Abstract
Borna disease virus (BDV) surface glycoprotein (GP) (p56) has a predicted molecular mass of 56 kDa. Due to extensive posttranslational glycosylation the protein migrates as a polypeptide of 84 kDa (gp84). The processing of gp84 by the cellular protease furin generates gp43, which corresponds to the C-terminal part of gp84. Both gp84 and gp43 have been implicated in viral entry involving receptor-mediated endocytosis and pH-dependent fusion. We have investigated the domains of BDV p56 involved in virus entry. For this, we used a pseudotype approach based on a recently developed recombinant vesicular stomatitis virus (VSV) in which the gene for green fluorescent protein was substituted for the VSV G protein gene (VSVΔG*). Complementation of VSVΔG* with BDV p56 resulted in infectious VSVΔG* pseudotypes that contained both BDV gp84 and gp43. BDV-VSV chimeric GPs that contained the N-terminal 244 amino acids of BDV p56 and amino acids 421 to 511 of VSV G protein were efficiently incorporated into VSVΔG* particles, and the resulting pseudotype virions were neutralized by BDV-specific antiserum. These findings indicate that the N-terminal part of BDV p56 is sufficient for receptor recognition and virus entry.
Borna disease virus (BDV) is the causal agent of Borna disease, a frequently fatal meningoencephalitis affecting mainly horses and sheep in certain regions of central Europe. Experimentally, BDV can infect a remarkably large number of vertebrate species. The infection is characterized by a variable period of incubation with diverse clinical and pathological manifestations (15, 25, 37), and behavioral disturbances are a hallmark of BDV infection. Serological and molecular-epidemiology data indicate that the host range, geographic distribution, and prevalence of BDV may be much broader than previously thought. There is also evidence that BDV can infect humans and might be associated with some neuropsychiatric disorders (1, 2, 9, 10, 18, 24, 28, 30, 38, 41). However, the prevalence and possible clinical significance of BDV in humans remain controversial (46).
BDV is an enveloped, nonsegmented, negative-strand (NNS) RNA virus (8, 43). BDV has the smallest genome size, 8.9 kb, among known mononegaviruses. Unlike what is found for all other NNS RNA animal viruses, transcription and replication of the BDV genome take place in the nucleus (3–5). BDV uses RNA splicing for the regulation of its genome expression, which is also unique among known mononegaviruses. Based on its unique biological and molecular-genetics features, BDV is now considered to be the prototypic member of a new family, Bornaviridae, within the order Mononegavirales.
The BDV genome contains six major open reading frames (ORFs). The product of BDV ORF IV is the counterpart of the type I surface glycoproteins (GPs) found in other members of the Mononegavirales. BDV GP (p56) has a predicted molecular mass of 56 kDa, but due to extensive N glycosylation the protein migrates with an apparent molecular mass of 84 kDa (gp84). BDV GP is processed posttranslationally by the subtilisin-like protease furin, resulting in two products (13, 31). The C-terminal portion has an apparent molecular mass of 43 kDa (gp43), whereas the N-terminal fragment (gp41) has not been detected yet in BDV-infected cells. gp84 accumulates in the endoplasmic reticulum and perinuclear region, but it is typically not detected on the plasma membrane. In contrast, gp43 accumulates at the cell surface (14).
Both gp84 and gp43 are associated with infectious BDV particles. Moreover, antibodies to BDV p56 exhibit neutralizing activity, suggesting that gp84 or gp43 or both participate in viral entry. BDV entry is via receptor-mediated endocytosis, and the fusion between viral and cellular membranes occurs in the acidic environment of the late endosome (13). Consistent with this, BDV-infected cells exhibit massive syncytium formation upon exposure to low-pH medium (13). This pH-dependent cell fusion event is likely mediated by gp43 since it is the only membrane-anchored GP species found on the plasma membrane. In addition, gp43 contains a hydrophobic sequence near its N terminus that has features similar to those of the fusogenic peptide sequences described for paramyxoviruses and influenza virus (42). However, the domains of BDV GP involved in receptor recognition and cell entry have not been defined. Studies to identify functional domains of BDV GP have been impaired by the inability to grow cell-free infectious BDV to high titers (6, 7). To overcome this problem, we took advantage of the potential that enveloped viruses have of incorporating heterologous surface GPs into their lipidic envelopes. This process, known as pseudotyping, allows a rapid analysis of the GP of interest.
Vesicular stomatitis virus (VSV), the prototype rhabdovirus, has been widely used as a model system to study replication and assembly of enveloped RNA virus due to its ability to grow to high titers in many different cell lines. Furthermore, the availability of a reverse genetic system for VSV (22, 51) has allowed the generation of recombinant viruses that express foreign proteins, and several heterologous GPs have been successfully incorporated into VSV particles (17, 20, 32, 33, 48, 53). In this report we have used recombinant VSVΔG* (47), in which the VSV G gene has been replaced by the green fluorescent protein (GFP) gene, to investigate the role of BDV p56 polypeptides in viral entry. Particles released from VSVΔG*-infected cells are not infectious unless an envelope protein responsible for receptor binding and fusion is provided in trans. We show here that both gp84 and gp43 can be incorporated into VSV virions. Furthermore, using BDV-VSV chimeric glycoproteins, we provide evidence that the amino-terminal region (amino acids 1 to 244) of the BDV p56 glycoprotein is sufficient for receptor recognition and for the promotion of entry of BDV into cells.
MATERIALS AND METHODS
Cells and viruses.
Baby hamster kidney BHK-21 cells (ATCC CCL 10) were maintained in high-glucose Dulbecco's modified Eagle medium (DMEM) supplemented with 10% heat-inactivated fetal calf serum (FCS; Life Technologies, Grand Island, N.Y.), 2 mM glutamine, 1× tryptose phosphate broth (Life Technologies), 1 mM sodium pyruvate (Life Technologies), and 0.5% glucose. Human embryonic kidney 293T cells were maintained in DMEM supplemented with 2 mM glutamine and 10% heat-inactivated FCS. Uninfected Vero cells and Vero cells persistently infected with BDV strain He80 (44), designated VeroNKBV, were maintained in 199 medium (Life Technologies) supplemented with 2 mM glutamine, 0.6% sodium bicarbonate, and 10% heat-inactivated FCS.
Stocks of VSVΔG* (47) were grown in BHK-21 cells transfected with pCAGGS (29) expressing VSV G. Thirty-two hours after transfection cells were infected with VSVΔG* at a multiplicity of infection of 3 PFU/cell for 1 h at 37°C. Virus-containing culture supernatant was harvested after 20 h.
Antibodies.
