Abstract
Mandrillus sphinx, a large primate living in Cameroon and Gabon and belonging to the Papionini tribe, was reported to be infected by a simian immunodeficiency virus (SIV) (SIVmndGB1) as early as 1988. Here, we have identified a second, highly divergent SIVmnd (designated SIVmnd-2). Genomic organization differs between the two viral types; SIVmnd-2 has the additional vpx gene, like other SIVs naturally infecting the Papionini tribe (SIVsm and SIVrcm) and in contrast to the other SIVmnd type (here designated SIVmnd-1), which is more closely related to SIVs infecting l'hoest (Cercopithecus lhoesti lhoesti) and sun-tailed (Cercopithecus lhoesti solatus) monkeys. Importantly, our epidemiological studies indicate a high prevalence of both types of SIVmnd; all 10 sexually mature wild-living monkeys and 3 out of 17 wild-born juveniles tested were infected. The geographic distribution of SIVmnd seems to be distinct for the two types: SIVmnd-1 viruses were exclusively identified in mandrills from central and southern Gabon, whereas SIVmnd-2 viruses were identified in monkeys from northern and western Gabon, as well as in Cameroon. SIVmnd-2 full-length sequence analysis, together with analysis of partial sequences from SIVmnd-1 and SIVmnd-2 from wild-born or wild-living mandrills, shows that the gag and pol regions of SIVmnd-2 are closest to those of SIVrcm, isolated from red-capped mangabeys (Cercocebus torquatus), while the env gene is closest to that of SIVmnd-1. pol and env sequence analyses of SIV from a related Papionini species, the drill (Mandrillus leucophaeus), shows a closer relationship of SIVdrl to SIVmnd-2 than to SIVmnd-1. Epidemiological surveys of human immunodeficiency virus revealed a case in Cameroon of a human infected by a virus serologically related to SIVmnd, raising the possibility that mandrills represent a viral reservoir for humans similar to sooty mangabeys in Western Africa and chimpanzees in Central Africa.
Studies of the origin of human immunodeficiency viruses (HIV) indicate that these viruses have entered the human population as a result of zoonotic transmissions of simian immunodeficiency viruses (SIV) (16). To date, SIV infections have been detected in more than 20 species of African nonhuman primates. Complete sequences of these viruses are now available for 13. These fully characterized SIV can be classified into at least six approximately equidistant lineages. Five of these are represented by (i) SIVcpz from chimpanzees (Pan troglodytes), which groups with HIV type 1 (HIV-1) (7, 16, 35); (ii) SIVsm from sooty mangabeys (Cercocebus atys), which groups with HIV-2 (5, 22, 27); (iii) SIVagm from the four African green monkey species (genus Chlorocebus) (1, 11, 21, 30); (iv) SIVsyk from Sykes' monkeys (Cercopithecus mitis albogularis) (9, 20); and (v) SIVlhoest from L'hoest monkeys (Cercopithecus lhoesti lhoesti) (2, 19), which groups with SIVsun from sun-tailed monkeys (Cercopithecus lhoesti solatus) (3). Note that prior to the identification of SIVlhoest, this fifth SIV lineage was represented by SIVmnd from mandrills (Mandrillus sphinx) (38). The sixth lineage, represented by the SIV infecting colobus monkeys (Colobus guereza), has been described recently (8).
Based on their genome organization, primate lentiviruses form three groups. Viruses from the SIVagm, SIVsyk, SIVlhoest/SIVsun, and SIVcol lineages have a common structure comprising the gag, pol, vif, vpr, tat, rev, env, and nef genes. Viruses from the SIVcpz/HIV-1 lineage have an additional gene, vpu, in the central part of the genome, whereas viruses from the SIVsm/HIV-2 lineage have a vpx gene in addition.
The divergence of SIV lineages often matches the divergence of their primate species host lineages, underscoring the ancient nature of these lentiviruses (1, 3, 18, 30). In addition to this apparent host-dependent evolution, different cross-species or cross-subspecies transmissions have occurred frequently between wild-living or captive primates (4, 7, 23, 39). African green monkeys have thus apparently transmitted their virus occasionally to patas monkeys and baboons (4, 23). And it has been suggested recently that the SIVmnd described in mandrills is the result of cross-species transmission to mandrills of a virus related to SIVlhoest (3, 19, 38).
The mandrill is a large semiterrestrial primate belonging to the Papionini tribe, living in the tropical rain forests of Cameroon and Gabon (15). SIVmnd was first isolated from mandrills in Gabon in 1988, and from one isolate (SIVmndGB1) a molecular clone was derived (38) that was the only representative of SIVmnd until now. The genetic divergences observed between SIVmndGB1 and other SIV from the Papionini tribe preclude an evolutionary history of purely host-dependent evolution (17, 18). The study of the evolution of SIV is helpful for the understanding of the origin and evolution of HIV in humans. SIV from sooty mangabeys belonging to the Papionini genus have already given rise to a human virus (HIV-2) (5, 13; R. Marlink, Editorial, AIDS 10:689–699, 1996). To elucidate the infection of M. sphinx by a SIV closely related to that infecting the Cercopithecini tribe, we investigated the nature of the SIVmnd in wild-born captive mandrills and in wild-living mandrills from Cameroon and Gabon using new serological and virological tools. Similarly, seropositive samples identified in an epidemiological study performed on the human populations living in these countries were tested in order to search for the presence of HIV closely related to SIVmnd.
MATERIALS AND METHODS
Simian samples.
Fifteen wild-born mandrills living in a large semi-free-ranging colony, established in 1983 at the International Center for Medical Research in Franceville, Gabon (CIRMF), were studied. The geographic origins of the mandrills are given in Table 1. Of the two viruses, SIVmndGB1 and SIVmndGB2, isolated from two founder animals in 1989, only SIVmndGB1 was completely sequenced, because SIVmndGB2 was considered very close (38). Viral transmission in the colony has occurred mostly via aggressive male-to-male conflicts (five males, designated M3, M9, M13, M14, and M15, were infected in the colony between 1985 and 1992) (31). A case of vertical transmission from a female, F17, to one of her offspring was suspected (31). The six SIVmnd-infected males and F17 died at the ages of 15 to 20 years of causes unrelated to immunodeficiency. Serum samples have been collected every year from all mandrills in the colony and stored at −80°C. All founder wild-born mandrills in the colony and their descendants were screened retrospectively for SIV using a new serological assay designed for specific SIV screening.
TABLE 1.
Description of mandrills and drills included in this study
| Animal and virus | Sexa | Ageb (yr) | Originc |
|---|---|---|---|
| Mandrills with SIVmnd type 1 | |||
| Mnd F17 | F | 2 | ? |
| Mnd 17B | F | <1 | C |
| Mnd 17D | F | <1 | C |
| Mnd 17G | M | <1 | C |
| Mnd 17D1 | M | <1 | C |
| Mnd 17D2 | F | <1 | C |
| Mnd Lop 4 | F | 7 | Lop |
| Mnd Lop 5 | F | 7 | Lop |
| Mnd Lop 6 | F | 10 | Lop |
| Mnd Lop 7 | M | 10 | Lop |
| Mnd Lop 8 | M | 10 | Lop |
| Mnd Lop 9 | F | 12 | Lop |
| Mnd Lop 10 | M | 10 | Lop |
| Mnd Lop 11 | M | 10 | Lop |
| Mnd Lop 12 | M | 10 | Lop |
| Mnd Lop 13 | M | 10 | Lop |
| Mandrills with SIVmnd type 2 | |||
| Mnd M3 | M | 6 | Mk |
| Mnd M7 | M | 2 | Mk |
| Mnd M9 | M | >14 | FCV |
| Mnd M13 | M | >12 | Mk |
| Mnd M14 | M | >8 | Mk |
| Mnd M15 | M | >8 | ? |
| Mnd PG13 | F | 3 | PG |
| Mnd 302 | F | 3 | CA |
| Mnd BK | F | 18 | Zoo |
| Drills with SIVdrl | |||
| Drill 207 | M | 4 | CA |
| Drill 006 | F | <10 | CA |
| Drill 007 | M | <10 | CA |
M, male; F, female.
