Abstract
Ovarian cancer (OC) presents a global health challenge, with well-documented genetic aspects. However, to the best of our knowledge, the role of human papillomavirus (HPV) types 16 and 18 in OC remains unclear. The present meta-analysis assessed the prevalence of HPV in OC across 43 studies and included a comparative meta-analysis of 19 case-control studies to determine the association of HPV with OC risk. Subgroup analyses were performed based on geographic regions and histopathological types to explore heterogeneity, and publication bias was evaluated using funnel plots and statistical tests of asymmetry. The pooled prevalence of HPV was found to be 10% (95% CI, 5–18) and 7% (95% CI, 3–15) specifically for HPV 16/18. Case-control studies indicated an odds ratio (OR) of 4.92 (95% CI, 1.96–12.53) for HPV 16/18, with higher pooled prevalence rates of 17% for all HPV genotypes and 13% for HPV 16/18. Notably, Asian countries exhibited the highest HPV prevalence and OR in OC. These findings support the involvement of HPV, particularly HPV 16 and 18, in increasing the risk of OC, emphasizing the need for further research to confirm these associations and explore potential mechanisms.
Keywords: ovarian neoplasms, HPV 16, HPV 18, meta-analysis
Introduction
Ovarian cancer (OC) presents a significant global health challenge, with >300,000 new cases and 200,000 deaths reported worldwide in 2020 (1). Despite advancements in medical research, there are still no effective tools for general population screening, which complicates early detection. This challenge is reflected economically, as the cost of treatment per patient with OC remains the highest among all cancer types, with initial treatment costs in the first year amounting to approximately USD 80,000, and potentially increasing to USD 100,000 in the final year (2). Cost-effective strategies for early detection and prevention of OC have thus been a significant focus of research over the last decade.
This disease often remains undetected until the advanced stages due to its elusive symptoms, leading to late diagnoses and less effective treatments (3,4). Currently, CA125 and HE4 are the only approved biomarkers for use in epithelial OC (EOC); however, these markers are not sufficient for early detection. To mitigate the limitations of single serum biomarkers in EOC, multivariate index assays have been developed, particularly for the pre-surgical evaluation of adnexal masses. The Risk of Malignancy Algorithm, which integrates menopausal status, CA125, and HE4 concentrations, is used to diagnose women with a pelvic mass. Furthermore, microRNAs have shown remarkable potential in EOC prediction, though further work is needed before they can be utilized as reliable clinical biomarkers (5).
Among OC types, EOC is particularly known for its invasive nature and predominance. However, it is important to note that 10% of OC cases are non-epithelial, including germ cell tumors, sex cord-stromal tumors, and some extremely rare tumors such as small cell carcinomas. Germ cell tumors, for instance, differ significantly from EOCs, with earlier age of incidence, faster growth rates, unilateral localization in 95% of cases, and generally better prognosis (6).
EOC encompasses a spectrum of histologic subtypes, including serous, mucinous, endometrioid, clear cell, or combinations of these subtypes (7). These subtypes exhibit unique molecular profiles, contributing to the differences in causes, epidemiology, treatments, and prognoses (8). Specifically, Type I EOCs are generally indolent and genetically stable tumors that typically arise from precursor lesions such as endometriosis or borderline tumors with low malignant potential. In contrast, Type II EOCs are biologically aggressive tumors from their outset, with a tendency to metastasize from small-volume primary lesions. High-grade serous OC, following the Type II pathway, is often associated with p53 and BRCA mutations (9).
Genetic and molecular pathway alterations play significant roles in OC initiation and development. For instance, serous OCs often exhibit BRCA1/2 and TP53 mutations and is involved in the amplification of G1/S-specific cyclin-E or defective homologous recombination DNA repair pathways (10), whereas non-serous OCs are associated with mutations in genes involved in pathways such as AT-rich interactive domain-containing protein, phosphatidylinositol 3-kinase (PI3K), K-Ras/B-Raf, Wnt, or protein phosphatase 2A (11). Within this context, it is critical to note that the PI3K pathway plays a pivotal role in chemoresistance and preservation of genomic stability, as it is implicated in numerous processes of DNA replication and cell cycle regulation. Inhibition of the PI3K pathway may lead to genomic instability and mitotic catastrophe through decreased activity of the spindle assembly checkpoint protein Aurora kinase B, consequently increasing the occurrence of lagging chromosomes during prometaphase (12). Moreover, emerging evidence suggests that high-grade serous OC, closely connected to the fimbriated ends of the fallopian tubes, may originate from the precursor lesions in the fallopian tubes rather than from the ovary itself (3,11,13,14).
In addition to genetic factors, chronic inflammation is emerging as a key risk factor in EOC development. Persistent infections that affect the female reproductive organs can trigger a pro-inflammatory response, exacerbate DNA damage, and contribute to cancer initiation (15–17). In line with this, the prolonged exposure of ovarian epithelial cells to inflammatory mediators, such as pro-inflammatory cytokines, chemokines, and hormones, may lead to DNA damage through oxidative stress and cause genetic and epigenetic alterations (18). However, the role of environmental factors in EOC initiation and progression, particularly viral infections, is still being investigated.
High-risk HPV (HR-HPV) plays a crucial role in the development of cervical cancer and head and neck squamous cell carcinoma (19,20). HR-HPV carries E6 and E7 oncoproteins that can inactivate tumor suppressors such as p53, which may be related to OC development (21,22). Therefore, researchers have also been attracted to the potential role of HPV in EOC. However, recent studies have yielded mixed results regarding the presence of HPV in EOC, with some studies confirming its presence, whereas others negating it. Notably, among HR-HPV types, HPV 16 and/or 18 (HPV 16/18) are most frequently associated with advanced-stage disease, while the others have not been. Despite two meta-analyses that have explored the link between HPV and EOC, a specific focus on HPV 16/18 in OC has yet to be reported (23,24). To address this gap and provide an updated perspective, emphasizing on HPV 16/18 investigation, this meta-analysis was conducted to deepen our understanding of this connection.
Materials and methods
Searching protocol and data collection
The meta-analysis followed the PRISMA criteria. Two researchers, TML and HDNT, independently conducted a systematic literature search covering the period from 1987 to August 2023. Searches were conducted in PubMed (on October 6, 2023), Embase (on October 8, 2023), and Web of Science (on October 8, 2023), using the following search terms with detailed Boolean logic: ((Human papillomavirus) OR (HPV)) AND ((ovarian malignancies) OR (ovarian neoplasm) OR (ovarian cancer)).
The initial search yielded 364 records in PubMed, 420 in Embase, and 350 in Web of Science, totaling 1,134 records. To ensure reproducibility, the search process was independently verified by both researchers. After eliminating duplicates, all abstracts (n=701) were independently reviewed to exclude studies irrelevant to the topic or lacking sufficient data on the association between HPV and OC. Subsequently, 646 papers were excluded based on abstract reviews. Full-text copies of potentially relevant papers were obtained and independently reviewed (n=55). In total, 43 papers met the inclusion criteria for the meta-analysis (25–67). Data from these identified studies were extracted independently, and any disagreements regarding inclusion or exclusion were resolved through discussion. Recorded data included the first author's details, publication country, publication year, detection method, histological type, specimen type, sample size, HPV genotype, and number of HPV-positive and HPV-negative OCs, as well as HPV-positive and HPV-negative ovarian benign tumors or normal ovaries.
To be eligible for inclusion in this meta-analysis, studies must meet the following criteria: (1) they were observational studies published between 1987 and August 2023 and provided data on the association between HPV and OC, (2) ovarian tissues were used in the study to identify HPV genotypes, and (3) they were written in English and published as full, peer-reviewed articles. The exclusion criteria were as follows: (1) studies not meeting the inclusion criteria and (2) studies conducted solely on animals.