Anti-VSV antibodies used included murine monoclonal antibodies (MAbs) against VSV nucleocapsid protein (10G4; anti-VSV N) and GP (I1; anti-VSV G) (23) and a hyperimmune rabbit serum against the carboxyl terminus of the VSV GP (anti-C tail [anti-CT]). The rabbit polyclonal antibody against VSV P was kindly provided by Jacques Perrault (San Diego State University, San Diego, Calif.). The anti-p56 rabbit serum was raised against a bacterially expressed truncated form of the BDV p56 glycoprotein (14).
For the preparation of pooled rat sera used in the neutralization assays, 5-week-old Lewis rats were inoculated intracerebrally with BDV He80 (1,000 focus-forming units [FFU]), and 30 days later blood samples were collected for serum preparation.
Construction of expression vectors.
All vectors for the expression of BDV p56 chimeras were derived from parental plasmid pCRIIp56 (14). Plasmid pCRIIp56 was obtained by cloning full-length ORF IV (BDV p56) into the pCRII vector (Invitrogen, La Jolla, Calif.). This plasmid was digested with EcoRI, and the resulting fragment containing the BDV p56 ORF was cloned into pCAGGS (29) to generate plasmid pCp56. Plasmid pCVSVG is the pCAGGS plasmid containing the full-length cDNA of the VSV G protein (Indiana serotype, San Juan strain) (36). Plasmid pCRIIp56N was obtained by cloning a DNA fragment corresponding to the first 244 amino acids of BDV p56 (p56N) into pCRII. This DNA fragment was produced by PCR using primers P56RIF (5′-GGAATTCCGCCATGGAGCTTTC-3′) and P56BstBIR (5′-GTTCGAACAACTTGGACCGGCAGG-3′). The underlined ATG sequence in primer P56RIF corresponds to the AUG of the BDV p56 ORF. To generate construct pCp56/GS, p56N was excised from pCRIIp56N by digestion with EcoRI and BstBI and cloned into pCVSVG digested with the same two enzymes. A similar strategy was used in the construction of pCp56/GlyGS, with the only difference that primer P56GlyR (5′-GTTCGAATCCTCCTCCCAACTTGGACCGGCAG-3′) was used instead of primer P56BstBIR. To generate plasmid p56N/G, a PCR fragment was made with primers P56RIF and P56RAvaII (5′-CAACTTGGACCGGCAGGACGAC-3′) using p56N/GS as a template. The amplified fragment (710 bp) was digested with NcoI and AvaII. A second PCR product (230 bp) corresponding to the transmembrane (TM) and CT domain sequences of VSV G was generated using the pair of primers VSVFAvaII (5′-GGGCGGTCCAAGTTGAGTAGTTGGAAAAGC-3′) and VSVRIR (5′-GGAATTCCCTCTTTGAGCATGGTATCAC-3′) and digested with AvaII and EcoRI. Plasmid pTM1 (27) was digested with NcoI and EcoRI, treated with alkaline phosphatase, and ligated to the 710- and 230-bp PCR products in a three-way ligation to generate plasmid pTMp56N/G. A PCR was carried over pTMp56N/G using primers P56RIF and VSVRIR to amplify gene p56N/G. The resulting product was digested with EcoRI and cloned into pCAGGS linearized with EcoRI. This plasmid was denominated pCp56N/G. Correct orientation of the inserts was determined by restriction analysis and sequencing through the junction sites. Plasmid DNAs were prepared using Qiagen technology.
Expression of wild-type and chimeric GPs.
The expression of the GPs was analyzed by indirect immunofluorescence and Western blotting. BHK-21 cells (105) were transfected with 0.4 μg of plasmid DNA and 1.2 μg of Lipofectamine (Life Technologies). After 48 h, cells were examined for protein expression. For immunofluorescence, transfected cells grown on coverslips were washed with phosphate-buffered saline (PBS) and fixed either with methanol-acetone (1:1) for 5 min for intracellular staining or with 4% paraformaldehyde in PBS for 20 min at room temperature for surface antigen staining and processed for immunofluorescence as described previously (5). Briefly, after being blocked with 10% normal goat serum for 45 min at room temperature, cells were stained with primary antibodies for 1 h. Secondary fluorescent antibodies used were Texas red-labeled anti-mouse immunoglobulin G (IgG) or anti-rabbit IgG (Cappel, West Chester, Pa.). The coverslips were mounted on microscope slides with Mowiol (Calbiochem, San Diego, Calif.) and viewed with a fluorescence microscope. For Western blot analysis, cells were harvested in sample buffer (50 mM Tris-HCl [pH 8], 62.5 mM EDTA, 1% NP-40, 0.4% deoxycholate). Cell lysates were separated by sodium dodecyl sulfate–8 to 16% polyacrylamide gel electrophoresis (SDS–8 to 16% PAGE) by using the buffer system of Laemmli, and the proteins were transferred onto Immobilon-P membranes (Millipore, Bedford, Mass.) as described previously (39). Immunodetection of the proteins was done by using the BM chemiluminescence kit (Boehringer Mannheim, Indianapolis, Ind.).
Production of VSV pseudotypes.
293T cell monolayers in six-well plates (80% confluent) were transfected with 2 μg of plasmid DNA by using Lipofectamine as recommended by the manufacturer. Thirty-two hours after transfection, cells were infected with VSVΔG* (47) at a multiplicity of infection of 3 PFU/cell for 1 h at 37°C. After a 1-h adsorption period, the inoculum was removed, cells were extensively washed with DMEM, and fresh culture medium was added. After 20 h of incubation at 37°C in a CO2 incubator, culture supernatant was collected, clarified by low-speed centrifugation, and stored at −70°C. Based on the GP provided in trans by transfection, pseudotyped viruses were designated VSVΔG*-G, VSVΔG*-p56, VSVΔG*-p56/GS, VSVΔG*-p56/GlyGS, VSVΔG*-p56/G, and VSVΔG*.
Titration of pseudotyped virus.
BHK-21 cells grown on 96-well plates were infected with 50 μl of serially diluted virus stock. After a 1-h adsorption period, the inoculum was removed and, after two washes, fresh culture medium was added and cells were incubated at 37°C in a CO2 incubator. At 20 h postinfection (p.i.), GFP-expressing cells were counted under an inverted fluorescence microscope. Doublets of GFP-expressing cells were counted as one infectious unit.
Neutralization of VSV pseudotypes.
VSV pseudotypes (500 PFU) and BDV He80 (100 FFU) were incubated (60 min at 37°C) with serial dilutions of pooled sera from BDV-infected rats. Samples were then diluted to a final volume of 300 μl in DMEM and used to infect BHK-21 cells (105) grown in M24 wells (2 cm2). Cells infected with VSV pseudotypes were examined at 24 h p.i. based on GFP expression, whereas BDV-infected cells were examined at 72 h p.i. by using an immunofocus assay (14).
Treatment with NH4Cl.