At date of seropositive identification.
?, place of birth in Gabon unknown; C, born in semifree colony in CIRMF; Lop, wild-living mandrills from Lopé reserve; Mk, Makokou; FCV, Franceville; PG, Port Gentil; CA, Cameroon (see Fig. 1); Zoo, San Diego, Calif.
Thirteen wild-living mandrills (Lop 1 through Lop 13) were captured, sampled, and released in 1998 (n = 6) and 1999 (n = 7) during ecological studies in central Gabon.
Fourteen wild-born but captive mandrills and three drills captured in the wild when juvenile and since then housed in sanctuaries in Cameroon or Gabon were tested on the date of capture, using the same serological assay. A female mandrill (BK) housed in the San Diego Wild Animal Park since 1984 was also studied.
Human sera.
A total of 19,762 human blood samples from Cameroon and Gabon were screened between 1994 and 1999. Of these, 6,515 and 15 were considered HIV-1 and HIV-2 positive, respectively (28; C. Tevi-Benissan, M. Okome, M. Makuwa, M. N. Nkoume, J. Lansoud-Soukate, A. Georges, M. C. Georges-Courbot, and L. Belec, Letter, Emerg. Infect. Dis. 4:130–131, 1998). These HIV-2 samples were further studied for their specific reactivities against SIV antigens.
V3 peptide EIA screening.
All the simian and human samples were tested by a new peptide-based enzyme-linked immunoassay (EIA) detecting and differentiating antibodies against the V3 regions representative of the different SIV/HIV lineages (34). Peptides corresponding to V3 loops of HIV-1, HIV-2, SIVcpz, SIVsm, SIVagm, SIVrcm, and SIVmnd were synthesized. Wells of polyvinyl microtiter plates (Falcon) were coated with 100 μl each of 2 μg of antigen per ml diluted in 0.05 M bicarbonate buffer, pH 9.6, by incubation for 20 h at 37°C. The wells were washed twice with phosphate-buffered saline (PBS) containing 0.5% Tween 20 (PBS-TW), and unoccupied sites were saturated with PBS containing 2% newborn calf serum (NBCS) by incubation for 45 min at 37°C, followed by washing in PBS-TW. Each serum sample was tested at a 1:100 dilution in 0.01 M sodium phosphate buffer, pH 7.4, containing 0.75 M NaCl, 10% NBCS, and 0.5% Tween 20 (PBS-TW-NBCS). The reactivity of each sample to all the peptides was tested. One hundred microliters of diluted serum was added to the wells and incubated for 30 min at room temperature. The wells were washed four times with PBS-TW, and peroxidase-conjugated goat F(ab′)2 anti-human immunoglobulin (Sigma; 100 μl of a 1:2,000 dilution in PBS-TW-NBCS) was added and incubated for 30 min at room temperature. The wells were washed four times with PBS-TW, and the reaction was revealed by incubation with hydrogen peroxide-o-phenylendiamine for 15 min at room temperature. Color development was stopped with 2 N H2SO4, and the absorbance (expressed as the optical density [OD]) was read at 492 nm. The cutoff was established at 0.20.
Virus isolation.
Frozen or fresh peripheral blood mononuclear cells (PBMC) from all infected adult mandrills but one (M3, for which no PBMC samples were available) were cocultured with human phytohemagglutinin (PHA)-stimulated PBMC as previously described (35). Cultures were performed separately in time to avoid cross-contamination. Viral replication was monitored by a reverse transcriptase (RT) assay (Lenti-RT Kit; Cavidi Tech AB, Uppsala, Sweden) and by measurement of p27 antigen with the SIV-monoclonal assay (SIV p27; Coulter, Hialeah, Fla.) and the HIV-1-polyclonal assay (Elavia p24; Sanofi-Pasteur, Paris, France).
PCR and sequences.
In order to amplify all SIVmnd, degenerate primers (DR1 and Hpol4538 for the first round and Hpol4235 and Hpol4538 for the second round) were used to amplify a 303-bp fragment of pol from total cellular DNA or plasma (7, 10, 35). The 303-bp PCR products were directly sequenced. For the full-length genome of SIVmnd-2 M14, two specific primers (N-OR1, CCAAAGGACATGAAAAATAGGCATC, and N-OF1, AAGGTAGCCACAGTGTGTTGGTGG) were generated to amplify the 5′ and 3′ parts of the genome by targeting unintegrated circular DNA from cultured cells (Expand high-fidelity Taq polymerase; Roche Diagnostics, Mannheim, Germany). Nested PCRs were performed with another specific primer (N-OIR1, 5′-GGCCACTGTTTAATTCTKGGKCCATC-3′) used with primer LPBS to amplify the 5′ part. We used Hpol4235 with LPBS reverse to obtain the 3′ part (10). The two fragments were cloned into the pGEM-T Easy Vector (Promega, Madison, Wis.). Serial SIVmnd-2 primers were then synthesized to walk along the cloned fragments.
The primers used for amplification of Gp41 were designed by alignment of SIVmnd-1 GB1 and SIVmnd-2 M14 sequences. In the first round we amplified a 2.2-kb fragment by using EnvF1 (5′-ATAGGAAAACAATRTGTRACAGT-3′) and EnvR1 (5′-GTTTAGGCAGGGCTATCGACC-3′), and for the second round we used Gp41F (5′-CAGTGTCGGTGGCACTGACTGTC-3′) and Gp41R (5′-CAGTGTCGGTGGCACTGAC TGTC-3′). PCR products of 540 bp were sequenced directly.
Sequence alignment.
Alignments were constructed for the partial pol gene (integrase region), the partial env gene (gp41 region), and the gag, pol, vif, vpr, vpx, tat, env, and nef genes using CLUSTAL W (37). Alignments were adjusted manually where necessary. Exons that are entirely overlapped by other genes (the second tat exon and both rev exons) were not included in the analyses. Regions of ambiguous alignment and all gap-containing sites were excluded.
SIVmnd type 2 diversity plot.
A concatenated amino acid alignment (proteome) was constructed that included the genes gag, pol, vif, and env. Note that the regions of pol that overlapped gag and vif were excluded from the concatenated alignment. The genetic distance between SIVmnd-2 M14 and representative SIV was calculated in 300-amino-acid windows that were incremented by 10 amino acids across the proteome alignment. For each pairwise comparison, amino acid sequence differences were plotted against the midpoint of each window.
Phylogenetic analyses.
Phylogenetic trees inferred from amino acid sequence alignments were constructed by the neighbor-joining method (33), implemented using NEIGHBOR from the Phylogeny Inference Package (PHYLIP), version 3.5c, and distance matrices were generated with the JTT model of amino acid substitution (24), implemented using TREE-PUZZLE (36). Phylogenetic clusterings were assessed by performing 1,000 bootstrap replicates (implemented using SEQBOOT, NEIGHBOR, and CONSENSE from the PHYLIP package), with the JTT distance matrices generated using PUZZLEBOOT.sh (available from TREE-PUZZLE's website at http://www.tree-puzzle.de). Phylogenetic trees inferred from nucleotide sequence alignments were constructed by the neighbor-joining method with the HKY model of nucleotide substitution, implemented by using PAUP (Phylogenetic Analysis Using Parsimony), version 4. Phylogenetic clusterings were assessed by performing 1,000 bootstrap replicates using PAUP to implement neighbor joining with the HKY model of nucleotide substitution. Maximum likelihood analysis, using 10 sequences from each alignment, was also implemented, by using PROTML (with the JTT model of amino acid substitution) or NUCML (with the HKY model of nucleotide substitution) from the MOLPHY package, version 2.2, to perform an exhaustive search of all possible tree topologies and to identify the 100 best topologies with approximate likelihoods; the maximum-likelihood tree was then identified among these.
Nucleotide sequence accession numbers.
All the SIVmnd sequences obtained in this study have been submitted to GenBank (accession numbers AF328276 to AF328295).
RESULTS
SIVmnd infects adult and juvenile mandrills in a semi-free-ranging colony.
