Abstract
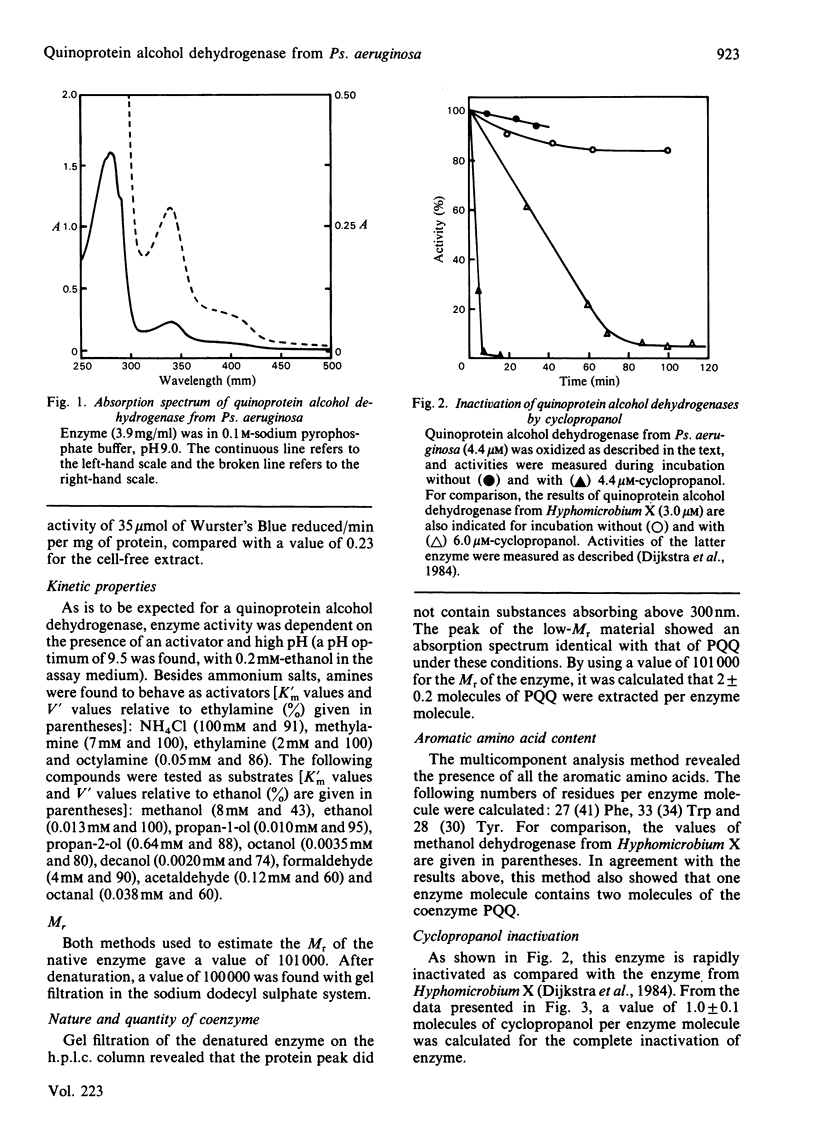
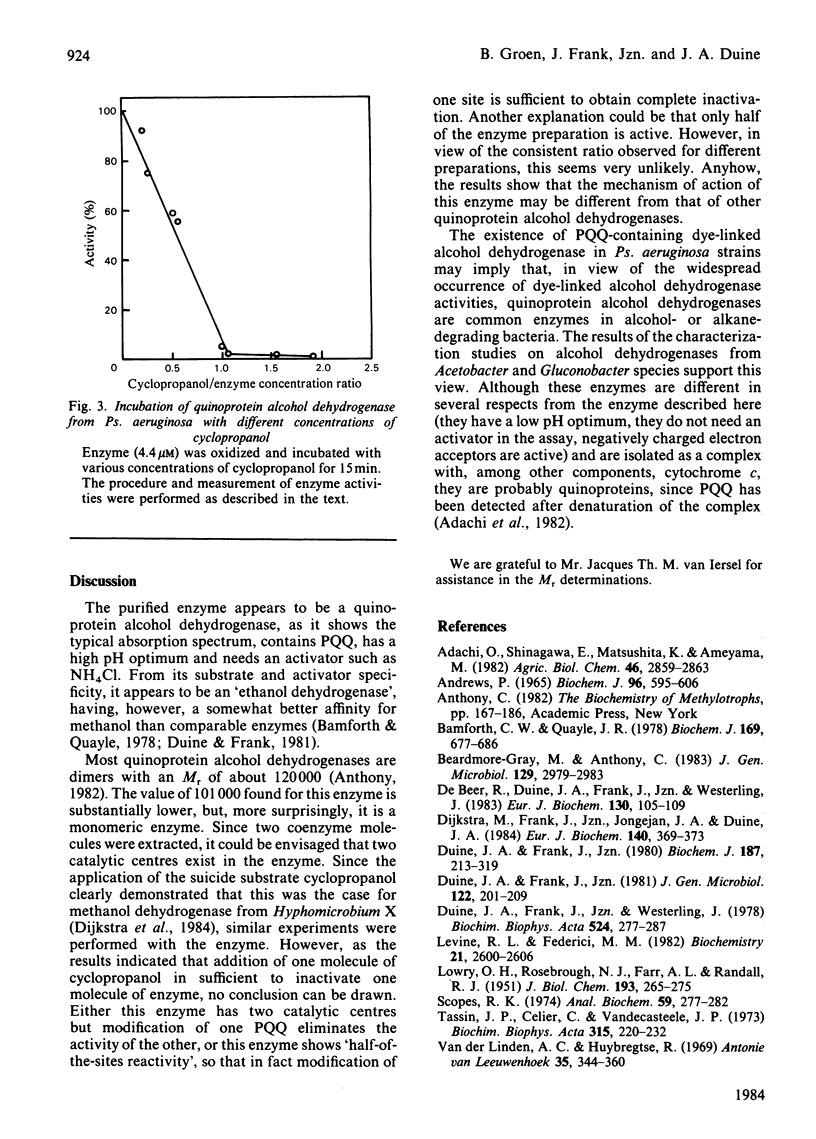
Cell-free extracts of Pseudomonas aeruginosa strains, grown on ethanol, showed dye-linked alcohol dehydrogenase activities. The enzyme responsible for this activity was purified to homogeneity. It appeared to contain two molecules of pyrroloquinoline quinone per enzyme molecule. In many respects, it resembled other quinoprotein alcohol dehydrogenases (EC 1.1.99.8), having a substrate specificity intermediate between that of methanol dehydrogenases and ethanol dehydrogenases in this group. On the other hand, it also showed dissimilarities: the enzyme was found to be a monomer (Mr 101 000), to need only one molecule of the suicide substrate cyclopropanol to become fully inactivated, and to have a different aromatic amino acid composition.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Andrews P. The gel-filtration behaviour of proteins related to their molecular weights over a wide range. Biochem J. 1965 Sep;96(3):595–606. doi: 10.1042/bj0960595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamforth C. W., Quayle J. R. The dye-linked alcohol dehydrogenase of Rhodopseudomonas acidophila. Comparison with dye-linked methanol dehydrogenases. Biochem J. 1978 Mar 1;169(3):677–686. doi: 10.1042/bj1690677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beardmore-Gray M., Anthony C. The absence of quinoprotein alcohol dehydrogenase in Acinetobacter calcoaceticus. J Gen Microbiol. 1983 Oct;129(10):2979–2983. doi: 10.1099/00221287-129-10-2979. [DOI] [PubMed] [Google Scholar]