BHK-21 cells (105) in M24 wells (2 cm2) were treated for 2 h with NH4Cl at various concentrations (0 to 20 mM; in DMEM at 37°C in a CO2 incubator) and then infected with the VSV pseudotypes or with BDV He80. Infections were done in the presence of NH4Cl at various concentrations. After 24 h, infected cells were determined based on GFP expression for the VSV pseudotypes or by using an immunofocus assay for BDV He80 (14).
RESULTS
Plasmid-mediated expression of wild-type and chimeric BDV GPs.
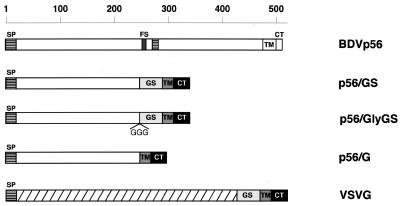
To analyze the role of BDV p56 in viral entry, we generated a series of constructs that included both wild-type BDV p56 and a number of chimeric GPs (Fig. 1). All the chimeric GPs contained the 20-amino-acid TM domain and the 29-amino-acid CT of VSV G. The TM domain and CT of VSV G protein were used to facilitate the expression of the chimeric GPs at the cell surface, which is essential for incorporation of GPs into VSV particles. Initially we cloned the full-length GP of BDV strain He80 into mammalian expression vector pCAGGS (29). As already mentioned, BDV GP (p56) is a type I integral membrane protein that is posttranslationally processed via cleavage at position 249 by the subtilisin-like protease furin. The fragment of BDV p56 chosen to generate the chimeric GPs comprised amino acids 1 to 244 of BDV p56. This fragment of BDV p56 contains the predicted signal peptide followed by the N-terminal part of the p56 ectodomain up to the start of the furin recognition signal. In construct p56/G, the N-terminal region of p56 was directly fused to the TM domain and CT of VSV G. Recently, the extracellular membrane-proximal stem region (GS) of VSV G has been reported to be required for efficient VSV budding (34). Hence, we decided to generate construct p56/GS, in which amino acids 1 to 244 were fused to amino acid F421 of GS. Plasmids p56/GS and p56/GlyGS differ only in the presence of a three-glycine hinge between the BDV p56 ectodomain and the GS of VSV G. All the constructs were characterized by restriction enzyme digestion and by nucleotide sequence analysis of the junction region for the chimeric genes and of the furin site for the wild-type gene.
FIG. 1.
Schematic representation of the wild-type and chimeric GPs. Open boxes, BDV sequences. BDV p56 is processed via cleavage by the protease furin. A canonical furin recognition sequence (FS) is found at amino acids 245 to 249 of BDV p56. Striped boxes, hydrophobic sequences corresponding to the predicted signal peptide (SP) and the putative fusion domain of BDV p56; hatched box, long ectodomain of VSV G. Boxes representing the GS, TM domain, and CT of VSV G are indicated. In the chimeric constructs the CT and TM domain of VSV G were exchanged for the CT and TM domain of BDV p56. GGG, addition of three glycine residues at the junction between BDV p56 sequences and VSV G sequences in construct p56/GlyGS.
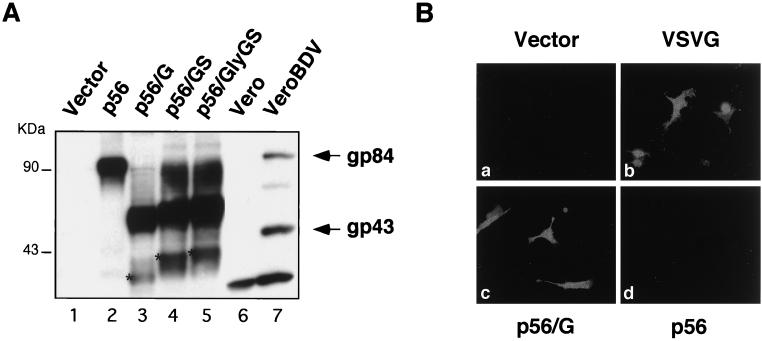
To characterize the polypeptides expressed by the different chimeric gene constructs, we performed Western blot assays using a polyclonal anti-BDV p56 rabbit serum and extracts from BHK-21 cells transfected with the corresponding plasmids (Fig. 2A). After transfection with plasmid p56, only one polypeptide (gp84) was detected by the antibody to p56 (Fig. 2A, lane 2), whereas in BDV-infected cells this antibody detected both gp84 and gp43 (Fig. 2A, lane 7). This apparent discrepancy is likely due to slower and less-efficient processing by furin of the transfected GP. The same result was observed in transfected cells when BDV p56 was expressed under the control of different promoters than the one present in pCAGGS (data not shown). The chimeric GPs had predicted molecular masses of 37.2 (p56/GS), 37.4 (p56/GlyGS), and 32.9 (p56/G) kDa. The predicted molecular masses of these polypeptides represent the nonglycosylated forms of the corresponding chimeric GPs. All three of the chimeric GPs were clearly detected by the rabbit polyclonal serum to BDV p56 (Fig. 2A, lanes 3 to 5). However, in all three cases the major polypeptide band migrated with an apparent molecular weight higher than that predicted. The domain of p56 present in the chimeric proteins still contains 9 of the 13 putative N-glycosylation sites present in full-length p56. Therefore, the observed differences were most likely due to posttranslational modifications involving the addition of carbohydrates.
FIG. 2.
Expression of wild-type and chimeric GPs. (A) Detection of wild-type BDV p56 and chimeric GPs in cell lysates. BHK-21 cells were transfected with the indicated plasmids. Forty-eight hours after transfection cells were lysed in sample buffer. Protein lysates were separated by SDS–8 to 16% PAGE and analyzed by Western blotting using an anti-BDV p56 rabbit serum. Vero cells mock infected (lane 6) or persistently infected with BDV (lane 7) were used as negative and positive controls, respectively. Arrows, migrations of the full-length (gp84) and processed (gp43) BDV GPs; asterisks, positions of the nonglycosylated precursors. (B) Intracellular staining of cells transiently expressing VSV G and BDV-VSV chimeric GPs. After 48 h, BHK-21 cells transfected with the indicated plasmid were fixed and permeabilized with methanol-acetone. Cells were stained with a rabbit serum that specifically recognizes the CT of the VSV GP. The results with p56/G (c) are shown as a representative example. Similar results were obtained with p56/GS and p56/GlyGS. The antibody anti-VSV CT did not cross-react with BDV p56 (d).