Statistical analysis
In this analysis, the pooled prevalence was estimated using both fixed- and random-effect models, based on the data from the included studies (68). Forest plots were used to display the prevalence for all the studies sorted by their publication year (69). The prevalence derived from individual studies and pooled proportions were presented with 95% confidence intervals (CIs). In addition, for case-control studies, the pooled odds ratio (OR) was computed for the presence of HPV in OC cases, along with the corresponding 95% CI. Both random- and fixed-effect models were employed for this analysis. The overall heterogeneity among the included studies was assessed using the I2 statistic, with I2 values >50% and/or P-value <0.05 indicating significant heterogeneity (70,71). To explore the potential sources of heterogeneity, subgroup analysis was conducted using meta-regression. This analysis included variables of geographic region.
To evaluate publication bias, a funnel plot, which illustrated the association between the logarithm of HPV prevalence and standard error, was generated (68,72). In the comparative (case-control) meta-analysis, potential asymmetry was examined using two methods: Egger's regression test and the Begg and Mazumdar adjusted rank correlation test (73,74). For the proportional meta-analysis, Peter's test, which is based on precision-effect estimates with standard errors, was employed (75). Statistical significance was defined as P-values <0.05.
Analyses were conducted using R v4.3.1 (R Core Team 2023) with packages including ‘meta,’ ‘metasens,’ and ‘metafor’ (69,72,76–78).
Results
Study description
This meta-analysis included 43 studies to investigate the prevalence of HPV in OC tumor tissue. The study selection process is illustrated in Fig. 1, and details of the additional study are available in Table I.
Figure 1.
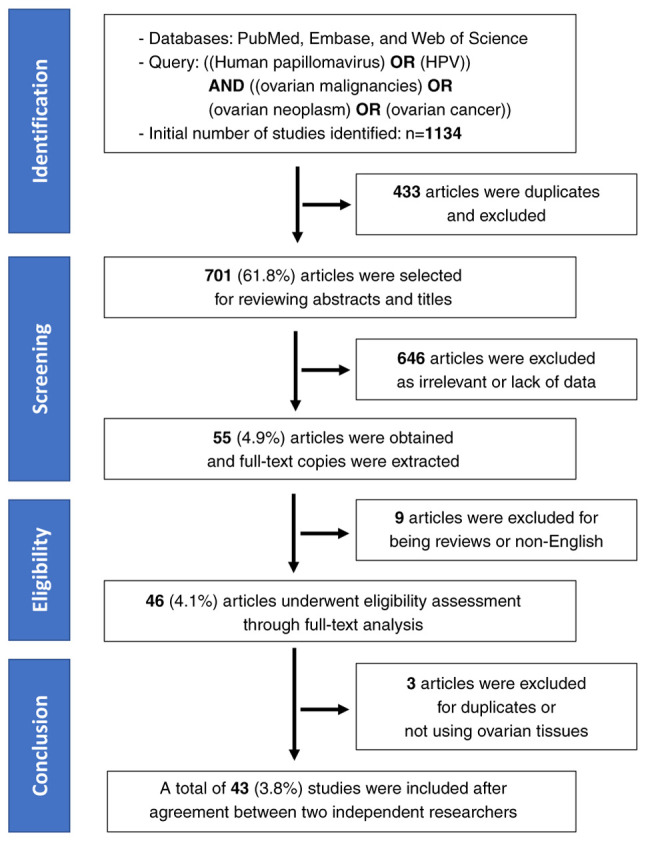
Preferred Reporting Items for Systematic reviews and Meta-Analyses flowchart. HPV, human papillomavirus.
Table I.
Overview of the included studies.
| No. of patients | ||||||||
|---|---|---|---|---|---|---|---|---|
|
|
||||||||
| Variable 1 | Variable 2 | No. of included studies | Serous POS | Serous total | Case POS | Case total | Control POS | Control total |
| Country | Africa | 2 | 6 | 53 | 18 | 170 | 0 | 0 |
| Asia | 21 | 45 | 563 | 426 | 1,691 | 32 | 677 | |
| Europe | 13 | 35 | 425 | 102 | 763 | 47 | 252 | |
| North America | 7 | 0 | 30 | 9 | 130 | 0 | 35 | |
| Method | PCR | 36 | 58 | 947 | 483 | 2,498 | 68 | 871 |
| IHC | 1 | 3 | 26 | 3 | 31 | 0 | 18 | |
| PCR/IHC | 1 | 9 | 53 | 9 | 53 | 0 | 0 | |
| ISH | 2 | 15 | 24 | 41 | 90 | 11 | 62 | |
| PCR/ISH | 2 | 1 | 6 | 17 | 31 | 0 | 8 | |
| PCR/Southern blot | 1 | 0 | 15 | 2 | 51 | 0 | 5 | |
| Sample type | FFPE | 29 | 46 | 768 | 466 | 2,218 | 77 | 805 |
| FFPE, frozen tissue | 1 | 7 | 14 | 10 | 17 | 0 | 0 | |
| Frozen tissue | 9 | 27 | 132 | 69 | 287 | 2 | 137 | |
| Fresh tissue | 4 | 6 | 157 | 10 | 232 | 0 | 22 | |
FFPE, formalin-fixed paraffin-embedded; IHC, Immunohistochemistry; ISH, in situ hybridization; POS, positive.
The analysis included a total of 2,754 patients from 43 selected studies (Tables I and SI). Of these studies, 21 were conducted in Asia, and the remaining 22 were carried out in other regions (Europe=13, North America=7, and Africa=2). Among the selected papers, 19 of 43 were case-control studies that involved normal ovarian or benign ovarian tumor tissues in the control group. These studies used formalin-fixed paraffin-embedded samples (n=30), frozen sections (n=11), and fresh tissues (n=4). These studies (n=36) predominantly used HPV detection methods based on polymerase chain reaction. Furthermore, 17 of 43 selected studies focused specifically only on the detection of HPV 16/18 in the analysis.
Prevalence of HPV in OC and subgroups
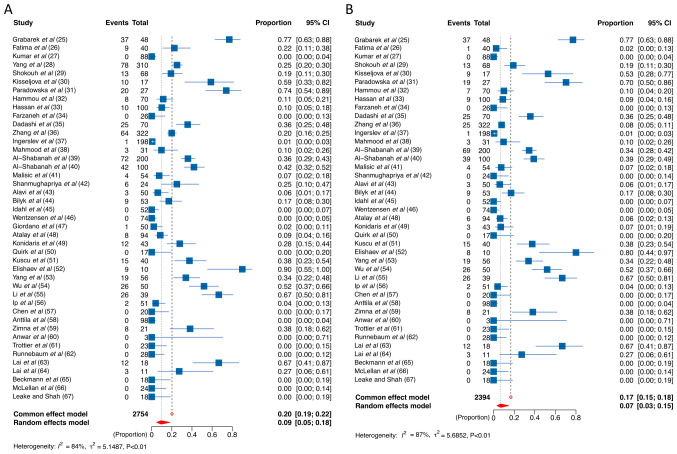
Forest plots of the 43 studies (Fig. 2) show the prevalence of HPV in OC. The pooled prevalence of all HPV genotypes and HPV 16/18 in OC were 20 and 17% (fixed-effect model) and 10 and 7% (random-effect model), respectively. To gain deeper insights into the relationship between HPV and OC, we conducted more subgroup meta-regression analyses, including histologic types and ethnic groups.
Figure 2.
