Abstract
Background
Human papillomavirus type 16 (HPV-16) infection is strongly associated with considerable parts of cervical, neck, and head cancers. Performed investigations have had moderate clinical success, so research to reach an efficient vaccine has been of great interest. In the present study, the immunization potential of a newly designed HPV-16 construct was evaluated in a mouse model.
Results
Initially, a construct containing HPV-16 mutant (m) E6/E7 fusion gene was designed and antigen produced in two platforms (i.e., DNA vaccine and recombinant protein). Subsequently, the immunogenicity of these platforms was investigated in five mice) C57BL/6 (groups based on several administration strategies. Three mice groups were immunized recombinant protein, DNA vaccine, and a combination of them, and two other groups were negative controls. The peripheral blood mononuclear cells (PBMCs) proliferation, Interleukin-5 (IL-5) and interferon-γ (IFN-γ) cytokines, IgG1 and IgG2a antibody levels were measured. After two weeks, TC-1 tumor cells were injected into all mice groups, and subsequently further analysis of tumor growth and metastasis and mice survival were performed according to the schedule.
Overall, the results obtained from in vitro immunology and tumor cells challenging assays indicated the potential of the mE6/E7 construct as an HPV16 therapeutic vaccine candidate. The results demonstrated a significant increase in IFN-γ cytokine (P value < 0.05) in the Protein/Protein (D) and DNA/Protein (E) groups. This finding was in agreement with in vivo assays. Control groups show a 10.5-fold increase (P value < 0.001) and (C) DNA/DNA group shows a 2.5-fold increase (P value < 0.01) in tumor growth compared to D and E groups. Also, a significant increase in survival of D and E (P value < 0.001) and C (P value < 0.01) groups were observed.
Conclusions
So, according to the findings, the recombinant protein could induce stronger protection compared to the DNA vaccine form. Protein/Protein and DNA/Protein are promising administration strategies for presenting this construct to develop an HPV-16 therapeutic vaccine candidate.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12896-024-00893-y.
Keywords: Recombinant protein, DNA vaccine, HPV-16, Therapeutic vaccine, Tumor-specific antigens (TSAs), Fusion gene, Prime-boost strategy
Background
The fourth most common female malignant disease is cervical cancer, resulting in 570,000 new cases and 311,000 deaths globally in 2018 as released by WHO [1]. Approximately 60% of all cases of cervical cancer have belonged to the most common carcinogenic type, HPV-16 [2–4].
Vaccination is the most preferred approach in the control of infectious diseases such as HPV [5]. Current HPV vaccines elicit antibody responses against the late viral capsid proteins of HPV (L1) and have been useful for preventive purposes in people who have not yet been infected [6]. Due to the critical role of cell-mediated immunity (CMI) in HPV infection clearance, there is an emergency need for developing therapeutic HPV vaccines to induce CMI responses [5, 7].
Therapeutic cancer vaccines mainly utilize oncogenic viral proteins as tumor-specific antigens (TSAs) to activate the patient’s CMI responses [3, 8]. For this purpose, amongst the early phase proteins of HPV, the E6 and E7 are ideal candidates because they have a constant, essential, and specific presence in cancerous cells. Moreover, cytotoxic T lymphocytes (CTLs) responses against HPV E6 and E7 are inherently more frequent than other HPV proteins and these proteins do not induce immune tolerance [9–11]. Also, E6 and E7 are the biomarkers that drive cancer progression, have been proven that approaches targeting them are highly efficient in terms of concentrated eliminating circulating malignant cells [12].
The E6 protein promotes degradation of the p53 protein, using the E6-associated protein (E6AP), and also induces cellular telomerase activity [13, 14]. On the other hand, the E7 protein binds to retinoblastoma protein (pRb) family members and subsequently inactivates them [15, 16]. Moreover, this protein activates Mi2β, which is a member of the histone deacetylase complex [17].
With meticulous attention to such details, further operations are therefore required for removing the oncogenicity of these proteins for safe use in vaccine formulation. Several studies reported that HPV-16 E6 and E7 proteins have their individual antigenic sites and incorporating both of them in a construct resulted in higher antitumor protection effects [18, 19].
Although efforts to develop vaccines with the ability to confer E6/E7 HPV-specific immune responses have shown good results in preclinical studies, there is also a burning need to develop new strategies with clinical efficacy [20]. To accomplish vaccine development strategies, vaccine formulation, platforms, and route of administration are the main objective areas in research.
Common platforms that were utilized in developing therapeutic vaccines are DNA vaccines, viral vectors, live attenuated, and subunit vaccines with or without adjuvant [21]. DNA vaccine platform has many advantages for cancer immune therapy but it suffers from insufficient intrinsic specificity for antigen-presenting cells (APCs) and limited ability to spread between cells in vivo [14, 16, 22]. As CTLs are activated efficiently by APCs through T helper (Th) 1 cells and its cytokines stimulation [23], it is hypothesized that the delivery of antigens through different administration routes can activate different parts of the immune system. Indeed, generally, DNA vaccines strongly instigate CD4 + T cell and recombinant proteins are more effective at stimulating antibody responses and CD4 + T cell activation. Thus DNA/protein strategy can lead to stronger, long-term CMI against intracellular pathogens [24].
This study aimed to establish an inclusive assessment of a new proposed construct through vaccine formulation, platforms, and delivery strategies to present a new HPV therapeutic vaccine candidate. For this purpose, a new fusion gene of HPV-16 was designed based on mutant forms of E6 and E7 (mE6/E7) that fused by using a helical linker. Then the construct was produced in two different platforms of DNA vaccine and recombinant protein using Escherichia coli (E. coli) strains. Subsequently, five mice) C57BL/6 (groups were organized and immunized with different vaccine platforms (i.e., recombinant protein, DNA vaccine) or negative control agents to accomplish the immunization schedule. After performing the immunization program, suitable in vitro immunology assays were used to assess immunological responses amongst all groups. Further challenging of all mice groups with TC-1 tumor cells, the protective efficacy of newly designed construct according to tumor growth, tumor metastasis, and mice survival was evaluated (Fig. 1).
Fig. 1.
The experimental path of the study
Results
The results of the bioinformatical assessment
Analysis in IEDB and SYFPEITHI servers demonstrated E6 and E7 proteins containing a lot of CTL epitopes throughout the entire sequences. Analysis of mutated proteins by these servers confirmed that variations in recognized epitopes exclusively occur around mutated regions (Additional file 1: Table S1 and Table 2 S).