To confirm that the chimeric GPs contained the corresponding sequences of VSV G protein, transfected cells were analyzed by immunofluorescence using a rabbit serum that specifically recognizes the CT of VSV G. Similar results were obtained with p56/G, p56/GS, and p56/GlyGS, and a representative example obtained with p56/G is shown in Fig. 2Bc. The pattern of expression for the chimeric GPs was identical to that seen with wild-type VSV G (Fig. 2Bb), which demonstrates that the chimeric proteins were not severely misfolded and were transported to the cell surface. The presence of the VSV G domains could have had an effect on the glycosylation and/or conformational folding of the chimeric GPs, which may have altered their expression and/or antigenicities. However, all the chimeric proteins were produced in large amounts, and they were recognized by the different antibodies used in this study. As predicted, the anti-tail serum did not stain cells transfected with either the empty control plasmid (Fig. 2Ba) or BDV p56 (Fig. 2Bd).
Production of VSV pseudotypes.
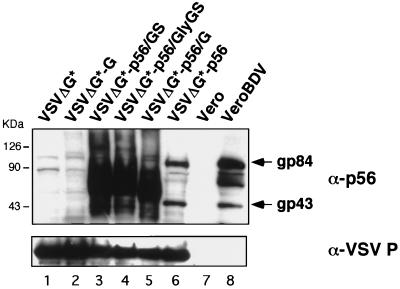
To determine if BDV p56 and the chimeric proteins were able to rescue the infectivity of the VSVΔG* virus, we first analyzed the efficiency of incorporation of these heterologous GPs into VSV particles. For this, we harvested culture medium supernatant from 293T cells transfected with the different plasmid-encoded chimeric GPs and subsequently infected with VSVΔG*. These supernatants were first clarified by centrifugation at low speed, and the viral particles were then pelleted by ultracentrifugation through a sucrose cushion. Viral particles in the pellet produced by high-speed centrifugation were analyzed by Western blotting using a polyclonal antibody specific for p56 (Fig. 3, top). All three chimeric GPs (Fig. 3, lanes 3 to 5) were incorporated with higher efficiency than p56 (Fig. 3, lane 6). VSV pseudotype particles rescued from cells transfected with p56 contained the BDV gp84 and gp43 polypeptides detected in cells persistently infected with BDV (Fig. 3, lane 8). The total amount of VSV particles produced did not appear to be affected by the incorporation of the heterologous GPs, as suggested by the Western blotting results with an antibody to VSV P (Fig. 3, bottom).
FIG. 3.
Incorporation of wild-type BDV p56 GP and BDV-VSV chimeric GPs into VSVΔG* particles. VSVΔG* particles complemented by transfection with the GPs indicated after each hyphen (lanes 2 to 6) or with empty control pCAGGS (lane 1) were partially purified by centrifugation through 20% sucrose and lysed in sample buffer. Proteins were separated by SDS-PAGE in 8 to 16% gels under reducing conditions and analyzed by Western blotting with an anti-p56 polyclonal rabbit serum (top). Vero cells mock infected (lane 7) or persistently infected with BDV (lane 8) were used as negative and positive controls, respectively. The same membrane was then stripped and reacted with an anti-VSV P rabbit serum (bottom).
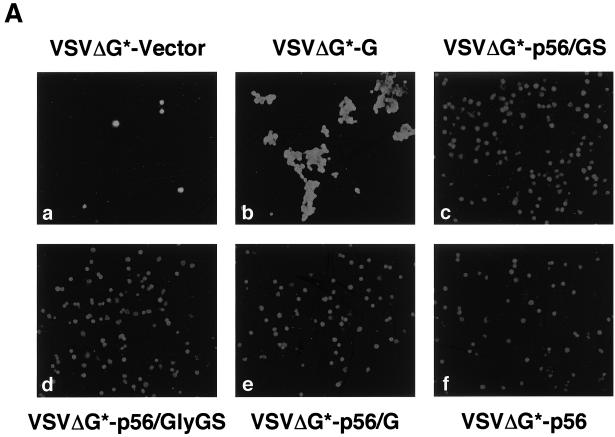
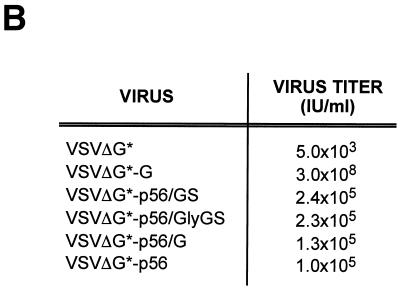
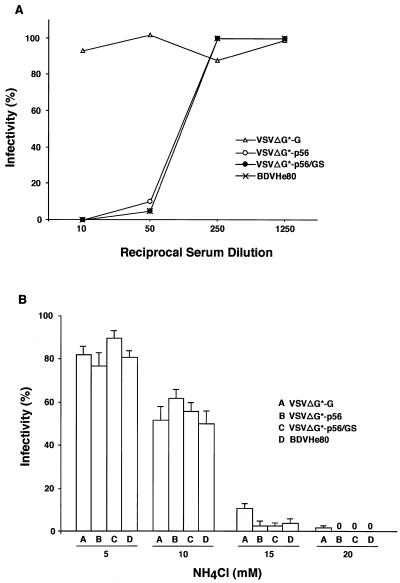
After verifying that the chimeric GPs could be efficiently incorporated into VSV particles, we tested if the corresponding pseudotyped particles were infectious. For this, we infected BHK-21 cells with serial dilutions of each pseudotype virus and 20 h later GFP-expressing living cells were directly counted using an inverted fluorescence microscope. Subsequently, cells were fixed with methanol-acetone (1:1) and stained with a monoclonal antibody against VSV N (Fig. 4A). VSVΔG* particles pseudotyped with the homologous VSV G had titers in the range of 107 to 108 IU/ml. VSVΔG* particles pseudotyped with BDV p56 or the chimeric GPs were also infectious and exhibited titers above 105 IU/ml (Fig. 4B). In contrast, VSVΔG* particles in the supernatant of cells transfected with the pCAGGS control plasmid had titers of approximately 103 IU/ml, which represents the residual inoculum remaining in the culture supernatant. Heat-treated pooled sera from BDV-infected rats neutralized the infectivity of VSVΔG* particles pseudotyped with BDV p56 or p56/GS but not those pseudotyped with VSV G (Fig. 5A). To assess whether the chimeric GP-pseudotyped viruses entered the cell by following the endocytic pathway described for BDV (13), we treated the cells with well-characterized lysosomotropic agent NH4Cl. Exposure of cells to NH4Cl prevents endosomal acidification, thus blocking the pH-dependant fusion between the virus envelope and the intracellular endosomal membranes (35). Treatment with 20 mM NH4Cl caused complete inhibition of infection by VSVΔG* pseudotypes complemented with either VSV G or BDV p56 or p56/GS (Fig. 5B).
FIG. 4.