The retrospective serological screening with the new SIV-specific EIA (34) confirmed that the two founder mandrills, M7 and F17, were already SIV positive upon their arrival at the CIRMF. EIA analysis also confirmed subsequent intracolony infection of five adult males (M3, M9, M13, M14, and M15). Five out of the 16 descendants of F17 (17B, 17D, 17D1, 17D2, and 17G) were revealed to be seropositive as well (Table 1). We thus confirmed the SIV infection in monkeys previously described as SIV infected (M7, F17, M3, M9, M13, M14, M15, and F17B) (31) and revealed four new cases of SIV seropositivity in recently born descendants of the female F17, suggesting a mother-to-child transmission in the colony (Fig. 1 and Table 1).
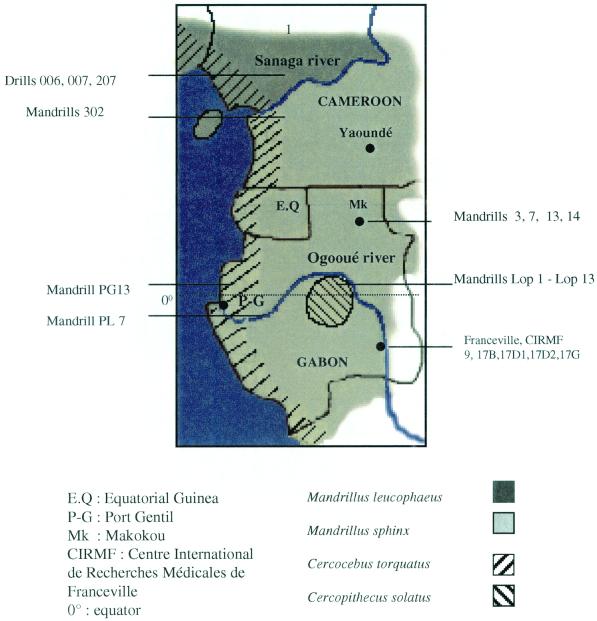
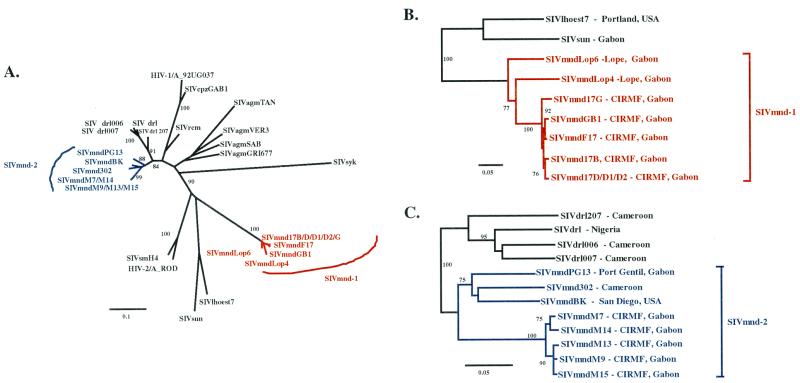
FIG. 1.
Geographic origins of the seropositive samples and range of M. sphinx, M. leucophaeus, C. solatus, and C. torquatus.
In order to isolate the virus from all animals, frozen or fresh PBMC from all seropositive mandrills except one (M3, for which PBMC samples were missing) were cocultured with human PHA-stimulated PBMC. Cultures were performed separately in time to avoid cross-contamination. Strong RT activity was detected in all culture supernatants after 7 to 14 days of culture. SIVmnd isolates from males were positive in both the anti-SIV p27 monoclonal and anti-HIV-1 p24 polyclonal assays, whereas the Gag antigen of the SIVmnd isolated from F17 and her offspring was detected only by the HIV-1 polyclonal assay (data not shown). This discrepancy between polyclonal and monoclonal antibody detection suggested strong antigenic differences between the strains.
Evidence for the cocirculation of a second SIVmnd strain within the colony.
In order to further examine the antigenic differences between the SIVmnd viruses derived from the founder animals M7 and F17, we tested the serum samples from the SIVmnd-infected animals with distinct commercial HIV-1/HIV-2 serological assays (Genscreen HIV1/2 [Bio-Rad–Sanofi] and HIV Determine [Abbott]). Interestingly, the sera from F17 and three of the five infected offspring remained negative in these tests, whereas the sera from the adult males were positive. To analyze whether these antigenic differences were associated with genetic differences between the M7- and F17-derived strains, we performed PCR amplification with DNA from uncultured PBMC from all seropositive mandrills. The PCRs were performed at different times for each strain, in addition to the usual precautions, in order to avoid contamination. By using primers specific for SIVmndGB1, we obtained positive results only for samples from F17 and her five seropositive descendants. In contrast, using these SIVmndGB1-related primers, we failed to amplify the DNA from the PBMC collected from the males. In order to exclude the possibility that the negative results for the adult males were due to a lower viral load, we amplified a region (pol integrase) of all SIVmnd in the colony by using degenerated pol primers for PCR. This pol region was sequenced for all viruses.
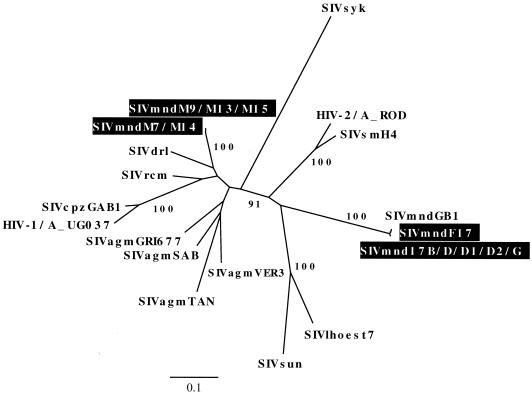
Phylogenetic analysis of these pol sequences showed a very close clustering of SIVmnd from F17 with that from her five descendants (17B, 17D, 17D1, 17D2, and 17G), confirming that they are epidemiologically linked infections (Fig. 2). The integrase fragment of SIVmndF17 was also very closely related to the sequence of the SIVmndGB1 clone. Pairwise sequence comparison indicated that SIVmndF17 shares high nucleotide identity with the SIVmndGB1 clone (99% identity), strongly suggesting that they are viruses from the same infected individual. Together with the SIVmndGB1 clone, SIVmndF17 sequences were more closely related to SIV from l'hoest monkeys than to other SIV. In contrast, SIVmnd from M7, M9, M13, M14, and M15 formed a group of highly related sequences that are more closely related to SIVdrl, a virus from the drill (Mandrillus leucophaeus) (6), than to SIVmndGB1 in this pol region (Fig. 2), confirming that the viruses from the males are epidemiologically related to each other (31) and indicating that the founder animal M7 was a carrier of a highly divergent SIV, distinct from SIVmndF17/GB1. These results show that two distinct SIVmnd are cocirculating in this colony, in contrast to what has been previously published (31, 38).
FIG. 2.
Phylogenetic relationships in a pol region of newly derived SIVmnd sequences from adult and juvenile mandrills living in a semi-free-ranging colony at CIRMF in Gabon to other primate lentiviruses. This unrooted neighbor-joining tree (see Materials and Methods for further details) was inferred from amino acid comparisons from the pol integrase region; 138 amino acid sites were included. SIVmndGB1 corresponds to the sequence of the molecular clone (38). The numbers correspond to bootstrap support for phylogenetic clusterings. Bootstrap values lower than 75% are not shown. Branch lengths are drawn to scale, with the scale bar indicating amino acid replacements per site.
In order to exclude the possibility of contamination producing these results, uncultured frozen PBMC sampled from M7 early after his arrival in the colony and stored since then were directly amplified using the same strategy. Resequencing confirmed that this animal was infected in the wild by a strain serologically and genetically different from SIVmndGB1, the sequence of which was previously published. The female F17, who was still alive, was resampled, her proviral and plasma viruses were directly sequenced, and these sequences confirmed that her virus corresponds to SIVmndGB1. These data correlate with the antigenic and serological discrepancies observed between SIVmndM7 and SIVmndF17.
Identification of a second SIVmnd type.