Construct structure and production
The chimeric protein sequence consists of 271 amino acids, in which the mutated E6 protein was linked to the mutated E7 protein through the HL2 linker (Fig. 2).
Fig. 2.
The amino acid sequence of the designed construct. The mutated amino acids are highlighted. (Additional file 1: Figure S1) shows the 3D structure of the designed construct in a solid ribbon model
The induction and purification bands of the mE6/E7 recombinant protein are shown in Additional file 1: Figure S2 and confirmation of mE6/E7 identity is represented in Additional file 1: Figure S3.
PBMCs proliferation assay
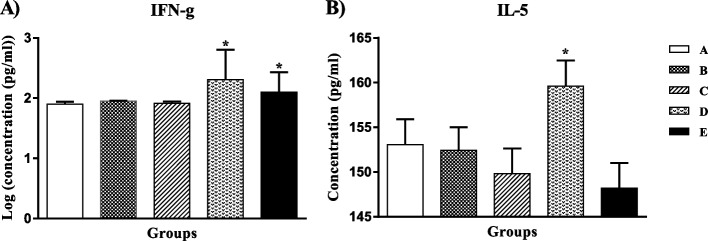
To evaluate the cell proliferation rate, lymphocytes were triggered using mE6/E7 recombinant antigens. Vaccinated groups (C, D E) showed a significant increase in the number of proliferated cells compared to the A and B groups (P value < 0.05). However, the results didn’t show any significant differences between the vaccinated groups (Fig. 3).
Fig. 3.
PBMCs proliferation in different groups. A PBS (SD=0.010), B pcDNA3 (SD=0.011), C mE6/E7 inserted pcDNA3 (SD=0.070), D mE6/E7 recombinant protein (SD=0.064), E mE6/E7 inserted pcDNA3 + mE6/E7 recombinant protein (SD=0.075). * P value < 0.05
Cytokine assay
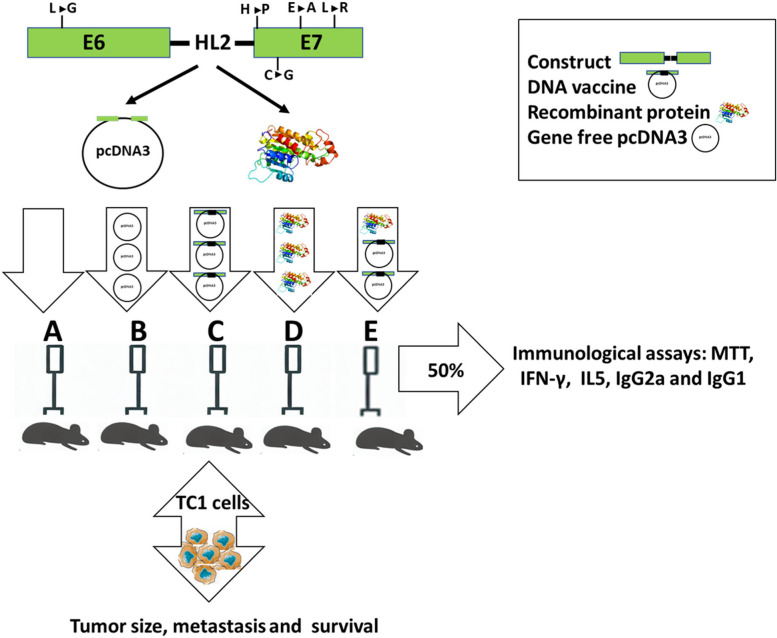
Th1 and Th2 cells produce IFN-γ and IL-5 cytokines respectively. A significant increase (* P value < 0.05) was observed in the levels of IFN-γ in D and E compared to the A and B groups. However, there was no significant difference between the D and E groups (Fig. 4A). Comparing IL-5 levels between different groups implies a significant increase in D compared to other groups (Fig. 4B).
Fig. 4.
The concentration of IFN-γ (A) and IL-5 (B) cytokines in different groups. A PBS, B pcDNA3, C mE6/E7 inserted pcDNA3, D mE6/E7 recombinant protein, E mE6/E7 inserted pcDNA3 + mE6/E7 recombinant protein. * P value < 0.05. SD of all groups respectively is: 0.039, 0.009, 0.034, 0.494, and 0.330 in IFNg and 2.8, 2.5, and 2.8 for other groups in IL5 cytokines
IgG1 and IgG2a antibody levels
IFN-γ and IL-5 cytokines support the generation of IgG2a and IgG1 in mice, respectively.
The pattern of IgG1 and IgG2a subclasses induced in different immunized groups was evaluated.
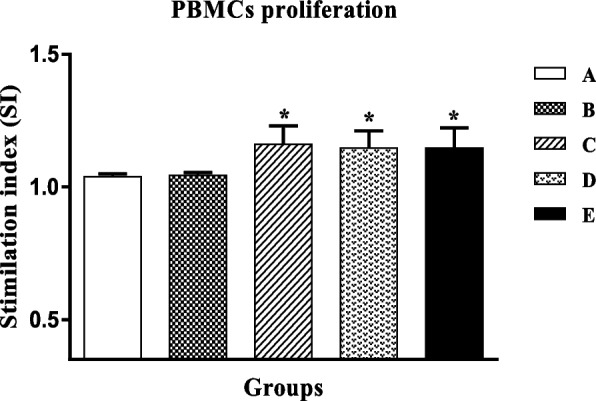
The D and E groups showed a significant increase in IgG1 and IgG2a antibody levels compared to the A, B, and C groups. However, there was no significant variation among the D and E groups.
Moreover, IgG1 antibody levels in all groups are more than IgG2a levels (Fig. 5).
Fig. 5.