Forest plot. HPV prevalence in ovarian cancer. (A) All HPV genotypes and (B) HPV 16/18. HPV, human papillomavirus; HPV 16/18, HPV 16 and/or 18.
Regarding histopathological types, serous OC, on the one hand, exhibited a significantly higher prevalence of HPV 16/18 positivity, approximately doubling that of non-serous OC (4% vs. 2%) as indicated by a random-effect model (Table II). On the other hand, positivity prevalence for all HPV genotypes was 6 and 5% in serous and non-serous OC, respectively (Table II).
Table II.
HPV prevalence in selected studies and subgroups.
| No. of patients | Pooled HPV prevalence | ||||||
|---|---|---|---|---|---|---|---|
|
|
|
||||||
| Variable | No. of included studies | Summed events | Summed total | Fixed effect model (95% CI) | Random effects model (95% CI) | I2 | P-value (Cochrane Q-test) |
| Ovarian cancer (all cases) | |||||||
| HPV 16/18 | 41 | 401 | 2,394 | 0.17 (0.15–0.18) | 0.07 (0.03–0.15) | 86.8 | <0.0001 |
| All genotypes | 43 | 555 | 2,754 | 0.20 (0.19–0.22) | 0.10 (0.05–0.18) | 83.8 | <0.0001 |
| Ovarian cancer by pathological types | |||||||
| HPV 16/18 | |||||||
| Serous | 25 | 75 | 791 | 0.09 (0.08–0.12) | 0.04 (0.01–0.13) | 72.9 | <0.0001 |
| Non-serous | 23 | 38 | 434 | 0.09 (0.06–0.12) | 0.02 (0.01–0.12) | 12.5 | <0.0001 |
| HPV (all genotypes) | |||||||
| Serous | 26 | 86 | 791 | 0.11 (0.09–0.13) | 0.06 (0.02–0.16) | 74.5 | <0.0001 |
| Non-serous | 24 | 47 | 459 | 0.10 (0.08–0.13) | 0.05 (0.01–0.16) | 24.0 | <0.0001 |
| Subgroup by region | |||||||
| HPV 16/18 | |||||||
| Asia | 20 | 287 | 1,381 | 0.21 (0.19–0.23) | 0.14 (0.07–0.26) | 88.6 | <0.0001 |
| Europe | 12 | 90 | 713 | 0.13 (0.10–0.15) | 0.05 (0.01–0.26) | 88.5 | <0.0001 |
| HPV (all genotypes) | |||||||
| Asia | 21 | 426 | 1,691 | 0.25 (0.23–0.27) | 0.20 (0.12–0.32) | 83.3 | <0.0001 |
| Europe | 13 | 102 | 763 | 0.13 (0.11–0.16) | 0.05 (0.01–0.26) | 88.0 | <0.0001 |
HPV, human papillomavirus; HPV 16/18, HPV 16 and/or 18.
When considering the effect of regional factors on HPV prevalence, variations in HPV status were observed across geographic regions. Although the test for subgroup difference using a meta-regression model did not yield statistical significance, Asian countries showed a considerably higher HPV prevalence in OC than European countries. Specifically, the prevalence of HPV across all genotypes in Asian countries was 20% compared with 5% in European countries. Moreover, the HPV 16/18 positivity was 14% in Asian countries compared with 5% in European countries (Table II; Figs. S1 and S2).
Unfortunately, owing to the limited number of studies from other regions (only two studies from Africa and seven from North America), a meta-regression could not be conducted. Nonetheless, these figures remain notably higher than the HPV prevalence in the control group, which stands at 3 and 2% for all HPV genotypes and HPV 16/18, respectively, as determined by the random-effect model (Table SII).
HPV status in the case-control analysis
In our case-control analysis, 19 studies, including 1,071 OC samples in contrast to 906 samples of normal or benign ovarian tissues, were obtained. The study by Li et al (55) was excluded from our analysis because it used blood as a control sample instead of ovarian tissues. The control groups in the selected studies ranged from women without ovarian diseases [Shokouh et al (29); Paradowska et al (31); Zhang et al (36); Shanmughapriya et al (42); Alavi et al (43); Konidaris et al (49); Ip et al (56); Trottier et al (61); Lai et al (63); Leake et al (67)], benign ovarian tumors [Grabarek et al (25); Farzaneh et al (34); Dadashi et al (35); Mahmood et al (38); Idahl et al (45); Kuscu et al (51); Quirk et al (50); Wu et al (54)], or adjacent normal ovarian tissues [Al-Shabanah et al (40)].
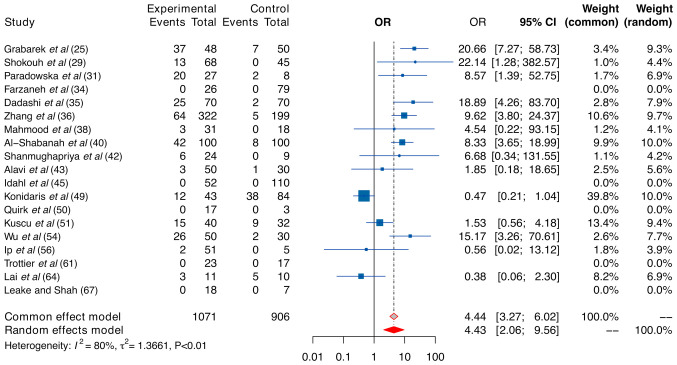
The findings of our case-control analysis regarding all HPV genotypes are presented in Fig. 3. In our investigation, a significant risk associated with OC was found in the context of HPV infection, reflected in fixed- and random-effect size ORs of 4.44 (95% CI, 3.27–6.02) and 4.43 (95% CI, 2.06–9.56), respectively. Importantly, owing to the high level of heterogeneity indicated by a sample I2 of 80% and P-value of <0.01, the random model should be considered a more appropriate choice for our case-control analysis. In addition, our analysis using the random-effect model revealed that the pooled proportion of all HPV genotypes in the case group was 18%.
Figure 3.
Forest plot. Comparative analysis of human papillomavirus positivity (any genotype) in the ovarian cancer and control groups. Data are presented as pooled ORs with 95% CIs. OR, odds ratio.
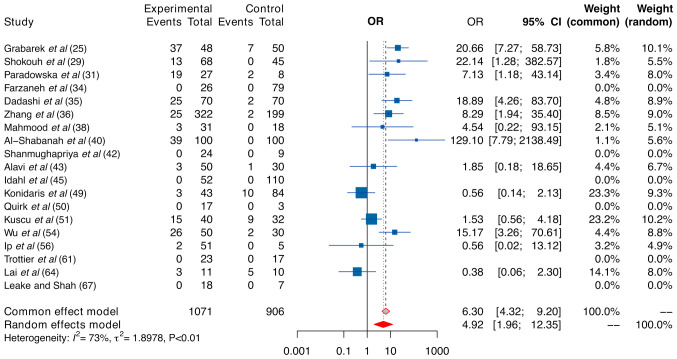
In light of exploring the sources of heterogeneity, a subgroup analysis was conducted by considering HPV 16/18 and different regions. When examining the subgroup related to HPV 16/18, HPV 16/18 was strongly linked to a heightened risk. Both the fixed- and random-effect models produced ORs of 6.30 and 4.92 (Fig. 4), respectively, which were higher than those of all HPV genotypes (4.44 and 4.43), respectively. As anticipated, the pooled prevalence of HPV 16/18 in the case group was considerably high, reaching 13% when using the random-effect model.
Figure 4.
Forest plot. Comparative analysis of human papillomavirus 16 and/or 18 statuses in ovarian cancer and control groups. Data are presented as pooled ORs with 95% CIs. OR, odds ratio.