We decided to analyze the full-length genome sequence from the divergent SIVmnd circulating in the males. Due to the limited volume of the original M7 samples, we sequenced the proviral genome from another male (M14) infected by M7 in 1988 (31). In contrast to SIVmndGB1, the genome organization of SIVmndM14 was similar to that of SIVsm and HIV-2, with the presence of an additional accessory gene, vpx.
SIVmndM14 also has an unusual long terminal repeat (LTR) structure, as the potential trans-activation response element (TAR) structure is different from that of the other SIV. Three stem-loop elements can be predicted. The first stem-loop is most similar to that found in SIVrcm, with a 2-base UU bulge, but is distinguished by the presence of a 7-base loop (CUGGGUU). The other two TAR elements found in SIVmndM14 were more similar to the first stem-loop structure found in SIVmndGB1, both presenting the characteristic 4-base bulge. The loop sequences were indistinguishable from the consensus CUGGGX. Two Sp1 and two NF-κB binding sites were predicted upstream of the TATA box.
Whatever the causes of the previous misidentification of SIVmnd, due to the distinct genome organizations and phylogenetic relationships we have identified, and for reasons of clarity, we propose to classify the SIVmndGB1-related viruses as SIVmnd type 1 (SIVmnd-1) and the SIVmndM14-related viruses as type 2 (SIVmnd-2).
SIVmnd-2 is a complex recombinant.
Genetic identities between the SIVmnd-2 representative SIVmnd-2 M14 and other representative SIV were determined (Table 2). Across gag, pol, vif, vpx, and tat, SIVmnd-2 M14 is more closely related to SIVrcm. In vpr SIVmnd-2 M14 is just as closely related to SIVsmH4 as to SIVrcm. Across env and nef, SIVmnd-2 M14 is more closely related to SIVmndGB1, the SIVmnd-1 representative. The env V3 loop sequences in SIVmnd-1 (NRSVVSTPSATGLLFYHGLEPGKNLKKG) and SIVmnd-2 (NRSIVSVPSASGLIFYHGLEPGRNLKKG) are highly conserved relative to each other, explaining why our V3-based peptide EIA strategy succeeded in detecting these different viruses in the CIRMF colony, regardless of the infecting type.
TABLE 2.
Amino acid sequence identities from pairwise comparisons of SIVmnd-2 M14 with other SIV representative of primate lentiviruses
| SIV | % Amino acid sequence identity of SIVmnd-2 M14 with the indicated SIV
|
|||||||
|---|---|---|---|---|---|---|---|---|
| gag | pol | vif | vpx | vpr | tat | env | nef | |
| SIVmndGB1 | 57.3 | 62.6 | 40.0 | 44.6 | 47.6 | 61.1 | 63.1 | |
| SIVlhoest7 | 56.4 | 61.8 | 36.9 | 44.6 | 42.9 | 50.7 | 53.4 | |
| SIVsun | 54.3 | 60.8 | 37.7 | 44.6 | 50 | 51.9 | 61.2 | |
| SIVrcm | 72.8 | 79.8 | 56.9 | 64.3 | 65.1 | 66.7 | 37.1 | 52.4 |
| SIVsmH4 | 69.9 | 63.2 | 47.7 | 43.9 | 65.1 | 52.4 | 42.3 | 49.5 |
| SIVagmVER3a | 66.4 | 66.3 | 46.1 | 40.8 | 39.8 | 57.1 | 39.7 | 51.5 |
| SIVcpzGAB1 | 59.4 | 71.0 | 45.4 | 53.0 | 52.4 | 36.4 | 45.6 | |
| SIVsyk | 62.4 | 57.7 | 46.9 | 22.9 | 59.5 | 36.4 | 41.7 | |
Included in both the vpr and vpx comparisons.
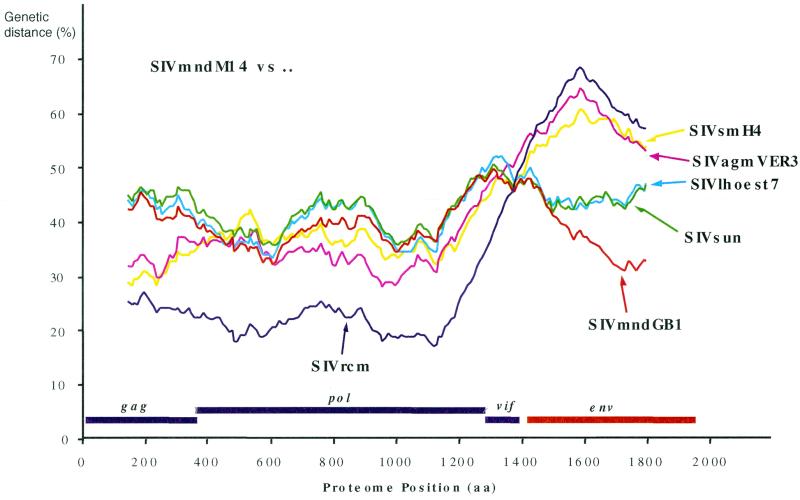
The distinct relationships of SIVmnd-2 M14 with other SIV according to the genomic region analyzed were further investigated by constructing a proteome including the genes gag, pol, vif, and env and performing diversity plotting comparing SIVmnd-2 M14 with representative SIV (Fig. 3). Across gag, pol, and vif, SIVmnd-2 M14 is more closely related to SIVrcm, while across env, SIVmnd-2 M14 is closest to SIVmnd-1 GB1. Such a switch in pairwise genetic distances between sequences is indicative of a recombinant ancestry of SIVmnd-2 M14.
FIG. 3.
Diversity plot comparing SIVmndM14 (the SIVmnd-2 representative) to SIVmndGB1 (the SIVmnd-1 representative), SIVrcm, SIVsun, SIVlhoest, SIVsm, and SIVagm. For each pairwise comparison, protein sequence difference was plotted against the midpoint of a 300-amino-acid window that was incremented by 10 amino acids across a proteome alignment including the genes gag, pol, vif, and env. The positions of these genes are shown beneath the plot.
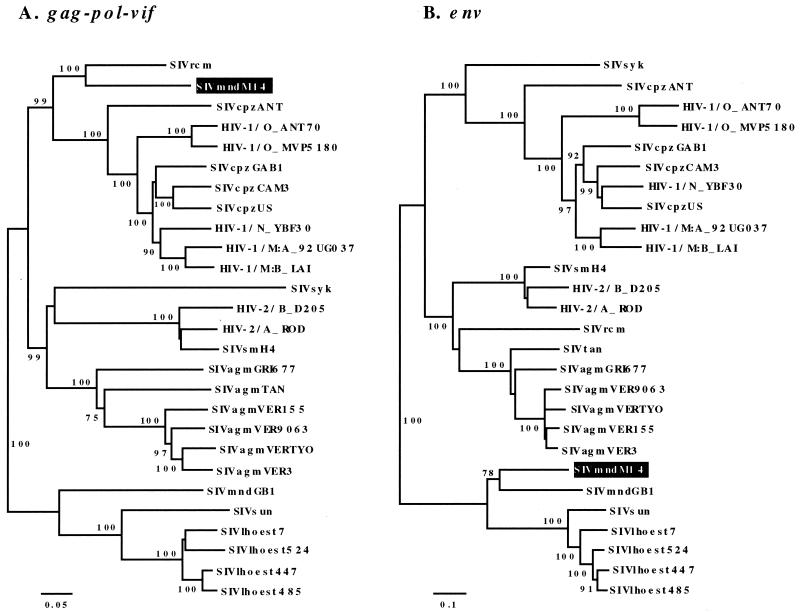
We further tested the hypothesis that SIVmnd-2 M14 is a recombinant strain by phylogenetic analysis. Phylogenetic trees constructed for each gene region confirmed the relationship of SIVmnd-2 M14 with SIVrcm in gag, pol, vif, vpr, and vpx and with SIVmnd-1 GB1 in env and nef. The discordant phylogenetic position of SIVmnd-2 M14 according to genomic region is shown in Fig. 4 and was inferred from the gag-pol-vif-env concatenated alignment. Note that both SIVrcm and SIVmnd-2 M14 show similarity with SIVcpz and HIV-1 in at least part of pol, while in env and nef, SIVmnd-2 M14 clusters with SIVmndGB1, close to SIVlhoest. The phylogenetic clustering of SIVmnd-2 M14 with either SIVrcm or SIVmndGB1 was highly significant (as assessed by bootstrap values), and the same discordant relationships were confirmed in tree topologies chosen using maximum likelihood.