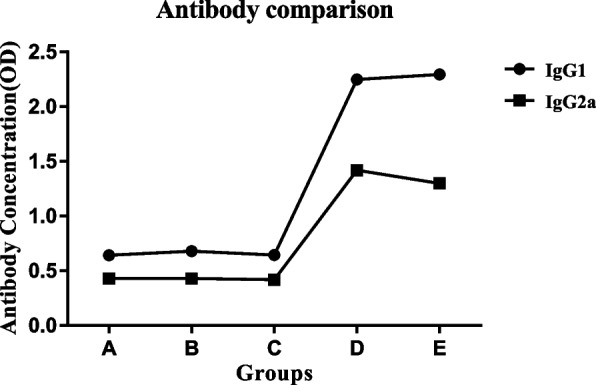
Comparison of IgG1 and IgG2a levels in different groups. A PBS, B pcDNA3, C mE6/E7 inserted pcDNA3, D mE6/E7 recombinant protein, E mE6/E7 inserted pcDNA3 + mE6/E7 recombinant protein. **P value < 0.01
Tumor growth, survival rate, and tumor metastasis assay
Tumor masses were examined by caliper measurement and analyzed using SPSS 16 software. The average of tumor size in the A and B groups showed significant increases (P value < 0.001) compared to the C, D, and E groups. D and E groups didn’t induce any tumor at all. The tumor growth in the C group was significant compared to A and B with a P value < 0.05. The tumor growth in the A and B groups was significantly higher than the C group with P value < 0.01 (Fig. 6).
Fig. 6.
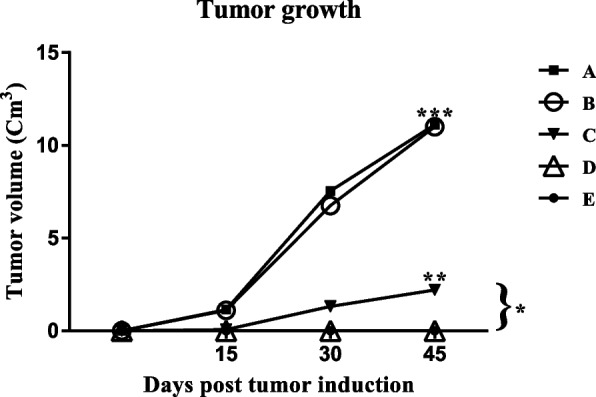
Tumor growth of different mice groups, in immunization by the mE6/mE7 construct. A PBS, B pcDNA3, C mE6/E7 inserted pcDNA3, D mE6/E7 recombinant protein, E mE6/E7 inserted pcDNA3 + mE6/E7 recombinant protein. *P value < 0.05, **P value < 0.01, ***P value < 0.001
The groups C, D, and E showed a significant increase in survival rate compared to the A and B groups (p < 0.001) (Fig. 7). The survival rate of the C group was not significant compared to D and E and was significantly higher than A and B groups with a P value < 0.01(Fig. 7).
Fig. 7.
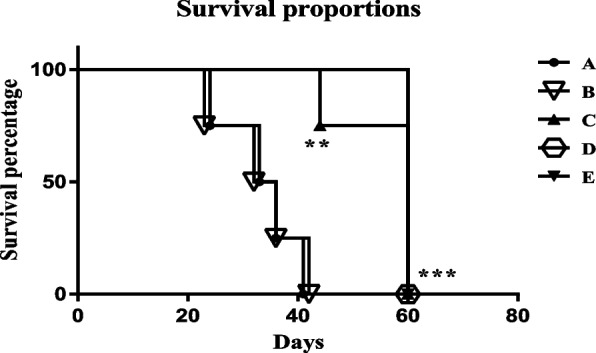
The survival rate of different mice groups, in immunization by the mE6/mE7 construct. A PBS, B pcDNA3, C mE6/E7 inserted pcDNA3, D mE6/E7 recombinant protein, E mE6/E7 inserted pcDNA3 + mE6/E7 recombinant protein. *P value < 0.05, **P value < 0.01, ***P value < 0.001
In terms of tumor metastasis, histological analysis implied the absence or minimal transformation of tissues in the D and E groups, and their tissues were almost normal. However, other groups showed completely abnormal tissues at the same time (Fig. 8).
Fig. 8.
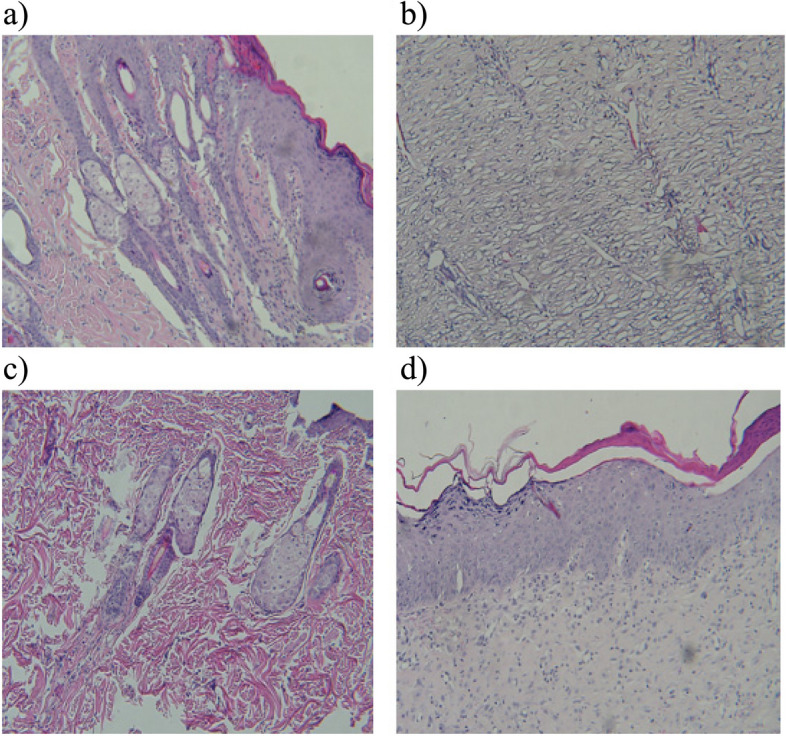
Histological examination of tissue's metastasis of a PBS (A group), b mE6/E7 inserted pcDNA3(C group), c mE6/E7 recombinant protein (D group) and d mE6/E7 inserted pcDNA3 + mE6/E7 recombinant protein (E group)
Discussion
Vaccination as the most effective way to prevent HPV has been performed for several years, but HPV-related disease is also bothersome for human societies because current prophylactic HPV vaccines are inefficient in the treatment of HPV persistent infections. Hence, developing a therapeutic vaccine is an emergency strategy to alleviate these difficulties. To the best of our knowledge, such research has not been promising up till now, and deliberated studies with thoughtful consideration of main vaccine development aspects including vaccine formulation, platform, and delivery method may be hopeful. In this study, the immunogenicity and protective efficacy of the new proposed construct (based on HPV16 as the major cause of HPV-related cancers) in three strategies of DNA/DNA (C), Protein/Protein (D), and DNA/protein (E) were evaluated.