Infectivity of the pseudotyped particles. (A) BHK-21 cells were infected with the same volume of the indicated supernatant-containing pseudotype virus. At 20 h p.i. cells were fixed and permeabilized with methanol-acetone and analyzed by immunofluorescence with MAb anti-VSV N. The apparent differences in staining between cells infected with VSVΔG*-G and those infected with pseudotypes bearing the chimeric GPs were due to the high fusogenic activity of VSV G. (B) Titers of the pseudotyped viruses were determined on BHK-21 cells by serial dilutions of viral stocks. Data from representative experiments are shown.
FIG. 5.
(A) Neutralization of VSV pseudotypes. Equal amounts (500 PFU) of VSVΔG*-G, VSVΔG*-p56, and VSVΔG*-p56/GS viruses, as well as BDV He80 (100 FFU), were incubated at 37°C for 60 min with the indicated dilution of pooled sera from BDV-infected rats. The infectivity of each sample was normalized to that obtained after incubation in the presence of the same dilutions of pooled normal rat sera. Infectivity was determined based on GFP expression for the VSV pseudotypes and, for BDV He80, by using an immunofocus assay (14). (B) Susceptibility to NH4Cl. Cells treated with indicated concentrations of NH4Cl were infected with 500 PFU of each VSV pseudotype or with 1,000 FFU of BDV He80. Infectivity of VSV pseudotypes was determined at 24 h p.i. based on their GFP expression. BDV He80 infectivity was determined at 72 h p.i. by using an immunofocus assay. The infectivity for each NH4Cl treatment was normalized to that obtained for each virus in untreated cells. Treatment with 20 mM NH4C1 abrogated completely the infectivity (0%) of VSVΔG*-p56, VSVΔG*-p56/GS, and BDV He80.
Cell surface expression of the BDV-VSV chimeric GPs.
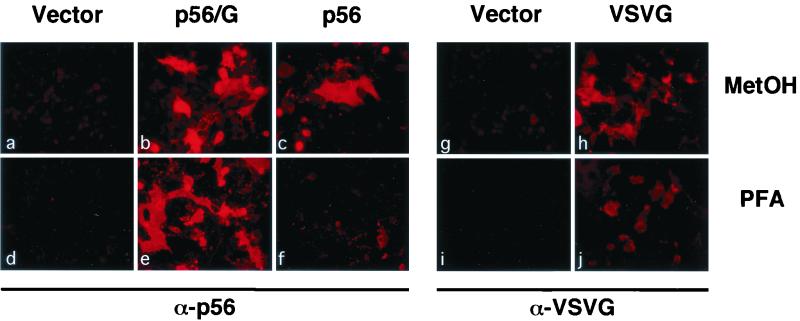
To investigate the reasons why the VSVΔG* particles seemed to incorporate significantly larger amounts of the chimeric GPs than of BDV p56, we used indirect immunofluorescence (IF) procedures to analyze the cell surface and intracellular expression of these GPs in transfected BHK-21 cells (Fig. 6). Transfected cells were fixed with 4% paraformaldehyde (surface staining; Fig. 6d to f, i, and j) or with methanol-acetone (intracellular staining; Fig. 6a to c, g, and h) and then stained with an anti-p56 rabbit serum (Fig. 6a to f) or a MAb to VSV G (Fig. 6g to j), followed by a Texas red-conjugated second antibody. Mock-transfected cells did not show any reactivity with the antibodies used (Fig. 6a, d, g, and i). The chimeric proteins were efficiently transported to the cell surface, as indicated by the staining of cells that were fixed with paraformaldehyde and not permeabilized (Fig. 6e). In contrast, wild-type BDV p56 remained mainly intracellular and was detected only in cells permeabilized with methanol (Fig. 6; compare panels c and f). This finding indicates that the presence of the TM domain and CT of VSV G enhanced the transport of the chimeric GPs to the plasma membrane. Therefore, the lack of detectable cell surface expression of BDV gp84 and gp43 in cells transfected with wt BDVp56 may account for the poor incorporation of BDV p56 polypeptides into VSVΔG* particles. Results obtained by IF were confirmed by flow cytometry analysis (data not shown).
FIG. 6.
Cell surface expression of p56/G chimeric GP. BHK-21 cells were transfected with empty pCAGGS (a, d, g, and i), pCAGGS expressing the full-length BDV p56 (c and f), VSV G (h and j), or chimeric protein p56/G (b and e). Forty-eight hours after transfection the cells were either fixed and permeabilized with methanol-acetone (MetOH) or fixed with 4% paraformaldehyde (PFA). Cells were analyzed by immunofluorescence using an anti-p56 rabbit serum (a to f) or a MAb to VSV G (g to j). Intracellular (a to c, g, and h) and cell surface (d to f, i, and j) expression of the proteins is shown.
DISCUSSION
BDV has a single surface GP (gp84), which is processed via cleavage by the cellular protease furin (13, 31). Among the known mononegaviruses, only the filoviruses have a similar situation in which a single envelope GP that is responsible for both binding and membrane fusion becomes cleaved by cellular proteases (40, 49, 50). However, for BDV the predicted N-terminal part of p56 (gp41), which is generated upon cleavage by furin, has not been detected yet in BDV-infected cells. Both gp84 and gp43 are associated with cell-free infectious BDV particles and appear to participate in virus entry. A plausible model for BDV cell entry is that the N-terminal part of the virion-associated gp84 is involved in BDV receptor binding and endocytosis, whereas gp43 would mediate the pH-dependent fusion event required for BDV infection. The role of gp43 in membrane fusion is supported by the observations that it is the only known BDV polypeptide present at the cell surface and that fusion of BDV-infected cells occurs upon exposure to low pH (13). Interestingly, near the N terminus of gp43 there is a highly hydrophobic sequence that is reminiscent of the fusogenic domain described for the surface GPs of other enveloped viruses. Nonetheless, we cannot rule out the possibility that other viral proteins not expressed at the cell surface could influence fusion, as has been described for the herpes simplex virus gK (16). Intriguingly, the BDV matrix protein has also been implicated in virus entry, which represents a unique case among known mononegaviruses (19).
BDV entry might occur through a hetero-oligomeric complex of gp84 and gp43, although the question as to the specific contribution of each GP to entry remains. To investigate the role of the N-terminal part of gp84 in virus entry, we used a pseudotype approach. We employed a recombinant VSV (VSVΔG*) lacking the G protein gene. Thus, the pseudotype viruses are expected to enter cells using the G protein provided in trans, with the subsequent intracellular steps being those characteristic of the VSV replication cycle. Therefore, the infectivity of these pseudotype viruses reflects the ability of trans-complementing G proteins to mediate entry. To facilitate expression of the BDV gp84 N-terminal part at the cell surface and hence its incorporation into VSV particles, we generated a BDV chimeric G comprising the N terminus of BDVgp84 and the TM domain and CT of VSV. Some of the chimeric GPs also contained 42 amino acids of GS of VSV G, which is a region that has been recently implicated in high-efficiency viral budding (34). However, we observed only a slight increase in the infectivity of the pseudotypes containing chimeric p56/GS molecules compared to that of a chimera containing just the VSV G TM domain and CT. Therefore, our results indicate that amino acids 1 to 244 of BDV p56 contain a domain that is sufficient, in the absence of gp43, for receptor recognition and cell entry.