FIG. 4.
Discordant phylogenetic relationships of SIVmnd-2 M14 (highlighted) in the gag-pol-vif (A) and env (B) regions. Rooted phylogenetic trees inferred by neighbor joining (see Materials and Methods for details) show the relationships of SIVmnd M14, the SIVmnd-2 representative, to other primate lentiviruses. The gag-pol-vif (A) and env (B) regions used to construct the trees correspond to positions 1 to 1403 and 1404 to 1959, respectively, in the proteome alignment (see Fig. 3). The numbers correspond to bootstrap support for the clusters to the right.
Both SIVmnd types are present in the wild.
If the recombination between SIVmnd-1 and SIVmnd-2 had happened in the colony, we should have detected a dually infected animal. Since this was not the case, we suspected that the SIVmnd-2 recombinant form was circulating in the wild and was introduced into the colony by the founder animal M7. We therefore screened wild animals for SIVmnd-2 and addressed in parallel the question whether SIVmnd-1 is equally distributed in the wild or rather resembles a “dead-end” infection. Thirteen free-living mandrills in a large colony of 700 individuals in the Lopé reserve (central Gabon [Fig. 1]), were tested first: all 10 adults (6 males and 4 females) were strongly reactive against SIVmnd V3 in the EIA, while the three juvenile mandrills were negative (Table 1). PCR amplification of short fragments of pol (integrase) or env (gp41) was successful in 4 out of 10 seropositive samples (Lop 4, 6, 7, and 12); cellular control DNA was unamplifiable in the others, suggesting a low quality of the DNA collected under field conditions. We first sequenced a region in pol because it allows discrimination between SIVmnd-1 and SIVmnd-2. We sequenced this region for SIVmndLop4 and SIVmndLop6. Phylogenetic analysis shows that these adult wild M. sphinx individuals were infected by viruses that cluster with SIVmnd-1 (Fig. 5A and B). Amino acid comparisons of SIVmndLop4 to SIVmndLop6 show that these strains have 95.6% identity with each other, SIVmndLop4 identities with SIVmnd-1 GB1 and SIVmnd-2 M7 being 96 and 63%, respectively. The sequence divergences between SIVmndLop4 and SIVmndLop6 were significantly higher than that among SIVmnd-1 viruses from the semi-free-ranging colony at CIRMF, suggesting that they are not directly epidemiologically linked, but rather that they correspond to distinct viral representatives of SIVmnd-1. For SIVmndLop7 and SIVmndLop12, we successfully sequenced a short fragment (339 nucleotides) of gp41 env. The length of the sequence did not allow us to perform a phylogenetic analysis, but we observed 95 and 70% amino acid identify with SIVmnd-1 GB1 and SIVmnd-2 M14, respectively. No SIVmnd-2-infected mandrills were found in this group. Thus, the first assessment of SIVmnd prevalence in wild-living monkeys indicates that SIVmnd-1 circulates in wild mandrills of central Gabon. In addition, the infection prevalence seems high in sexually mature mandrills, irrespective of sex, and raises the possibility of sexual transmission of SIVmnd-1 in the wild.
FIG. 5.
Phylogenetic relationships in a pol region of SIVmnd and SIVdrl strains isolated from adult and juvenile mandrills and drills of distinct geographic origins (Table 1). (A) Unrooted phylogenetic tree showing both the SIVmnd-1 and SIVmnd-2 clades; (B and C) rooted phylogenetic trees showing the SIVmnd-1 clade and the SIVmnd-2 clade, respectively. The three phylogenetic trees were inferred by neighbor joining. The unrooted phylogeny was inferred from amino acid sequence comparisons (138 amino acid sites) of pol integrase. The two rooted phylogenies were inferred from nucleotide sequence comparisons (414 nucleotide sites) from the same pol region.
We then screened mandrills originating from other geographic regions. The animals studied correspond to 14 mandrills that were captured at a young age in southwestern Cameroon and northern or western Gabon and were subsequently kept as pets or housed in rescue centers (Fig. 1; Table 1). These animals had been removed from the wild before the onset of sexual maturity and had had no further contact with infected animals. Three out of the 14 juvenile mandrills, one from Cameroon (female 302) and two from western Gabon (female PG13 and male PL7), were V3 EIA positive. Plasma amplification succeeded in two cases (animals 302 and PG13). Sequencing of the integrase confirmed that these two M. sphinx individuals were infected by SIVmnd (Fig. 5A and C). Both SIVmndPG13 and SIVmnd302 clustered together with SIVmnd-2 in this region. In order to study whether they have a mosaic genome similar to SIVmnd-2 M14, we analyzed the gp41 env sequence from SIVmnd302. This SIVmnd from Cameroon is closely related to the SIVmnd-2 found in Gabon, indicating that it is also a recombinant (Fig. 6). In addition it provides evidence for a widespread distribution of SIVmnd-2.
FIG. 6.
Phylogenetic relationships of SIVmnd-2 M14, SIVmnd302, and SIVdrl007 isolated from mandrills and drills from Cameroon and Gabon to other primate lentiviruses in an env region. This unrooted tree is inferred from amino acid comparisons (146 amino acid sites remained after gap stripping) from env (gp41 region).
Like type 1, SIVmnd-2 was found in both sexes. As the age range of the SIVmnd-2-infected mandrills resembles that of mandrills infected vertically by SIVmnd-1 in the CIRMF colony, mother-to-child transmission in the wild cannot be excluded.
We identified another case of SIVmnd-2 infection in the United States (Table 1). A female mandrill (BK) born in Sarasota, Fla., in 1971 and housed in the San Diego Wild Animal Park since 1984 was found to be seropositive in 1989, and the virus was isolated. Sequence comparision in gag, pol (Fig. 5A and C), and env indicated 90% identity with the SIVmnd-2 from mandrill M14. BK died at the age of 18 years from persistent diarrhea and weight loss, invasive Balantidium coli infection unresponsive to standard therapies, and disseminated atypical mycobacteriosis. However, despite the fact that such symptoms are indicative of immunodeficiency, one should note that 18 years represents the natural life span of a mandrill in captivity.
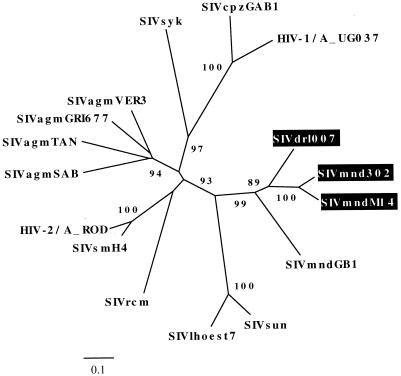
SIVdrl are more related to SIVmnd-2 than to SIVmnd-1.
Mandrills are closely related to drills (M. leucophaeus) and allopatric with them (13). Drills have recently been reported to carry SIV (6). In order to study the relationship between SIVmnd and SIVdrl (Fig. 2), we characterized SIVdrl further. We identified three drills (Drl 006, Drl 007, and Drl 207) whose sera reacted positively with the V3 SIVmnd peptide in an EIA. All three were wild born in Cameroon. Drl 207, a sexually immature 4-year-old monkey, was already positive at the date of its rescue, confirming that this SIVdrl strain represents a natural infection. We sequenced a pol region from all three SIVdrl (Fig. 5A and C). They are the most closely related to the formerly described SIVdrl (6), which in turn is much more closely related to SIVmnd-2 than to SIVmnd-1. A 339-bp sequence in gp41 of SIVdrl007 indicates that it has the same recombinant structure as SIVmnd-2 (Fig. 6). M. sphinx and M. leucophaeus are the two species of the Mandrillus genus, and the phylogenetic relationships between SIVdrl and SIVmnd-2 suggest that these related viruses might have codiverged in their respective host species.