To design the construct, two E6 and E7 oncogenes were selected because they are expressed permanently and specifically in cancer cells and previous studies showed that both E6 and E7 proteins elicit protective immunity, separately [25]. Based on this study and other studies, since both proteins have an immunization effect, both of them were used in the construct.
However, it has been proven that E6 and E7 antigens are oncogenes [2, 15], therefore vaccines composed of these antigens are not safe. Thus, according to studies five mutations [26] were performed and mutated sequences were evaluated using bioinformatical software. Limited variation of epitopes in mutated sequences produce this supposing that these mutations do not disrupt inherent epitopes of proteins significantly and therefore confirmed for presence in vaccine formulation.
Moreover, since, the bioinformatical analysis showed several epitopes in all E6 and E7 proteins lengths, the full length of the proteins was used in designing the construct.
Then designed construct was produced in two DNA vaccine and recombinant protein platforms and evaluated its potential in three delivery strategies of DNA/DNA(C), Protein/Protein (D), and DNA/protein (E).
Several immunological assays were used to analyse the immune response.
The results of the PBMCs proliferation test demonstrated the designed construct could induce proliferation of lymphocytes as one of the goals of cancer immunotherapy and construct platforms (i.e., DNA vaccine and recombinant protein) had limited and non-significant effects on cell proliferation.
Various factors such as the sequence of the peptide ligand type, amount of antigen dose, adjuvant type, and delivery affect naive CD4 + T cells differentiation into Th1 or Th2 [27]. To trace different parts of the immune system, two IFN-γ and IL5 cytokines were evaluated. IFN-γ is known to arrange innate and adaptive immune responses against viruses and tumors resulting in broad tumor removal in mouse models. It is produced by T cell and natural killer (NK) cells and can finally lead to the group of differentiated immune responses Th1 and CTL cells [28]. Th2 promotes humoral immunity by cytokines such as IL-5 [29]. As expected, the IL-5 levels showed that recombinant protein platforms induce humoral immunity while in DNA vaccine platforms such effects were not observed. Studies showed that peptide length, amino acid composition, and their arrangement affect IFN-γ inducing abilities of these peptides [30]. The high level of IFNγ in all platforms demonstrated that the construct sequence effect for induction of cellular immunity is promising even in protein platforms (D group). Moreover, complete Feround’s adjuvant (CFA) and incomplete Feround’s adjuvant (IFA) which can induce a strong CTL and local inflammatory response for activation of APCs, engrossed huge attention in immunotherapy by researchers. These adjuvants have been used for HPV therapeutic vaccines and inhibit tumor growth by induction of Th1/TC-1 [31]. Altogether, our other immunological assays, such as IFN-γ, IL-5, and antibodies imply a higher propensity to Th1 responses in the D, E, and C groups respectively.
Similar to above-mentioned details, high levels of IgG2a compared to control groups imply strong stimulation of IgG2a.
The generally lower level of IgG2a related to IgG1 in all groups originated from aspecific and elevated baseline response of C57BL/6 mouse strain, which this relation obvious in A and B groups as control groups. The absence of significant IgG2a levels in the C group related to the A and B groups is consistent with the fact that DNA vaccine mostly simulates cell-mediated immunity by nature [32, 33].
In this study, 5 × 105 TC1 cells were used for mouse challenge. Tumour growth, survival, and metastatic analysis present identical and strong effectiveness strategies in the D and E groups. That is, the results demonstrated that this construct induced strong immunogenicity thereby producing favourable responses and complete protection against the high number of injected cells in the D and E groups. The C group was also significantly protected in comparison with the A and B groups.
Indeed, the presentation of this vaccine candidate in the protein platform showed predominate immunity and protectiveness. This means that the inherent therapeutic features of the selected antigens and adjutants lead to this wonderful therapeutic effect in the protein platform.
Moreover, the strong protection effect in the D and E groups despite the elevated levels of both IgG1 and IgG2a antibodies, was in agreement with other studies.
For example, a study indicated that the up-regulation of both IgG1 and IgG2a isotypes has a significant role in the vaccine-induced protection [34]. Also, in patients with cervical cancer, supportive Th immunity, which is induced by the secretion of both Th1/Th2 cytokines, is protective [35, 36].
Previously, epitope vaccines containing E6 (49–57, 127–135 amino acids) and E7 (49–57, 67–75 amino acids) peptides were evaluated in mice, and then immunized mice were challenged by 7.5 × 104 TC-1 cells [37]. Results implied limited anti-tumor protection effects of E6E7 peptides compared to the construct used in our study (challenging with 5 × 105 TC1 cells) which can be due to removing many epitopes that naturally exist in these proteins (STable 2).
Recently, Xuechao Han et al. used an adenoviral vector encoding a mutant HPV16 E7 which is the deletion of amino acids 21–24 to remove the capability to bind to pRb. The study demonstrated that Ad-E7 was an effective candidate vaccine and upon combination with PD-1/PD-L1 antibody, induced a strong immune response and inhibited tumor growth in mouse cervical cancer models. However, our bioinformatical evaluation using IEDB software showed that by deletion of these amino acids, at least one of the epitopes of the native gene (Allele: HLA-DRB3*02:02, 16–30) is destroyed while in the mutated construct of our study, it conserved. Moreover, any frameshift can increase the risk of changing antigen presentation and formation of neoantigens. As declared in the previous studies, several mechanisms are involved in E7 oncogenicity. It seems deletion of oncogenic sites restrains the ability to modify all of them. However more deliberate in silico, in vitro, and in vivo studies are needed to introduce a modified construct for further clinical evaluations [38].
In another study DNA vaccine with mutations in the LYCYE motif of E7 induced higher levels of tumor protection compared to wild-type E7, this is because of increasing proteasomal degradation and antigen processing through MHC class I [39].
In agreement with the last-mentioned study, we supposed that such efficient responses could also be induced by designing of construct. Maybe even the insertion of the linker or performed mutations improved proteasomal degradations. High protection efficacy in challenge study with this newly designed construct, implies that epitopes presented as the same as natural antigens.
Certainly, the method of administration, vaccine doses, and adjuvant and administration schedule strengthen this candidate vaccine effect.