As heterogeneity persisted after subgrouping by HPV 16/18 (I2=73%, P-value <0.01), further subgrouping by regions was conducted. Due to the limited sample size from North America and Africa, patients were categorized into Asian (n=12 studies) and non-Asian (n=8 studies) groups. Heterogeneity decreased to less than 67% in Asian subgroups (Table III). HPV infection, particularly HPV 16/18, emerged as a more substantial risk factor for OC among Asian women compared to those from other regions (Asia vs. non-Asia: 4.75 vs. 4.13 for all HPV genotypes, and 5.12 vs. 4.40 for HPV 16/18). Despite the observed trend, the test for differences was not statistically significant.
Table III.
Regional subgroup analysis: Asian vs. non-Asian countries.
| Case group | Control group | ||||||||
|---|---|---|---|---|---|---|---|---|---|
|
|
|
Random effects model OR (95% CI) | P-value (meta-regression) | I2 | P-value (Cochrane Q-test) | ||||
| Subgroups | No. of studies | Summed events | Summed total | Summed events | Summed total | ||||
| HPV (all genotypes) | |||||||||
| Asia | 12 | 202 | 422 | 32 | 176 | 4.75 (2.15–10.49) | 0.91 | 60.2 | 0.0051 |
| Non-Asia | 7 | 69 | 228 | 47 | 279 | 4.13 (0.42–40.90) | 94.2 | <0.0001 | |
| HPV 16/18 | |||||||||
| Asia | 12 | 154 | 843 | 21 | 627 | 5.12 (1.74–15.06) | 0.90 | 66.9 | 0.0013 |
| Non-Asia | 7 | 59 | 228 | 19 | 279 | 4.40 (0.52–37.19) | 88.5 | 0.0002 | |
Pooled odds ratios and 95% CIs for random effects models. HPV, human papillomavirus; HPV 16/18, HPV 16 and/or 18; OR, odds ratio.
Publication bias
In our meta-analysis, publication bias was assessed using funnel plots and tests for asymmetry (Figs. 5 and 6; Tables SIII and SIV). Accordingly, the selected studies that were used to investigate the prevalence of all HPV genotypes and HPV 16/18 in OC displayed no substantial indications of significant asymmetry. Moreover, the results from Begg's, Egger's, and Peter's tests collectively confirmed the absence of significant publication bias.
Figure 5.
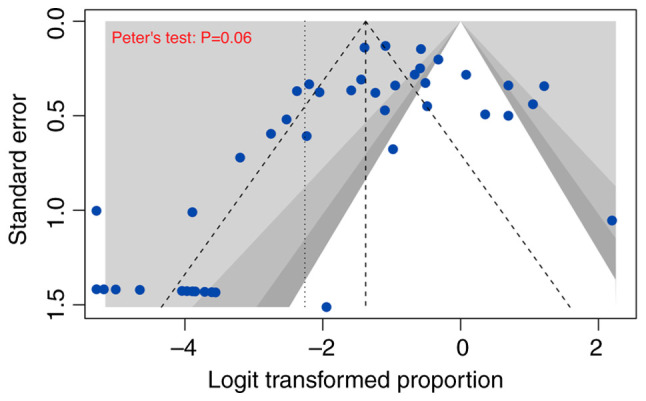
Contour-enhanced funnel plot. Evaluation of publication bias across all 43 included studies.
Figure 6.
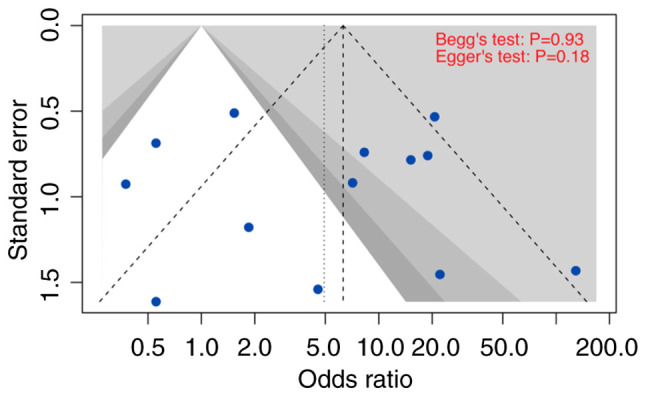
Contour-enhanced funnel plot. Evaluation of publication bias in Asian studies providing human papillomavirus 16 and/or 18 status.
Discussion
HR-HPV is a well-known etiological factor in anogenital and oropharyngeal cancers (79–82). Recently, interest in the prevalence of HPV, particularly HPV 16/18, and its potential role in OC has grown. HPV may reach EOC through: (1) anatomical continuity with endocervical glands, where the endometrium and fallopian tubes extend from the endocervical glands, allowing infection to spread. The fallopian tube's fimbriae are close to the ovarian surface (27); 2) sperm-mediated transmission, where sperm facilitate this by absorbing HPV DNA and transmitting the virus to reproductive system cells, and also serve as virus carriers during their passage through the endocervical canal, potentially reaching the ovarian cortex after ovulation (83).
In the context of HPV infection, integration into the human genome produces oncoproteins such as E6 and E7, crucial in initiating cancer, including EOC. E6 degrades p53, suppressing viral DNA synthesis and enhancing telomerase activity to evade senescence (21), while E7 disrupts retinoblastoma protein function, overriding p21-mediated growth arrest and increasing p16 for cell immortalization (22,84). Remarkably, over half of EOCs exhibit p53 and RB pathway mutations, with serous carcinomas representing 40% of cases (85). HPV infection leads to the emergence of HPV-related lesions over time. Interestingly, the precursor lesions for EOC, including serous tubal intraepithelial carcinoma and p53 signature, are widely accepted to originate within the fallopian tubes at the fimbriated end because of their proximity to these structures (10). Naturally, while some lesions transition into a quiescent state, others continue to proliferate, contributing to cancer development through genomic instability, telomere maintenance, and immune cell responses (10,11,13).
Given these considerations, HPV prevalence in OC may exhibit a substantial effect, akin to what has been observed in cervical cancer. In line with this, our meta-analysis, which consolidated data from 43 studies involving a total of 2,754 patients, unveiled a significant correlation between the prevalence of HPV (specifically HPV 16/18) and the risk of OC.
In our proportional meta-analysis, the overall pooled HPV prevalence was approximately 20% for all genotypes and 17% for the high-risk genotypes HPV 16/18 using a fixed-effect model. However, when employing the random-effect model, lower prevalence, with all HPV genotypes at 10% and HPV 16/18 at 7%, was obtained. In studies designed as case-control investigations, a higher pooled prevalence of HPV for all genotypes was found at 17% and HPV 16/18 at 13% using the random-effect model. Owing to the substantial heterogeneity across the studies, the random-effect model was considered more appropriate.
In addition, our meta-analysis considered variations in HPV prevalence among different regions. Asian countries exhibited higher HPV prevalence in OC than European countries. This regional disparity may be due to differences in healthcare practices, genetic factors, or environmental influences. The limited number of studies from Africa and North America prevented a comprehensive regional analysis, highlighting the need for more research in these regions to understand the disparities better.
This study further substantiated the role of HPV in OC risk through a comparative analysis. Accordingly, HPV 16/18 demonstrated a stronger association with OC than all HPV genotypes, particularly in Asian countries (with respective ORs of 5.12 (95% CI, 1.74–15.06) and 4.75 (95% CI, 2.15–10.49)). This finding emphasized the importance of recognizing specific HPV genotypes that may carry a higher risk for OC development and the role of geographical and host genetic factors in influencing susceptibility to HPV infection.