In our experiments we observed a relatively large, yet consistent, amount of “background” infectivity in the supernatants of cells that were transfected with the empty pCAGGS plasmid and infected with VSV G-complemented VSVΔG* (VSVΔG*-G) particles. In the absence of a complementing GP, the supernatant of VSVΔG*-G-infected cells contains primarily noncomplemented (bald) VSVΔG* particles but it can also contain some amount of residual input VSVΔG*-G virus that was not removed by washing. VSV G-mediated infectivity contributed by the residual input VSVΔG*-G virus, together with some possible level of G-independent virus entry, likely explains the infectivity associated with the negative-control supernatants. This infectivity was considered the background level and is unrelated to the properties of the complementing GP provided by transfection.
Two lines of evidence support our conclusion that the N-terminal part of gp84 (amino acids 1 to 244 of BDV p56) is capable of mediating virus entry. First, the infectivity of pseudotypes complemented with either BDV p56 or any of the chimeric BDV-VSV GPs was 2 to 3 log units higher than that of mock-complemented VSVΔG* particles. Therefore, the background infectivity represents only 1% of the infectivity present in the supernatant of cells transfected with any of the BDV GP constructs. Second, the infectivity of VSVΔG* pseudotypes containing either BDV p56 or any of the BDV-VSV chimeric GPs was neutralized with a pooled sera from BDV-infected rats. Therefore, the infectivity associated with pseudotyped particles was BDV specific. In addition, the infectivity of pseudotype viruses complemented with any of the BDV-VSV chimeric GPs exhibited a pattern of sensitivity to NH4Cl similar to that seen with BDV He80 (13).
Both VSV G and the chimeric BDV-VSV GPs were expressed to similar levels in cell extracts from transfected cells, but supernatants containing pseudotypes complemented with the homologous VSV G protein had significantly higher titers than those found in the BDV-VSV pseudotype supernatants. This difference in titer is likely due to better incorporation of the homologous G protein into the VSV particles (data not shown). Another contributing factor could be that the VSV G protein may have a fusogenic activity stronger than that associated with the chimeric GPs.
One unexpected result obtained from our studies was that we detected gp84, but not gp43, in lysates from p56-transfected cells (Fig. 2A). However, both gp84 and gp43 were readily detected at similar levels on concentrated pseudotyped particle resulting from complementation with p56 (Fig. 3). The lack of gp43 detection was also observed in cells transfected with plasmids that used other RNA polymerase II (pol II) promoters to drive BDV p56 expression. In contrast, gp43 was detected at levels similar to those of gp84 in lysates of cells infected with a recombinant vaccinia virus expressing p56 (14). The reasons for these differences remain unknown, but it is likely that they reflect a slow, and perhaps inefficient, processing of gp84 when expressed under the control of a pol II promoter. The detection of similar amounts of both gp84 and gp43 in pseudotype particles complemented with p56 may be explained by cleavage of gp84 once it is incorporated into viral particles. Alternatively, it might reflect the requirement of a physical interaction between gp84 and gp43 in such a way that the incorporation of gp84 into virions is dependent on the incorporation of gp43.
Although gp43 likely accounts for most of the fusogenic activity associated with BDV G, it is possible that the N-terminal part of gp84 also has some fusion capability that would be manifested in the chimeric GPs. For hepatitis C virus, both E1 and E2 GPs have been reported to be independently competent in virus entry and fusion (21, 26). For VSV G, the fusion peptide appears to reside at an internal location, between amino acids 117 and 139, which is in the N-terminal one-third of the ectodomain (11, 12, 52, 54). In addition, a region near the TM domain is highly conserved in all vesiculovirus G proteins and appears to be important for both VSV G-mediated fusion (45) and efficient virus budding (34). However, this domain was not present in the p56/G chimeric GP, whose incorporation into the VSVΔG* pseudotypes resulted in titers similar to those produced by the p56/GS chimeras that did contain this domain. Nevertheless, we cannot exclude the possibility that the TM domain and CT of VSV G could also influence the fusogenic behavior of the chimeric GPs and contribute to the infectivity observed.
The pseudotype approach described here may permit the investigation of the functional domains of BDV G that are responsible for virus entry. The use of VSVΔG* pseudotypes complemented with BDV GP may also allow the identification of cell lines resistant to BDV infection due to a blockade in virus entry, thus facilitating the search for cellular receptors for BDV.
ACKNOWLEDGMENTS
We thank Beatrice Cubitt and Calvin Ly for technical assistance and Diana Frye for editorial assistance.
This work was supported by NIH grant RO1 NS32355 to J.C.T. and GM-53726 to M.A.W. M.P. was supported by a postdoctoral fellowship from the Ministerio de Educacion y Ciencia of Spain.
Footnotes
Publication 13774-NP from The Scripps Research Institute.