Based on immunological assays, the D group showed the best effectiveness in the stimulation of cellular immunity but results of in vivo challenging demonstrated that the D group is as effective as the E group. These two groups showed the strongest immunity among all groups.
However, delivery of DNA vaccines using a more efficient method or construct delivery using different platforms could also be very promising. Moreover, the absence of any abnormality or tumor formation in the injection sites of the construct (as a result of oncogenicity of E6 and E7) means mutations are probably effective in the destruction of oncogenicity along with preserving immunogenicity. However, to assure the safety of vaccine candidates more and deeper in silico, in vivo and in vitro studies are recommended.
Conclusion
This construct is a promising vaccine candidate for HPV therapeutic vaccine. It was concluded that the construct with these sequences, mutations, linker, and immunization strategies induced such a strong immunity. High protection in challenging TC1 cells implies the ability of the protein platform to present this construct as a therapeutic vaccine. So, the Protein/Protein (D) and DNA/Protein (E) groups had the strongest responses. In terms of evaluating the effects of combining two delivery routes, this study doesn’t show significant differences between D and E groups. Thus in vivo test showed the identical efficacy of vaccines in these two groups, despite the discrepancy of their in vitro responses. Probably by challenging with a higher number of TC1 cells, we will be able to discriminate responses between Protein/Protein and DNA/Protein strategies in vaccination with this construct. Delivery of the designed DNA vaccine using a more efficient method or various platforms could also be very promising for better evaluating the potential of the designed construct in DNA/DNA and DNA/ protein strategy.
Methods
Construct designing
Amino acid sequences of HPV-16 E6 (Accession No.: NP_041325.1) and E7 proteins (Accession No.: NP_041326.1) were obtained from NCBI’s (National Center for Biotechnology Information) GenBank database. To predict continuous T cell epitopes of E6 and E7 proteins, the sequences were analyzed by the Immune Epitope Database (IEDB) (http://www.iedb.org) and SYFPEITHI (http://www.syfpeithi.de/bin/MHCServer.dll/EpitopePrediction.htm) databases. The mE6/E7 protein sequence was mutated in silico as follows: In the E6 protein, Leucine at position 57 changed to Glycine, and also four-point mutations in HPV-16 E7 were performed by replacement of Histidine (position 2) to Proline, Cysteine (position 24) to Glycine, Glutamic acid (position 46) to Alanine, and Leucine to Arginine at residue 67 [26]. To evaluate the effects of mutations on E6 E7 epitopes, they were reanalyzed using mentioned software. Finally, mutated forms of proteins were selected and a construct was designed in which the stop codon of the E6 protein (the first gene in the construct) was omitted and the helical linker 2 (HL2; LAEAAAKEAAAKAAA) was inserted between E6 and E7 sequences. The fusion protein sequence is converted to the nucleic acid by Oligo-5 software. For the prediction of the 3D structure of the designed construct, the ab initio online program of 3Dpro (http://scratch.proteomics.ics.uci.edu) was used. Then by PyMOL1.3 software, different segments of computed structure in a solid ribbon model were determined.
DNA vaccine preparation
The construct sequence was optimized based on mice codons and inserted in BamHI and XhoI restriction sites of the pcDNA3 expression vector (Biomatic, Canada). The pcDNA3-mE6/E7 was transferred into E. coli DH5α competent cells in Luria-Bertani medium. Large-scale purification of the plasmids was performed using Endo free plasmid mega kit (Qiagen, USA).
Recombinant protein production and purification
The codons of the construct were optimized for expression in bacteria and cloned in the pBluescript SK vector (Biomatic, Canada). The mE6/E7 gene was subcloned into the pET-32a expression vector (Novagene, USA) using BamHI and XhoI restriction enzymes (Fermentas, Lithuania) and T4 Ligase (Fermentas, Lithuania). Then the E.coli BL21 (DE3) competent cells were transformed with the recombinant pET-32a - mE6/E7 vector. The E.coli BL21 (DE3) cells containing pET-32a -mE6/E7 were cultured in 100 ml nutrient broth supplemented with 100 µg/ml of ampicillin and incubated at 37 °C and 220 rpm. The protein production was induced by adding a 1mM concentration of Isopropylthio-β-D-galactoside (IPTG) and purifying using the Ni-NTA protein purification kit (Qiagen, USA) [40, 41]. The quantitative and qualitative evaluation of the protein was performed using measuring absorbance at 280 nm and SDS-polyacrylamide gel electrophoresis (SDS-PAGE) on 12% gels [42–44].
Western blotting
Recombinant mE6/E7 was separated from other host cell proteins using SDS-PAGE and transferred to polyvinylidene difluoride (PVDF) membrane (Roche, Germany) [45]. The identity of recombinant mE6/E7 protein was verified using mouse anti-E7 monoclonal antibody (0.5 µg/ml) (Abcam, USA) and goat anti-mouse (0.25 µg/ml) antibody conjugated with horseradish peroxidase (HRP) enzyme (Abcam, USA). Finally, the detection was completed by 3, 3’-Diaminobenzidine (DAB) (Sigma, USA) substrate solution.
Mice
Male C57BL/6 mice (5–7 weeks old) were purchased from the Pasteur Institute of Iran (n = 40). The mice were housed for one week before the experiment in a standard condition.
Immunization
Finally, the animals were organized into five groups with eight mice in each group. The immunization strategy was scheduled as shown in Table 1.
Table 1.