In comparison to two existing meta-analyses, Cherif et al (23) analyzed 29 studies involving 2,280 OC cases and reported a pooled HPV proportion of 15.9% (95% CI, 11–22), while Ibragimova et al (24) included 14 case-control studies with 1,163 ovarian tumor samples and 738 normal ovarian tissue samples, showing a relative risk of 2.68 (95% CI, 1.97–3.64). Despite similarities in search strategy with the previous study (23), our study's larger and more recent dataset (43 studies vs. 29 studies, 2,754 patients vs. 2,280 patients) allows for more comprehensive subgroup analyses, revealing significant variations in HPV 16/18 prevalence across histologic types and ethnic groups. This approach offers a deeper understanding of HPV's impact on OC, addressing gaps in previous research. We observed a similar OC risk related to HPV infection with a random-effects size OR of 4.43, but a lower pooled prevalence of HPV (10%). However, the HPV proportion in case-control studies was comparable (17% vs. 15.9%). In the case of HPV 16/18, these two meta-analyses calculated the proportion among HPV-positive studies, which differed from our approach of calculating HPV 16/18 prevalence across all studies. Our approach not only allows us to better demonstrate the predominance of HPV 16/18 among HPV genotypes in OC but also to mitigate the potential bias that arises from certain studies focusing solely on HPV 16/18, which can inflate the proportion of these two genotypes. As expected, this study effectively highlighted the association of HPV 16/18 in OC. Although the prevalence of HPV 16/18 (7%) was lower than that of all HPV genotypes (10%), the OR for HPV 16/18 was higher than that for all HPV genotypes (4.92 vs. 4.43).
The prevalence of HPV 16/18 in OC (7.0%) was lower than that of HPV 16/18 in women with uterine cervical lesions. According to the latest data from the ICO/IARC Information Centre on HPV and Cancer (2023), the worldwide incidence rates of HPV are 3.9, 25.8, 51.9, and 69.4% in normal cytology, low-grade lesions, high-grade lesions, and cervical cancer, respectively (86). This difference may arise from variations in sample type, as our data exclusively assessed the presence of HPV in OC tissue.
Publication bias is a potential concern in meta-analyses. However, the lack of significant asymmetry in the funnel plot and the results of Begg's, Egger's, and Peter's tests suggest that publication bias is not a significant issue in this study, thereby strengthening the credibility of the findings.
This study has several limitations. Firstly, the data availability was limited to regions outside Asia and Europe, which may restrict the comprehensiveness of our meta-analysis. Secondly, we only included studies published in English, potentially excluding important findings from non-English literature. Moreover, the absence of data on high-grade serous OC, especially in case-control studies, highlights a significant knowledge gap regarding HPV's role in this particular subtype. Additionally, further research, including well-designed and multi-ethnic epidemiological studies or prospective cohort studies, can provide stronger evidence of the causal relationship and underlying mechanisms between HPV and ovarian cancer.
In conclusion, this meta-analysis provides evidence of a significant association between HPV infection and OC risk, particularly in Asian countries. The high prevalence of HPV, particularly HPV 16/18, in OC cases highlights the imperative need for further in-depth research to elucidate the underlying mechanisms governing this association. Furthermore, a comprehensive understanding of the role and intricate interplay between HPV infections and this cancer can be crucial for early detection and prevention efforts.
Supplementary Material
Acknowledgements
Not applicable.
Glossary
Abbreviations
- HPV
human papillomavirus
- OC
ovarian cancer
- EOC
epithelial ovarian cancer
- HR-HPV
high-risk human papillomavirus
- OR
odds ratio
Funding Statement
This research was supported by a grant of the Korea Health Technology R&D Project through the Korea Health Industry Development Institute, funded by the Ministry of Health & Welfare, Republic of Korea (grant no. HI23C0515), and the National Research Foundation of Korea (grant no. NRF-2021R1A5A202161).
Availability of data and materials
The data generated in the present study may be requested from the corresponding author.
Authors' contributions
IS, GOC and HSH contributed to conception and design. IS, TML and HDTN were involved in the acquisition and analysis of data. HSH, GOC, IS, JC and NJYP were involved in the interpretation of data. IS, TML and HDTN were involved in visualization. TML wrote the original draft. GOC, IS, HSH, HDTN, JC and NJYP revised the manuscript critically for important intellectual content. IS and GOC confirm the authenticity of all the raw data. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Ghose A, Bolina A, Mahajan I, Raza SA, Clarke M, Pal A, Sanchez E, Rallis KS, Boussios S. Hereditary ovarian cancer: Towards a cost-effective prevention strategy. Int J Environ Res Public Health. 2022;19:12057. doi: 10.3390/ijerph191912057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lõhmussaar K, Kopper O, Korving J, Begthel H, Vreuls CPH, van Es JH, Clevers H. Assessing the origin of high-grade serous ovarian cancer using CRISPR-modification of mouse organoids. Nat Commun. 2020;11:2660. doi: 10.1038/s41467-020-16432-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maringe C, Walters S, Butler J, Coleman MP, Hacker N, Hanna L, Mosgaard BJ, Nordin A, Rosen B, Engholm G, et al. Stage at diagnosis and ovarian cancer survival: Evidence from the international cancer benchmarking partnership. Gynecol Oncol. 2012;127:75–82. doi: 10.1016/j.ygyno.2012.06.033. [DOI] [PubMed] [Google Scholar]
- 5.Ghose A, McCann L, Makker S, Mukherjee U, Gullapalli SVN, Erekkath J, Shih S, Mahajan I, Sanchez E, Uccello M, et al. Diagnostic biomarkers in ovarian cancer: Advances beyond CA125 and HE4. Ther Adv Med Oncol. 2024;16:17588359241233225. doi: 10.1177/17588359241233225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saani I, Raj N, Sood R, Ansari S, Mandviwala HA, Sanchez E, Boussios S. Clinical challenges in the management of malignant ovarian germ cell tumours. Int J Environ Res Public Health. 2023;20:6089. doi: 10.3390/ijerph20126089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Singh N. WHO classification of tumors of the ovary. In: Encyclopedia of Pathology. In: Van Krieken JHJM, editor. Springer International Publishing; Cham: 2022. pp. 1–4. [Google Scholar]