- Dijkstra M., Frank J., Jongejan J. A., Duine J. A. Inactivation of quinoprotein alcohol dehydrogenases with cyclopropane-derived suicide substrates. . Eur J Biochem. 1984 Apr 16;140(2):369–373. doi: 10.1111/j.1432-1033.1984.tb08111.x. [DOI] [PubMed] [Google Scholar]
- Duine J. A., Frank J. Quinoprotein alcohol dehydrogenase from a non-methylotroph, Acinetobacter calcoaceticus. J Gen Microbiol. 1981 Feb;122(2):201–209. doi: 10.1099/00221287-122-2-201. [DOI] [PubMed] [Google Scholar]
- Duine J. A., Frank J., Westerling J. Purification and properties of methanol dehydrogenase from Hyphomicrobium x. Biochim Biophys Acta. 1978 Jun 9;524(2):277–287. doi: 10.1016/0005-2744(78)90164-x. [DOI] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Levine R. L., Federici M. M. Quantitation of aromatic residues in proteins: model compounds for second-derivative spectroscopy. Biochemistry. 1982 May 25;21(11):2600–2606. doi: 10.1021/bi00540a004. [DOI] [PubMed] [Google Scholar]
- Scopes R. K. Measurement of protein by spectrophotometry at 205 nm. Anal Biochem. 1974 May;59(1):277–282. doi: 10.1016/0003-2697(74)90034-7. [DOI] [PubMed] [Google Scholar]
- Van der Linden A. C., Huybregtse R. Occurrence of inducible and NAD(P)-independent primary alcohol dehydrogenases in an alkane-oxidizing Pseudomonas. Antonie Van Leeuwenhoek. 1969;35(3):344–360. doi: 10.1007/BF02219154. [DOI] [PubMed] [Google Scholar]
- de Beer R., Duine J. A., Frank J., Westerling J. The role of pyrrolo-quinoline semiquinone forms in the mechanism of action of methanol dehydrogenase. Eur J Biochem. 1983 Jan 17;130(1):105–109. doi: 10.1111/j.1432-1033.1983.tb07123.x. [DOI] [PubMed] [Google Scholar]