A human virus serologically related to V3 of SIVmnd.
Our results show that SIVmnd-2 is related to SIV from the Papionini tribe. Indeed, it has the same genomic organization as SIVsm, and it is phylogenetically related to SIVrcm and SIVdrl. Viruses from the Papionini, such as SIVsm, seem to be transmissible to humans. We addressed the question whether humans could be at risk for SIVmnd-2 by looking for SIVmnd-related viruses in humans. We screened samples collected during a large survey of HIV diversity in Cameroon and Gabon performed between 1994 and 1999 (28; Tevi-Benissan et al., letter, 1998). Among the 6,515 HIV-positive sera, 15 were positive by HIV-2 Western blotting (WB). By our HIV and SIV V3-specific EIA, 14 out of the 15 sera reacted specifically with the SIVsm/HIV-2 V3 loop. The remaining subject (patient 97-6178) was a 65-year-old symptom-free man attending a clinic in South Cameroon. His HIV-2 WB was positive for transmembrane and core antigens but not for pol products. Conversely, his HIV-1 WB was positive only for pol products. On a commercial dot test using separate HIV-1 recombinant protein and HIV-2 transmembrane peptide as antigens (Multispot; Bio-Rad–Pasteur), the serum reacted only against the HIV-1 recombinant spot. These unusual serological profiles prompted us to further characterize this sample. We observed a high reactivity directed solely and strongly against the SIVmnd V3 loop (Table 3). We then compared these results to those obtained for 164 HIV-2-infected patients (150 living in France and 14 from Cameroon and Gabon) and to those of our SIVmnd-infected mandrills. Table 3 summarizes the specificity of our V3 loop peptide assay. The HIV-2 samples reacted strongly against the HIV-2/SIVsm V3 loop and displayed a low cross-reactivity against the SIVagm V3 loop. None of these HIV-2 samples reacted against the SIVmnd loop. All the samples from seropositive mandrills reacted only against their specific peptide, with no cross-reactivity against the heterologous V3 peptides (Table 3).
TABLE 3.
Reactivity of a human sample against SIVmnd V3 loop peptidea
| Sample source | No. of samples | OD ± SD for:
|
||
|---|---|---|---|---|
| V3 SIVagmb | V3 SIVsm/HIV-2c | V3 SIVmndd | ||
| HIV-2 infected patients | 164 | 0.12 ± 0.2 | 0.98 ± 0.46 | 0.03 ± 0.02 |
| SIVmnd-infected mandrills and drills | 28 | 0.06 ± 0.05 | 0.09 ± 0.08 | 1.8 ± 0.53 |
| Patient 97-6178 | 1 | 0.01 | 0.02 | 1.7 |
Determined by a V3 loop peptide-based EIA (34). The reactivity of the sample (from patient 97-6178) was compared with those for HIV-2-positive samples and SIVmnd-positive samples.
Peptide corresponding to the V3 loop from SIV of African green monkeys (C. sabaeus).
Peptide corresponding to the V3 loop consensus sequence from SIV of sooty mangabeys and the HIV-2 Rod prototype strain.
Peptide mimicking the V3 loop of SIVmnd-1 GB1.
All molecular investigations of this atypical human serum sample, stored under local conditions before shipping to our laboratory, were unsuccessful. An attempt to isolate the strain was also negative. A poorly replicating virus could explain these failures. The patient has since been lost to follow-up, precluding firm diagnosis. However, given the high V3 loop sequence divergence between SIV and HIV, the accuracy and the high specificity of our peptide EIA, and the strong and specific reactivity of this human sample against the SIVmnd V3 region, this result can only be explained by infection with a virus that is SIVmnd-like, at least in this region of Env.
DISCUSSION
SIVmnd classification.
Our results show that two distinct SIV types infect wild-living mandrills. These two SIVmnd are different with respect to (i) their phylogenetic relationships, clustering in different SIV lineages in phylogenies inferred from different genomic regions; (ii) genome structures, as SIVmnd-1 (represented by GB1) lacks the vpx gene whereas the second virus, SIVmnd-2 (represented by M14), includes it; and (iii) antigenic properties, as shown by a commercial EIA and a p24 antigen assay. These data are analogous to the situation encountered in humans. The human population is infected by two different lentiviral types, HIV-1 and HIV-2, which have different origins, different genome structures, and different antigenic properties (Marlink, editorial, 1996). These features, together with epidemiological evidence for the circulation of these two mandrill viruses in the wild, call for a classification system for SIVmnd. We propose, by analogy with HIV classification, that these viruses be considered different “types” of SIVmnd. We suggest that the original lineage of SIVmnd, which includes SIVmndGB1, be named SIVmnd type 1 (SIVmnd-1) and that the second lineage, identified here, be classified as SIVmnd type 2 (SIVmnd-2).
SIVmnd, an SIV diversity paradigm: host-dependent evolution versus cross-species transmission.
The two types of SIVmnd naturally infecting M. sphinx can be considered a model for the complexity of HIV/SIV evolution. Both SIVmnd types circulate in the wild, but they might have a distinct geographic distribution. All SIVmnd-2-infected mandrills originated from Cameroon (the region south of the Sanaga River) and the neighboring region of north Gabon (north of the Ogooué River), (Fig. 1). Conversely, the SIVmnd-1-infected mandrills were found south of the Ogooué River. Studies of mitochondrial DNA from infected mandrills will determine if a differential haplotype distribution coincides with the SIVmnd type distribution.
SIVmnd-1 most likely originates from an ancient cross-species transmission from a Cercopithecinae ancestor (2, 3, 19). Our data demonstrate that this cross-transmission between two different genera is not a dead-end, as this virus is apparently spreading among wild mandrills. According to our data, however, SIVmnd-2 might have a more host-specific history than SIVmnd-1. SIVmnd-2 shares a high degree of homology in the 5′ end of the genome with SIVdrl and SIVrcm, which have been isolated from other species belonging to the Papionini tribe, favoring a common SIV ancestor for these strains. The ancestor of the current SIVmnd-2 is probably a recombinant, as SIVmnd-2 is related to SIVrcm in gag and pol, and SIVrcm itself is a recombinant form, sharing homology in pol with SIVcpz and a relationship with SIVsm in gag (7, 14). Interestingly, C. torquatus is the closest relative of the Mandrillus genus (17). Our data indicate that the prevalence of both types seems sufficiently high in nature to allow dual infection. An overlapping geographic range in the past between mandrills infected by the different types would explain how recombination could occur between the SIVmnd-2 parental virus and SIVmnd-1, but although possible, this remains to be demonstrated.
These apparently successful recombination events between strains infecting different genera illustrate the potential of lentiviral diversification. Recent evidence suggests not only that recombination between distinct lentiviruses is possible but also that this phenomenon is responsible for the emergence of viruses which have succeeded in dominating the epidemics in both human and nonhuman primates. HIV-1 group M recombinant forms are successfully spreading in the world (12, 25, 29, 32). Also, at least two nonhuman primate species are infected with recombinant forms of lentiviruses: Cercopithecus aethiops sabaeus (23) and red-capped mangabeys (14). Taken together, the characterization of two SIVmnd types epitomizes our knowledge of SIV diversity, including codivergence of SIV and host species, SIV cross-species transmission between primates, and, finally, recombination. These data further demonstrate that recombinant viral forms have a great capacity to contribute to epidemics and might explain why the circulation of the parental, nonrecombinant SIVmnd-2 ancestor, which might be rare or is now extinct, went undetected in epidemiological studies.
Zoonotic transmission of SIVmnd?
A case of retrovirus transmission from mandrills to humans has already been documented. Simian T-cell lymphotrophic virus type 1 (STLV-1) from M. sphinx has been described as the simian counterpart of human T-cell lymphotrophic virus type 1 (HTLV-1) subtype D (26). Indeed, a close molecular and phylogenetic relationship has been reported between STLV-1 subtype D from mandrills in Gabon and HTLV-1 strains obtained from pygmies living in Cameroon and the Central African Republic and from a healthy nonpygmy carrier in Gabon.