The animal grouping and immunization strategy
| Groups | Received Component | Route of delivery | Immunization | Adjuvant |
|---|---|---|---|---|
| A | PBS (100 µl) | IM | Three times at 0, 21, 42 days | No |
| B | pcDNA3 (100 µg/100 µl) | IM | Three times at 0, 21, 42 days | No |
| C | mE6/E7 inserted pcDNA3 (100 µg/100 µl) | IM | Three times at 0, 21, 42 days | No |
| D | mE6/E7 recombinant protein (100 µg/100 µl) | SC | Three times at 0, 14, 21 days | CFA (Sigma, USA) in the first injection and IFA in the second and third injections. |
| E | mE6/E7 inserted pcDNA3 (100 µg/100 µl) + mE6/E7 recombinant protein (100 µg/100 µl) | IM + SC | Three times at 0, 21, 42 days | IFA in the third injection |
IM Intramuscularly, SC Subcutaneous, PBS Phosphate buffered saline, CFA Complete Feround’s adjuvant, IFA Incomplete Feround’s adjuvant
PBMCs proliferation assay
About 30 µl ketamine and 10 µl xylazine (or even more quantity with this ratio, if needed) were used for anesthesia. The mice were sacrificed in the home cage so that other mice couldn’t be able to see or even sense this process. The peripheral blood mononuclear cells (PBMC) or splenocytes of the immunized mice were harvested and dispensed into a 96-well culture plate (Nunc, Denmark) at cell densities of 1 × 105 in 10% Fetal bovine serum (FBS) (Sigma, USA) supplemented RPMI-1640 (Sigma, USA). The splenocytes were treated with mE6/E7 recombinant protein (10 µg/ml) and Phytohaemagglutinin (PHA) (5 µg/ml) separately and incubated at 37 °C for 72 h in a humidified atmosphere containing 5% CO2. Then the splenocytes were separated and incubated for 4 h with 20 µl of a 5 mg/ml solution of 3-(4, 5-Dimethylthiazol-2-yl)-2, 5-Diphenyltetrazolium Bromide (MTT). Subsequently, the plates were incubated for an additional 30 min in the presence of 100 µl of Dimethyl Sulfoxide (DMSO). Optical density was measured at the wavelength of 560 nm and the stimulation index (SI) was calculated as follows:
Cytokine assay
The splenocytes (2 × 106 cells/ml) were cultured in a 24-well tissue culture plate in the presence of mE6/E7 recombinant protein (10 µg/ml) and PHA (5 µg/ml). The supernatants were harvested and assayed for interferon-gamma (IFNγ) and interleukin 5 (IL-5) using ELISA kits (Biosciences, USA) according to the manufacturer’s instructions.
Determination of antigen-specific antibody responses
an antigen sandwich enzyme immunoassay to evaluate IgG1 and IgG2a specific antibodies against mE6/E7, serum samples were analyzed by ELISA. Briefly, 96-well Maxisorp plates (Nunc, Denmark) were coated with mE6/E7 recombinant protein (10 µg/ml) overnight at 4 °C. To prevent nonspecific binding, plates were blocked with PBS containing 1% skim milk at 25 °C for 2 h. The mice sera were added (with the dilution of 1:5 by PBS) and incubated for 2 h at 25 °C. After three times washing (PBS + 0.05% Tween20), Anti-mouse IgG1 (0.5 µg/ml) and IgG2a (0.5 µg/ml) (Abcam, USA) were added for 1 h at 25 °C. After washing, the plates were incubated for 1 h at 25 ◦C with goat anti-mouse IgG conjugated to horseradish peroxidase (Abcam, USA). Detection was evaluated by tetramethylbenzidine (TMB) as enzyme-substrate and then the reactions were stopped and signals were measured at 450 nm.
In vivo tumor challenge and metastasis assay
The TC-1 cells, containing E6 and E7 genes were purchased from the Pasteur Institute of Iran. The cells were cultured in supplemented RPMI-1640. Two weeks after immunization, a suspension containing 1 × 105 TC-1 cells in 70 µl PBS was prepared and injected in the right flank of mice. The mice followed up for 60 days to analyze tumor growth and mice survival. Tumor sizes were measured every two weeks after the challenge, with calipers, and the tumor volumes were calculated by this formula (width × length × (width + length) /2) [46]. It needs to be mentioned if the mice died before the day of measuring tumor size, the size of tumors was measured on death day. These sizes of tumors are the mean of all mice tumors in a group. To evaluate metastasis, after 60 days samples from sensitive organs such as the lungs, kidneys, and liver of mice were prepared, and a histological assay was performed.
Statistical analysis
Statistical analysis of data was performed by SPSS16 software. For this purpose, the Mann-Whitney U analysis test and One-Way ANOVA were used. We considered P ≤ 0.001, P ≤ 0.01, and P ≤ 0.05 as our statistical threshold.
Supplementary Information
Additional file 1: Figure S1. Three-dimensional structure of the mE6/E7 construct. The gray HL2 linker between E6 and E7 with black color clearly observed. Figure S2. Induction and purification of recombinant mE6/mE7. Line M,1,2, 3, and 4 respectively are protein marker, before induction, 2 h after induction, 4 h after induction, and purified mE6/mE7 samples. Figure S3. Verification of recombinant mE6/mE7 by western blotting. M is marker and line 1 is mE6/mE7. Table S1: T-cell epitopes of the E6 protein in SYFPEITHI and IEDB servers. Table S2: T-cell epitopes of the E7 protein in SYFPEITHI and IEDB servers.
Acknowledgements
The authors would like to thank the deputy of Research and Technology, and educational and research specialists of Arak University of Medical Sciences for equipment support.
Abbreviations
- HPV
Human papillomavirus
- mE6/E7
Mutant E6/E7
- TSAs
tumor-specific antigens
- PBMCs
peripheral blood mononuclear cells
- IL5
interleukin 5
- IFNγ
interferonγ
- IgG1
immunoglobulin G1
- IgG2a
immunoglobulin G2a
- CMI
cell-mediated immunity
- CTLs
Cytotoxic T lymphocytes
- pRb
Retinoblastoma protein
- APCs
Antigen-presenting cells
- Th1
T helper 1
- Th2
T helper 2
- E.Coli
Escherichia coli
- MHC
Major histocompatibility complex
- HL2
helical linker 2
- IEDB
Immune Epitope Database
- PVDF
polyvinylidinedifuoride
Authors’ contributions
Conceptualization; HA, MMG; Formal analysis: MMG; Funding acquisition: HA, GM; Investigation: MMG; Methodology; HA, MMG, GM, AG, ER, SS, SB; Project administration; HA, MMG, AG; Resources: HA, GM, AG; Visualization; MMG; Supervision: HA, AG, GM; Roles/Writing- original draft: MMG; Writing- review & editing: HA, AG, GM.
Funding
This study is part of the thesis proposal of M.Sc. (N: 1296). This study was conducted with financial assistance from the Molecular and Medicine Research Center and Arak University of Medical Sciences, Arak, Iran, and we are grateful for their invaluable contribution to this study.
Data availability
To request data, please contact MMG.
Declarations
Ethics approval and consent to participate
All experiments were done according to the care and use of laboratory animals’ guidelines of the Ethical Committee of the Arak University of Medical Science (No.IR.ARAKMU.REC.1394.202).
Competing interests
The authors declare no competing interests.