REFERENCES
- 1.Bode L. Human infections with Borna disease virus and potential pathogenic implications. Curr Top Microbiol Immunol. 1995;190:103–130. doi: 10.1007/978-3-642-78618-1_7. [DOI] [PubMed] [Google Scholar]
- 2.Bode L, Zimmermann W, Ferszt R, Steinbach F, Ludwig H. Borna disease virus genome transcribed and expressed in psychiatric patients. Nat Med. 1995;1:232–236. doi: 10.1038/nm0395-232. [DOI] [PubMed] [Google Scholar]
- 3.Briese T, de la Torre J C, Lewis A, Ludwig H, Lipkin W I. Borna disease virus, a negative-strand RNA virus, transcribes in the nucleus of infected cells. Proc Natl Acad Sci USA. 1992;89:11486–11489. doi: 10.1073/pnas.89.23.11486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carbone K M, Moench T R, Lipkin W I. Borna disease virus replicates in astrocytes, Schwann cells and ependymal cells in persistently infected rats: location of viral genomic and messenger RNAs by in situ hybridization. J Neuropathol Exp Neurol. 1991;50:205–214. doi: 10.1097/00005072-199105000-00003. [DOI] [PubMed] [Google Scholar]
- 5.Cubitt B, de la Torre J C. Borna disease virus (BDV), a nonsegmented RNA virus, replicates in the nuclei of infected cells where infectious BDV ribonucleoproteins are present. J Virol. 1994;68:1371–1381. doi: 10.1128/jvi.68.3.1371-1381.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Danner K, Heubeck D, Mayr A. In vitro studies on Borna virus. I. The use of cell cultures for the demonstration, titration and production of Borna virus. Arch Virol. 1978;57:63–75. doi: 10.1007/BF01315638. [DOI] [PubMed] [Google Scholar]
- 7.Danner K, Mayr A. In vitro studies on Borna virus. II. Properties of the virus. Arch Virol. 1979;61:261–271. doi: 10.1007/BF01315012. [DOI] [PubMed] [Google Scholar]
- 8.de la Torre J C. Molecular biology of Borna disease virus: prototype of a new group of animal viruses. J Virol. 1994;68:7669–7675. doi: 10.1128/jvi.68.12.7669-7675.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de la Torre J C, Bode L, Durrwald R, Cubitt B, Ludwig H. Sequence characterization of human Borna disease virus. Virus Res. 1996;44:33–44. doi: 10.1016/0168-1702(96)01338-x. [DOI] [PubMed] [Google Scholar]
- 10.de la Torre J C, Gonzalez-Dunia D, Cubitt B, Mallory M, Mueller-Lantzsch N, Grasser F A, Hansen L A, Masliah E. Detection of Borna disease virus antigen and RNA in human autopsy brain samples from neuropsychiatric patients. Virology. 1996;223:272–282. doi: 10.1006/viro.1996.0479. [DOI] [PubMed] [Google Scholar]
- 11.Fredericksen B L, Whitt M A. Attenuation of recombinant vesicular stomatitis viruses encoding mutant glycoproteins demonstrate (sic) a critical role for maintaining a high pH threshold for membrane fusion in viral fitness. Virology. 1998;240:349–358. doi: 10.1006/viro.1997.8921. [DOI] [PubMed] [Google Scholar]
- 12.Fredericksen B L, Whitt M A. Vesicular stomatitis virus glycoprotein mutations that affect membrane fusion activity and abolish virus infectivity. J Virol. 1995;69:1435–1443. doi: 10.1128/jvi.69.3.1435-1443.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gonzalez-Dunia D, Cubitt B, de la Torre J C. Mechanism of Borna disease virus entry into cells. J Virol. 1998;72:783–788. doi: 10.1128/jvi.72.1.783-788.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gonzalez-Dunia D, Cubitt B, Grasser F A, de la Torre J C. Characterization of Borna disease virus p56 protein, a surface glycoprotein involved in virus entry. J Virol. 1997;71:3208–3218. doi: 10.1128/jvi.71.4.3208-3218.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gosztonyi G, Ludwig H. Borna disease—neuropathology and pathogenesis. Curr Top Microbiol Immunol. 1995;190:39–73. [PubMed] [Google Scholar]
- 16.Hutchinson L, Roop-Beauchamp C, Johnson D C. Herpes simplex virus glycoprotein K is known to influence fusion of infected cells, yet is not on the cell surface. J Virol. 1995;69:4556–4563. doi: 10.1128/jvi.69.7.4556-4563.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kahn J S, Schnell M J, Buonocore L, Rose J K. Recombinant vesicular stomatitis virus expressing respiratory syncytial virus (RSV) glycoproteins: RSV fusion protein can mediate infection and cell fusion. Virology. 1999;254:81–91. doi: 10.1006/viro.1998.9535. [DOI] [PubMed] [Google Scholar]
- 18.Kishi M, Nakaya T, Nakamura Y, Zhong Q, Ikeda K, Senjo M, Kakinuma M, Kato S, Ikuta K. Demonstration of human Borna disease virus RNA in human peripheral blood mononuclear cells. FEBS Lett. 1995;364:293–297. doi: 10.1016/0014-5793(95)00406-y. [DOI] [PubMed] [Google Scholar]
- 19.Kliche S, Briese T, Henschen A H, Stitz L, Lipkin W I. Characterization of a Borna disease virus glycoprotein, gp18. J Virol. 1994;68:6918–6923. doi: 10.1128/jvi.68.11.6918-6923.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kretzschmar E, Buonocore L, Schnell M J, Rose J K. High-efficiency incorporation of functional influenza virus glycoproteins into recombinant vesicular stomatitis viruses. J Virol. 1997;71:5982–5989. doi: 10.1128/jvi.71.8.5982-5989.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lagging L M, Meyer K, Owens R J, Ray R. Functional role of hepatitis C virus chimeric glycoproteins in the infectivity of pseudotyped virus. J Virol. 1998;72:3539–3546. doi: 10.1128/jvi.72.5.3539-3546.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lawson N D, Stillman E A, Whitt M A, Rose J K. Recombinant vesicular stomatitis viruses from DNA. Proc Natl Acad Sci USA. 1995;92:4477–4481. doi: 10.1073/pnas.92.10.4477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lefrancois L, Lyles D S. The interaction of antibody with the major surface glycoprotein of vesicular stomatitis virus. II. Monoclonal antibodies of nonneutralizing and cross-reactive epitopes of Indiana and New Jersey serotypes. Virology. 1982;121:168–174. doi: 10.1016/0042-6822(82)90126-x. [DOI] [PubMed] [Google Scholar]
- 24.Lipkin W I, Schneemann A, Solbrig M V. Borna disease virus: implications for human neuropsychiatric illness. Trends Microbiol. 1995;3:64–69. doi: 10.1016/s0966-842x(00)88877-0. [DOI] [PubMed] [Google Scholar]
- 25.Ludwig H, Bode L, Gosztonyi G. Borna disease: a persistent virus infection of the central nervous system. Prog Med Virol. 1988;35:107–151. [PubMed] [Google Scholar]
- 26.Meyer K, Basu A, Ray R. Functional features of hepatitis C virus glycoproteins for pseudotype virus entry into mammalian cells. Virology. 2000;276:214–226. doi: 10.1006/viro.2000.0547. [DOI] [PubMed] [Google Scholar]
- 27.Moss B, Elroy-Stein O, Mizukami T, Alexander W A, Fuerst T R. Product review. New mammalian expression vectors. Nature. 1990;348:91–92. doi: 10.1038/348091a0. [DOI] [PubMed] [Google Scholar]