The atypical serological reactivities observed with commercial assays for a serum from an HIV-infected human in Cameroon already indicated that the patient could be infected by a virus which is different from known HIV-1 or HIV-2. Our peptide-based serological test, which displays a high discriminatory capacity between HIV and SIV from different lineages (34), revealed a human case of lentiviral infection serologically reactive to the SIVmnd V3 peptide. The high rate of SIV seropositivity in wild mandrills favors the probability of exposure of the population to infected blood during hunting or food preparation, as the wild troops of mandrills in Cameroon and Gabon are heavily hunted. Moreover, juvenile mandrills are often kept as pets. As SIVsm was able to jump to the human population, the possibility that SIVmnd-infected mandrills could also represent a reservoir posing a risk for humans cannot be excluded. This single case of human infection by a strain serologically related to SIVmnd V3 may represent a dead-end infection, similar to those observed for subtypes D and E of HIV-2 infection (13). However, this needs confirmation by sequence identification. This case, nonetheless, does illustrate the potential for currently unrecognized zoonotic reservoirs of AIDS viruses for humans.
In conclusion, we have identified a second SIV type naturally infecting M. sphinx. Mandrills, naturally infected by two distinct SIV lineages, could be a useful model for coinfection studies. The observation of SIVmnd mother-to-child transmission opens up research opportunities for better understanding of one major public health problem of HIV/AIDS. Finally, our epidemiological study of humans illustrates that the HIV pandemic still calls for large-scale and longitudinal worldwide epidemiological surveys.
ACKNOWLEDGMENTS
This work was supported by the French National Agency on AIDS Research (ANRS), grant 2000/038, and grants NO1 AI 85338 (to B.H.H.) and RO1 AI 44596 (to P.M. and B.H.H.) from the National Institutes of Health. D.L.R. is supported by a Wellcome Trust Biodiversity Fellowship, and C.K. is supported by the Daimler-Benz Foundation.
We thank John Clewley for permission to use an unpublished SIVdrl sequence. Blood sampling from mandrills was performed by Pierre Rouquet (CIRMF), Jack Allen, Kent Osbom, April Gorow (SDZ/WAP, San Diego, Calif.), Stacey Hoffman, Myra Jennings, Nicholas W. Lerche (California Regional Primate Research Center, University of California, Davis), William Karesh, and John Lewis.
REFERENCES
- 1.Allan J S, Short M, Taylor M E, Su S, Hirsch V M, Johnson P R, Shaw G M, Hahn B H. Species-specific diversity among simian immunodeficiency viruses from African green monkeys. J Virol. 1991;65:2816–2828. doi: 10.1128/jvi.65.6.2816-2828.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beer B E, Bailes E, Dapolito G, Campbell B J, Goeken R M, Axthelm M K, Markham P D, Bernard J, Zagury D, Franchini G, Sharp P M, Hirsch V M. Patterns of genomic sequence diversity among their simian immunodeficiency viruses suggest that L'Hoest monkeys (Cercopithecus lhoesti) are a natural lentivirus reservoir. J Virol. 2000;74:3892–3898. doi: 10.1128/jvi.74.8.3892-3898.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beer B E, Bailes E, Goeken R, Dapolito G, Coulibaly C, Norley S G, Kurth R, Gautier J P, Gautier-Hion A, Vallet D, Sharp P M, Hirsch V M. Simian immunodeficiency virus (SIV) from sun-tailed monkeys (Cercopithecus solatus): evidence for host-dependent evolution of SIV within the C. lhoesti superspecies. J Virol. 1999;73:7734–7744. doi: 10.1128/jvi.73.9.7734-7744.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bibollet-Ruche F, Galat-Luong A, Cuny G, Sarni-Manchado P, Galat G, Durand J P, Pourrut X, Veas F. Simian immunodeficiency virus infection in a patas monkey (Erythrocebus patas): evidence for cross-species transmission from African green monkeys (Cercopithecus aethiops sabaeus) in the wild. J Gen Virol. 1996;77:773–781. doi: 10.1099/0022-1317-77-4-773. [DOI] [PubMed] [Google Scholar]
- 5.Chen Z, Telfer P, Gettie A, Reed P, Zhang L, Ho D D, Marx P A. Genetic characterization of new West African simian immunodeficiency virus SIVsm: geographic clustering of household-derived SIV strains with human immunodeficiency virus type 2 subtypes and genetically diverse viruses from a single feral sooty mangabey troop. J Virol. 1996;70:3617–3627. doi: 10.1128/jvi.70.6.3617-3627.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clewley J P, Lewis J C, Brown D W, Gadsby E L. A novel simian immunodeficiency virus (SIVdrl) pol sequence from the drill monkey, Mandrillus leucophaeus. J Virol. 1998;72:10305–10309. doi: 10.1128/jvi.72.12.10305-10309.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Corbet S, Müller-Trutwin M C, Versmisse P, Delarue S, Ayouba A, Lewis J, Brunak S, Martin P, Brun-Vezinet F, Simon F, Barre-Sinoussi F, Mauclere P. env sequences of simian immunodeficiency viruses from chimpanzees in Cameroon are strongly related to those of human immunodeficiency virus group N from the same geographic area. J Virol. 2000;74:529–534. doi: 10.1128/jvi.74.1.529-534.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Courgnaud V, Pourrut X, Bibollet-Ruche F, Mpoudi-Ngole E, Bourgeois A, Delaporte E, Peeters M. Characterization of a novel simian immunodeficiency virus from guereza colobus monkeys (Colobus guereza) in Cameroon: a new lineage in the nonhuman primate lentivirus family. J Virol. 2001;75:857–866. doi: 10.1128/JVI.75.2.857-866.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Emau P, McClure H M, Isahakia M, Else J G, Fultz P N. Isolation from African Sykes' monkeys (Cercopithecus mitis) of a lentivirus related to human and simian immunodeficiency viruses. J Virol. 1991;65:2135–2140. doi: 10.1128/jvi.65.4.2135-2140.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fransen K, Zhong P, De Beenhouwer H, Carpels G, Peeters M, Louwagie J, Janssens W, Piot P, van der Groen G. Design and evaluation of new, highly sensitive and specific primers for polymerase chain reaction detection of HIV-1 infected primary lymphocytes. Mol Cell Probes. 1994;8:317–322. doi: 10.1006/mcpr.1994.1043. . (Erratum, 9:373, 1995.) [DOI] [PubMed] [Google Scholar]
- 11.Fukasawa M, Miura T, Hasegawa A, Morikawa S, Tsujimoto H, Miki K, Kitamura T, Hayami M. Sequence of simian immunodeficiency virus from African green monkey, a new member of the HIV/SIV group. Nature. 1988;333:457–461. doi: 10.1038/333457a0. [DOI] [PubMed] [Google Scholar]
- 12.Gao F, Robertson D L, Morrison S G, Hui H, Craig S, Decker J, Fultz P N, Girard M, Shaw G M, Hahn B H, Sharp P M. The heterosexual human immunodeficiency virus type 1 epidemic in Thailand is caused by an intersubtype (A/E) recombinant of African origin. J Virol. 1996;70:7013–7029. doi: 10.1128/jvi.70.10.7013-7029.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao F, Yue L, Robertson D L, Hill S C, Hui H, Biggar R J, Neequaye A E, Whelan T M, Ho D D, Shaw G M, et al. Genetic diversity of human immunodeficiency virus type 2: evidence for distinct sequence subtypes with differences in virus biology. J Virol. 1994;68:7433–7447. doi: 10.1128/jvi.68.11.7433-7447.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Georges-Courbot M C, Lu C Y, Makuwa M, Telfer P, Onanga R, Dubreuil G, Chen Z, Smith S M, Georges A, Gao F, Hahn B H, Marx P A. Natural infection of a household pet red-capped mangabey (Cercocebus torquatus torquatus) with a new simian immunodeficiency virus. J Virol. 1998;72:600–608. doi: 10.1128/jvi.72.1.600-608.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grubb P. Distribution, divergence and speciation of the drill and mandrill. Fol Primatol. 1973;20:161–177. doi: 10.1159/000155574. [DOI] [PubMed] [Google Scholar]