- 8.Matulonis UA, Sood AK, Fallowfield L, Howitt BE, Sehouli J, Karlan BY. Ovarian cancer. Nat Rev Dis Primers. 2016;2:16061. doi: 10.1038/nrdp.2016.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pavlidis N, Rassy E, Vermorken JB, Assi T, Kattan J, Boussios S, Smith-Gagen J. The outcome of patients with serous papillary peritoneal cancer, fallopian tube cancer, and epithelial ovarian cancer by treatment eras: 27 years data from the SEER registry. Cancer Epidemiol. 2021;75:102045. doi: 10.1016/j.canep.2021.102045. [DOI] [PubMed] [Google Scholar]
- 10.Kindelberger DW, Lee Y, Miron A, Hirsch MS, Feltmate C, Medeiros F, Callahan MJ, Garner EO, Gordon RW, Birch C, et al. Intraepithelial carcinoma of the fimbria and pelvic serous carcinoma: Evidence for a causal relationship. Am J Surg Pathol. 2007;31:161–169. doi: 10.1097/01.pas.0000213335.40358.47. [DOI] [PubMed] [Google Scholar]
- 11.Shih IM, Wang Y, Wang TL. The origin of ovarian cancer species and precancerous landscape. Am J Pathol. 2021;191:26–39. doi: 10.1016/j.ajpath.2020.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aliyuda F, Moschetta M, Ghose A, Sofia Rallis K, Sheriff M, Sanchez E, Rassy E, Boussios S. Advances in ovarian cancer treatment beyond PARP Inhibitors. Curr Cancer Drug Targets. 2023;23:433–446. doi: 10.2174/1568009623666230209121732. [DOI] [PubMed] [Google Scholar]
- 13.Lee Y, Miron A, Drapkin R, Nucci MR, Medeiros F, Saleemuddin A, Garber J, Birch C, Mou H, Gordon RW, et al. A candidate precursor to serous carcinoma that originates in the distal fallopian tube. J Pathol. 2007;211:26–35. doi: 10.1002/path.2091. [DOI] [PubMed] [Google Scholar]
- 14.Colvin EK, Howell VM. Why the dual origins of high grade serous ovarian cancer matter. Nat Commun. 2020;11:1200. doi: 10.1038/s41467-020-15089-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Greten FR, Grivennikov SI. Inflammation and cancer: Triggers, mechanisms, and consequences. Immunity. 2019;51:27–41. doi: 10.1016/j.immuni.2019.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Łaniewski P, Ilhan ZE, Herbst-Kralovetz MM. The microbiome and gynaecological cancer development, prevention and therapy. Nat Rev Urol. 2020;17:232–250. doi: 10.1038/s41585-020-0286-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chambers LM, Bussies P, Vargas R, Esakov E, Tewari S, Reizes O, Michener C. The microbiome and gynecologic cancer: Current evidence and future opportunities. Curr Oncol Rep. 2021;23:92. doi: 10.1007/s11912-021-01079-x. [DOI] [PubMed] [Google Scholar]
- 18.Jia D, Nagaoka Y, Katsumata M, Orsulic S. Inflammation is a key contributor to ovarian cancer cell seeding. Sci Rep. 2018;8:12394. doi: 10.1038/s41598-018-30261-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sabatini ME, Chiocca S. Human papillomavirus as a driver of head and neck cancers. Br J Cancer. 2020;122:306–314. doi: 10.1038/s41416-019-0602-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Demarco M, Hyun N, Carter-Pokras O, Raine-Bennett TR, Cheung L, Chen X, Hammer A, Campos N, Kinney W, Gage JC, et al. A study of type-specific HPV natural history and implications for contemporary cervical cancer screening programs. EClinicalMedicine. 2020;22:100293. doi: 10.1016/j.eclinm.2020.100293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scheffner M, Werness BA, Huibregtse JM, Levine AJ, Howley PM. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell. 1990;63:1129–1136. doi: 10.1016/0092-8674(90)90409-8. [DOI] [PubMed] [Google Scholar]
- 22.Fischer M, Uxa S, Stanko C, Magin TM, Engeland K. Human papilloma virus E7 oncoprotein abrogates the p53-p21-DREAM pathway. Sci Rep. 2017;7:2603. doi: 10.1038/s41598-017-02831-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cherif S, Amine A, Thies S, Taube ET, Braicu EI, Sehouli J, Kaufmann AM. Prevalence of human papillomavirus detection in ovarian cancer: A meta-analysis. Eur J Clin Microbiol Infect Dis. 2021;40:1791–1802. doi: 10.1007/s10096-021-04282-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ibragimova MK, Kokorina EV, Tsyganov MM, Churuksaeva ON, Litviakov NV. Human papillomavirus and ovarian cancer (review of literature and meta-analysis) Infect Genet Evol. 2021;95:105086. doi: 10.1016/j.meegid.2021.105086. [DOI] [PubMed] [Google Scholar]
- 25.Grabarek BO, Ossowski P, Czarniecka J, Ożóg M, Prucnal J, Dziuba I, Ostenda A, Dziobek K, Boroń D, Peszek W, et al. Detection and genotyping of human papillomavirus (HPV16/18), Epstein-barr virus (EBV), and human cytomegalovirus (HCMV) in endometrial endometroid and ovarian cancers. Pathogens. 2023;12:397. doi: 10.3390/pathogens12030397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fatima B, Masud R, Sultana N, Javed A, Justin S. Detection of human papillomavirus in archival bladder and ovarian cancer samples. Clin Epidemiology Glob Health. 2023;22:101339. doi: 10.1016/j.cegh.2023.101339. [DOI] [Google Scholar]
- 27.Kumar P, Ranmale S, Tongaonkar H, Mehta S, Mania-Pramanik J. Human papillomavirus infection: Is it associated with epithelial ovarian cancer? Indian J Med Microbiol. 2021;39:311–314. doi: 10.1016/j.ijmmb.2021.05.001. [DOI] [PubMed] [Google Scholar]
- 28.Yang X, You Q, Yao G, Geng J, Ma R, Meng H. Evaluation of p16 in epithelial ovarian cancer for a 10-year study in northeast China: Significance of HPV in correlation with PD-L1 expression. Cancer Manag Res. 2020;12:6747–6753. doi: 10.2147/CMAR.S262678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shokouh MR, Safaei A, Moattari A, Sarvari J. Association of human papilloma virus and Epstein-barr virus with ovarian cancer in Shiraz, southwestern Iran. Iran J Pathol. 2020;15:292–298. doi: 10.30699/ijp.2020.119681.2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kisseljova N, Zhordania K, Fedorova M, Katargin A, Valeeva A, Pajanidi J, Pavlova L, Khvan O, Vinokurova S. Detection of human papillomavirus prevalence in ovarian cancer by different test systems. Intervirology. 2019;62:198–204. doi: 10.1159/000506050. [DOI] [PubMed] [Google Scholar]
- 31.Paradowska E, Jabłońska A, Studzińska M, Wilczyński M, Wilczyński JR. Detection and genotyping of CMV and HPV in tumors and fallopian tubes from epithelial ovarian cancer patients. Sci Rep. 2019;9:19935. doi: 10.1038/s41598-019-56448-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hammou RA, Benhessou M, Bouziyane A, Hassou N, Benhchekroun MN, Bessi H, Ennaji MM. Oncogenic human papillomavirus involvement in epithelial ovarian carcinoma among women in Morocco. Bioinformation. 2019;15:55–60. doi: 10.6026/97320630015055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hassan ZK, Hafez MM, Kamel MM, Zekri ARN. Human Papillomavirus genotypes and methylation of CADM1, PAX1, MAL and ADCYAP1 genes in epithelial ovarian cancer patients. Asian Pac J Cancer Prev. 2017;18:169–176. doi: 10.22034/APJCP.2017.18.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Farzaneh F, Nadji SA, Khosravi D, Hosseini MS, Hashemi Bahremani M, Chehrazi M, Bagheri G, Sigaroodi A, Haghighatian Z. Lack of HPV in benign and malignant epithelial ovarian tumors in Iran. Asian Pac J Cancer Prev. 2017;18:1233–1236. doi: 10.22034/APJCP.2017.18.5.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dadashi M, Eslami G, Faghihloo E, Pourmohammad A, Hosseini J, Taheripanah R, Arab-Mazar Z. Detection of human papilloma virus type 16 in epithelial ovarian tumors samples. Arch Clin Infect Dis. 2017;12:e39666. [Google Scholar]