- 28.Nakamura Y, Takahashi H, Shoya Y, Nakaya T, Watanabe M, Tomonaga K, Iwahashi K, Ameno K, Momiyama N, Taniyama H, Sata T, Kurata T J, de la Torre C, Ikuta K. Isolation of Borna disease virus from human brain tissue. J Virol. 2000;74:4601–4611. doi: 10.1128/jvi.74.10.4601-4611.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Niwa H, Yamamura K, Miyazaki J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene. 1991;108:193–199. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- 30.Planz O, Rentzsch C, Batra A, Winkler T, Buttner M, Rziha H J, Stitz L. Pathogenesis of Borna disease virus: granulocyte fractions of psychiatric patients harbor infectious virus in the absence of antiviral antibodies. J Virol. 1999;73:6251–6256. doi: 10.1128/jvi.73.8.6251-6256.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Richt J A, Furbringer T, Koch A, Pfeuffer I, Herden C, Bause-Niedrig I, Garten W. Processing of the Borna disease virus glycoprotein gp94 by the subtilisin-like endoprotease furin. J Virol. 1998;72:4528–4533. doi: 10.1128/jvi.72.5.4528-4533.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roberts A, Buonocore L, Price R, Forman J, Rose J K. Attenuated vesicular stomatitis viruses as vaccine vectors. J Virol. 1999;73:3723–3732. doi: 10.1128/jvi.73.5.3723-3732.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roberts A, Kretzschmar E, Perkins A S, Forman J, Price R, Buonocore L, Kawaoka Y, Rose J K. Vaccination with a recombinant vesicular stomatitis virus expressing an influenza virus hemagglutinin provides complete protection from influenza virus challenge. J Virol. 1998;72:4704–4711. doi: 10.1128/jvi.72.6.4704-4711.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Robison C S, Whitt M A. The membrane-proximal stem region of vesicular stomatitis virus G protein confers efficient virus assembly. J Virol. 2000;74:2239–2246. doi: 10.1128/jvi.74.5.2239-2246.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rodriguez E, Everitt E. Adenovirus uncoating and nuclear establishment are not affected by weak base amines. J Virol. 1996;70:3470–3477. doi: 10.1128/jvi.70.6.3470-3477.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rose J K, Bergmann J E. Expression from cloned cDNA of cell-surface secreted forms of the glycoprotein of vesicular stomatitis virus in eucaryotic cells. Cell. 1982;30:753–762. doi: 10.1016/0092-8674(82)90280-x. [DOI] [PubMed] [Google Scholar]
- 37.Rott R, Becht H. Natural and experimental Borna disease in animals. Curr Top Microbiol Immunol. 1995;190:17–30. doi: 10.1007/978-3-642-78618-1_2. [DOI] [PubMed] [Google Scholar]
- 38.Salvatore M, Morzunov S, Schwemmle M, Lipkin W I. Borna disease virus in brains of North American and European people with schizophrenia and bipolar disorder. Bornavirus Study Group. Lancet. 1997;349:1813–1814. doi: 10.1016/s0140-6736(05)61693-5. [DOI] [PubMed] [Google Scholar]
- 39.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 40.Sanchez A, Yang Z Y, Xu L, Nabel G J, Crews T, Peters C J. Biochemical analysis of the secreted and virion glycoproteins of Ebola virus. J Virol. 1998;72:6442–6447. doi: 10.1128/jvi.72.8.6442-6447.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sauder C, Muller A, Cubitt B, Mayer J, Steinmetz J, Trabert W, Ziegler B, Wanke K, Mueller-Lantzsch N, de la Torre J C, Grasser F A. Detection of Borna disease virus (BDV) antibodies and BDV RNA in psychiatric patients: evidence for high sequence conservation of human blood-derived BDV RNA. J Virol. 1996;70:7713–7724. doi: 10.1128/jvi.70.11.7713-7724.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schlesinger M J, Schlesinger S. Domains of virus glycoproteins. Adv Virus Res. 1987;33:1–44. doi: 10.1016/S0065-3527(08)60315-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schneemann A, Schneider P A, Lamb R A, Lipkin W I. The remarkable coding strategy of Borna disease virus: a new member of the nonsegmented negative strand RNA viruses. Virology. 1995;210:1–8. doi: 10.1006/viro.1995.1311. [DOI] [PubMed] [Google Scholar]
- 44.Schneider P A, Briese T, Zimmermann W, Ludwig H, Lipkin W I. Sequence conservation in field and experimental isolates of Borna disease virus. J Virol. 1994;68:63–68. doi: 10.1128/jvi.68.1.63-68.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shokralla S, Chernish R, Ghosh H P. Effects of double-site mutations of vesicular stomatitis virus glycoprotein G on membrane fusion activity. Virology. 1999;256:119–129. doi: 10.1006/viro.1999.9606. [DOI] [PubMed] [Google Scholar]
- 46.Staeheli P, Sauder C, Hausmann J, Ehrensperger F, Schwemmle M. Epidemiology of Borna disease virus. J Gen Virol. 2000;81:2123–2135. doi: 10.1099/0022-1317-81-9-2123. [DOI] [PubMed] [Google Scholar]
- 47.Takada A, Robison C, Goto H, Sanchez A, Murti K G, Whitt M A, Kawaoka Y. A system for functional analysis of Ebola virus glycoprotein. Proc Natl Acad Sci USA. 1997;94:14764–14769. doi: 10.1073/pnas.94.26.14764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tatsuo H, Okuma K, Tanaka K, Ono N, Minagawa H, Takade A, Matsuura Y, Yanagi Y. Virus entry is a major determinant of cell tropism of Edmonston and wild-type strains of measles virus as revealed by vesicular stomatitis virus pseudotypes bearing their envelope proteins. J Virol. 2000;74:4139–4145. doi: 10.1128/jvi.74.9.4139-4145.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Volchkov V E, Feldmann H, Volchkova V A, Klenk H D. Processing of the Ebola virus glycoprotein by the proprotein convertase furin. Proc Natl Acad Sci USA. 1998;95:5762–5767. doi: 10.1073/pnas.95.10.5762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Volchkov V E, Volchkova V A, Stroher U, Becker S, Dolnik O, Cieplik M, Garten W, Klenk H D, Feldmann H. Proteolytic processing of Marburg virus glycoprotein. Virology. 2000;268:1–6. doi: 10.1006/viro.1999.0110. [DOI] [PubMed] [Google Scholar]
- 51.Whelan S P, Ball L A, Barr J N, Wertz G T. Efficient recovery of infectious vesicular stomatitis virus entirely from cDNA clones. Proc Natl Acad Sci USA. 1995;92:8388–8392. doi: 10.1073/pnas.92.18.8388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Whitt M A, Zagouras P, Crise B, Rose J K. A fusion-defective mutant of the vesicular stomatitis virus glycoprotein. J Virol. 1990;64:4907–4913. doi: 10.1128/jvi.64.10.4907-4913.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wool-Lewis R J, Bates P. Characterization of Ebola virus entry by using pseudotyped viruses: identification of receptor-deficient cell lines. J Virol. 1998;72:3155–3160. doi: 10.1128/jvi.72.4.3155-3160.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang L, Ghosh H P. Characterization of the putative fusogenic domain in vesicular stomatitis virus glycoprotein G. J Virol. 1994;68:2186–2193. doi: 10.1128/jvi.68.4.2186-2193.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]