamani E, Morris H P. Ribonucleotide reductase and cell proliferation. I. Variations of ribonucleotide reductase activity with tumor growth rate in a series of rat hepatomas. J Biol Chem. 1970;245:5228–5233. [PubMed] [Google Scholar]
- 17.Engstöm Y, Eriksson S, Jildevik I, Skog S, Thelander L, Tribukai B. Cell cycle-dependent expression of mammalian ribonucleotide reductase. J Biol Chem. 1985;260:9114–9116. [PubMed] [Google Scholar]
- 18.Engström Y, Rozell B, Hansson H A, Stemme S, Thelander L. Localization of ribonucleotide reductase in mammalian cells. EMBO J. 1984;3:863–867. doi: 10.1002/j.1460-2075.1984.tb01897.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Filatov D, Thelander L. Role of a proximal NF-Y binding promoter element in S phase-specific expression of mouse ribonucleotide reductase R2. J Biol Chem. 1995;270:25239–25243. doi: 10.1074/jbc.270.42.25239. [DOI] [PubMed] [Google Scholar]
- 20.Goldstein D J, Weller S K. Factor(s) present in herpes simplex virus type 1-infected cells can compensate for the loss of the large subunit of the viral ribonucleotide reductase: characterization of an ICP6 deletion mutant. Virology. 1988;166:41–51. doi: 10.1016/0042-6822(88)90144-4. [DOI] [PubMed] [Google Scholar]
- 21.Goldstein D J, Weller S K. Herpes simplex virus type 1-induced ribonucleotide reductase activity is dispensable for virus growth and DNA synthesis: isolation and characterization of an ICP6 lacZ insertion mutant. J Virol. 1988;62:196–205. doi: 10.1128/jvi.62.1.196-205.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goulian M, Bleile B M, Dickey L M, Grafstrom R H, Ingraham H A, Neynaber S A, Peterson M S, Tseng B Y. Mechanism of thymineless death. Adv Exp Med Biol. 1986;195:89–95. doi: 10.1007/978-1-4684-1248-2_15. [DOI] [PubMed] [Google Scholar]
- 23.Gräslund A, Ehrenberg A, Thelander L. Characterization of the free radical of mammalian ribonucleotide reductase. J Biol Chem. 1982;257:5711–5715. [PubMed] [Google Scholar]
- 24.He B, Gross M, Roizman B. The γ134.5 protein of herpes simplex virus 1 complexes with protein phosphatase 1alpha to dephosphorylate the alpha subunit of the eukaryotic translation initiation factor 2 and preclude the shutoff of protein synthesis by double-stranded RNA-activated protein kinase. Proc Natl Acad Sci USA. 1997;94:843–848. doi: 10.1073/pnas.94.3.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hollander M C, Zhan Q, Bae I, Fornace A J., Jr Mammalian GADD34, an apoptosis- and DNA damage-inducible gene. J Biol Chem. 1997;272:13731–13737. doi: 10.1074/jbc.272.21.13731. [DOI] [PubMed] [Google Scholar]
- 26.Ingraham H A, Dickey L, Goulian M. DNA fragmentation and cytotoxicity from increased cellular deoxyuridylate. Biochemistry. 1986;25:3225–3230. doi: 10.1021/bi00359a022. [DOI] [PubMed] [Google Scholar]
- 27.Jackson R C. The regulation of thymidylate biosynthesis in Novikoff hepatoma cells and the effects of amethopterin, 5-fluorodeoxyuridine, and 3-deazauridine. J Biol Chem. 1978;253:7440–7446. [PubMed] [Google Scholar]
- 28.Kooby D A, Carew J F, Halterman M W, Mack J E, Bertino J R, Blumgart L H, Federoff H J, Fong Y. Oncolytic viral therapy for human colorectal cancer and liver metastases using a multi-mutated herpes simplex virus type-1 (G207) FASEB J. 1999;13:1325–1334. doi: 10.1096/fasebj.13.11.1325. [DOI] [PubMed] [Google Scholar]