- 36.Zhang PP, Zhou L, Cao JS, Li YP, Zeng Z, Sun N, Shen L, Zhu HY, Ruan Y, Zha WT, et al. Possible epithelial ovarian cancer association with HPV18 or HPV33 infection. Asian Pac J Cancer Prev. 2016;17:2959–2964. [PubMed] [Google Scholar]
- 37.Ingerslev K, Hogdall E, Skovrider-Ruminski W, Schnack TH, Karlsen MA, Nedergaard L, Hogdall C, Blaakær J. High-risk HPV is not associated with epithelial ovarian cancer in a Caucasian population. Infect Agent Cancer. 2016;11:39. doi: 10.1186/s13027-016-0087-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mahmood FM, Kadhim HS, Mousa Al Khuzaee LR. Detection of human papillomavirus-16 E6-oncoprotein in epithelial ovarian tumors samples of Iraqi patients. Jundishapur J Microbiol. 2014;7:e11945. doi: 10.5812/jjm.11945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Al-Shabanah OA, Hafez MM, Hassan ZK, Sayed-Ahmed MM, Abozeed WN, Alsheikh A, Al-Rejaie SS. Methylation of SFRPs and APC genes in ovarian cancer infected with high risk human papillomavirus. Asian Pac J Cancer Prev. 2014;15:2719–2725. doi: 10.7314/APJCP.2014.15.6.2719. [DOI] [PubMed] [Google Scholar]
- 40.Al-Shabanah OA, Hafez MM, Hassan ZK, Sayed-Ahmed MM, Abozeed WN, Al-Rejaie SS, Alsheikh AA. Human papillomavirus genotyping and integration in ovarian cancer Saudi patients. Virol J. 2013;10:343. doi: 10.1186/1743-422X-10-343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Malisic E, Jankovic R, Jakovljevic K. Detection and genotyping of human papillomaviruses and their role in the development of ovarian carcinomas. Arch Gynecol Obstet. 2012;286:723–728. doi: 10.1007/s00404-012-2367-6. [DOI] [PubMed] [Google Scholar]
- 42.Shanmughapriya S, SenthilKumar G, Vinodhini K, Das BC, Vasanthi N, Natarajaseenivasan K. Viral and bacterial aetiologies of epithelial ovarian cancer. Eur J Clin Microbiol Infect Dis. 2012;31:2311–2317. doi: 10.1007/s10096-012-1570-5. [DOI] [PubMed] [Google Scholar]
- 43.Alavi G, Sharifi N, Sadeghian A, Rezaei A, Shidaee H. Failure to demonstrate the role of high risk human papilloma virus in epithelial ovarian cancer. Iran J Pathol. 2012;7:151–156. [Google Scholar]
- 44.Bilyk OO, Pande NT, Buchynska LG. Analysis of P53, P16INK4A, PRB and Cyclin D1 expression and human papillomavirus in primary ovarian serous carcinomas. Exp Oncol. 2011;33:150–156. [PubMed] [Google Scholar]
- 45.Idahl A, Lundin E, Elgh F, Jurstrand M, Møller JK, Marklund I, Lindgren P, Ottander U. Chlamydia trachomatis, Mycoplasma genitalium, Neisseria gonorrhoeae, human papillomavirus, and polyomavirus are not detectable in human tissue with epithelial ovarian cancer, borderline tumor, or benign conditions. Am J Obstet Gynecol. 2010;202:71.e1–e6. doi: 10.1016/j.ajog.2009.07.042. [DOI] [PubMed] [Google Scholar]
- 46.Wentzensen N, du Bois A, Kommoss S, Pfisterer J, Von Knebel Doeberitz M, Schmidt D, Kommoss F. No metastatic cervical adenocarcinomas in a series of p16INK4a-positive mucinous or endometrioid advanced ovarian carcinomas: An analysis of the AGO ovarian cancer study group. Int J Gynecol Pathol. 2008;27:18–23. doi: 10.1097/pgp.0b013e318074b83f. [DOI] [PubMed] [Google Scholar]
- 47.Giordano G, D'Adda T, Gnetti L, Froio E, Merisio C, Melpignano M. Role of human papillomavirus in the development of epithelial ovarian neoplasms in Italian women. J Obstet Gynaecol. 2008;34:210–217. doi: 10.1111/j.1447-0756.2008.00759.x. [DOI] [PubMed] [Google Scholar]
- 48.Atalay F, Taskiran C, Taner MZ, Pak I, Or M, Tuncer S. Detection of human papillomavirus DNA and genotyping in patients with epithelial ovarian carcinoma. J Obstet Gynaecol. 2007;33:823–828. doi: 10.1111/j.1447-0756.2007.00663.x. [DOI] [PubMed] [Google Scholar]
- 49.Konidaris S, Kouskouni EE, Panoskaltsis T, Kreatsas G, Patsouris ES, Sarivalassis A, Nonni A, Lazaris AC. Human papillomavirus infection in malignant and benign gynaecological conditions: A study in Greek women. Health Care Women Int. 2007;28:182–191. doi: 10.1080/07399330601128627. [DOI] [PubMed] [Google Scholar]
- 50.Quirk JT, Kupinski JM, DiCioccio RA. Analysis of ovarian tumors for the presence of human papillomavirus DNA. J Obstet Gynaecol. 2006;32:202–205. doi: 10.1111/j.1447-0756.2006.00376.x. [DOI] [PubMed] [Google Scholar]
- 51.Kuscu E, Ozdemir BH, Erkanli S, Haberal A. HPV and p53 expression in epithelial ovarian carcinoma. Eur J Gynaecol Oncol. 2005;26:642–645. [PubMed] [Google Scholar]
- 52.Elishaev E, Gilks CB, Miller D, Srodon M, Kurman RJ, Ronnett BM. Synchronous and metachronous endocervical and ovarian neoplasms: Evidence supporting interpretation of the ovarian neoplasms as metastatic endocervical adenocarcinomas simulating primary ovarian surface epithelial neoplasms. Am J Surg Pathol. 2005;29:281–294. doi: 10.1097/01.pas.0000152136.81771.12. [DOI] [PubMed] [Google Scholar]
- 53.Yang HJ, Liu VW, Tsang PC, Yip AM, Ng TY, Cheung AN, Ngan HY. Comparison of human papillomavirus DNA levels in gynecological cancers: Implication for cancer development. Tumor Biol. 2003;24:310–316. doi: 10.1159/000076463. [DOI] [PubMed] [Google Scholar]
- 54.Wu QJ, Guo M, Lu ZM, Li T, Qiao HZ, Ke Y. Detection of human papillomavirus-16 in ovarian malignancy. Br J Cancer. 2003;89:672–675. doi: 10.1038/sj.bjc.6601172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li T, Lu ZM, Guo M, Wu QJ, Chen KN, Xing HP, Mei Q, Ke Y. p53 codon 72 polymorphism (C/G) and the risk of human papillomavirus-associated carcinomas in China. Cancer. 2002;95:2571–2576. doi: 10.1002/cncr.11008. [DOI] [PubMed] [Google Scholar]
- 56.Ip SM, Wong LC, Xu CM, Cheung AN, Tsang PC, Ngan HY. Detection of human papillomavirus DNA in malignant lesions from Chinese women with carcinomas of the upper genital tract. Gynecol Oncol. 2002;87:104–111. doi: 10.1006/gyno.2002.6784. [DOI] [PubMed] [Google Scholar]
- 57.Chen TR, Chan PJ, Seraj IM, King A. Absence of human papillomavirus E6-E7 transforming genes from HPV 16 and 18 in malignant ovarian carcinoma. Gynecol Oncol. 1999;72:180–182. doi: 10.1006/gyno.1998.5255. [DOI] [PubMed] [Google Scholar]