- 16.Hahn B H, Shaw G M, De Cock K M, Sharp P M. AIDS as a zoonosis: scientific and public health implications. Science. 2000;287:607–614. doi: 10.1126/science.287.5453.607. [DOI] [PubMed] [Google Scholar]
- 17.Harris E E, Disotell T R. Nuclear gene trees and the phylogenetic relationships of the mangabeys (Primates: Papionini) Mol Biol Evol. 1998;15:892–900. doi: 10.1093/oxfordjournals.molbev.a025993. [DOI] [PubMed] [Google Scholar]
- 18.Hasegawa M, Kishino H, Yano T. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J Mol Evol. 1985;22:160–174. doi: 10.1007/BF02101694. [DOI] [PubMed] [Google Scholar]
- 19.Hirsch V M, Campbell B J, Bailes E, Goeken R, Brown C, Elkins W R, Axthelm M, Murphey-Corb M, Sharp P M. Characterization of a novel simian immunodeficiency virus (SIV) from L'Hoest monkeys (Cercopithecus l'hoesti): implications for the origins of SIVmnd and other primate lentiviruses. J Virol. 1999;73:1036–1045. doi: 10.1128/jvi.73.2.1036-1045.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hirsch V M, Dapolito G A, Goldstein S, McClure H, Emau P, Fultz P N, Isahakia M, Lenroot R, Myers G, Johnson P R. A distinct African lentivirus from Sykes' monkeys. J Virol. 1993;67:1517–1528. doi: 10.1128/jvi.67.3.1517-1528.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hirsch V M, McGann C, Dapolito G, Goldstein S, Ogen-Odoi A, Biryawaho B, Lakwo T, Johnson P R. Identification of a new subgroup of SIVagm in tantalus monkeys. Virology. 1993;197:426–430. doi: 10.1006/viro.1993.1606. [DOI] [PubMed] [Google Scholar]
- 22.Hirsch V M, Olmsted R A, Murphey-Corb M, Purcell R H, Johnson P R. An African primate lentivirus (SIVsm) closely related to HIV-2. Nature. 1989;339:389–392. doi: 10.1038/339389a0. [DOI] [PubMed] [Google Scholar]
- 23.Jin J M, Hui H, Robertson D L, Müller M C, Barré-Sinoussi F, Hirsch V M, Allan J F, Shaw G M, Sharp P M, Hahn B H. Mosaic genome structure of simian immunodeficiency viruses from West African green monkeys. EMBO J. 1994;13:2935–2947. doi: 10.1002/j.1460-2075.1994.tb06588.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jones D T, Taylor W R, Thomton J M. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- 25.Liitsola K, Tashkinova I, Laukkanen T, Korovina G, Smolskaja T, Momot O, Mashkilleyson N, Chaplinskas S, Brummer-Korvenkontio H, Vanhatalo J, Leinikki P, Salminen M O. HIV-1 genetic subtype A/B recombinant strain causing an explosive epidemic in injecting drug users in Kaliningrad. AIDS. 1998;12:1907–1919. doi: 10.1097/00002030-199814000-00023. [DOI] [PubMed] [Google Scholar]
- 26.Mahieux R, Chappey C, Georges-Courbot M C, Dubreuil G, Mauclere P, Georges A, Gessain A. Simian T-cell lymphotropic virus type 1 from Mandrillus sphinx as a simian counterpart of human T-cell lymphotropic virus type 1 subtype D. J Virol. 1998;72:10316–10322. doi: 10.1128/jvi.72.12.10316-10322.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marx P A, Li Y, Lerche N W, Sutjipto S, Gettie A, Yee J A, Brotman B H, Prince A M, Hanson A, Webster R G, et al. Isolation of a simian immunodeficiency virus related to human immunodeficiency virus type 2 from a West African pet sooty mangabey. J Virol. 1991;65:4480–4485. doi: 10.1128/jvi.65.8.4480-4485.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mauclere P, Loussert-Ajaka I, Damond F, Fagot P, Souquiere S, Monny Lobe M, Mbopi Keou F X, Barre-Sinoussi F, Saragosti S, Brun-Vezinet F, Simon F. Serological and virological characterization of HIV-1 group O infection in Cameroon. AIDS. 1997;11:445–453. doi: 10.1097/00002030-199704000-00007. [DOI] [PubMed] [Google Scholar]
- 29.McCutchan F E, Carr J K, Bajani M, Sanders-Buell E, Harry T O, Stoeckli T C, Robbins K E, Gashau W, Nasidi A, Janssens W, Kalish M L. Subtype G and multiple forms of A/G intersubtype recombinant human immunodeficiency virus type 1 in Nigeria. Virology. 1999;254:226–234. doi: 10.1006/viro.1998.9505. [DOI] [PubMed] [Google Scholar]
- 30.Müller M C, Saksena N K, Nerrienet E, Chappey C, Herve V M, Durand J P, Legal-Campodonico P, Lang M C, Digoutte J P, Georges A J, Sonigo P, Barré-Sinoussi F. Simian immunodeficiency viruses from central and western Africa: evidence for a new species-specific lentivirus in tantalus monkeys. J Virol. 1993;67:1227–1235. doi: 10.1128/jvi.67.3.1227-1235.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nerrienet E, Amouretti X, Müller-Trutwin M C, Poaty-Mavoungou V, Bedjebaga I, Nguyen H T, Dubreuil G, Corbet S, Wickings E J, Barre-Sinoussi F, Georges A J, Georges-Courbot M C. Phylogenetic analysis of SIV and STLV type 1 in mandrills (Mandrillus sphinx): indications that intracolony transmissions are predominantly the result of male-to-male aggressive contacts. AIDS Res Hum Retrovir. 1998;14:785–796. doi: 10.1089/aid.1998.14.785. [DOI] [PubMed] [Google Scholar]
- 32.Peeters M, Liegeois F, Torimiro N, Bourgeois A, Mpoudi E, Vergne L, Saman E, Delaporte E, Saragosti S. Characterization of a highly replicative intergroup M/O human immunodeficiency virus type 1 recombinant isolated from a Cameroonian patient. J Virol. 1999;73:7368–7375. doi: 10.1128/jvi.73.9.7368-7375.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 34.Simon F, Souquière S, Damond F, Kfutwah A, Makuwa M, Leroy E, Rouquet P, Berthier J L, Rigoulet J, Lecu A, Telfer P T, Pandrea I, Plantier J C, Marx P A, Barré-Sinoussi F, Müller-Trutwin M C, Apetrei C. A synthetic peptide strategy for the detection of and discrimination between highly divergent primate lentiviruses. AIDS Res Hum Retrovir. 2001;17:937–952. doi: 10.1089/088922201750290050. [DOI] [PubMed] [Google Scholar]
- 35.Simon F, Mauclere P, Roques P, Loussert-Ajaka I, Müller-Trutwin M C, Saragosti S, Georges-Courbot M C, Barré-Sinoussi F, Brun-Vézinet F. Identification of a new human immunodeficiency virus type 1 distinct from group M and group O. Nat Med. 1998;4:1032–1037. doi: 10.1038/2017. [DOI] [PubMed] [Google Scholar]
- 36.Strimmer K, von Haeseler A. Quartet puzzling: a quartet maximum likelihood method for reconstructing tree topologies. Mol Biol Evol. 1996;13:964–969. [Google Scholar]
- 37.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W—improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tsujimoto H, Hasegawa A, Maki N, Fukasawa M, Miura T, Speidel S, Cooper R W, Moriyama E N, Gojobori T, Hayami M. Sequence of a novel simian immunodeficiency virus from a wild-caught African mandrill. Nature. 1989;341:539–541. doi: 10.1038/341539a0. [DOI] [PubMed] [Google Scholar]
- 39.van Rensburg E J, Engelbrecht S, Mwenda J, Laten J D, Robson B A, Stander T, Chege G K. Simian immunodeficiency viruses (SIVs) from eastern and southern Africa: detection of a SIVagm variant from a chacma baboon. J Gen Virol. 1998;79:1809–1814. doi: 10.1099/0022-1317-79-7-1809. [DOI] [PubMed] [Google Scholar]