- 29.Markert J M, Medlock M D, Rabkin S D, Gillespie G Y, Todo T, Hunter W D, Palmer C A, Feigenbaum F, Tornatore C, Tufaro F, Martuza R L. Conditionally replicating herpes simplex virus mutant, G207 for the treatment of malignant glioma: results of a phase I trial. Gene Ther. 2000;7:867–874. doi: 10.1038/sj.gt.3301205. [DOI] [PubMed] [Google Scholar]
- 30.McGeoch D J, Barnett B C. Neurovirulence factor. Nature. 1991;353:609. doi: 10.1038/353609b0. [DOI] [PubMed] [Google Scholar]
- 31.Mineta T, Rabkin S D, Yazaki T, Hunter W D, Martuza R L. Attenuated multi-mutated herpes simplex virus-1 for the treatment of malignant gliomas. Nat Med. 1995;1:938–943. doi: 10.1038/nm0995-938. [DOI] [PubMed] [Google Scholar]
- 32.Nüsse M, Beisker W, Hoffmann C, Tarnok A. Flow cytometric analysis of G1- and G2/M-phase subpopulations in mammalian cell nuclei using side scatter and DNA content measurements. Cytometry. 1990;11:813–821. doi: 10.1002/cyto.990110707. [DOI] [PubMed] [Google Scholar]
- 33.Proud C G. Protein phosphorylation in translational control. Curr Top Cell Regul. 1992;32:243–369. doi: 10.1016/b978-0-12-152832-4.50008-2. [DOI] [PubMed] [Google Scholar]
- 34.Proud C G. PKR: a new name and new roles. Trends Biochem Sci. 1995;20:241–246. doi: 10.1016/s0968-0004(00)89025-8. [DOI] [PubMed] [Google Scholar]
- 35.Roizman B, Seras A E. Herpes simplex virus and their replication. In: Fields B N, Knipe D M, Howley P, Chanock R M, Hirsch M S, Melnick J L, Monath T P, Roizman B, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2231–2295. [Google Scholar]
- 36.Roy B, Decout J L, Beguin C, Fontecave M, Allard P, Kuprin S, Ehrenberg A. NMR studies of binding of 5-FdUDP and dCDP to ribonucleoside-diphosphate reductase from Escherichia coli. Biochim Biophys Acta. 1995;1247:284–292. doi: 10.1016/0167-4838(94)00241-8. [DOI] [PubMed] [Google Scholar]
- 37.Samuel C E. The eIF-2 alpha protein kinases, regulators of translation in eukaryotes from yeast to humans. J Biol Chem. 1993;268:7603–7606. [PubMed] [Google Scholar]
- 38.Steeper J P, Stuart C D. A rapid assay for CDP reductase activity in mammalian cell extracts. Anal Biochem. 1970;34:123–130. doi: 10.1016/0003-2697(70)90092-8. [DOI] [PubMed] [Google Scholar]
- 39.Tanaka H, Arakawa H, Yamaguchi T, Shiraishi K, Fukuda S, Matsui K, Takei Y, Nakamura Y. A ribonucleotide reductase gene involved in a p53-dependent cell-cycle checkpoint for DNA damage. Nature. 2000;404:42–49. doi: 10.1038/35003506. [DOI] [PubMed] [Google Scholar]
- 40.Thelander L, Reichard P. Reduction of ribonucleotides. Annu Rev Biochem. 1979;48:133–158. doi: 10.1146/annurev.bi.48.070179.001025. [DOI] [PubMed] [Google Scholar]
- 41.Whitley R J, Kern E R, Catterjee S, Chou J, Roizman B. Replication, establishment of latency, and induced reactivation of herpes simplex virus γ134.5 deletion mutants in rodent models. J Clin Investig. 1993;91:2837–2843. doi: 10.1172/JCI116527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yoshioka A, Tanaka S, Hiraoka S, Koyama Y, Hirota Y, Ayusawa D, Seno T, Garrett C, Wataya Y. Deoxyribonucleoside triphosphate imbalance. 5-fluorodeoxyuridine-induced DNA double strand breaks in mouse FM3A cells and the mechanism of cell death. J Biol Chem. 1987;262:8235–8241. [PubMed] [Google Scholar]
- 43.Zhan Q, Lord K A, Alamo I, Jr, Hollander C, Carrier F, Ron D, Kohn K W, Hoffman B, Liebermann D A, Foranace A J., Jr The gadd and MyD genes define a novel set of mammalian genes encoding acidic proteins that synergistically suppress cell growth. Mol Cell Biol. 1994;14:2361–2371. doi: 10.1128/mcb.14.4.2361. [DOI] [PMC free article] [PubMed] [Google Scholar]