Declaration of competing interest
All the authors of the manuscript declare no scientific and financial conflict of interest.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.WHO. Cervical cancer estimated incidence, mortality and prevalence worldwide in 2018. 2018. [Google Scholar]
- 2.Sarabia-Vega V, Banks L. Acquisition of a phospho-acceptor site enhances HPV E6 PDZ-binding motif functional promiscuity. J Gen Virol. 2020;101:954–62. [DOI] [PubMed] [Google Scholar]
- 3.Ibrahim Khalil A, Zhang L, Muwonge R, Sauvaget C, Basu P. Efficacy and safety of therapeutic HPV vaccines to treat CIN 2/CIN 3 lesions: a systematic review and meta-analysis of phase II/III clinical trials. BMJ Open. 2023;13:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen S, Mei C, Huang W, Liu P, Wang H, Lin W, et al. Human papillomavirus vaccination related knowledge, and recommendations among healthcare providers in Southern China: a cross-sectional survey. BMC Womens Health. 2022;22:169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barrios K, Celis E. TriVax-HPV: an improved peptide-based therapeutic vaccination strategy against human papillomavirus-induced cancers. Cancer Immunol Immunother. 2012;61:1307–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bosch FX, Castellsague X, De Sanjosé S. HPV and cervical cancer: screening or vaccination? Br J Cancer. 2008;98:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moazamigoodarzi M, Fotouhi Ardakani R, Afgar A. Insilco new aspects of peptide-based vaccine designing for human papilloma virus infection. Int J Bioinform Res Appl. 2017;13:223 http://www.inderscience.com/link.php?id=85857. [Google Scholar]
- 8.Mougel A, Terme M, Tanchot C. Therapeutic Cancer Vaccine and Combinations With Antiangiogenic Therapies and Immune Checkpoint Blockade. Front Immunol. 2019;10:476. 10.3389/fimmu.2019.00467. [DOI] [PMC free article] [PubMed]
- 9.Brehm A, Nielsen SJ, Miska EA, McCance DJ, Reid JL, Bannister AJ, et al. The E7 oncoprotein associates with Mi2 and histone deacetylase activity to promote cell growth. EMBO J. 1999;18:2449–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zeng Q, Peng S, Monie A, Yang M, Pang X, Hung CF, et al. Control of cervicovaginal HPV-16 E7-expressing tumors by the combination of therapeutic HPV vaccination and vascular disrupting agents. Hum Gene Ther. 2010;22:809–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Whitehead M, Öhlschläger P, Almajhdi FN, Alloza L, Marzábal P, Meyers AE, et al. Human papillomavirus (HPV) type 16 E7 protein bodies cause tumour regression in mice. BMC Cancer. 2014;14:367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pal A, Kundu R. Human papillomavirus E6 and E7: the cervical cancer hallmarks and targets for therapy. Front Microbiol. 2020;10:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thomas M, Tomaic V, Pim D, Myers MP, Tommasino M, Banks L. Interactions between E6AP and E6 proteins from alpha and beta HPV types. Virology. 2013;435:357–62. [DOI] [PubMed] [Google Scholar]
- 14.Steele JC, Mann CH, Rookes S, Rollason T, Murphy D, Freeth MG, et al. T-cell responses to human papillomavirus type 16 among women with different grades of cervical neoplasia. Br J Cancer. 2005;93:248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vats A, Thatte J, Banks L. Identification of E6AP-independent degradation targets of HPV E6. J Gen Virol. 2019;100:1674–9. [DOI] [PubMed] [Google Scholar]
- 16.Lin K, Roosinovich E, Ma B, Hung CF, Wu TC. Therapeutic HPV DNA vaccines. Immunol Res. 2010;47:86–112. http://link.springer.com/, 10.1007/s12026-009-8141-6. [DOI] [PMC free article] [PubMed]
- 17.Veldman T, Liu X, Yuan H, Schlegel R. Human papillomavirus E6 and Myc proteins associate in vivo and bind to and cooperatively activate the telomerase reverse transcriptase promoter. Proc Natl Acad Sci U S A. 2003;100:8211–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gupta S, Gupta S. Role of human papillomavirus in oral squamous cell carcinoma and oral potentially malignant disorders: a review of the literature. Indian J Dent. 2015;6:91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheng WF, Chang MC, Sun WZ, Jen YW, Liao CW, Chen YY, et al. Fusion protein vaccines targeting two tumor antigens generate synergistic anti-tumor effects. PLoS ONE. 2013;8:e71216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xie Q, Zhou ZX, Li ZL, Zeng Y. Transforming activity of a novel mutant of HPV16 E6E7 fusion gene. Virol Sin. 2011;26:206–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Griffiths KL, Khader SA. Novel vaccine approaches for protection against intracellular pathogens. Curr Opin Immunol. 2014;28:58–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seo SH, Jin HT, Park SH, Youn JI, Sung YC. Optimal induction of HPV DNA vaccine-induced CD8 + T cell responses and therapeutic antitumor effect by antigen engineering and electroporation. Vaccine. 2009;27:5906–12. [DOI] [PubMed] [Google Scholar]
- 23.Asiaf A, Ahmad ST, Mohammad SO, Zargar MA. Review of the current knowledge on the epidemiology, pathogenesis, and prevention of human papillomavirus infection. Eur J Cancer Prev. 2014;23:206–24. [DOI] [PubMed] [Google Scholar]
- 24.Gupta S, Garg NJ. Prophylactic efficacy of TcVac2 against trypanosoma Cruzi in mice. PLoS Negl Trop Dis. 2010;4: e797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li YL, Qiu XH, Shen C, Liu JN, Zhang J. Vaccination of full-length HPV16 E6 or E7 protein inhibits the growth of HPV16 associated tumors. Oncol Rep. 2010;24:1323. [DOI] [PubMed] [Google Scholar]
- 26.Wieking BG, Vermeer DW, Spanos WC, Lee KM, Vermeer P, Lee WT, et al. A non-oncogenic HPV 16 E6/E7 vaccine enhances treatment of HPV expressing tumors. Cancer Gene Ther. 2012;19:667–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.De Maria A, Ferraris A, Guastella A. A role for adaptive immunity V I E W P O I N T How the MHC selects Th1/Th2 immunity. 1977. [Google Scholar]