- 58.Anttila M, Syrjänen S, Ji H, Saarikoski S, Syrjänen K. Failure to demonstrate human papillomavirus DNA in epithelial ovarian cancer by general primer PCR. Gynecol Oncol. 1999;72:337–341. doi: 10.1006/gyno.1998.5264. [DOI] [PubMed] [Google Scholar]
- 59.Zimna K, Poreba E, Kedzia W, Gozdzicka-Józefiak A, Kezia H. Human papillomavirus (HPV) in upper genital tract carcinomas of women. Eur J Gynaecol Oncol. 1997;18:415–417. [PubMed] [Google Scholar]
- 60.Anwar K, Nakakuki K, Imai H, Shiraishi T, Inuzuka M. Infection of human papillomavirus (HPV) and p53 over-expression in human female genital tract carcinoma. J Pak Med Assoc. 1996;46:220–224. [PubMed] [Google Scholar]
- 61.Trottier AM, Provencher D, Mes-Masson AM, Vauclair R, Coutlée F. Absence of human papillomavirus sequences in ovarian pathologies. J Clin Microbiol. 1995;33:1011–1013. doi: 10.1128/jcm.33.4.1011-1013.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Runnebaum IB, Maier S, Tong XW, Rosenthal HE, Möbus VJ, Kieback DG, Kreienberg R. Human papillomavirus integration is not associated with advanced epithelial ovarian cancer in German patients. Cancer Epidemiol Biomarkers Prev. 1995;4:573–575. [PubMed] [Google Scholar]
- 63.Lai CH, Wang CY, Lin CY, Pao CC. Detection of human papillomavirus RNA in ovarian and endometrial carcinomas by reverse transcription/polymerase chain reaction. Gynecol Obstet Invest. 1994;38:276–280. doi: 10.1159/000292496. [DOI] [PubMed] [Google Scholar]
- 64.Lai CH, Hsueh S, Lin CY, Huang MY, You GB, Chang HC, Pao CC. Human papillomavirus in benign and malignant ovarian and endometrial tissues. Int J Gynecol Pathol. 1992;11:210–215. doi: 10.1097/00004347-199207000-00007. [DOI] [PubMed] [Google Scholar]
- 65.Beckmann AM, Sherman KJ, Saran L, Weiss NS. Genital-type human papillomavirus infection is not associated with surface epithelial ovarian carcinoma. Gynecol Oncol. 1991;43:247–251. doi: 10.1016/0090-8258(91)90029-5. [DOI] [PubMed] [Google Scholar]
- 66.McLellan R, Buscema J, Guerrero E, Shah KV, Woodruff JD, Currie JL. Investigation of ovarian neoplasia of low malignant potential for human papillomavirus. Gynecol Oncol. 1990;38:383–385. doi: 10.1016/0090-8258(90)90078-Y. [DOI] [PubMed] [Google Scholar]
- 67.Leake JF, Woodruff JD, Searle C, Daniel R, Shah KV, Currie JL. Human papillomavirus and epithelial ovarian neoplasia. Gynecol Oncol. 1989;34:263–273. doi: 10.1016/0090-8258(89)90158-3. [DOI] [PubMed] [Google Scholar]
- 68.Nikolakopoulou A, Mavridis D, Salanti G. Demystifying fixed and random effects meta-analysis. BMJ Ment Health. 2014;17:53–57. doi: 10.1136/eb-2014-101795. [DOI] [PubMed] [Google Scholar]
- 69.Schwarzer G. meta: An R package for meta-analysis. R News. 2007;7:40–45. [Google Scholar]
- 70.Higgins JP, Green S. Wiley; 2019. Cochrane handbook for systematic reviews of interventions. In: Cochrane Handbook for Systematic Reviews of Interventions. [Google Scholar]
- 71.Paule RC, Mandel J. Consensus values and weighting factors. J Res Natl Bur Stand (1977) 1982;87:377–385. doi: 10.6028/jres.087.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Balduzzi S, Rücker G, Schwarzer G. How to perform a meta-analysis with R: A practical tutorial. Evid Based Ment Health. 2019;22:153–160. doi: 10.1136/ebmental-2019-300117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sterne JA, Sutton AJ, Ioannidis JP, Terrin N, Jones DR, Lau J, Carpenter J, Rücker G, Harbord RM, Schmid CH, et al. Recommendations for examining and interpreting funnel plot asymmetry in meta-analyses of randomised controlled trials. BMJ. 2011;343:d4002. doi: 10.1136/bmj.d4002. [DOI] [PubMed] [Google Scholar]
- 74.Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50:1088–1101. doi: 10.2307/2533446. [DOI] [PubMed] [Google Scholar]
- 75.Peters JL, Sutton AJ, Jones DR, Abrams KR, Rushton L. Comparison of two methods to detect publication bias in meta-analysis. JAMA. 2006;295:676–680. doi: 10.1001/jama.295.6.676. [DOI] [PubMed] [Google Scholar]
- 76.Viechtbauer W. Conducting meta-analyses in R with the metafor package. J Stat Soft. 2010;36:1–48. doi: 10.18637/jss.v036.i03. [DOI] [Google Scholar]
- 77.R Core Team, corp-author. R Foundation for Statistical Computing; Vienna: 2023. R: A language and environment for statistical computing. [Google Scholar]
- 78.Schwarzer G, Carpenter JR, Rücker G. metasens: Statistical methods for sensitivity analysis in meta-analysis. https://CRAN.R-project.org/package=metasens R package version 1.5–2. [Google Scholar]
- 79.Halec G, Alemany L, Quiros B, Clavero O, Höfler D, Alejo M, Quint W, Pawlita M, Bosch FX, de Sanjose S. Biological relevance of human papillomaviruses in vulvar cancer. Mod Pathol. 2017;30:549–562. doi: 10.1038/modpathol.2016.197. [DOI] [PubMed] [Google Scholar]
- 80.Lechner M, Liu J, Masterson L, Fenton TR. HPV-associated oropharyngeal cancer: Epidemiology, molecular biology and clinical management. Nat Rev Clin Oncol. 2022;19:306–327. doi: 10.1038/s41571-022-00603-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bjørge T, Engeland A, Luostarinen T, Mork J, Gislefoss RE, Jellum E, Koskela P, Lehtinen M, Pukkala E, Thoresen SO, Dillner J. Human papillomavirus infection as a risk factor for anal and perianal skin cancer in a prospective study. Br J Cancer. 2002;87:61–64. doi: 10.1038/sj.bjc.6600350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cohen PA, Jhingran A, Oaknin A, Denny L. Cervical cancer. Lancet. 2019;393:169–182. doi: 10.1016/S0140-6736(18)32470-X. [DOI] [PubMed] [Google Scholar]
- 83.Chan PJ, Seraj IM, Kalugdan TH, King A. Evidence for ease of transmission of human papillomavirus DNA from sperm to cells of the uterus and embryo. J Assist Reprod Genet. 1996;13:516–519. doi: 10.1007/BF02066536. [DOI] [PubMed] [Google Scholar]
- 84.Dyson N, Howley PM, Münger K, Harlow E. The human papilloma virus-16 E7 oncoprotein is able to bind to the retinoblastoma gene product. Science. 1989;243:934–937. doi: 10.1126/science.2537532. [DOI] [PubMed] [Google Scholar]
- 85.Hashiguchi Y, Tsuda H, Yamamoto K, Inoue T, Ishiko O, Ogita S. Combined analysis of p53 and RB pathways in epithelial ovarian cancer. Hum Pathol. 2001;32:988–996. doi: 10.1053/hupa.2001.27115. [DOI] [PubMed] [Google Scholar]
- 86.Bruni L, Albero G, Serrano B, Mena M, Collado J, Gómez D, Muñoz J, Bosch F, de Sanjosé S. ICO/IARC Information Centre on HPV and Cancer (HPV Information Centre). Human papillomavirus and related diseases in the world. Summary Report 10 March 2023. https://hpvcentre.net/statistics/reports/XWX.pdf. [ October 8; 2023 ]; [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data generated in the present study may be requested from the corresponding author.