- 28.Alspach E, Lussier DM, Schreiber RD. Interferon γ and its important roles in promoting and inhibiting spontaneous and therapeutic cancer immunity. Cold Spring Harb Perspect Biol. 2019;11:a028480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Storcksdieck genannt Bonsmann M, Niezold T, Temchura V, Pissani F, Ehrhardt K, Brown EP, et al. Enhancing the quality of antibodies to HIV-1 envelope by GagPol-Specific Th cells. J Immunol. 2015;195:4861–72. [DOI] [PubMed] [Google Scholar]
- 30.Dhanda DK, Vir P, Raghava GP. Designing of interferon-gamma inducing MHC class-II binders. 2013. Available from: http://crdd.osdd.net/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mousavi T, Sattari Saravi S, Valadan R, Haghshenas MR, Rafiei A, Jafarpour H, et al. Different types of adjuvants in prophylactic and therapeutic human papillomavirus vaccines in laboratory animals: a systematic review. Arch Virol. 2020;165:263–84. [DOI] [PubMed] [Google Scholar]
- 32.Silveira MM, Conceição FR, Mendonça M, Moreira GMSG, Da Cunha CEP, Conrad NL, et al. Saccharomyces boulardii improves humoral immune response to DNA vaccines against leptospirosis. J Med Microbiol. 2017;66:184–90. [DOI] [PubMed] [Google Scholar]
- 33.Illiano E, Bissa M, Paolini F, Zanotto C, Morghen CDG, Franconi R, et al. Prime–boost therapeutic vaccination in mice with DNA/DNA or DNA/Fowlpox virus recombinants expressing the human papilloma virus type 16 E6 and E7 mutated proteins fused to the coat protein of Potato virus X. Virus Res. 2016;225:82–90. [DOI] [PubMed] [Google Scholar]
- 34.Gupta J, Pathak M, Misra S, Misra-Bhattacharya S. Immunogenicity and protective efficacy of Brugia malayi Heavy Chain myosin as homologous DNA, protein and heterologous DNA/protein prime boost vaccine in rodent model. PLoS ONE. 2015;10: e0142548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.de Jong A, van Poelgeest MIE, van der Hulst JM, Drijfhout JW, Fleuren GJ, Melief CJM, et al. Human papillomavirus type 16-positive cervical cancer is associated with impaired CD4 + T-cell immunity against early antigens E2 and E6. Cancer Res. 2004;64:5449–55. [DOI] [PubMed] [Google Scholar]
- 36.van Poelgeest MIE, van Seters M, van Beurden M, Kwappenberg KMC, Heijmans-Antonissen C, Drijfhout JW, et al. Detection of human papillomavirus (HPV) 16-specific CD4 + T-cell immunity in patients with persistent HPV16-induced vulvar intraepithelial neoplasia in relation to clinical impact of imiquimod treatment. Clin Cancer Res. 2005;11:5273–80. [DOI] [PubMed] [Google Scholar]
- 37.de Oliveira LMF, Morale MG, Chaves AAM, Cavalher AM, Lopes AS, de Oliveira Diniz M, et al. Design, Immune responses and Anti-tumor potential of an HPV16 E6E7 Multi-epitope Vaccine. PLoS ONE. 2015;10:e0138686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Han X, Gao Z, Cheng Y, Wu S, Chen J, Zhang W. A therapeutic DNA vaccine targeting HPV16 E7 in combination with anti-PD-1/PD-L1 enhanced tumor regression and cytotoxic immune responses. Int J Mol Sci. 2023;24(20):15469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bahrami AA, Ghaemi A, Tabarraei A, Sajadian A, Gorji A, Soleimanjahi H. DNA vaccine encoding HPV-16 E7 with mutation in LYCYE pRb-binding motif induces potent anti-tumor responses in mice. J Virol Methods. 2014;206:12–8. [DOI] [PubMed] [Google Scholar]
- 40.Farhangnia L, Ghaznavi-Rad E, Mollaee N, Abtahi H. Cloning, expression, and purification of recombinant lysostaphin from Staphylococcus simulans. Jundishapur J Microbiol. 2014;7:e10009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mahmoudi S, Abtahi H, Bahador A, Mosayebi G, Salmanian AH. Production of recombinant Streptokinase in E. Coli and reactivity with immunized mice. Pak J Biol Sci. 2010;13:380–4. [DOI] [PubMed] [Google Scholar]
- 42.Mirjamali NAS, Soufian S, Molaee N, Sadoogh Abbasian S, Abtahi H. Cloning and expression of the enzymatic region of streptococcal hyaluronidase. Iran J Basic Med Sci. 2014;17:667–72. [PMC free article] [PubMed] [Google Scholar]
- 43.Hasanzadeh L, Ghaznavi-Rad E, Soufian S, Farjadi V, Abtahi H. Expression and antigenic evaluation of vacA antigenic fragment of helicobacter pylori. Iran J Basic Med Sci. 2013;16:835–40. [PMC free article] [PubMed] [Google Scholar]
- 44.Moazami Goodarzi M, Jalalirad R, Doroud D, Hozouri H, Aghasadeghi M, Paryan M. Determining buffer conditions for downstream processing of VLP-based recombinant hepatitis B surface antigen using multimodal resins in bind-elute and flow-through purification modes. Sci Rep. 2023;13:10745. 10.1038/s41598-023-37614-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Farjadi V, Abtahi H, Zolfaghari MR, Soufian S, Hasanzadeh L. Expression, purification and evaluation of antigenicity of caga antigenic fragment of helicobacter pylori. Jundishapur J Microbiol 2013;6.
- 46.Radaelli A, Morghen CDG, Zanotto C, Pacchioni S, Bissa M, Franconi R, et al. A prime/boost strategy by DNA/fowlpox recombinants expressing a mutant E7 protein for the immunotherapy of HPV-associated cancers. Virus Res. 2012;170:44–52. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. Three-dimensional structure of the mE6/E7 construct. The gray HL2 linker between E6 and E7 with black color clearly observed. Figure S2. Induction and purification of recombinant mE6/mE7. Line M,1,2, 3, and 4 respectively are protein marker, before induction, 2 h after induction, 4 h after induction, and purified mE6/mE7 samples. Figure S3. Verification of recombinant mE6/mE7 by western blotting. M is marker and line 1 is mE6/mE7. Table S1: T-cell epitopes of the E6 protein in SYFPEITHI and IEDB servers. Table S2: T-cell epitopes of the E7 protein in SYFPEITHI and IEDB servers.
Data Availability Statement
To request data, please contact MMG.